Abstract
The current study examined the role of PLD2 in the maintenance of mast cell structure. Phospholipase D (PLD) catalyzes hydrolysis of phosphatidylcholine to produce choline and phosphatidic acid (PA). PLD has two isoforms, PLD1 and PLD2, which vary in expression and localization depending on the cell type. The mast cell line RBL-2H3 was transfected to overexpress catalytically active (PLD2CA) and inactive (PLD2CI) forms of PLD2. The results of this study show that PLD2CI cells have a distinct star-shaped morphology, whereas PLD2CA and RBL-2H3 cells are spindle shaped. In PLD2CI cells, the Golgi complex was also disorganized with dilated cisternae, and more Golgi-associated vesicles were present as compared with the PLD2CA and RBL-2H3 cells. Treatment with exogenous PA led to the restoration of the wild-type Golgi complex phenotype in PLD2CI cells. Conversely, treatment of RBL-2H3 and PLD2CA cells with 1% 1-Butanol led to a disruption of the Golgi complex. The distribution of acidic compartments, including secretory granules and lysosomes, was also modified in PLD2CI cells, where they concentrated in the perinuclear region. These results suggest that the PA produced by PLD2 plays an important role in regulating cell morphology in mast cells.
Keywords: mast cell, phospholipase D2, Golgi complex, morphology, secretory granules, lysosomes
Phospholipase D (PLD) is a phospholipid-modifying enzyme that plays a pivotal role in many cellular functions, such as rearrangement of the actin cytoskeleton, vesicle trafficking, mitogenesis, and cell survival (Fang et al. 2001; Kam and Exton 2001; Cazzolli et al. 2006; Oude Weernink et al. 2007). Two major mammalian isoforms of PLD have been identified, PLD1 (Hammond et al. 1995) and PLD2 (Colley, Sung, et al. 1997).
Both isoforms are widely expressed in a variety of tissues and cells (Gibbs and Meier 2000; Vorland et al. 2008). PLD1 and PLD2 share approximately 50% homology in the conserved catalytic core and are more variable at the N- and C-termini (Liscovitch et al. 2000; Exton 2002). PLD2 has a higher basal activity than PLD1 (Chen and Exton 2004). PLD hydrolyzes the terminal phosphodiester bond of phosphatidylcholine, the predominant membrane phospholipid, to produce phosphatidic acid (PA) and choline. The production of PA is highly regulated by a number of intracellular factors, including phosphoinositides, Rho, Arf, and a variety of GTPases. Furthermore, PA can be converted to other potentially bioactive lipids, such as diacylglycerol (DAG) and lysophosphatidic acid (LPA), which act as second messengers in many cellular responses (Jenkins and Frohman 2005). PLD2 is reported to localize to the plasma membrane (Colley, Altshuller, et al. 1997; Du et al. 2004) and to the Golgi complex (Freyberg et al. 2002) and to be associated with β-actin (Lee et al. 2001).
Mast cells are involved in a variety of allergic and inflammatory disorders (Galli et al. 2008). These cells bind IgE to the high-affinity receptor for IgE (FcϵRI) on the cell surface. In the presence of specific antigen, FcϵRI is cross-linked, thus activating the cell and releasing the preformed inflammatory mediators contained in secretory granules and generating other inflammatory lipids and cytokines. PLD is thought to play an essential role in mast cell degranulation (Dinh and Kennerly 1991; Way et al. 2000) and can be activated in isolated mast cells as well as in cultured RBL-2H3 mast cells by a variety of stimulants, including antigen (Lin and Gilfillan 1992; Kumada et al. 1994; Hitomi et al. 2004).
Most previous investigations on PLD in mast cells have focused on its role in signal transduction via FcϵRI. Previous studies have shown that PLD1, but not PLD2, is a negative regulator of mast cell degranulation (Hitomi et al. 2004). PLD2 had a higher basal activity than PLD1 but did not respond to antigenic stimulation. During the course of the previous investigation (Hitomi et al. 2004), it was noted that there were morphological changes in the cells that overexpressed the dominant-negative catalytically inactive form of PLD1b. Therefore, it was of interest to determine if PLD2 also plays a role in maintaining the structure of RBL-2H3 mast cells. Cell lines overexpressing catalytically active and catalytically inactive forms of PLD2 showed increases in both forms of PLD2 (Hitomi et al. 2004). The results of the present study indicate that the production of PA by PLD2 contributes to the basic cell morphology, maintenance of the Golgi complex, and distribution of lysosomes and secretory granules in mast cells.
Materials and Methods
Cells
RBL-2H3 cells, a rat mast cell line (Barsumian et al. 1981), as well as RBL-2H3 cells transfected to overexpress catalytically active PLD2 (PLD2CA; clone D2-WT-1) or catalytically inactive PLD2 (PLD2CI; clone D2/K758R-1) (generously provided by Reuben P. Siraganian, MD, PhD, National Institutes of Health, Bethesda, MD) were used in this study. All cells were grown as monolayers in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen, GIBCO, Carlsbad, CA) supplemented with 15% fetal calf serum (Invitrogen), 0.434 mg/ml glutamine, and an antibiotic-antimycotic mixture containing 100 units/ml penicillin, 100 µg/ml streptomycin, and 0.25 µg/ml amphotericin B (Invitrogen). Transfected cells were selected with geneticin (0.4 mg/ml) (Sigma-Aldrich; St. Louis, MO).
Antibodies, Fluorescent Markers, and Stains
The following primary antibodies were used: mouse mAb anti-GM-130 (4 µg/ml, Clone 35/GM130; BD Transduction Laboratories, San Jose, CA), mouse mAb anti-β-tubulin (0.75 µg/ml; Chemicon International, Billerica, MA), Phalloidin–Alexa 488 (1:100; Invitrogen), and rabbit polyclonal anti-PLD2 Internal (0.5 µg/ml, produced against a peptide corresponding to residues 476–486 derived from a mouse PLD2; Invitrogen). The lower concentration was used for Western blots and the higher concentration for immunofluorescence. The following secondary antibody used for Western blot was purchased from Jackson ImmunoResearch Laboratories (Port Washington, PA): goat anti-rabbit IgG–horseradish peroxidase (HRP) (1:20,000). The following secondary antibodies were used for immunofluorescence: goat anti-mouse IgG F(ab′)2–Alexa 488/594 (1:300 in PBS; Invitrogen, Molecular Probes) and goat anti-rabbit IgG F(ab′)2–Alexa 488 (1:300 in PBS; Invitrogen, Molecular Probes). The tracer LysoTracker red DND99 (Invitrogen, Molecular Probes) was used at 75 nM. Alcian Blue (1% Alcian Blue in 120 mM hydrochloric acid, pH 1.0) was used to stain secretory granules.
