Abstract
The Lpin1 gene encodes the phosphatidate phosphatase (PAP1) enzyme Lipin 1, which plays a critical role in lipid metabolism. In this study we describe the identification and characterization of a rat model with a mutated Lpin1 gene (Lpin11Hubr), generated by N-ethyl-N-nitrosourea mutagenesis. Lpin11Hubr rats are characterized by hindlimb paralysis and mild lipodystrophy that are detectable from the second postnatal week. Sequencing of Lpin1 identified a point mutation in the 5′-end splice site of intron 18 resulting in mis-splicing, a reading frameshift, and a premature stop codon. As this mutation does not induce nonsense-mediated decay, it allows the production of a truncated Lipin 1 protein lacking PAP1 activity. Lpin11Hubr rats developed hypomyelination and mild lipodystrophy rather than the pronounced demyelination and adipocyte defects characteristic of Lpin1fld/fld mice, which carry a null allele for Lpin1. Furthermore, biochemical, histological, and molecular analyses revealed that these lesions improve in older Lpin11Hubr rats as compared with young Lpin11Hubr rats and Lpin1fld/fld mice. We observed activation of compensatory biochemical pathways substituting for missing PAP1 activity that, in combination with a possible non-enzymatic Lipin 1 function residing outside of its PAP1 domain, may contribute to the less severe phenotypes observed in Lpin11Hubr rats as compared with Lpin1fld/fld mice. Although we are cautious in making a direct parallel between the presented rodent model and human disease, our data may provide new insight into the pathogenicity of recently identified human LPIN1 mutations.
Keywords: Enzyme Inactivation, Lipodystrophy, Mutant, Neurological Diseases, Rat, Lipin 1, Myelin, Peripheral Neuropathy, Phosphatidate Phosphatase, Schwann Cells
Introduction
Lipin 1 plays a crucial role in lipid metabolism in multiple cell types including glia, adipocytes, myocytes, and hepatocytes (1–4). Previous studies have shown that Lipin 1 is a Mg2+-dependent phosphatidate phosphatase (PAP1)5 enzyme catalyzing the dephosphorylation of phosphatidic acid (PA), yielding inorganic phosphate and diacylglycerol needed for the synthesis of triacylglycerol, phosphatidylcholine, and phosphatidylethanolamine (5–8). The mammalian Lipin protein family consists of three members, Lipin 1, Lipin 2, and Lipin 3 (9), which are encoded by transcripts with specific tissue expression patterns (5). Lpin1 is expressed at high levels in white adipose tissue (WAT) and muscle (5), and is also present in other tissues including peripheral nerve (2). Alternative splicing of Lpin1 generates two Lipin 1 isoforms that play distinct, but complementary, roles in adipogenesis: Lipin 1α, which affects adipocyte differentiation, and Lipin 1β, which induces lipogenic gene expression (10). The amino-terminal and carboxyl-terminal regions of Lipin 1 (NLIP and CLIP, respectively), and a predicted nuclear localization signal are highly conserved among the three mammalian Lipin family members and among species (9). The CLIP domain contains multiple key protein functional domains: four haloacid dehalogenase motifs and a transcription factor-binding motif (LXXIL) (1, 6, 11). Moreover, Lipin 1 and Lipin 2 have been predicted to possess the same structural organization as previously characterized HAD protein family members (11). In addition to its enzymatic function, Lipin 1 also has the ability to regulate gene expression, and it is likely that this ability is shared by Lipin 2 and Lipin 3 (1, 12, 13).
Overexpression of Lpin1 in either adipose tissue or skeletal muscle promotes obesity when mice are fed a high-fat (adipose and muscle tissue) or chow diet (muscle tissue) (4). On the other hand, a spontaneous null mutation of Lpin1 present in fatty liver dystrophy (Lpin1fld/fld) mice results in lipodystrophy, a progressive demyelinating neuropathy affecting peripheral nerves, and a neonatal fatty liver that resolves at weaning (9, 14). Recently, mice with a Schwann cell (SC)-specific deletion of Lpin1 were created resulting in SC abnormalities and a neuropathy similar to the full Lpin1-knock-out Lpin1fld/fld mice (2). This indicated that the neuropathy present in Lpin1fld/fld mice is a direct consequence of the absence of Lipin 1 within SCs. Moreover, SC-specific deletion of Lpin1 also revealed an interaction between PA and the MEK-ERK signaling pathway mediating SC de-differentiation and proliferation (2). In addition, mutant mice (Lpin120884) bearing a tyrosine to asparagine missense mutation (Y873N) were recently described (15). Lpin120884 mice develop adult-onset transitory hindlimb paralysis, a floppy gait, and a tendency to clench the hind limbs in toward the body when suspended by the tail. These phenotypes are, however, less severe as compared with Lpin1fld/fld mice, probably due to partial retention of PAP1 activity in Lpin120884 mice.
Here we describe a Lpin1 mutant allele in the rat (hereafter Lpin11Hubr) characterized by early-onset transitory peripheral neuropathy, muscle wasting, and lipodystrophy. The Lpin11Hubr mutation resulted in out-of-frame transcription leading to disruption of the Lipin 1 HAD domain IV. This resulted in complete inactivation of PAP1 activity, whereas preserving the function of the remainder of the Lipin 1 protein. Our data suggest that the age-related improvement of the Lpin11Hubr phenotypes may relate to Lipin 1 function that is independent of its PAP1 activity and/or to the activation of compensatory biochemical pathways partially substituting for the lack of PAP1 activity. In addition to its importance for the understanding of the role of Lipin 1 in lipid metabolism, the generated Lpin11Hubr rat model may provide an interesting model to study the pathophysiology of recently described human LPIN1 mutations.
EXPERIMENTAL PROCEDURES
Animal Housing
The Animal Care Committee of the Royal Dutch Academy of Science and the University of Lausanne approved all experiments according to Dutch and Swiss legal ethical guidelines. Lpin11Hubr pups were obtained at the expected Mendelian frequency, and the phenotype was not gender specific. If more than 8 littermates were present and survival of Lpin11Hubr pups could be compromised, wild-type/heterozygous pups were removed as soon as the Lpin11Hubr phenotype could be observed, until a total of 8 pups remained. Chow pellets were also added to the maternal cage floor to increase feeding chances of Lpin11Hubr pups. After weaning (around postnatal day (PND) 21), two rats were housed together, unless noted otherwise, under controlled experimental conditions (12 h light/dark cycle, light period 0600–1800, 21 ± 1 °C, ∼60% relative humidity). The standard fed diet in our animal facility (semi-high protein chow: RM3, 26.9% protein, 11.5% fat, and 61.6% carbohydrates; 3.33 kcal/g of AFE; SDS, Witham, United Kingdom) was provided ad libitum together with water.
Identification of Lpin11Hubr Rats
Following an N-ethyl-N-nitrosourea (ENU) mutagenesis screen (16), an accidental brother-sister mating between two F3 animals (Wistar/Crl background; backcrossed twice to wild-type Wistar background) revealed offspring that could be identified approximately between PND 7 and 14 by hindlimb paralysis and hindquarter muscle wasting.
Forward Screen, Sequencing, and Animals
For the mapping cross, the two initial F3 carriers of the Lpin11Hubr mutation were outcrossed with Brown Norway (BN/Crl) rats. F4 Wistar/BN rats were intercrossed to identify them as carriers of the mutation and to generate offspring homozygous for the mutation. After onset of the phenotype, DNA was isolated from F5 wild-type/heterozygous and homozygous Wistar/BN offspring. DNA was processed as described before (17), and a previous described Wistar/BN SNP panel (17) was used to map the phenotype in F5 Wistar/BN rats. In total, the SNP distribution pattern for 67 mutant Wistar/BN F5 rats was determined. Primers were designed to cover each exon with partial intron overhang, and sequencing of Lpin1 in 4 wild-type/heterozygous and 4 mutant pups revealed a point mutation (Thr > Ala) in the conserved 5′-end splice site of intron 18. In addition to the cross of BN/Crl animals, the initial F3 carriers of the Lpin11Hubr mutation were also backcrossed to wild-type Wistar/Crl animals for 4 additional generations to eliminate confounding effects from background mutations induced by ENU. F7 Wistar/Crl rats were used in all described experiments except for the mapping screen.
Genotyping
Genotyping was done using the KASPar SNP Genotyping System (KBiosciences, Hoddesdon, United Kingdom; as described by Ref. 18) using gene-specific primers (forward common, GAGCC CTTTT ATGCT GCTTT TGGGA A; reverse wild-type, GAAGG TGACC AAGTT CATGC TCCAC TGACC TCCTC GGGTA; reverse homozygous, GAAGG TCGGA GTCAA CGGAT TCTCC ACTGA CCTCC TCGGG TT). All pups were genotyped around PND 21. Genotypes were reconfirmed when experimental procedures were completed.
