Abstract
Polymerase (RNA) II (DNA directed) polypeptide A (RPB1) is the largest subunit of RNA polymerase II (RNAPII), and phosphorylation of its C-terminal domain (CTD) is required for transcription initiation, elongation and RNA processing. Little is known about the CTD phosphorylation pattern and potential function during cell division when transcription is silenced. In this study, we assessed the protein expression and subcellular distribution of RPB1 during mouse oocyte meiotic division. Western blot analysis revealed that the RPB1 CTD was highly phosphorylated on Ser2 (pRPB1Ser2), Ser5 (pRPB1Ser5) and Ser7 (pRPB1Ser7). High and stable expression of pRPB1Ser2 and pRPB1Ser5 was detected from germinal vesicle (GV) to Metaphase II (MII) stage. In contrast, pRPB1Ser7 only emerged after germinal vesicle breakdown (GVBD) and gradually increased to its peak level at metaphase I (MI) and MII. Immunofluorescence demonstrated that pRPB1Ser2, pRPB1Ser5 and pRPB1Ser7 were pronouncedly aggregated within the nucleus of GV oocytes with a non-surrounded nucleolus (NSN) but very faintly labeled in oocytes with a surrounded nucleolus (SN). After meiotic resumption, pRPB1Ser2 was again detected at spindle poles and co-localized with key microtubule organizing center (MTOC) components, pericentrin and γ-tubulin. pRPB1Ser5 and pRPB1Ser7 were assembled as filamentous aggregates and co-localized with microtubules throughout the spindle structure, responding to spindle-disturbing drugs, nocodazole or taxol, in pattern strongly similar to microtubules. pRPB1Ser2 and pRPB1Ser5 were constantly localized on chromosomes, with a relatively high concentration in centromere areas. Taken together, our data suggest that the CTD is highly phosphorylated and may be required for accurate chromosome segregation in mouse oocytes during meiosis.
Keywords: Chromosome separation, C-terminal domain (CTD) phosphorylation, Oocyte, Subcellular distribution
In somatic cells, transcription activity is silenced as chromatin is condensed when the cell cycle progresses to mitosis and is reactivated when the cells enter the next Gap I (G1) phase after mitosis completion. Similarly, in mammalian oocytes, genome transcription is shut down upon resumption of meiosis, remaining quiescent during the following meiotic progression and even several cycles of early cleavage after fertilization [1, 2]. Transcription is driven by a dynamic functional complex composed of RNA polymerase and various regulating factors, and this complex is detached from DNA when the transcription process is arrested during mitosis or meiosis; however, but its form, distribution and possible function are not yet entirely clear.
Up to now, only a few studies have been carried out to explore the expression and potential function of transcription factors during cell division. Transcription factor II H (TFIIH), a general transcription factor, is required for the formation of the RNA polymerase II (RNAPII) pre-initiation complex [3]. Two subunits of TFIIH, XPD (Xeroderma pigmentosum D ortholog) and XPB (Xeroderma pigmentosum B ortholog), are localized to the mitotic spindle during mitosis and required for proper chromosome segregation [4,5,6]. Early growth response 3 (Egr3), a transcription factor, has recently been verified to be a microtubule organizing center (MTOC)-associated protein in mouse oocytes, and it might play a role during meiotic spindle formation [7]. This evidence suggests transcription-associated factors may play a role in molecular mechanisms governing chromosome separation during transcription silencing, yet whether such an involvement is a universal feature for other transcription factors requires further study.
Polymerase (RNA) II (DNA directed) polypeptide A (RPB1) is the largest and catalytic subunit of RNAP II and contains a carboxy terminal domain (CTD) composed of 52 heptapeptide repeats of Tyr1-Ser2-Pro3-Thr4-Ser5-Pro6-Ser7, essential for polymerase activity [8]. CTD phosphorylation at Ser5 is required for transcriptional initiation [9], Ser7 phosphorylation is essential for transcription elongation [10] and Ser2 phosphorylation plays a critical role in mRNA 3’-end processing, splicing, transcriptional elongation and termination factor recruitment to RNAP II in vivo [9, 11]. Other modifications, including phosphorylation of Tyr1 and Thr4, also contribute to CTD functions during transcription [12, 13]. Therefore, CTD phosphorylation exerts a central role in transcription initiation and orderly completion. Whether or not CTD phosphorylation occurs in mitotic or meiotic cells is rarely investigated. Abe et al. (2010) reported that CTD is dephosphorylated in mouse oocytes approaching meiotic resumption [14]. An early investigation showed that CTD is obviously phosphorylated after resumption of meiosis and dephosphorylated upon the completion of meiosis after fertilization in Xenopus oocytes [15]. It has not yet been determined which amino acid residues are phosphorylated in CTD sequence nor have the intracellular localization and potential function of RPB1 been determined in relation to the phosphorylated CTD during cell division.