Microscopy
For all microscopic assays, except transmission electron microscopy, 3 × 104 cells were plated onto 13-mm coverslips and cultured overnight prior to use.
Differential Interference Contrast Microscopy
Cells fixed in 2% glutaraldehyde (Electron Microscopy Sciences; Hatfield, PA) were examined using an Olympus BX50 microscope (Olympus America; Melville, NY) equipped for differential interference contrast (DIC) microscopy. Images were collected using a Nikon DXM 1200 digital camera (Nikon USA; Melville, NY).
Fluorescence Microscopy
Cells were rinsed in PBS, fixed for 20 min with 2% formaldehyde (EM Sciences) in PBS, rinsed again, and permeabilized with 0.01% saponin (Sigma-Aldrich) in PBS for 20 min. Next, cells were rinsed twice in PBS and incubated for 30 min at room temperature in PBS containing 1% BSA (Sigma-Aldrich) and 5 µg/ml donkey-IgG (Jackson ImmunoResearch). Cells were then labeled with primary antibodies diluted in PBS containing 1% BSA for 1 hr at room temperature. They were rinsed in PBS, followed by incubation for 45 min at room temperature with the secondary antibodies diluted in PBS. Cells were then rinsed in PBS and mounted with Fluoromount-G (EM Sciences). Cells incubated without primary antibody served as controls. All controls were negative. For experiments with LysoTracker, unfixed cells were incubated for 1 hr at 37C with LysoTracker in DMEM. All samples were analyzed using a LEICA TCS-NT scanning confocal microscope (Leica Microsystems; Heidelberg, Germany).
Alcian Blue Staining
Cells were fixed with Carnoy’s solution (3% chloroform, 1% acetic acid, and 6% ethanol) for 15 min at room temperature and stained with Alcian blue for 15 min at room temperature. Cells were then rinsed twice in 70% ethanol and once in Milli-Q water (Millipore; Billerica, MA). Cells were counterstained with Weigert’s Fucsin-Resorcin (1% basic fucsin, 2% resorcin, 90% ethanol, 240 mM hydrochloric acid, and 30% FeCl3) for 15 min at room temperature; dehydrated in a graded ethanol series; cleared in xylol:ethanol, xylol; and mounted with Permount (Fisher Scientific; Hanover Park, IL). Cells were examined with an Olympus BX50 microscope (Olympus America) equipped with a Spot RT3 digital camera (Spot Imaging Solutions; Sterling Heights, MI).
Scanning Electron Microscopy (SEM)
Cells were rinsed in warm PBS (37C) and fixed in 2% glutaraldehyde (Ladd Research Industries; Burlington, VT) in warm PBS for 2 hr at room temperature. Cells were postfixed in 1% OsO4 (EM Sciences) for 2 hr, rinsed in Milli-Q water, and incubated with a saturated solution of thiocarbohydrazide (EM Sciences), followed by 1% OsO4. This step was repeated once. The cells were dehydrated in a graded ethanol series and critically point-dried with liquid CO2 in a BAL-TEC CPD 030 Critical-Point Dryer (BAL-TEC AG; Balzers, Liechtenstein), mounted on aluminum stubs with silver paint (EM Sciences), and coated with gold in a BAL-TEC SCD 050 Sputter Coater (BAL-TEC AG). Samples were examined with a JEOL JSM-5200 scanning electron microscope (JEOL, Ltd.; Tokyo, Japan).
Transmission Electron Microscopy (TEM)
Cells were plated at 2 × 105 cells/well in 6-well tissue culture plates and cultured for 3 days before fixation. Media were changed 16 to 24 hr before fixation. In some experiments, cells were treated with phosphatidic acid (10 mg/ml; Sigma-Aldrich) for 24 hr at 37C and in other experiments with 1% 1-Butanol (Merck KGaA; Darmstadt, Germany) for 20 min at 37C. Cells were fixed in 2% glutaraldehyde (Ladd Research) plus 2% formaldehyde (EM Sciences) in 0.1 M cacodylate buffer (pH 7.4), containing 0.05% CaCl2 for 1 hr at room temperature. Cells were postfixed in 1% OsO4 (EM Sciences) in 0.1 M cacodylate buffer (pH 7.4) for 2 hr, rinsed in Milli-Q water, and dehydrated in a graded ethanol series. Cells were removed from the tissue culture plates with propylene oxide and embedded in EMBED 812 (EM Sciences). Thin sections were cut with a diamond knife, mounted on copper grids, and stained for 10 min each in Reynolds’s lead citrate (Reynolds 1963) and 0.5% aqueous uranyl acetate, then examined with a Philips EM 208 transmission electron microscope (Fei Company; Eindhoven, Holland).
SDS-PAGE and Immunoblotting
For immunoblotting, whole-cell lysates of 106 cells from each cell line were washed twice with ice-cold PBS and immediately lysed with hot sample buffer (125 mM Tris-HCl [pH 6.8], 4% SDS, 10% glycerol, 0.006% bromophenol blue, and 1.8% β-mercaptoethanol) for 5 min at 70C. The samples were then sonicated for 10 sec on ice, and protease inhibitor cocktail (Sigma-Aldrich) was added to a final concentration of 1%, 30 µl/lane was loaded, and the proteins were separated electrophoretically on 10% polyacrylamide gels and transferred to Hybond membranes (GE Healthcare Life Sciences; Piscataway, NJ). After transfer, the membranes were blocked for 1 hr at room temperature in TBS (0.05 M Tris-HCl, 0.15 M NaCl [pH7.5], with 0.05% Tween 20) containing 5% BSA. After blocking, the membranes were incubated for 1 hr at room temperature with anti-PLD2 diluted in TBS. After washing with TBS, the membranes were incubated with secondary antibody for 45 min at room temperature. Membranes were then washed in TBS and developed using chemiluminescence (ECL-GE Healthcare). Mean optical density of the blots was determined using Adobe Photoshop 7.0 (Adobe Systems; San Jose, CA).