Quantitive RT-PCR
For all analyses, total RNA from complete sciatic nerve, endoneurium, and WAT were isolated using the Qiagen RNeasy lipid tissue kit (Qiagen) following the manufacturer's instructions. Total RNA from muscle, brain, liver, and rat primary Schwann cells was isolated in TRIzol (Invitrogen) reagent and purified with the RNeasy kit (Qiagen). RNA quality was verified by agarose gel and/or by the Qiaxcel capillary electrophoresis device (Qiagen), and the concentration was determined by an ND-1000 spectrophotometer (NanoDrop). Total RNA (250–500 ng) was subjected to reverse transcription using the SuperScript III First-strand Synthesis System for RT-PCR (Invitrogen) following the manufacturer's instructions. The resulting cDNA was used as a template for relative quantitative RT-PCR as described previously (19). Results were normalized using the reference gene Ubiquitin. See supplemental Table S1 for a complete list of oligonucleotides used for relative gene expression.
Immunohistochemistry
The sciatic nerves were dissected and embedded in OCT medium (Sakura) and longitudinal or cross-sections were prepared. Prior to specific staining, the slides were post-fixed in 4% paraformaldehyde for 10 min. For immunohistochemistry, slides were briefly (30 s) washed in PBS and blocked for 1 h using 10% normal donkey or goat serum (DAKO) and 0.3% Triton X-100 (Merck) in PBS (PBS-T). Slides were incubated overnight at 4 °C with primary antibodies diluted in PBS-T. The following primary antibodies were used: Lipin 1 (rabbit; 1:100; described below), Krox20 IgG (rabbit, 1:100; Covance), and Oct6 IgG (goat, 1:100; Abcam). The slides were then washed in PBS-T and hybridized with the appropriate secondary fluorescent antibodies (Alexa Fluor 594 or 488 conjugated anti-rabbit and mouse; all at a dilution of 1:200; Invitrogen) for 2 h at room temperature. Slides were washed in PBS-T and mounted with Vectashield mounting medium containing DAPI to counterstain cell nuclei (Vector Laboratories). Sections were visualized as described above.
BrdU Incorporation Assay
At PND 21, rats were injected intraperitoneally with 100 μg of BrdU/g of body weight. Six hours later, the sciatic nerves were dissected, fixed in 4% paraformaldehyde for 24 h, and embedded in OCT medium. Longitudinal cryosections were prepared, post-fixed in 4% paraformaldehyde (10 min), denatured with 2 m HCl for 20 min at 37 °C, and neutralized in 0.1 m sodium borate (pH 8.5) for 10 min. Sections were incubated with rat anti-BrdU (at a 1:200 dilution; Abcam) in 0.3% Triton X-100 overnight at 4 °C. The next day, the sections were incubated with anti-rat secondary antibody conjugated to Alexa Fluor 594 (at a 1:200 dilution; Invitrogen) and visualized by fluorescence microscopy. The nuclei were counterstained with DAPI.
WAT Morphology
Rats of both genders were sacrificed at PND 4 or 10, and male rats were sacrificed at PND 21 or 90 by decapitation after measuring body weight. The left and right dorsal subcutaneous WAT fat pads were subsequently harvested and weighed. WAT samples were then collected in 4% formaldehyde, rotated overnight at 4 °C, rinsed twice with 100% ethanol for 2 h, left in xylene overnight at RT, and embedded in paraffin. The tissue was then cut into 5-μm sections and stained with hematoxylin and eosin. Average adipocyte cell diameter was measured using NIH ImageJ freeware.
Western Blotting
Cells were lysed in ice-cold lysis buffer (20 mm Na2H2PO4, 250 mm NaCl, Triton X-100 1%, SDS 0.1%) supplemented with Complete protease inhibitors (Roche Applied Science). Protein levels were quantified using the Bio-Rad protein assay with BSA as a standard. Equal amounts of protein extracts were resolved by 10% SDS-PAGE and electrotransferred onto a polyvinylidene difluoride (PVDF) membrane (Amersham Biosciences). Blots were blocked in Tris-buffered saline containing 0.1% Tween (TBS-T) supplemented with 4% milk powder and subsequently incubated overnight at 4 °C in the same buffer supplemented with antibodies against the HA tag (Cell Signaling) and Tubulin (Sigma). After washing in TBS-T, blots were exposed to the appropriate horseradish peroxidase-conjugated secondary antibodies (DAKO) in TBS-T for 1 h at room temperature. Finally, the blots were developed using ECL reagents (Pierce) and Kodak Scientific Imaging Films (Kodak).
Microdissection of Sciatic Nerve
Sciatic nerves were harvested at PND 16, 21, 90, or 200–300 and placed in ice-cold PBS (pH 7.4). The perineurium and epineurium were gently dissected away from the endoneurium along the whole length of the nerve as previously described (3).
PAP Activity Measurement
PAP activity measurement was performed as previously described (2). Tissue samples were disrupted using a Dounce homogenizer at 4 °C in 50 mm Tris-HCl (pH 7.5) buffer containing 0.25 m sucrose, 1 mm EDTA, 10 mm β-mercaptoethanol, 1 mm benzamidine, 0.5 mm PMSF, 5 μg/ml of aprotinin, leupeptin, and pepstatin. The lysed cells were centrifuged at 1,000 × g for 10 min at 4 °C, and the supernatant was used as cell extract. Total PA phosphatase activity (Mg2+-dependent and Mg2+-independent) was measured at 37 °C for 20 min in the reaction mixture (total volume of 100 μl) containing 50 mm Tris-HCl (pH 7.5), 1 mm MgCl2, 10 mm β-mercaptoethanol, 0.2 mm [32P]PA (5,000 cpm/nmol), 2 mm Triton X-100, and enzyme protein. The Mg2+-independent PA phosphatase activity was measured in the same reaction mixture except that 2 mm EDTA was substituted for 1 mm MgCl2. The Mg2+-dependent PA phosphatase activity was calculated by subtracting the Mg2+-independent enzyme activity from total enzyme activity. One unit (U) of PA phosphatase activity was defined as the amount of enzyme that catalyzed the formation of 1 nmol of product/min. Specific activity was defined as units/mg protein.
Phosphatidic Acid Quantitation
PA was quantified as previously described (2). Tissues (the endoneurium from two nerves and about 300 mg for adipose tissue) were homogenized in 1.5 ml of PBS containing 0.5 mm sodium orthovanadate (Sigma) and extracted twice with 1 volume of butanol. After evaporation, phospholipids were solubilized in 1 ml of PBS containing 1% BSA and 0.5 mm sodium orthovanadate. An aliquot of the solution was incubated for 90 min at 37 °C with or without bovine pancreatic phospholipase A2 (3.8 units/ml, from Sigma). At the end of the incubation, phospholipids were extracted with butanol, dried, and LPA was quantified. The amount of PA corresponds to the amount of LPA detected after treatment with phospholipase A2 after subtraction of the amount of LPA detected without phospholipase A2 treatment. The assay was performed in triplicate for each sample.
Lipid Analysis
Lipids were extracted from sciatic nerve endoneurium derived from rats at PND 16 or PND 200–300 rats according to the method of Bligh and Dyer (20). Phospholipids were separated on silica gel high performance TLC plates (Merck, Darmstadt, Germany) using a solvent system consisting of chloroform, methanol, acetic acid, formic acid, and water (75:32:12.5:4:1, v/v) (21). Lipid spots were visualized by exposure to iodine vapor, and phosphorus was determined according to Rouser et al. (22). Because of incomplete resolution of phosphatidylinositol (PI) and phosphatidylserine (PS), the phosphorus content of the combined spots was measured and the contribution of each of the two lipid classes was determined using negative mode precursor scanning of m/z 241 atomic mass units (PI) and neutral loss scanning of 87 atomic mass units (PS), respectively, on an AB Sciex (Foster City, CA) 4000 Qtrap MS instrument. A mixture of authentic PI and PS standards was used to correct for ionization and fragmentation efficiencies (23). Cholesterol was quantified as described previously, using deuterated cholesterol (Avanti Polar Lipids) as an internal standard (24).
Toluidine Blue Staining and Morphometric Analysis
Semi-thin nerve cross-sections were stained with 1% toluidine blue and digitalized using AxioVision release 4.5 software (Zeiss). For each myelinated axon present, both an axonal area (defined by the inner limit of the myelin sheath) and a total fiber area (defined by the outer limit of the myelin sheath) were automatically measured using image analysis software (G-ratio calculator 1.0, Image J plug-in, Yannick Krempp, Cellular Imaging Facility, Lausanne, Switzerland). The g-ratio was calculated by dividing the axon area by the total fiber area. Each experimental group consisted of two rats.
Nile Red Staining
Sciatic nerve cross-sections were mounted in Nile Red (Sigma) solution (0.5 mg/ml in acetone) diluted 1,000 times in 75% glycerol, and visualized using a Zeiss Axioplan 2 microscope with an AxioCam MRc camera and AxioVision release 4.5 software (Zeiss).
Lipin 1 Antibody
A rabbit anti-Lipin 1β antibody was generated with a peptide (PSGSRPSTPKSDSEL) specific for the murine Lipin 1β isoform (Eurogentec) and used at 1:100 for immunohistochemistry.
cDNA Constructs
Constructs used in this study were generated using murine and rat Lipin 1α and 1β cDNAs with an N-terminal hemagglutinin (HA) epitope tag. The original mouse constructs were provided by Dr. Thurl Harris (University of Virginia, Charlottesville, VA) and the rat constructs (wild-type and truncated forms) were purchased as mini-genes in pcDNA3.1 from Integrated DNA Technologies (IDT, Coralville, IA).