In the present study, RPB1 was found to be highly phosphorylated on Ser2, Ser5 and Ser7 in the CTD sequence in mouse oocytes during meiosis. Phosphorylated RPB1s were localized to the MTOC, spindle microtubules and chromosome centromeres, implying a possible involvement of CTD phosphorylation in meiotic spindle formation and chromosome segregation during oocyte meiotic division.
Materials and Methods
Oocyte collection and culture
All experimental procedures were carried out in accordance with the policies for the Care and Use of Animals in Research and Teaching and approved by the Animal Care Commission of Capital Medical University. The oocytes were obtained from CB6F1 (female BALB/C × male C57BL/6) mice. Twenty-one-day-old females were euthanized with CO2 44–48 h after injection with 10 IU of pregnant mare serum gonadotropin (PMSG) (Beijing XinHuiZeAo Science and Technology), and cumulus-oocyte complexes (COCs) were harvested from ovaries and cultured in Minimal Essential Medium (MEM) with 3 mg/ml bovine serum albumin (BSA, Sigma) and 10% fetal bovine serum (FBS, Gibco, Grand Island, NY, USA) at 37 C in an incubator with 5% CO2 and 100% humidity. Oocytes at the germinal vesicle (GV), germinal vesicle breakdown (GVBD), prometaphase I (pro-MI), metaphase I (MI) and metaphase II (MII) stages of meiosis were obtained after cultures of 0, 2, 4, 8 and 17 h, respectively. After appropriate culture, oocytes were collected for further drug treatment experiments or immunofluorescence staining and western blot analysis.
Immunofluorescence analysis
Oocytes were fixed in 2% paraformaldehyde with 0.5% Triton X-100 in PEM Buffer (100 mM Pipes, pH 6.9, 1 mM MgCl2, 1 mM EGTA) for 45 min at room temperature. After being washed three times in phosphate-buffered saline (PBS) with 0.2% Triton X-100 (PBST) for 5 min each, the oocytes were blocked in 10% normal goat serum in PBS for 1 h at room temperature and then incubated in diluted primary antibodies: rabbit anti-RPB1(1:200, Novus, Littleton, CO, USA), rabbit anti-RNA polymerase II CTD repeat YSPTSPS (phospho Ser2) (1:500, Abcam, Hong Kong, China), rabbit anti-RNA polymerase II CTD repeat YSPTSPS (phospho Ser5) (1:1000, Abcam), rabbit anti-RNA polymerase II subunit B1 (phosphor-CTD Ser7) (1:250, Millipore, Temecula, CA, USA), mouse anti-acetylated tubulin (1:10000, Sigma, St. Louis, MO, USA), mouse anti-γ-tubulin (1:1000, Sigma) and mouse anti-pericentrin (1:3000, BD Transduction Laboratories, San Jose, CA, USA). After being washed in PBST, the oocytes were labeled with secondary antibodies for 45 min at room temperature and then mounted on microscope slides in mounting medium with DAPI (Vector Laboratories, Burlingame, CA, USA) and examined with a fluorescent microscope (Olympus Microsystems, Tokyo, Japan). A negative control was included to demonstrate the specificity of the primary antibodies used above, and oocyte samples were processed for the immunostaining procedure with no primary antibody incubation.
In order to determine the accurate localization of phosphorylated RPB1 on chromosomes, immunofluorescence was carried out on oocyte chromosome spreads as previously reported [16]. Briefly, the denuded oocytes were incubated in acid Tyrode’s solution (Sigma) at 37 C for 2 min to eliminate the zona pellucida and then recovered in MEM/BSA at 37 C within a short time. The oocytes were transferred onto glass slides and fixed in 1% paraformaldehyde with 0.1% Triton X-100. The air-dried slides were stored at –20 C for future use. Prior to immunolabeling, the slides were thoroughly washed in a sufficient volume of PBS to remove excess salts. The chromosome samples were incubated in primary antibody solution: Centromere antiserum (1:1000, Fitzgerald, Acton, MA, USA), anti-RNA polymerase II CTD repeat YSPTSPS (phospho Ser2) (1:500, Abcam) or anti-RNA polymerase II CTD repeat YSPTSPS (phospho Ser5) (1:500, Abcam) at 4 C overnight and then visualized with fluorescein-labeled goat anti-human and goat anti-rabbit IgG. Chromosomes were counterstained with DAPI in mounting medium (Vector Laboratories) and then assessed with a fluorescent microscope (Olympus Microsystems).