Detection of PLD2 mRNAs by Real-Time PCR
Total RNA was purified from each cell line using the RNeasy Mini kit (Qiagen; Valencia, CA) according to the manufacturer’s instructions. For mRNA analysis, 20 ng RNA was subjected to real-time RT-PCR using QuantiFast SYBR Green RT-PCR (Qiagen). For all RT-PCR analysis, actin mRNA was used to normalize RNA inputs. Primer sequences are as follows: mouse PLD2 forward (5′-ATGA CTGTAACCCAGACGGCACTC-3′) and reverse (5′-CAG CTCCTGAAAGTGTCGGAATTT-3′); actin forward (5′-T TGTCGACGACGAGCG-3′) and reverse (5′-GCACAGA GCCTCGCCTT-3′).
Image Analysis
PLD2 fluorescence intensity was quantified using Image-Pro Plus, ver. 7.0 (Media Cybernetics; Silver Spring, MD). To normalize fluorescence intensity, individual cells were selected with the freehand tool, and the mean fluorescence intensity was divided by the mean area of the cells. A minimum of 5 fields/cell line were analyzed. Volume renderings were obtained by analysis of Z-series of GM130-stained confocal images using ImageJ software (http://rsb.info.nih.gov/ij/; National Institutes of Health, Bethesda, MD). The analysis was expressed as 3D images of the Golgi complex. For the measurement of Golgi areas, the Golgi complexes were immunostained with anti-GM130. The boundary of the Golgi complex was delineated using the drawing tool in the Image-Pro Plus and the area of the Golgi complex determined. A minimum of 60 cells/cell line was analyzed.
For the measurement of the granule size, the diameter and the area of each individual granule were measured on TEM images using Image-Pro Plus. A minimum of 25 granules/cell line were analyzed.
Statistics
Results were expressed as means ± SD unless indicated otherwise. Differences between groups were assessed by unpaired, two-tailed Student’s t-test; p<0.05 was considered significant.
Results
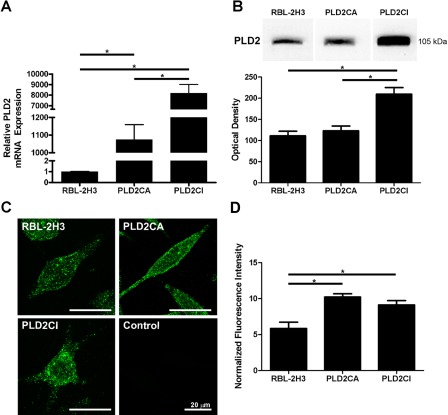
The expression and distribution of PLD2 varied among the three cell lines (Fig. 1). PLD2 mRNA was quantified by real-time qPCR, and the total amount of transcripts was significantly higher in PLD2CA and PLD2CI cells when compared with RBL-2H3 cells (Fig. 1A). The protein expression of PLD2 among the three cell lines generally reflected the levels of the transcripts (Fig. 1B). The expression of PLD2 was significantly higher in PLD2CI cells as compared with RBL-2H3 cells, whereas PLD2CA cells expressed only slightly more PLD2 than did the wild-type cells. The differences in expression may be a reflection of turnover rates. The three cell lines also showed differences in the distribution of PLD2 (Fig. 1C). In RBL-2H3 and PLD2CA cells, PLD2 was dispersed in a punctate pattern throughout the cytoplasm and followed the outline of the plasma membrane. In contrast, in PLD2CI cells, PLD2 was concentrated in the cell body. The normalized fluorescence intensity showed that PLD2CA and PLD2CI cells both had similar levels of fluorescence that were significantly higher than in the RBL-2H3 cells (Fig. 1D).
Figure 1.
Expression and distribution of PLD2. The levels of PLD2 transcripts were measured in RBL-2H3, PLD2CA, and PLD2CI cells. PLD2CA and PLD2CI cells had significantly higher levels of transcripts than did RBL-2H3 cells (*=p<0.05) (A). Lysates of RBL-2H3, PLD2CA, and PLD2CI cells were immunoblotted for PLD2 using an antibody against the internal sequence of PLD2 (B) and the resulting bands quantified. The PLDCI cells expressed significantly higher levels of PLD2 (*=p<0.05). Differences in the intracellular distribution of PLD2 were also seen (C). In RBL-2H3 and PLD2CA cell lines, PLD2 was localized throughout the cell cytoplasm, whereas in PLD2CI cells, PLD2 appeared to be localized mainly in the cell body. PLD2, green. The fluorescence intensity seen in PLD2CA and PLD2CI cells was significantly higher than that of the RBL-2H3 cells (*=p<0.05).