Cell Culture and Transfections
HEK293 cells were maintained in DMEM supplemented with 10% fetal bovine serum, 100 units/ml of penicillin, 100 μg/ml of streptomycin, and 2 mm l-glutamine (all reagents were from Invitrogen) cultured in a humidified 37 °C incubator with 5% CO2. Plasmid transfections were performed using Effectene (Qiagen), according to the manufacturer's instructions.
For the luciferase assay, rat Schwann cells were seeded in 6-well PRIMARIA (BD Biosciences) dishes. At 70–80% confluence, the cells were transfected in triplicate using FuGENE (Roche Applied Science) in the presence of 3% FCS, 5% Neu-differentiation factor β-conditioned medium and 2 μm forskolin. The next day, the cells were switched to serum-free medium (DMEM/F-12, 1× N2, 1× PS, 10 ng/ml of NGF, 1× BSA, 5% Neu-differentiation factor β conditioned medium) and incubated for 18 h. The next day, the cells were induced to differentiate for 36 h in induction medium (100 μm cAMP, DMEM/F-12, 1× N2, 1× PS, 10 ng/ml of NGF, 1× BSA, 5% Neu-differentiation factor β-conditioned medium) and used for luciferase assays. HeLa cells were plated in 12-well dishes (Greiner-bio one) containing DMEM, 10% FCS, and 1% PS. At 90% confluence they were transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. DNA quantity was kept constant using pBS empty vector. 36 h post-transfection, the cells were washed and lysed in 1× reporter lysis buffer (Promega). The extracts were measured for β-galactosidase (β-gal) activity using 2-nitro-phenyl-galactopyranoside as substrate. Luciferase activity was measured on a luminometer (PerkinElmer Life Sciences) using the Steady Glo luciferase assay substrate (Promega) and values were normalized to β-gal activity.
Immunofluorescence
HEK293 cells were grown as described above and transfected with plasmids for expression of rat Lipin 1 proteins (wild-type and truncated forms). One day after transfection, the cells were fixed with 4% formaldehyde for 30 min and permeabilized for 15 min with 0.1% Triton X-100. The cells were blocked with PBS containing 10% normal goat serum for 1 h before incubation with a primary antibody against the HA tag (Cell Signaling, 1:200) overnight at 4 °C. After washing in PBS-T, the cells were incubated with the appropriate secondary fluorescent antibody (Alexa Fluor 594) for 1 h in the dark followed by washing with PBS-T. The cells were mounted with Vectashield mounting medium containing DAPI to counterstain cell nuclei (Vector Laboratories) and visualized by confocal microscopy (Leica TCS SP5).
Data Analysis
All data are shown as mean ± S.E. All data were analyzed using a commercially available statistical program (SPSS for Macintosh, version 16.0) and were controlled for normality and homogeneity. Differences in longitudinal body weight measurements were assessed using repeated measure analysis, followed by Bonferroni post hoc analyses if significant overall interactions were observed. The statistical analysis for the lipid analysis was performed using the Prism GraphPad version 5.00 (GraphPad Software, San Diego, CA), and lipid analysis was assessed by two-way analysis of variance, using age and mutation as factors, followed by a Bonferroni post hoc analysis if a significant overall interaction was observed. The null hypothesis was rejected at the 0.05 level.
RESULTS
The Identification of the Lpin11Hubr Mutation in Rat
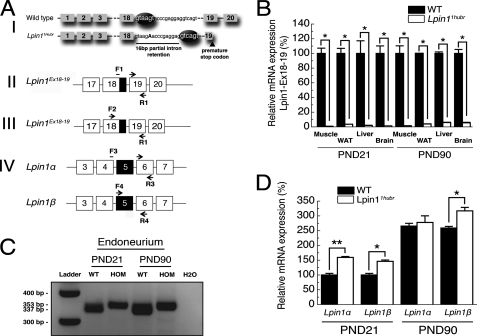
After an ENU mutagenesis screen on a Wistar/Crl background (16), a brother-sister mating (F3 generation) revealed offspring that could be identified approximately between PND 7 and 14 by difficulty walking, muscular atrophy, and a reduced amount of body fat (supplemental Fig. S1, A and B, and supplemental Video S1). Further evaluation of older rats (PND 28) revealed, in addition to the above mentioned phenotypes, presence of loss of hindlimb joint mobility, the absence of characteristic retraction of the hindlimbs when picked up by the neck, and the inability to splay hindlimbs when suspended by the tail (supplemental Fig. S1, C and D). The Wistar/Crl F3 rats heterozygous for the mutation were outcrossed with BN/Crl rats to generate an F4 Wistar/BN population for mapping purposes. On PND 21, DNA was collected from wild-type/heterozygous and mutant Wistar/BN F5 rats, and a forward screen was performed using a Wistar/BN-specific SNP marker panel (17). This resulted in the identification of a 3.6 Mb region on chromosome 6 between 37.1 and 40.7 Mb, resulting in the identification of Lpin1 (Chr.6; 40.3 Mb) as a prime candidate gene (supplemental Fig. S2). Sequencing of Lpin1 revealed a mutation (T>A) in the conserved 5′-end splice site of intron 18 potentially leading to the use of an alternative splice site 16 bp downstream of the original site. The new splice site was predicted to result in a change in reading frame introducing a premature stop codon in exon 19 (Fig. 1A, panel I). We have named this mutated allele of the rat Lpin1 gene Lpin11Hubr.
FIGURE 1.
The Lpin11Hubr mutation disturbs exon 18–19 splicing. A, panel I, ENU mutagenesis introduced a T>A mutation (indicated in capital bold) in intron 18 of Lpin11Hubr rats. The mutation disrupted a splice site motif (GTXAGT), creating a new splice site downstream and resulting in a 16-bp partial intron retention. Black ellipses indicate a splice motif. The new reading frame resulted in a premature stop codon in exon 19. B, relative quantification of the expression of the wild-type transcript in Lpin11Hubr rats at PND 21 and PND 90 (*, p < 0.001; n = 2 per group; primers are shown in A, panel II; black part in primer overview indicates the 16-bp intron retention). C, qualitative analysis of Lpin1 expression revealed the expression of a mutated allele (353 bp) without any residual wild-type allele (337 bp) expression in sciatic nerve tissue of Lpin11Hubr rats as compared with wild-type rats at PND 21 and PND 90 (primers are shown in A, panel III; black part in the primer overview indicates a 16-bp intron retention). H2O was used as a control. D, relative expression of Lpin1α and Lpin1β is increased in sciatic nerve tissue of Lpin11Hubr rats as compared with wild-type rats at PND 21. At PND 90, relative expression of Lpin1α and Lpin1β is unchanged and increased (respectively) in sciatic nerve tissue of Lpin11Hubr rats as compared with wild-type rats (*, p < 0.05; **, p < 0.001; n = 2 per group; primers are shown in A, panel IV). Exon 5 (shown in black in primer overview) is included in Lpin1β, but is absent in Lpin1α. Data are expressed as mean ± S.E.
Consequences of Lpin11Hubr Mutation on Lipin 1 Expression and Function
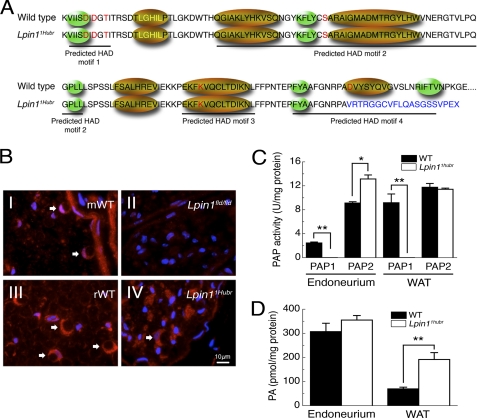
To verify these predictions, we evaluated the correct splicing of intron 18 using primers spanning the wild-type exon 18–19 boundary (Lpin1Ex18–19; Fig. 1A, panel II). The expression of correctly spliced Lpin1Ex18–19 was almost undetectable in muscle, WAT, liver, and brain samples of Lpin11Hubr rats at PND 21 or 90 (Fig. 1B). RT-PCR using primers spanning the exon 18–19 border (primers shown in Fig. 1A, panel III) failed to show any residual wild-type expression in mutant sciatic nerve endoneurium (Fig. 1C). Furthermore, expression of the Lpin1α and Lpin1β isoforms (10) was investigated in sciatic nerve endoneurium (Fig. 1D, primers shown in Fig. 1A, panel IV). The expression of both Lpin1 isoforms was increased in Lpin11Hubr rats as compared with wild-type rats at PND 21. Later in development, at PND 90, only Lpin1β expression was increased in Lpin11Hubr rats as compared with wild-type rats (Fig. 1D). Taken together these data therefore suggested the absence of nonsense-mediated decay (NMD) of the mutated Lpin11Hubr transcript. We further validated the absence of NMD by immunofluorescence analysis of the localization of Lipin 1 in sciatic nerves of wild-type and Lpin11Hubr rats. Using a Lipin 1 antibody we detected the presence of Lipin 1 in sciatic nerve endoneurium of both adult (PND 90) wild-type and Lpin11Hubr rats (Fig. 2B). Importantly, this staining was lost in Lpin1fld/fld mice in which the Lipin 1 protein is absent (Fig. 2B). In silico analysis suggested that the Lpin11Hubr mutation disrupts the conserved amino acid in the predicted HAD motif IV (wild-type, GNRPAD; Lpin11Hubr, GNRPAV, Fig. 2A). Importantly, the terminal acidic residue, aspartic acid of motif IV, along with those in motif I, are required for positioning of the Mg2+ ion into the active site (reviewed in Ref. 25). The disruption of the Mg2+ binding site therefore potentially leads to inactivation of Lipin 1 PAP activity. We have investigated PAP activity in Lpin11Hubr rats and observed that although we can detect the presence of mutated Lipin 1 protein in Lpin11Hubr rats, its PAP1 activity was completely lost in both sciatic nerve endoneurium and WAT (Fig. 2C). Interestingly, the level of PAP2, which has a similar enzymatic activity (26), was up-regulated in endoneurial samples from Lpin11Hubr rats. We then assessed the level of the substrate of the PAP1 enzyme, PA, in sciatic nerve and WAT isolated from Lpin11Hubr rats as compared with wild-type rats. Unlike in Lpin1fld/fld sciatic nerve (2), we found no significant differences in the concentration of PA in Lpin11Hubr sciatic nerves. However, the amount of PA in the WAT from Lpin11Hubr was three times higher as compared with wild-type rats (Fig. 2D).