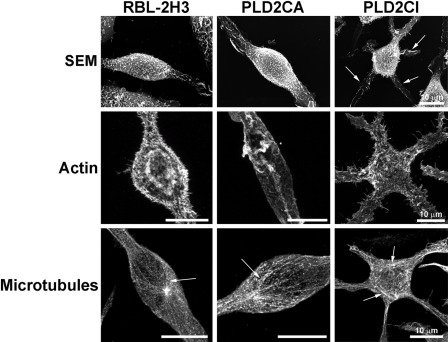
It then became of interest to examine if the differences in PLD2 expression were reflected in alterations in the morphology of these cells (Fig. 2). Wild-type RBL-2H3, PLD2CA, and PLD2CI cells were initially characterized by SEM. RBL-2H3 cells were fusiform (spindle shaped), and by SEM, it could be seen that the surface was covered with fine microvilli. The PLD2CA cells were also fusiform but with long cytoplasmatic extensions. By SEM, the PLD2CA cells showed the same fusiform morphology as the RBL-2H3 cells. In contrast, the PLD2CI cells differed from the other cell lines due to the presence of numerous cytoplasmatic extensions resulting in a characteristic star-shaped morphology. The distribution of cytoskeletal components in these cell lines was also examined. In RBL-2H3 and PLD2CA cells, the actin filaments followed the plasma membrane and the microvilli on the cell surface. In the PLD2CI cells, the actin filaments were also found under the plasma membrane, following the star-shaped form of the cell and in the microvilli. The microtubules in RBL-2H3 and PLD2CA cells were organized into long, straight bundles, giving the cell its fusiform shape. The microtubule distribution in the PLD2CI cells was altered when compared with the other cell lines. In PLD2CI cells, the microtubules were arranged in a basket-like form in the cell body and radiated throughout the numerous cytoplasmatic extensions in these star-shaped cells. Thus, overexpression of the catalytically inactive form of PLD2 appears to have a dramatic effect on the morphology of these cells.
Figure 2.
PLD2 is important for maintaining cell structure and organization. By scanning electron microscopy (SEM), RBL-2H3 cells were spindle shaped and their surface was covered with short microvilli. PLD2CA cells had morphology similar to that of RBL-2H3 cells. The majority of PLD2CI cells were star shaped with multiple cytoplasmic extensions (arrows), and their surface was also covered with short microvilli. Analysis of the cytoskeleton following 3D reconstruction of fluorescent images showed that there were differences in the distribution of the cytoskeleton in the PLD2CI cells when compared with the RBL-2H3 and PLD2CA cells. Actin filaments in RBL-2H3 and PLD2CA cells were under the plasma membrane following the spindle shape of the cells and in association with microvilli. Actin filaments were also seen in occasional ruffles. In PLD2CI cells, actin filaments followed the star shape of the cells and extended through the cytoplasmic projections. There were also numerous small actin containing projections on the surface of these cells. In RBL-2H3 and PLD2CA cells, microtubules (arrows) were organized into bundles that followed the fusiform shape of the cells, whereas in PLD2CI cells, the microtubules (arrows) in the cell body were more disorganized and appeared basket-like in the cell body.
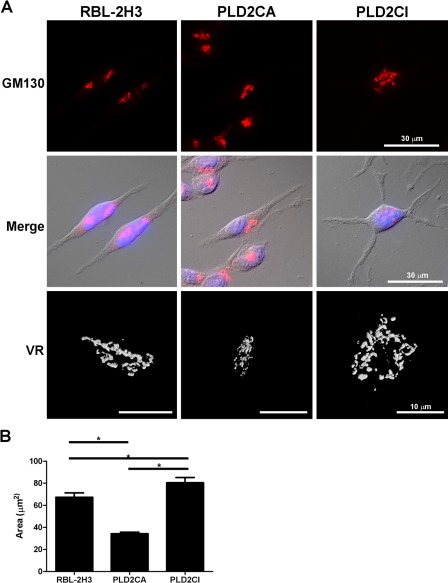
Because the organization of the Golgi complex is dependent on the cytoskeleton (Thyberg and Moskalewski 1999), the Golgi complex was also analyzed (Fig. 3A). In the RBL-2H3 and PLD2CA cells, the Golgi complex was compact and juxtanuclear in location. In PLD2CI cells, the Golgi complex was disorganized and dispersed. Volume rendering of the Golgi complex verified the observations made by fluorescence microscopy. This observation was further confirmed by quantifying the area of the Golgi complex in the various cell lines (Fig. 3B). The area occupied by the Golgi complex in the PLD2CA cells was significantly less than the RBL-2H3 cells, whereas the area occupied by the Golgi complex in the PLD2CI cells was significantly greater than the other two cell lines.
Figure 3.
PLD2CI cells have alterations in the Golgi complex. In RBL-2H3 and PLD2CA cells, the Golgi complex was compact and localized juxtanuclearly, but in PLD2CI cells, the Golgi complex was more diffuse and spread throughout the cytoplasm. Volume rendering (VR) of the Golgi complex confirmed the differences in the size of the Golgi complex among the three cell lines. (B) Quantification of the area of the Golgi complex showed that PLD2CA cells had a smaller Golgi complex than the RBL-2H3 cells (*=p<0.05). The PLD2CI cells had a larger Golgi area when compared with the RBL-2H3 cells (*=p<0.05). GM130, red; DAPI, blue.
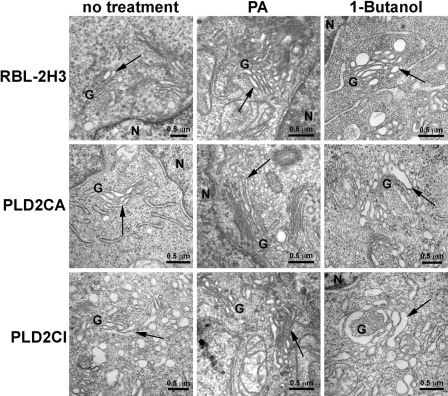
By TEM (Fig. 4), the Golgi complex in RBL-2H3 and PLD2CA cells was seen to be compact and close to the nucleus, with thin parallel cisternae. In PLD2CI cells, the Golgi complex was dispersed throughout the cytoplasm, with dilated cisternae and numerous dilated Golgi-associated vesicles that were absent in the other cell lines. Because of the differences seen in the morphology of the Golgi complex in the PLD2CI cells, it was of interest to investigate whether PA produced by PLD was involved in maintaining the structure of the Golgi complex. By TEM, it was possible to characterize the effects of treatment with PA or 1% 1-Butanol (Fig. 4). Treatment with PA in RBL-2H3 and PLD2CA cells did not alter the organization of the Golgi complex. The Golgi complex in these cell lines remained well organized, with thin and flattened cisternae, whereas in PLD2CI cells, the treatment with PA led to a more organized Golgi complex. The Golgi complex recovered its structural organization with parallel cisternae, and the number of Golgi-associated vesicles was reduced. To confirm that the lack of PA was responsible for the effects seen in the Golgi complex of the PLD2CI cells, the cell lines were treated with 1% 1-Butanol, which blocks the production of PA synthesized from PLD. In RBL-2H3 and PLD2CA cells, the treatment with 1% 1-Butanol led to the dispersion of the Golgi complex and the Golgi cisternae, and some vesicles were dilated. It was also observed that the cisternae were no longer parallel in the PLD2CA cells but were curved in a horseshoe shape. The treatment of PLD2CI cells with 1% 1-Butanol led to dilation of the Golgi cisternae and an increase in the number of vesicles.