FIGURE 2.
The Lpin1 PAP1 activity is disrupted in Lpin11Hubr rats. A, amino acid sequence showing the four predicted HAD motifs, the conserved amino acids from the HAD family of proteins are indicated in red letters, the transcription coactivator motif LXXIL in yellow letters, green ellipses indicate predicted β-strands, and gold ellipses indicate predicted α-helices (11). The partial intron retention disrupts the conserved amino acid in the predicted HAD motif IV (WT, GNRPAD; Lpin11Hubr, GNRPAV). Moreover, the out-of-frame translation (indicated in blue) disrupts the predicted α-helix and the second predicted β-strand of the predicted HAD motif IV. B, immunolabeling of Lipin 1 (red) and 4′,6-diamidino-2-phenylindole (DAPI; blue) in cross-sections of sciatic nerve derived from wild-type (mWT; B, panel I) or Lpin1fld/fld mice at PND 56 (B, panel II), and derived from wild-type (rWT; B, panel III), or Lpin11Hubr rats (B, panel IV) at PND 90. The endoneurial part of the control nerves shows the expression of Lipin 1 in myelinating Schwann cells (red-stained croissant-shaped cells indicated by white arrows). This labeling is completely absent in Lpin1fld/fld mice but partially preserved in Lpin11Hubr rats. C, PAP1 activity is substantially decreased in sciatic nerve endoneurium and WAT of Lpin11Hubr rats as compared with wild-type rats at PND 21. PAP2 activity is increased in sciatic nerve endoneurium of Lpin11Hubr rats as compared with wild-type rats, whereas no significant difference in WAT PAP2 activity was observed between genotypes (*, p < 0.005; **, p < 0.001; n = 5–6 per group). D, at PND 21, PA levels were unchanged in sciatic nerve and increased in WAT tissue of Lpin11Hubr rats as compared with wild-type rats (*, p < 0.001; n = 6 per group). Data are expressed as mean ± S.E.
Sciatic Nerve Abnormalities in Lpin11Hubr Rats
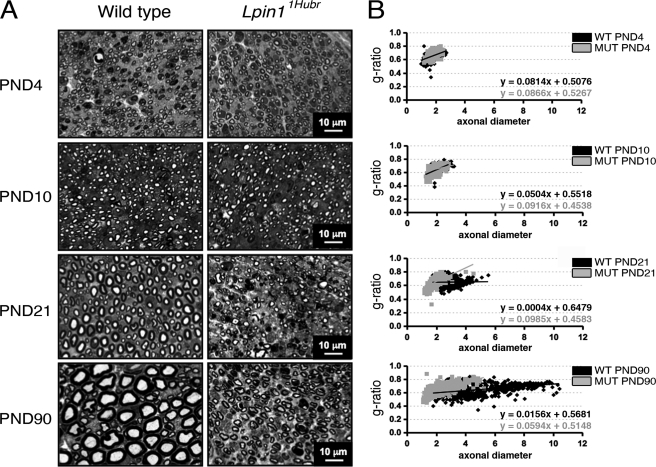
Null deletions of Lpin1 in mice result in severe sciatic nerve myelin abnormalities (2, 14), whereas Lpin120884 mutant mice show a less severe demyelination phenotype (15). We therefore investigated sciatic nerve morphology using toluidine blue-stained semi-thin nerve sections at PND 4, 10, 21, and 90. At PND 4 and 10, no clear sciatic nerve myelin abnormalities were observed between genotypes (Fig. 3A). This observation was strengthened by an equal axon diameter and g-ratio at PND 4 and 10 (Fig. 3B). At PND 21, Lpin11Hubr rats showed hypomyelination, a decreased axon diameter, and the presence of dark-colored debris (Fig. 3, A and B). Although all these defects were still present at PND 90, overall sciatic nerve morphology in Lpin11Hubr rats at this late developmental time point showed an improved phenotype resembling sciatic nerve morphology of wild-type rats at PND 10 or 21. Thus, sciatic nerve morphology was perturbed in Lpin11Hubr rats consistent with the finding that Lpin1 is critical for normal SC function (2). However, contrary to mice with a null mutation of Lpin1, in Lpin11Hubr rats the aberrant SC phenotype seemed to attenuate over time. The fact that Lpin11Hubr rats regained the ability to use their hind limbs when walking, although still with a floppy gait, and regained the ability to splay hindlimbs when picked up by the tail supported this observation (supplemental Fig. S1 and Videos S1 and S2).
FIGURE 3.
Progression of myelination in Lpin11Hubr rats. A, toluidine blue-stained semi-thin sections from the medial region of the sciatic nerve of Lpin11Hubr and wild-type rats at PND 4, 10, 21, and 90. At PND 4 and 10, the level of myelination is similar in wild-type and mutant nerve. At PND 21 and 90, hypomyelination is visible in sciatic nerves of Lpin11Hubr rats compared with wild-type nerves. At PND 90, the general morphology of Lpin11Hubr sciatic nerves is improved as compared with the general morphology of sciatic nerves isolated from Lpin11Hubr rats at PND 21. B, at PND 4 and 10, average g-ratio, g-ratio trend line, and average axonal diameter of medial sciatic nerve tissue is equal between genotypes. At PND 21 and PND 90, axons of Lpin11Hubr rats (MUT) show a decreased axonal diameter as compared with wild-type (WT) axons, and an aberrant g-ratio trend line indicating hypomyelination. Axons of Lpin11Hubr rats, however, show an improvement in trend line at PND 90 as compared with PND 21, indicative of partial myelination recovery (PND 4, 10, 21, and 90; n = 2, n = 2, n = 2, and n = 1 per group, respectively; n = 107–374 axons per genotype).
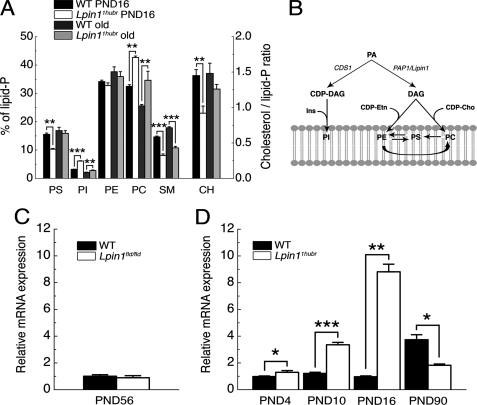
Next, we examined the amplitude of myelin perturbation in Lpin11Hubr rats as compared with Lpin1fld/fld mice, which completely lack Lipin 1 function. Similar to previously reported changes in Lpin1fld/fld sciatic nerves (14), thin-layer chromatography analysis performed on lipids extracted from sciatic nerves isolated from young (PND 16) and adult (between PND 200 and 300) Lpin11Hubr rats revealed major changes in PS, PI, phosphatidylcholine, sphingomyelin, and cholesterol (Fig. 4A). But in contrast to observations in Lpin1fld/fld mice, Lpin11Hubr rats exhibited age-dependent increases in PS levels (time × genotype interaction: F(1,12) = 7.0; p < 0.05), age-dependent decreases in PI levels (time × genotype interaction: F(1,12) = 61.0; p < 0.001), and a tendency for cholesterol levels to increase over time (time × genotype interaction: F(1,12) = 2.3; p = 0.16). This suggested that the sciatic nerve myelin lipid composition of Lpin11Hubr rats improved with age.