Figure 4.
By transmission electron microscopy (TEM), the RBL-2H3 and PLD2CA cells had a well-organized Golgi complex, with their cisternae showing the typical arrangement of flattened saccules (arrows). The Golgi complex of the PLD2CI cells was dispersed in the cytoplasm, and the cisternae were disorganized and dilated (arrow). There was also an increase in the number of Golgi-associated vesicles in the PLD2CI cells. By TEM, treatment of RBL-2H3 cells with 10 mg/ml PA for 24 hr did not alter the morphology of Golgi complex. However, treatment with 1% 1-Butanol for 20 min led to the disorganization of the Golgi complex and dilated cisternae. Treatment of PLD2CA cells with phosphatidic acid (PA) did not alter the organization of the Golgi complex. In these cells, treatment with1% 1-Butanol also resulted in the disorganization of the Golgi complex and dilation of the cisternae. Treatment with PA in PLD2CI cells led to a decrease in the size of the Golgi complex cisternae and number of Golgi-associated vesicles (arrows), but treatment with 1% 1-Butanol did not alter the structure of the Golgi complex (arrows). G, Golgi complex; N, nucleus.
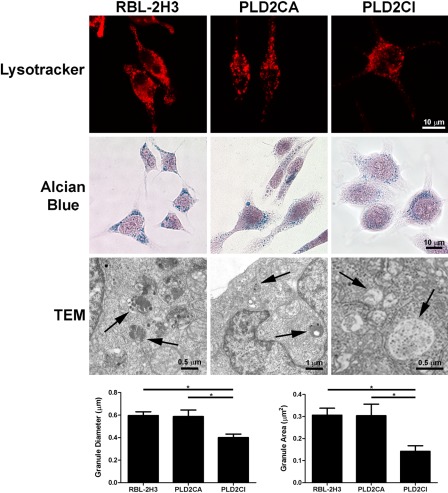
Because the Golgi complex is responsible for the production of lysosomes, secretory granules, and their contents, the distribution and morphology of acidic compartments, including secretory granules and lysosomes, were evaluated with LysoTracker. RBL-2H3 and PLD2CA cells presented numerous acidic vesicles spread throughout the cytoplasm and cell projections (Fig. 5). In PLD2CI cells, the acidic vesicles were concentrated in the perinuclear region with few vesicles in the cell extensions. Staining with Alcian blue, which stains the glycosaminoglycans present in the granules, showed that, in RBL-2H3 cells, the granules are present throughout the cytoplasm. In PLD2CA cells, the granules had the same distribution but appeared larger. The PLD2CI cells appear to have fewer, smaller granules that are concentrated near the nucleus. The morphology of the secretory granules was further characterized by TEM. In RBL-2H3 and PLD2CA cells, the secretory granules were heterogeneous, whereas in PLD2CI cells, the granules had an electron lucid content. Quantification of granule diameter and area showed that cytoplamatic granules in the RBL-2H3 and PLD2CA cells had the same overall size, whereas PLD2CI cells had significantly smaller granules when compared with the other cell lines.
Figure 5.
In the PLD2CI cells, secretory granules are altered. By LysoTracker staining, RBL-2H3 and PLD2CA cells have acidic compartments distributed in the cellular body and the cellular extensions. In the PLD2CI cells, the acidic vesicles are concentrated in the perinuclear region with few vesicles in the cell extensions. In RBL-2H3 cells, the granules are dispersed through the cytoplasm. Staining with Alcian blue shows that in PLD2CA cells, the granules are larger and PLD2CI cells have smaller granules in comparison with RBL-2H3 cells. By transmission electron microscopy (TEM), RBL-2H3 and PLD2CA cells contain electron-dense granules (arrows). However, in the PLD2CI cells, the granules are electron-lucid (arrows). The measurement of the granules size shows that in the PLD2CI cells, the granules are significantly smaller than those in RBL-2H3 and PLD2CA cells (*=p<0.05).
Discussion
In the present study, the importance of PLD2 in maintaining cell structure and function was examined in stable mast cell lines (Hitomi et al. 2004) that overexpress the wild-type or a dominant-negative form of PLD2. Overexpression of the catalytically inactive form of PLD2 appears to have a dramatic effect on the morphology of these cells, suggesting that the production of PA by PLD2 contributes to structural maintenance of the cytoskeleton and the Golgi complex as well as influencing the distribution of lysosomes and secretory granules in mast cells. The morphological changes observed in the present study are most likely the result of low levels of PA in the PLD2CI cells as well as the interaction of PLD2 with the cytoskeleton.
One of the striking features of the PLD2CI cells was the alteration in their shape. In contrast to the fusiform shape of the wild-type RBL-2H3 cells and the PLD2CA cells, most of the PLD2CI cells were star shaped with long cytoplasmic extensions. The shape of the cells correlated with the distribution of actin fibers and microtubules. Rat embryo fibroblasts overexpressing PLD2 also exhibited numerous cell projections and filopodia (Colley, Sung, et al. 1997). PLD2 is also known to contribute to stimulus-induced cytoskeletal reorganization (Iyer and Kusner 1999; Liscovitch et al. 1999). In RBL-2H3 cells, activation of ARF6 by antigen and continual PLD2 activity were both necessary for actin cytoskeletal rearrangements (O’Luanaigh et al. 2002). The reorganization of the actin cytoskeleton following PLD stimulation was first demonstrated in cultured fibroblasts stimulated by LPA or by the addition of exogenous PA (Ha and Exton 1993; Ha et al. 1994).