FIGURE 4.
Sciatic nerve lipid composition in the Lpin11Hubr rat. A, the amounts of different lipid classes (see B for legend) as percentage of the total amount in lipid extracts derived from control (WT) and Lpin11Hubr rats at PND 16 or between PND 200 and 300 (“old”; n = 4). B, schematic overview of one part of membrane lipid synthesis in mammals (adapted from Ref. 27). DAG, diacylglycerol; Ins, inositol; CDP-Etn, cytidine diphosphate ethanolamine; CDP-Cho, cytidine diphosphate choline; Cds1, CDP-diacylglycerol synthase gene; CH, cholesterol. C, relative gene expression of Cds1 in sciatic nerve tissue of Lpin1fld/fld mice as compared with wild-type mice at PND 56 (n = 4). D, relative gene expression of Cds1 in sciatic nerve tissue of Lpin11Hubr rats as compared with wild-type rats at PND 4, 10, 16, and 90 (n = 3, n = 3, n = 3, and n = 2 per group, respectively, *, p < 0.05; **, p < 0.01; ***, p < 0.0001). Data are expressed as mean ± S.E.
Phosphatidic acid in mammalian cells is metabolized via two pathways: the cytidine diphosphate-diacylglycerol pathway and the Kennedy pathway (Fig. 4B) (8, 27). In the cytidine diphosphate-diacylglycerol pathway, CDS1 catalyzes the synthesis of cytidine diphosphate-diacylglycerol, which serves as a precursor of PI. Consistent with the observed accumulation of PI especially at PND 16, we observed elevated levels of Cds1 mRNA in sciatic nerves derived from Lpin11Hubr rats between PND 4 and 16, whereas Cds1 expression was unchanged between wild-type and Lpin1fld/fld mice at PND 56 or even decreased in adult Lpin11Hubr rats (PND 90) as compared with adult wild-type rats (Fig. 4, C and D).
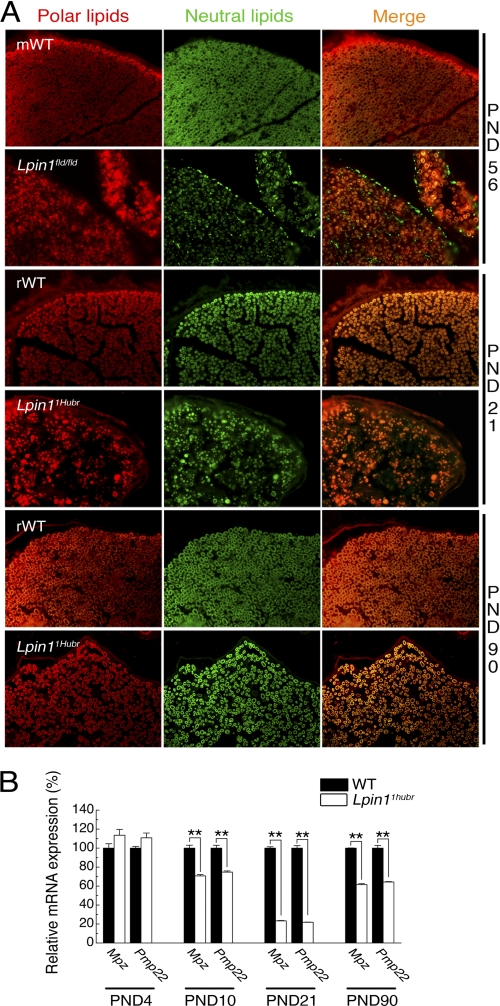
Nile Red staining, emitting a green fluorescence in the presence of neutral lipids (cholesterol, lipoproteins, and triglycerides) and a red fluorescence in the presence of polar lipids (phospholipids) (28) allowed us to determine that demyelinated sciatic nerves from Lpin1fld/fld mice accumulated neutral lipids in their perineurial and endoneurial compartments, which was rarely observed at PND 21 and not observed at PND 90 in sciatic nerves from Lpin1Hubr rats (Fig. 5A). In addition, the Nile Red staining confirmed the presence of hypomyelination at PND 21 in Lpin11Hubr rats, a phenotype that substantially improved in Lpin11Hubr rats at PND 90 (Fig. 5A).
FIGURE 5.
PNS nerves are hypomyelinated in Lpin11Hubr rats. A, Nile Red staining of sciatic nerve sections from wild-type (mWT) and Lpin1fld/fld mice at PND 56, and wild-type (rWT) and Lpin11Hubr rats at PND 21 and 90. Nile Red staining demonstrated that Lpin1fld/fld sciatic nerve specifically accumulates neutral lipids in the perineurium and endoneurium consecutive to demyelination. In the rat, at PND 21, the typical “donut”-like myelin structures present in wild-type sciatic nerve are replaced in Lpin11Hubr nerves by “dot”-like structures probably corresponding to the accumulation of neutral and polar lipids in SCs. Lpin11Hubr nerves partially recover the donut-like myelin staining at PND 90 suggesting the presence of hypomyelination rather than demyelination. B, relative gene expression analysis of Mpz and Pmp22 (mature SC markers) in sciatic nerve tissue of Lpin11Hubr rats as compared with wild-type rats at PND 4, 10, 21 and 90 (n = 3, n = 3, n = 3, and n = 2 per group, respectively, **, p < 0.001). Data are expressed as mean ± S.E.
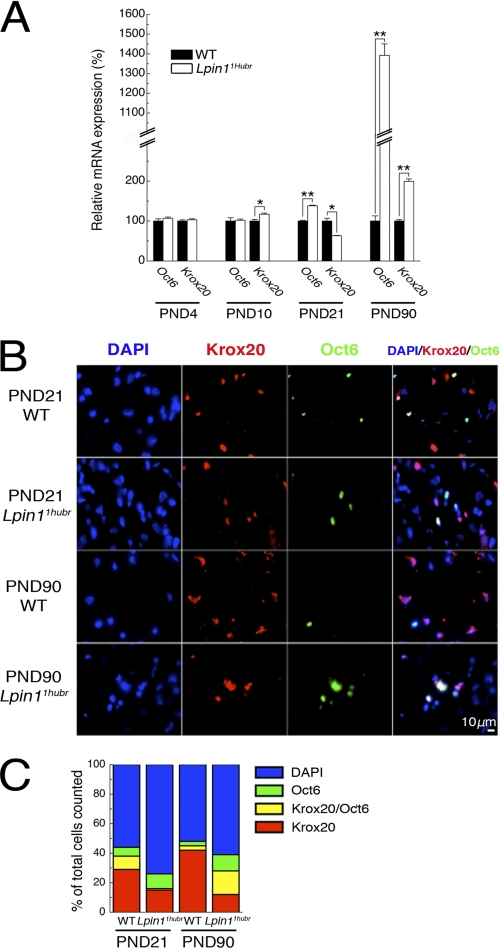
We further evaluated the level of myelin gene expression using a set of molecular markers. Mpz (also designated P0) and Pmp22, two transcripts encoding key myelin structural proteins, were expressed at equal levels between genotypes at PND 4, but were decreased in Lpin11Hubr rats as compared with wild-type rats at PND 10, 21, and 90 (Fig. 5B). These results were in agreement with the observed myelin phenotypes at PND 21 and 90 (Figs. 3 and 5A), and confirmed a relatively early onset (∼PND 10) of the myelin phenotype in Lpin11Hubr rats on a molecular level. Also, the relative increase in expression of both Mpz and Pmp22 at PND 90 as compared with PND 21 further confirmed partial recovery of the myelin phenotype in Lpin11Hubr rats. These observations led us to investigate expression of the POU domain transcription factor Pou3f1/Oct6/Scip/Tst-1 (hereafter Oct6), a marker of immature promyelinating SCs (29, 30), and the zinc finger transcription factor Krox20/Egr2 (hereafter Krox20), a marker of mature SCs (29), and an important controller of myelination. The level of expression of both of these markers was not significantly different between genotypes at PND 4 (Fig. 6A). Starting at PND 10, deregulation of the level of relative Krox20 expression became visible and was followed by an up-regulation of relative Oct6 expression starting at PND 21. At PND 90, both relative Oct6 and Krox20 expression was increased in Lpin11Hubr rats as compared with wild-type rats (Fig. 6A). This suggested the presence of a mixed SC population in PND 90 nerves. Immunostaining of wild-type and Lpin11Hubr sciatic nerves indeed revealed that wild-type and mutant nerves contained SCs at different stages. At PND 21, wild-type nerves contained a high number of myelinated (Krox20+, Oct6−) SCs in addition to early myelinating (Krox20+, Oct6+) and immature (Krox20−, Oct6+) SCs, whereas the large proportion of SCs in Lpin11Hubr nerves were immature (Krox20−, Oct6+) SCs. At PND 90, a substantial increase in the number of myelinated (Krox20+, Oct6−) cells was observed in wild-type nerves, whereas most SCs in Lpin11Hubr nerves were early myelinating (Krox20+, Oct6+) SCs (Fig. 6, B and C). Additionally, we observed an increased cellularity and increased number of replicating cells in sciatic nerve of Lpin11Hubr rats as compared with wild-type sciatic nerve at PND 90 (supplemental Fig. S3A). This, together with a maintained elevated level of Cyclin D1 expression in Lpin11Hubr rats as compared with wild-type rats at PND 21 and 90 (supplemental Fig. S3B), further confirmed that even at PND 90, Lpin11Hubr endoneurium contains immature and pro-myelinating SCs.