Although the roles of PLD in general and PLD1 specifically in cytoskeletal reorganization have been extensively studied, little is known about the specific role of PLD2 in this process (McDermott 2004; Morris 2007; Oude Weernink et al. 2007). In human umbilical vein endothelial cells (HUV-EC), PLD2 has recently been shown to have an important role in stress fiber formation and in the permeability of endothelial cell layers. This change in endothelial permeability appears to be linked to PA-induced cytoskeletal rearrangement as well as activation of the Raf-MEK-ERK1/2 signaling pathway. Activation of this pathway then downregulates the expression of occludin, a tight junction structural protein (Zeiller et al. 2009). The activity of PLD2 itself may be controlled by binding to cytoskeletal components. β-Actin binds directly to PLD2 at amino acids 613 to 723 and inhibits its activity (Lee et al. 2001). PLD2 also directly interacts with tubulin by binding PLD2 at amino acids 476 to 612. Binding of tubulin to PLD2 also inhibits PLD2 activity, and this activity is inversely proportional to the concentration of monomeric tubulin (Chae et al. 2005). Thus, the cytoskeleton itself can regulate the production of PA by PLD, controlling the level and the cellular location of PA necessary to modify cytoskeleton organization (Exton 2002; Rudge and Wakelam 2009).
In the current study, it was observed that the presence of the catalytically inactive form of PLD2 also resulted in changes in the Golgi complex, which can be explained by the low level of PA in these cells (Hitomi et al. 2004). PLD is known to be associated with the Golgi complex in many cell types (Freyberg et al. 2002; McDermott et al. 2004; Riebeling et al. 2009) and is involved with the transport of Golgi-associated vesicles. The results of the present study agree with previous studies showing that PA and phosphatidylinositol 4,5-bisphosphate are required to maintain the structural integrity of the Golgi complex (Siddhanta et al. 2000; Sweeney et al. 2002). In rat pituitary GH3 cells, >90% of the Golgi-associated PLD2 is localized to the rims of the Golgi cisternae and to Golgi-associated vesicles (Freyberg et al. 2002). The budding of COPI vesicles from the Golgi complex depends on PA produced by PLD2. PA is known to play a critical role in vesicle formation. Activation of PLD and the resultant PA production promotes a negative curvature of membranes, facilitating vesicle fission (McMahon and Gallop 2005; Cazzolli et al. 2006). In PLD2-depleted cells, budding COPI vesicles accumulate at the lateral rims of the Golgi cisternae, and the cis-Golgi is reduced and disorganized. COPI vesicle fission can be divided into two stages. In the early stage BARS (Brefeldin-A ADP-ribosylated substrate), PA and other COPI components are required for bud-neck constriction. The second step, in which the bud neck is cut, is dependent on PA generated by PLD2 (Yang et al. 2008). Inhibition of PLD2 also results in a mild disruption in the Golgi complex, similar to that reported in the current study.
PLD is an important regulator of secretion in mast cells (Choi, Chahdi, et al. 2002; Choi, Kim, et al. 2002). The distribution and morphology of lysosomes and secretory granules in the various cell lines can be related to the basal levels of PA produced in these cells (Hitomi et al. 2004). The low levels of PA in the PLD2CI cells affect the distribution of secretory granules and lysosomes. One possible explanation for this change in distribution may be the effect of the catalytically inactive form of PLD2 on the actin cytoskeleton, as the actin cytoskeleton is known to play a role in vesicle transport (Pfeiffer et al. 1985; Kuznetsov et al. 1992). In mast cells, activation via FcϵRI stimulates actin polymerization (Sahara et al. 1990; Oka et al. 2002; Farquhar et al. 2007) and subsequent fusion of secretory granules with the plasma membrane and mediator release.
Another explanation for the change in distribution of acidic compartments seen in the PLD2CI cells may relate to the requirement for PA in vesicle formation. PLD2 has been shown to regulate the transport of vesicles between the trans Golgi network and the apical plasma membrane in human intestinal HT29-c119A cells. Although the exact mechanism of action is unclear, it is most likely related to the ability of PA to influence membrane curvature. Therefore, PLD2 may be controlling secretory vesicle formation at the Trans Golgi Network (TGN) level (Denmat-Ouisse et al. 2001) in a manner similar to that seen for COPI vesicle formation (Yang et al. 2008). PA is considered a fusogenic lipid that participates in membrane fission and fusion (Kooijman et al. 2003; Cazzolli et al. 2006). These processes require a transitory reorganization of bilipid membranes that do not occur spontaneously (Lentz et al. 2000). Therefore, the local concentration of PA produced by PLD2 may be the determining factor in vesicle formation. The present study shows that the production of PA by PLD2 is essential for the regulation of the structure and organization of RBL-2H3 mast cells. The exact mechanism by which PLD2 exerts its effects is not clearly understood but seems to be related, at least in part, to intracellular PA levels.
Acknowledgments
We would like to thank Anderson Roberto de Souza for technical assistance, Márcia Sirlene Graeff and Lenaldo Branco Rocha for assistance with the confocal microscopy, and Maria Dolores S. Ferreira, Tereza P. Maglia, and José Augusto Moulin for assistance with the electron microscopy, all from the Department of Cell and Molecular Biology and Pathogenic Bioagents, FMRP-USP, Ribeirão Preto, SP. We also thank CAPES (Coordenação de Aperfeiçoamento de Pessoa de Nível Superior), FAEPA (Fundação de Apoio ao Ensino, Pesquisa e Assistência) and FAPESP (Fundação de Amparo à Pesquisa do Estado de São Paulo) (Individual Grant 01/10752–2 to CO) for the financial support.
Footnotes
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: Institutional grants to support research: CAPES (Coordenação de Aperfeiçoamento de Pessoa de Nível Superior), FAEPA (Fundação de Apoio ao Ensino, Pesquisa e Assistência) and FAPESP (Fundação de Amparo à Pesquisa do Estado de São Paulo) (Individual Grant 01/10752–2 to CO).