FIGURE 6.
Myelination status of Schwann cells in Lpin11Hubr peripheral nerve. A, relative gene expression analysis of Oct6 and Krox20 (SC markers) in sciatic nerve tissue of Lpin11Hubr rats as compared with wild-type rats at PND 4, 10, 21, and 90 (n = 3, n = 3, n = 3, and n = 2 per group, respectively; *, p < 0.05; **, p < 0.001). B, sciatic nerve cross-sections of wild-type (WT) and Lpin11Hubr rats at PND 21 and 90 were immunostained with antibodies against transcription factors Krox20 (red) and Oct6 (green). The nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI; blue). C, the percentage of DAPI+ (blue), Krox20+ (red), Krox20+ and Oct6+ (yellow), and only Oct6+ (green) cells is shown (n = 2 per group). Data are expressed as mean ± S.E.
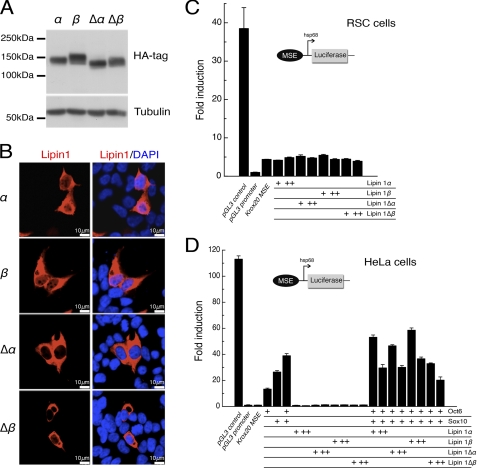
Lipin 1 was previously described to have a transcriptional coactivator activity (1), which could potentially participate in the observed increase of Krox20 gene expression in Lpin11Hubr sciatic nerve tissue at PND 90. We first examined the subcellular localization of wild-type and mutated forms of rat Lipin 1α and β in HEK293 cells (Figs. 7, A and B). Although rat Lipin 1α and Lipin 1β wild-type forms localized to the cytoplasm and the nucleus, the mutated rat Lipin 1Δα and Δβ forms preferentially localized to the cytoplasm (Fig. 7B). Using luciferase gene reporter assays, we found that both wild-type and mutated forms of mouse Lipin 1α and Lipin 1β did not influence Krox20 myelinating Schwann cell enhancer (MSE) (31) mediated activation of the hsp68 promoter in differentiating Schwann cells (Fig. 7C). In addition, whereas overexpression of Oct6 and Sox10 strongly activated the Krox20 MSE in HeLa cells, wild-type and mutated forms of Lipin 1α and/or Lipin 1β showed no or minimal effect (Fig. 7D). These data indicate that Lipin 1α or Lipin 1β (either alone or through Sox10 or Oct6) does not activate Krox20 transcription.
FIGURE 7.
Lipin 1α or Lipin 1β do not activate expression through the Krox20 MSE. A, HEK293 cells were transfected with vectors expressing rat Lipin 1α, 1β, or its mutated truncated forms, (Δα) and (Δβ), which were detected by Western blot using an anti-HA antibody. B, subcellular localization of HA-tagged rat Lipin 1 variants (red) was examined by confocal microscopy to compare the localization with the nuclear marker DAPI (blue). C and D, luciferase constructs containing the Krox20 myelination-associated enhancer (MSE), and driven by the hsp68 promoter, were co-transfected with expression plasmids encoding mouse Lipin 1α or Lipin 1β or its mutated truncated forms, Lipin 1Δα and Lipin1 Δβ (C), and/or Oct6 and Sox10 (D), in different cell types as indicated. Fold-induction was represented as the luciferase activity over the minimal hsp68 promoter. CMV enhancer/hsp68 promoter luciferase construct (pGL3 control) and the hsp68 promoter luciferase construct (pGL3 promoter) served as controls in these experiments (+, 100 ng/well; ++, 500 ng/well of Lipin 1α, Lipin 1β, Lipin 1Δα or Lipin1 Δβ; +, 200 ng/well of Oct6 and Sox10). Data are expressed as mean ± S.E.
WAT Morphology in Lpin11Hubr Rats
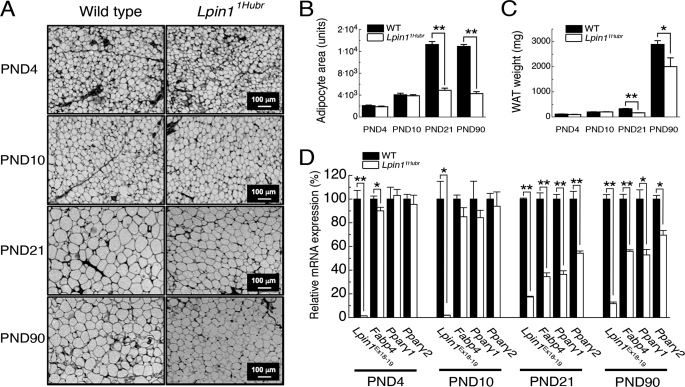
Lipodystrophy is one of the major phenotypes observed in models with null deletions of Lpin1 (9, 14). We have, therefore, evaluated the WAT phenotype in Lpin11Hubr rats. The body weight of Lpin11Hubr and wild-type rats was not significantly different until PND 10, but at PND 21 and onward, Lpin11Hubr rats showed a reduction in body weight as compared with wild-type rats (supplemental Fig. S4, A–E). WAT tissue analysis revealed a decreased adipocyte size, both at PND 21 and 90, whereas adipocyte cell size was equal between genotypes at PND 4 or 10 (Fig. 8, A and B). The decreased ability to store lipids at a later age was reflected by a reduction in adipose mass at PND 21 and 90 (Fig. 8C). To evaluate the WAT phenotype on a molecular level, we measured the relative expression of peroxisome proliferator-activated receptor-γ 1 and 2 (Pparγ1 and Pparγ2), key markers of adipocyte differentiation (32), and fatty acid-binding protein 4 (Fabp4, also designated aP2), which reversibly bind fatty acids and other lipids (33). In WAT derived from Lpin11Hubr and wild-type rats at PND 4 and 10, the expression level of these markers was not significantly different between genotypes (Fig. 8D). However, relative expression of Pparγ1, Pparγ2, and Fabp4 was decreased in Lpin11Hubr as compared with wild-type rats at PND 21 and 90 (Fig. 8D). As a control, we also evaluated the relative expression of the wild-type transcript Lpin1Ex18–19 in WAT of Lpin11Hubr rats as compared with wild-type rats at PND 4, 10, 21, and 90, and observed that the presence of normal splicing in Lpin11Hubr samples was dramatically reduced (Fig. 8D). Together, these data demonstrated an aberrant WAT phenotype in Lpin11Hubr rats, consistent with the finding that Lpin1 is strongly expressed in adipose tissue (9). However, similar to the above described phenotype in peripheral nerves, the WAT phenotype in Lpin11Hubr rats seems to attenuate over time, as reflected by higher expression of adipogenic factors at PND 90 as compared with PND 21 (Fig. 8D), and relatively increased adipose mass at PND 90 (51% at PND 21 and 69% at PND 90 in Lpin11Hubr rats as compared with wild-type rats).
FIGURE 8.
Lipodystrophy phenotype in Lpin11Hubr rats. A, dorsal subcutaneous adipose tissue sections prepared from wild-type and Lpin11Hubr rats at PND 4, 10, 21, and 90 stained with hematoxylin and eosin. Starting from PND 21 the reduced size of adipocytes became visible in Lpin11Hubr samples. B, average adipocyte area is not significantly different between genotypes at PND 4 and 10, but is decreased in Lpin11Hubr rats as compared with wild-type rats at PND 21 and 90 (n = 2 per group; n = 41–83 adipocytes per group; **, p < 0.001). C, dorsal subcutaneous WAT weight is equal between genotypes at PND 4 and 10 (n = 2 per group), but is decreased in Lpin11Hubr rats compared with wild-type rats at PND 21 (n = 2 per group) and PND 90 (n = 2–4 per group; *, p < 0.05; **, p < 0.001). D, relative gene expression analysis in WAT of Lpin1 (Lpin1Ex18–19) and genes involved in adipocyte differentiation (Fabp4, Pparγ1, and Pparγ2) in WAT isolated from wild-type and Lpin11Hubr rats (PND 4, n = 3; PND 10, n = 3; PND 21, n = 5; and PND 90, n = 2 per group, *, p < 0.05; **, p < 0.001). Data are expressed as mean ± S.E.
DISCUSSION
Here we describe a new Lpin11Hubr rat model that is characterized by a peripheral neuropathy, muscle wasting, and lipodystrophy. Importantly, the Lpin11Hubr mutation does not prevent the production of a truncated protein that lacks PAP1 enzymatic activity. Moreover, in contrast to the models with complete loss of Lipin 1 function, the deficits present in Lpin11Hubr rats become attenuated during adulthood, resulting in improvement of its phenotypes.
The Lpin11Hubr phenotype was initially discovered in F3 rats that were already outcrossed twice to wild-type Wistar/Crl rats. This highlights the importance of outcrossing ENU-mutagenized rats to clear the background of unwanted mutations. The F3 Wistar/Crl Lpin11Hubr mutation was backcrossed to the Wistar background for an additional 4 generations (6 outcrosses in total), which should theoretically decrease the total number of random background mutations to 1 (34). However, we cannot formally exclude the presence of tightly linked confounding mutations in Lpin11Hubr rats.
The Lpin11Hubr mutation disrupted the 5′-end splice site of intron 18. Although potentially deleterious for correct splicing, this mutation led to a small amount (1–6%) of Lpin1 transcript with a correctly spliced exon 18–19 boundary (Lpin1Ex18–19) in various tissues isolated from Lpin11Hubr rats at either PND 21 or 90. This is probably a consequence of the capacity of the splicing machinery to occasionally use the mutated splicing site (see for example, Ref. 35). Our data, however, clearly demonstrate that the attenuation of the different Lpin11Hubr phenotypes is not a consequence of an age-related increase of the presence of correctly spliced exon 18–19 boundary in Lpin11Hubr rats. Importantly, by using three different sets of primers (covering the boundary region between exons 18 and 19, and the region around exons 4–6, which can distinguish Lpin1α and Lpin1β isoforms), we detected the presence of mutated Lpin1 transcript in Lpin11Hubr rats. We even observed increased expression of both Lpin1α and Lpin1β isoforms in Lpin11Hubr rats at PND 21, suggesting a functional feedback loop affecting their expression. These data, together with our observation that the stop codon is induced in the 19th exon of 20 in total and close to the last exon-exon junction, suggested that the Lpin11Hubr mutation does not induce NMD. This is in agreement with previous observations suggesting that a mutation close to the 3′-end of a transcript does not necessarily lead to NMD (36).
The Lpin11Hubr mutation completely inactivated the Lipin 1 PAP1 activity in WAT and nerve tissue. Thus, whereas Lpin1 mRNA appears to be present in Lpin11Hubr rats, the Lpin11Hubr mutation disrupts the last HAD motif resulting in complete lack of PAP1 activity in the corresponding protein. This observation clearly distinguishes Lpin11Hubr mutant rats from the recently published Lpin120884 mouse mutant bearing a Y873N missense mutation in Lpin1, which results in partial reduction of PAP1 activity underlying a milder transient peripheral neuropathy (15). In addition, the murine mutant line 20884 also harbors a mutation in Nrcam, which is not the case for Lpin11Hubr rats (data not shown).
In our previous study (2), we have demonstrated that a decrease in PAP1 activity was followed by significant accumulation of PA in the endoneurium of both Lpin1fld/fld and Lpin1-SC conditional (MPZCre/+/LpfE2–3/fE2–3) mutant mice. Surprisingly, the Lpin11Hubr mutation does not provoke the same effect. We found no significant differences in the levels of PA in the sciatic nerve tissue derived from Lpin11Hubr. However, similar to the observation in Lpin1fld/fld mice the amount of PA in the Lpin11Hubr WAT was three times higher as compared with wild-type rats. This difference between Lpin1 null mice (Lpin1fld/fld, MPZCre/+/LpfE2–3/fE2–3) and the Lpin11Hubr rat model could be explained by a process of compensation that is tissue- and species-dependent. SCs may compensate the loss of PAP1 activity by PAP2 (26). Indeed, we observed that PAP2 activity is only increased in sciatic nerve endoneurium of Lpin11Hubr rats, whereas no significant difference was observed in WAT. Interestingly, the Lpin1fld/fld endoneurium and WAT showed no significant differences in PAP2 activity (2). Furthermore, the accumulation of PI, especially in young Lpin11Hubr rats, which is associated with the increased expression of Cds1 in sciatic nerves of these rats, supports the idea of a compensatory pathway activation in these animals. Thus, both elevated PAP2 activity and increased production of PI may underlie the absence of PA accumulation, and consequently milder myelin phenotype in Lpin11Hubr rats as compared with Lpin1fld/fld mice. In line with this hypothesis, we observed partial restoration (e.g. relatively elevated or increased levels of cholesterol and phosphatidylserine, respectively) of lipid levels in peripheral nerves of adult Lpin11Hubr rats, which is in parallel with the observed improvement in myelination in adult Lpin11Hubr rats.
In the PNS, partially recovered Mpz and Pmp22 expression levels accompanied the progression of myelination in Lpin11Hubr sciatic nerves at PND 90. In contrast to Lpin1fld/fld and MPZCre/+/LpfE2–3/fE2–3 mice, expression of Krox20 was decreased but not abolished at PND 21, whereas it was increased at PND 90. Krox20 is an important controller of myelination and a marker of mature SCs (29), thus indicating that myelination processes are down-regulated in Lpin11Hubr sciatic nerve tissue at PND 21 but up-regulated at PND 90, resulting in a relative increase in myelination at PND 90, a process that was never observed in Lpin1fld/fld and MPZCre/+/LpfE2–3/fE2–3 mice (2). Oct6, a marker of immature and promyelinating SCs (29, 30), is up-regulated at PND 21, thus indicative of an increased number of non-differentiated SCs. Interestingly, at PND 90, Oct6 levels are even further increased, which correlates with the observed elevated level of cell proliferation at this stage. Indeed, we observed the presence of a mixed population of three different SC lineages in Lpin11Hubr PND 90 nerves: immature promyelinating (Krox20−, Oct6+), early myelinating (Krox20+, Oct6+), and mature myelinated (Krox20+, Oct6−) SCs. In contrast, the main SC population observed in adult Lpin1fld/fld and MPZCre/+/LpfE2–3/fE2–3 mice were immature (Krox20−, Oct6+) SCs. These observations, which are further strengthened by our Nile Red staining data, reveal that Lpin11Hubr animals predominantly develop characteristics of hypomyelination rather than characteristics of demyelination, which was observed in Lpin1fld/fld and MPZCre/+/LpfE2–3/fE2–3 mice.
Lipin 1 was previously reported to play a role as a transcriptional coactivator (1). However, our observations that both Lipin 1Δα and 1Δβ forms preferentially localized to the cytoplasm and that both wild-type and mutated forms of Lipin 1α and 1β failed to activate the Krox20 MSE do not support the idea that the truncated form of Lipin 1 in Lpin11Hubr rats acts as a transcriptional regulator involved in PNS myelination and suggest that Lipin 1 may have additional yet to be discovered function(s).
The transitory lipodystrophy phenotype in Lpin11Hubr rats might be the result of immobility-induced hypophagia and a subsequent decreased lipid accumulation at a young age, whereas at an older age rats become more able to feed correctly due to increased mobility. This explanation is, however, unlikely, as most Lpin11Hubr pups retained partial ability to move around after the onset of the Lpin11Hubr phenotype. In addition, chow pellets were provided to the maternal cage floor to enable feeding under competitive conditions. An alternative explanation could come from the previously observed regulatory role of Lipin 1 during initial phases of adipogenesis, for the induction of adipogenic factors including PPARγ and CCAAT enhancer-binding protein (C/EBP) α (9, 37). Our observation that relative expression levels of Fabp4, PPARγ1, and PPARγ2 are increased in WAT derived from Lpin11Hubr rats at PND 90 as compared with PND 21, mirrored by a relative increase in WAT amount in Lpin11Hubr rats, potentially indicates that a regulatory function of Lipin 1 in Lpin11Hubr rats remained functional in WAT, thus enabling partial stimulation of adipogenesis. Additional studies are required to corroborate the suggested Lipin 1 involvement in regulation of nerve myelination and WAT function.
In humans, mutations in LPIN1 were recently described to result in recurrent myoglobinuria (i.e. the presence of myoglobin in urine, which is usually linked to destruction of skeletal muscle fibers, the latter also known as rhabdomyolysis) or statin-induced myopathy (38). An additional study reported severe early childhood myoglobinuria due to an in-frame intragenic Lpin1 deletion, removing a large part of the Lipin 1 CLIP domain (exons 18 and 19) but preserving its transcriptional LXXIL motif (39). Unfortunately, the PNS status of these patients was not evaluated and these patients may be too young to clearly detect a potentially mild WAT-related phenotype. The cause of the difference between so far described human phenotypes and the phenotypes of rodents with mutated Lipin 1 forms remains largely unknown.
Interestingly, a combination of episodic rhabdomyolysis and peripheral neuropathy occurred in patients with mitochondrial trifunctional protein deficiency (40). Thus, it remains possible that so far undetected peripheral neuropathies might indeed affect patients carrying a LPIN1 mutation.