References
- Barsumian EL, Isersky C, Petrino MG, Siraganian RP. 1981. IgE-induced histamine release from rat basophilic leukemia cell lines: isolation of releasing and nonreleasing clones. Eur J Immunol. 11:317–323 [DOI] [PubMed] [Google Scholar]
- Cazzolli R, Shemon AN, Fang MQ, Hughes WE. 2006. Phospholipid signalling through phospholipase D and phosphatidic acid. IUBMB Life. 58:457–461 [DOI] [PubMed] [Google Scholar]
- Chae YC, Lee S, Lee HY, Heo K, Kim JH, Kim JH, Suh PG, Ryu SH. 2005. Inhibition of muscarinic receptor-linked phospholipase D activation by association with tubulin. J Biol Chem. 280:3723–3730 [DOI] [PubMed] [Google Scholar]
- Chen JS, Exton JH. 2004. Regulation of phospholipase D2 activity by protein kinase C alpha. J Biol Chem. 279:22076–22083 [DOI] [PubMed] [Google Scholar]
- Choi WS, Chahdi A, Kim YM, Fraundorfer PF, Beaven MA. 2002. Regulation of phospholipase D and secretion in mast cells by protein kinase A and other protein kinases. Ann N Y Acad Sci. 968:198–212 [DOI] [PubMed] [Google Scholar]
- Choi WS, Kim YM, Combs C, Frohman MA, Beaven MA. 2002. Phospholipases D1 and D2 regulate different phases of exocytosis in mast cells. J Immunol. 168:5682–5689 [DOI] [PubMed] [Google Scholar]
- Colley WC, Altshuller YM, Sue-Ling CK, Copeland NG, Gilbert DJ, Jenkins NA, Branch KD, Tsirka SE, Bollag RJ, Bollag WB, et al. 1997. Cloning and expression analysis of murine phospholipase D1. Biochem J. 15;326(Pt 3):745–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colley WC, Sung TC, Roll R, Jenco J, Hammond SM, Altshuller Y, Bar-Sagi D, Morris AJ, Frohman MA. 1997. Phospholipase D2, a distinct phospholipase D isoform with novel regulatory properties that provokes cytoskeletal reorganization. Curr Biol. 7:191–201 [DOI] [PubMed] [Google Scholar]
- Denmat-Ouisse LA, Phebidias C, Honkavaara P, Robin P, Geny B, Min DS, Bourgoin S, Frohman MA, Raymond MN. 2001. Regulation of constitutive protein transit by phospholipase D in HT29-c119A cells. J Biol Chem. 276:48840–48846 [DOI] [PubMed] [Google Scholar]
- Dinh TT, Kennerly DA. 1991. Assessment of receptor-dependent activation of phosphatidylcholine hydrolysis by both phospholipase D and phospholipase C. Cell Regul. 2:299–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du G, Huang P, Liang BT, Frohman MA. 2004. Phospholipase D2 localizes to the plasma membrane and regulates angiotensin II receptor endocytosis. Mol Biol Cell. 15:1024–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exton JH. 2002. Regulation of phospholipase D. FEBS Lett. 531:58–61 [DOI] [PubMed] [Google Scholar]
- Fang Y, Vilella-Bach M, Bachmann R, Flanigan A, Chen J. 2001. Phosphatidic acid–mediated mitogenic activation of mTOR signaling. Science. 294:1942–1945 [DOI] [PubMed] [Google Scholar]
- Farquhar MJ, Powner DJ, Levine BA, Wright MH, Ladds G, Hodgkin MN. 2007. Interaction of PLD1b with actin in antigen-stimulated mast cells. Cell Signal. 19:349–358 [DOI] [PubMed] [Google Scholar]
- Freyberg Z, Bourgoin S, Shields D. 2002. Phospholipase D2 is localized to the rims of the Golgi apparatus in mammalian cells. Mol Biol Cell. 13:3930–3942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli SJ, Grimbaldeston M, Tsai M. 2008. Immunomodulatory mast cells: negative, as well as positive, regulators of immunity. Nat Rev Immunol. 8:478–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs TC, Meier KE. 2000. Expression and regulation of phospholipase D isoforms in mammalian cell lines. J Cell Physiol. 182:77–87 [DOI] [PubMed] [Google Scholar]
- Ha KS, Exton JH. 1993. Activation of actin polymerization by phosphatidic acid derived from phosphatidylcholine in IIC9 fibroblasts. J Cell Biol. 123:1789–1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha KS, Yeo EJ, Exton JH. 1994. Lysophosphatidic acid activation of phosphatidylcholine-hydrolysing phospholipase D and actin polymerization by a pertussis toxin–sensitive mechanism. Biochem J. 303(Pt 1):55–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SM, Altshuller YM, Sung TC, Rudge SA, Rose K, Engebrecht J, Morris AJ, Frohman MA. 1995. Human ADP-ribosylation factor-activated phosphatidylcholine-specific phospholipase D defines a new and highly conserved gene family. J Biol Chem. 270:29640–29643 [DOI] [PubMed] [Google Scholar]
- Hitomi T, Zhang J, Nicoletti LM, Grodzki AC, Jamur MC, Oliver C, Siraganian RP. 2004. Phospholipase D1 regulates high-affinity IgE receptor-induced mast cell degranulation. Blood. 104:4122–4128 [DOI] [PubMed] [Google Scholar]
- Iyer SS, Kusner DJ. 1999. Association of phospholipase D activity with the detergent-insoluble cytoskeleton of U937 promonocytic leukocytes. J Biol Chem. 274:2350–2359 [DOI] [PubMed] [Google Scholar]
- Jenkins GM, Frohman MA. 2005. Phospholipase D: a centric review. Cell Mol Life Sci. 62(19–20):2305–2316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kam Y, Exton JH. 2001. Phospholipase D activity is required for actin stress fiber formation in fibroblasts. Mol Cell Biol. 21:4055–4066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooijman EE, Chupin V, de Kruijff B, Burger KN. 2003. Modulation of membrane curvature by phosphatidic acid and lysophosphatidic acid. Traffic. 4:162–174 [DOI] [PubMed] [Google Scholar]