Supplementary Material
Acknowledgments
We gratefully acknowledge Magdalena Harakolova and Raoul Kuiper for performing initial studies, Jeroen Korving for support with histological experiments, Mark Verheul for help with mutation mapping, and Goranka Tanackovic and Carlo Rivolta for helpful discussions about the splicing mutations.
This work was supported, in whole or in part, by National Institutes of Health Grant GM-28140 (to G. M. C.), Swiss National Science Foundation Grant PP00P3_124833/1 (to R. C.), INSERM and Fondation de la Recherche Médicale Grant DRM20101220459 (to J. S. S. B.), a grant from the European Science Foundation EURYI (to E. C.), and Dutch government and European community Grants NWO 918.66.616 and FP7/2007-2013 (to D. M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4, Table S1, and Videos S1 and S2.
- PAP1/2
- phosphatidate phosphatase 1/2
- WAT
- white adipose tissue
- SC
- Schwann cell
- HAD
- haloacid dehalogenase
- PND
- postnatal day
- ENU
- N-ethyl-N-nitrosourea
- BN
- Brown Norway
- PPARγ
- peroxisome proliferator-activated receptor-γ
- PA
- phosphatidic acid
- PI
- phosphatidylinositol
- PS
- phosphatidylserine
- Cds1
- CDP-diacylglycerol synthase gene
- NMD
- nonsense-mediated decay
- MSE
- myelinating Schwann cell enhancer
- FABP
- fatty acid-binding protein.
REFERENCES
- 1. Finck B. N., Gropler M. C., Chen Z., Leone T. C., Croce M. A., Harris T. E., Lawrence J. C., Jr., Kelly D. P. (2006) Cell Metab. 4, 199–210 [DOI] [PubMed] [Google Scholar]
- 2. Nadra K., de Preux Charles A. S., Médard J. J., Hendriks W. T., Han G. S., Grès S., Carman G. M., Saulnier-Blache J. S., Verheijen M. H., Chrast R. (2008) Genes Dev. 22, 1647–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Verheijen M. H., Chrast R., Burrola P., Lemke G. (2003) Genes Dev. 17, 2450–2464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Phan J., Reue K. (2005) Cell Metab. 1, 73–83 [DOI] [PubMed] [Google Scholar]
- 5. Donkor J., Sariahmetoglu M., Dewald J., Brindley D. N., Reue K. (2007) J. Biol. Chem. 282, 3450–3457 [DOI] [PubMed] [Google Scholar]
- 6. Han G. S., Wu W. I., Carman G. M. (2006) J. Biol. Chem. 281, 9210–9218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Harris T. E., Huffman T. A., Chi A., Shabanowitz J., Hunt D. F., Kumar A., Lawrence J. C., Jr. (2007) J. Biol. Chem. 282, 277–286 [DOI] [PubMed] [Google Scholar]
- 8. Carman G. M., Han G. S. (2009) J. Biol. Chem. 284, 2593–2597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Péterfy M., Phan J., Xu P., Reue K. (2001) Nat. Genet. 27, 121–124 [DOI] [PubMed] [Google Scholar]
- 10. Péterfy M., Phan J., Reue K. (2005) J. Biol. Chem. 280, 32883–32889 [DOI] [PubMed] [Google Scholar]
- 11. Donkor J., Zhang P., Wong S., O'Loughlin L., Dewald J., Kok B. P., Brindley D. N., Reue K. (2009) J. Biol. Chem. 284, 29968–29978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Reue K., Zhang P. (2008) FEBS Lett. 582, 90–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kim H. B., Kumar A., Wang L., Liu G. H., Keller S. R., Lawrence J. C., Jr., Finck B. N., Harris T. E. (2010) Mol. Cell. Biol. 30, 3126–3139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Langner C. A., Birkenmeier E. H., Roth K. A., Bronson R. T., Gordon J. I. (1991) J. Biol. Chem. 266, 11955–11964 [PubMed] [Google Scholar]
- 15. Douglas D. S., Moran J. L., Bermingham J. R., Jr., Chen X. J., Brindley D. N., Soliven B., Beier D. R., Popko B. (2009) J. Neurosci. 29, 12089–12100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Smits B. M., Mudde J. B., van de Belt J., Verheul M., Olivier J., Homberg J., Guryev V., Cools A. R., Ellenbroek B. A., Plasterk R. H., Cuppen E. (2006) Pharmacogen. Genomics 16, 159–169 [DOI] [PubMed] [Google Scholar]
- 17. Nijman I. J., Kuipers S., Verheul M., Guryev V., Cuppen E. (2008) BMC Genomics 9, 95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. van Boxtel R., Toonen P. W., van Roekel H. S., Verheul M., Smits B. M., Korving J., de Bruin A., Cuppen E. (2008) Carcinogenesis 29, 1290–1297 [DOI] [PubMed] [Google Scholar]
- 19. de Preux A. S., Goosen K., Zhang W., Sima A. A., Shimano H., Ouwens D. M., Diamant M., Hillebrands J. L., Rozing J., Lemke G., Beckmann J. S., Smit A. B., Verheijen M. H., Chrast R. (2007) Mol. Cell. Neurosci. 35, 525–534 [DOI] [PubMed] [Google Scholar]
- 20. Bligh E. G., Dyer W. J. (1959) Can. J. Biochem. Physiol. 37, 911–917 [DOI] [PubMed] [Google Scholar]
- 21. Cui Z., Vance D. E. (1996) J. Biol. Chem. 271, 2839–2843 [DOI] [PubMed] [Google Scholar]
- 22. Rouser G., Siakotos A. N., Fleischer S. (1966) Lipids 1, 85–86 [DOI] [PubMed] [Google Scholar]
- 23. Brügger B., Erben G., Sandhoff R., Wieland F. T., Lehmann W. D. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 2339–2344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Brouwers J. F., Boerke A., Silva P. F., Garcia-Gil N., van Gestel R. A., Helms J. B., van de Lest C. H., Gadella B. M. (2011) Biol. Reprod., in press [DOI] [PubMed] [Google Scholar]
- 25. Burroughs A. M., Allen K. N., Dunaway-Mariano D., Aravind L. (2006) J. Mol. Biol. 361, 1003–1034 [DOI] [PubMed] [Google Scholar]
- 26. Roberts R. Z., Morris A. J. (2000) Biochim. Biophys. Acta 1487, 33–49 [DOI] [PubMed] [Google Scholar]
- 27. Nohturfft A., Zhang S. C. (2009) Annu. Rev. Cell Dev. Biol. 25, 539–566 [DOI] [PubMed] [Google Scholar]
- 28. Fowler S. D., Greenspan P. (1985) J. Histochem. Cytochem. 33, 833–836 [DOI] [PubMed] [Google Scholar]
- 29. Zorick T. S., Syroid D. E., Arroyo E., Scherer S. S., Lemke G. (1996) Mol. Cell. Neurosci. 8, 129–145 [DOI] [PubMed] [Google Scholar]
- 30. Jaegle M., Mandemakers W., Broos L., Zwart R., Karis A., Visser P., Grosveld F., Meijer D. (1996) Science 273, 507–510 [DOI] [PubMed] [Google Scholar]
- 31. Ghislain J., Charnay P. (2006) EMBO Rep. 7, 52–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rosen E. D., Spiegelman B. M. (2006) Nature 444, 847–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Makowski L., Hotamisligil G. S. (2005) Curr. Opin. Lipidol. 16, 543–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mul J. D., Yi C. X., van den Berg S. A., Ruiter M., Toonen P. W., van der Elst M. C., Voshol P. J., Ellenbroek B. A., Kalsbeek A., la Fleur S. E., Cuppen E. (2010) Am. J. Physiol. Endocrinol. Metab. 298, E477–488 [DOI] [PubMed] [Google Scholar]
- 35. Rio Frio T., McGee T. L., Wade N. M., Iseli C., Beckmann J. S., Berson E. L., Rivolta C. (2009) Hum. Mutat. 30, 1340–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nicholson P., Yepiskoposyan H., Metze S., Zamudio Orozco R., Kleinschmidt N., Mühlemann O. (2010) Cell Mol. Life Sci. 67, 677–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Phan J., Péterfy M., Reue K. (2004) J. Biol. Chem. 279, 29558–29564 [DOI] [PubMed] [Google Scholar]
- 38. Zeharia A., Shaag A., Houtkooper R. H., Hindi T., de Lonlay P., Erez G., Hubert L., Saada A., de Keyzer Y., Eshel G., Vaz F. M., Pines O., Elpeleg O. (2008) Am. J. Hum. Genet. 83, 489–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Michot C., Hubert L., Brivet M., De Meirleir L., Valayannopoulos V., Müller-Felber W., Venkateswaran R., Ogier H., Desguerre I., Altuzarra C., Thompson E., Smitka M., Huebner A., Husson M., Horvath R., Chinnery P., Vaz F. M., Munnich A., Elpeleg O., Delahodde A., de Keyzer Y., de Lonlay P. (2010) Hum. Mutat. 31, E1564–1573 [DOI] [PubMed] [Google Scholar]
- 40. Spiekerkoetter U., Bennett M. J., Ben-Zeev B., Strauss A. W., Tein I. (2004) Muscle Nerve 29, 66–72 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.