- Kumada T, Nakashima S, Miyata H, Nozawa Y. 1994. Potent activation of phospholipase D by phenylarsine oxide in rat basophilic leukemia (RBL-2H3) cells. Biochem Biophys Res Commun. 199:792–798 [DOI] [PubMed] [Google Scholar]
- Kuznetsov SA, Langford GM, Weiss DG. 1992. Actin-dependent organelle movement in squid axoplasm. Nature. 356:722–725 [DOI] [PubMed] [Google Scholar]
- Lee S, Park JB, Kim JH, Kim Y, Kim JH, Shin KJ, Lee JS, Ha SH, Suh PG, Ryu SH. 2001. Actin directly interacts with phospholipase D, inhibiting its activity. J Biol Chem. 276:28252–28260 [DOI] [PubMed] [Google Scholar]
- Lentz BR, Malinin V, Haque ME, Evans K. 2000. Protein machines and lipid assemblies: current views of cell membrane fusion. Curr Opin Struct Biol. 10:607–615 [DOI] [PubMed] [Google Scholar]
- Lin P, Gilfillan AM. 1992. The role of calcium and protein kinase C in the IgE-dependent activation of phosphatidylcholine-specific phospholipase D in a rat mast (RBL 2H3) cell line. Eur J Biochem. 207:163–168 [DOI] [PubMed] [Google Scholar]
- Liscovitch M, Czarny M, Fiucci G, Lavie Y, Tang X. 1999. Localization and possible functions of phospholipase D isozymes. Biochim Biophys Acta. 1439:245–263 [DOI] [PubMed] [Google Scholar]
- Liscovitch M, Czarny M, Fiucci G, Tang X. 2000. Phospholipase D: molecular and cell biology of a novel gene family. Biochem J. 345(Pt 3):401–415 [PMC free article] [PubMed] [Google Scholar]
- McDermott M, Wakelam MJ, Morris AJ. 2004. Phospholipase D. Biochem Cell Biol. 82:225–253 [DOI] [PubMed] [Google Scholar]
- McMahon HT, Gallop JL. 2005. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 438:590–596 [DOI] [PubMed] [Google Scholar]
- Morris AJ. 2007. Regulation of phospholipase D activity, membrane targeting and intracellular trafficking by phosphoinositides. Biochem Soc Symp. (74):247–257 [DOI] [PubMed] [Google Scholar]
- Oka T, Sato K, Hori M, Ozaki H, Karaki H. 2002. FcepsilonRI cross-linking-induced actin assembly mediates calcium signalling in RBL-2H3 mast cells. Br J Pharmacol. 136:837–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Luanaigh N, Pardo R, Fensome A, Allen-Baume V, Jones D, Holt MR, Cockcroft S. 2002. Continual production of phosphatidic acid by phospholipase D is essential for antigen-stimulated membrane ruffling in cultured mast cells. Mol Biol Cell. 13:3730–3746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oude Weernink PA, Han L, Jakobs KH, Schmidt M. 2007. Dynamic phospholipid signaling by G protein-coupled receptors. Biochim Biophys Acta. 1768:888–900 [DOI] [PubMed] [Google Scholar]
- Pfeiffer JR, Seagrave JC, Davis BH, Deanin GG, Oliver JM. 1985. Membrane and cytoskeletal changes associated with IgE-mediated serotonin release from rat basophilic leukemia cells. J Cell Biol. 101:2145–2155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds ES. 1963. The use of the lead citrate at high pH as an electron-opaque stain in electron microscopy. J Cell Biol. 17:208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riebeling C, Morris AJ, Shields D. 2009. Phospholipase D in the Golgi apparatus. Biochim Biophys Acta. 1791:876–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudge SA, Wakelam MJ. 2009. Inter-regulatory dynamics of phospholipase D and the actin cytoskeleton. Biochim Biophys Acta. 1791:856–861 [DOI] [PubMed] [Google Scholar]
- Sahara N, Siraganian RP, Oliver C. 1990. Morphological changes induced by the calcium ionophore A23187 in rat basophilic leukemia (2H3) cells. J Histochem Cytochem. 38:975–983 [DOI] [PubMed] [Google Scholar]
- Siddhanta A, Backer JM, Shields D. 2000. Inhibition of phosphatidic acid synthesis alters the structure of the Golgi apparatus and inhibits secretion in endocrine cells. J Biol Chem. 275:12023–12031 [DOI] [PubMed] [Google Scholar]
- Sweeney DA, Siddhanta A, Shields D. 2002. Fragmentation and re-assembly of the Golgi apparatus in vitro: a requirement for phosphatidic acid and phosphatidylinositol 4,5-bisphosphate synthesis. J Biol Chem. 277:3030–3039 [DOI] [PubMed] [Google Scholar]
- Thyberg J, Moskalewski S. 1999. Role of microtubules in the organization of the Golgi complex. Exp Cell Res. 246:263–279 [DOI] [PubMed] [Google Scholar]
- Vorland M, Thorsen VA, Holmsen H. 2008. Phospholipase D in platelets and other cells. Platelets. 19:582–594 [DOI] [PubMed] [Google Scholar]
- Yang JS, Gad H, Lee SY, Mironov A, Zhang L, Beznoussenko GV, Valente C, Turacchio G, Bonsra AN, Du G, et al. 2008. A role for phosphatidic acid in COPI vesicle fission yields insights into Golgi maintenance. Nat Cell Biol. 10(10):1146–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Way G, O’Luanaigh N, Cockcroft S. 2000. Activation of exocytosis by cross-linking of the IgE receptor is dependent on ADP-ribosylation factor 1-regulated phospholipase D in RBL-2H3 mast cells: evidence that the mechanism of activation is via regulation of phosphatidylinositol 4,5-bisphosphate synthesis. Biochem J. 346(Pt 1):63–70 [PMC free article] [PubMed] [Google Scholar]
- Zeiller C, Mebarek S, Jaafar R, Pirola L, Lagarde M, Prigent AF, Nemoz G. 2009. Phospholipase D2 regulates endothelial permeability through cytoskeleton reorganization and occluding downregulation. Biochim Biophys Acta. 1793:1236–1249 [DOI] [PubMed] [Google Scholar]