Western blot analysis
A total of 100 oocytes were collected in Laemmli Sample Buffer (Bio-Rad, Hercules, CA, USA) with protease inhibitor cocktail (Sigma) and stored at –80 C. Prior to analysis, the samples were thawed and heated at 100 C for 5 min. The proteins were separated by SDS-PAGE on 7.5% polyacrylamide gel with 0.1% SDS and electroblotted onto polyvinylidene fluoride membranes (PVDF) (Amersham Biosciences, Buckinghamshire, UK). After blocking in 1% BSA in Tris-buffered saline (TBS) with 0.1% Tween-20 (TBST) at 4 C overnight, the membranes were incubated at 37 C for 2 h with diluted primary antibodies, including rabbit anti-RPB1 antibody (1:250, Novus), rabbit anti-RNA polymerase II CTD repeat YSPTSPS (phospho Ser2) (1:500, Abcam), rabbit anti-RNA polymerase II CTD repeat YSPTSPS (phospho Ser5) (1:2000, Abcam) and rat anti-RNA polymerase II subunit B1 (phosphor-CTD Ser7) (1:100, Millipore), and thereafter, they were thoroughly washed in TBST and incubated in peroxidase-conjugated secondary antibodies (ZSGB-BIO, Beijing, China) for 1 h at room temperature. The protein bands on membranes were visualized with enhanced chemiluminescence detection reagents (Applygen Technologies Inc, Beijing, China), according to the manufacturer’s protocol.
Treatment with nocodazole and taxol
Nocodazole is a microtubule depolymerization agent, and taxol is a microtubule stabilizer. The stock solution of nocodazole (Sigma) was prepared at a concentration of 20 mM in dimethyl sulfoxide (DMSO, Sigma), and taxol (Sigma) stock solution was made at a concentration of 10 mM in DMSO. Just before use, the stock solutions were diluted to suitable concentrations in culture medium. The final concentration of DMSO in the medium was not more than 0.1% (v/v). For nocodazole treatment, MI-stage oocytes were incubated in MEM/BSA containing 10 μM nocodazole for 5, 10 or 15 min at 37 C in an atmosphere of 5% CO2 in air. For taxol treatment, the oocytes were treated with 10 μM taxol in MEM/BSA for 45 min. Control oocytes were incubated in MEM/BSA supplemented with the same concentration of DMSO. After drug treatment, the oocytes were fixed for immunofluorescence analysis.
Results
CTD phosphorylation on Ser2, Ser5 and Ser7 occurred in mouse oocytes during meiotic division
To explore whether RPB1 CTD is phosphorylated in oocytes during meiotic division, western blot analysis was conducted in mouse oocytes at the GV, GVBD, MI and MII stages. As shown in Fig. 1, CTD phosphorylation on Ser2 (pRPB1Ser2), Ser5 (pRPB1Ser5) and Ser7 (pRPB1Ser7) was obviously detected in meiotic oocytes. In brief, stable expression of pRPB1Ser2 and pRPB1Ser5 was consistently probed from the GV to MII stage. In contrast, pRPB1Ser7 was very faintly detected at the GV stage but increased gradually from GVBD and reach its peak level in MI and MII. As an additional control, the level of total RPB1 was detected in stable level throughout meiosis. These data strongly indicate RNAP II CTD phosphorylation definitely occurs in mammalian oocytes after meiotic resumption and that such modification is not required for transcription but might be involved in meiotic cycle regulation.
Fig. 1.
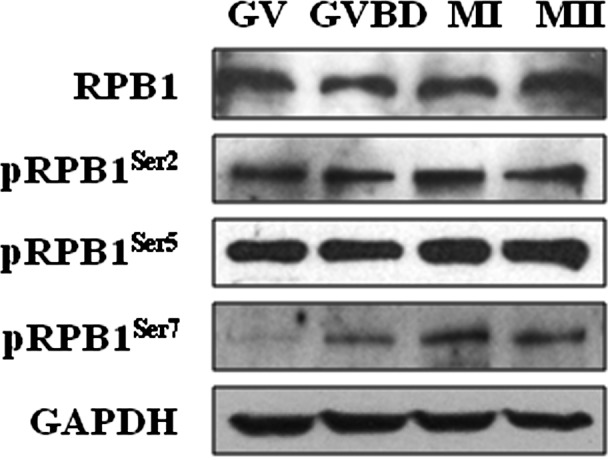
Protein expression of RPB1, pRPB1Ser2, pRPB1Ser5 and pRPB1Ser7 in mouse oocytes during meiotic division. Western blot analysis showed the protein levels of RPB1 and phosphorylated RPB1s in oocytes during meiotic maturation. The experiments were repeated at least three times.
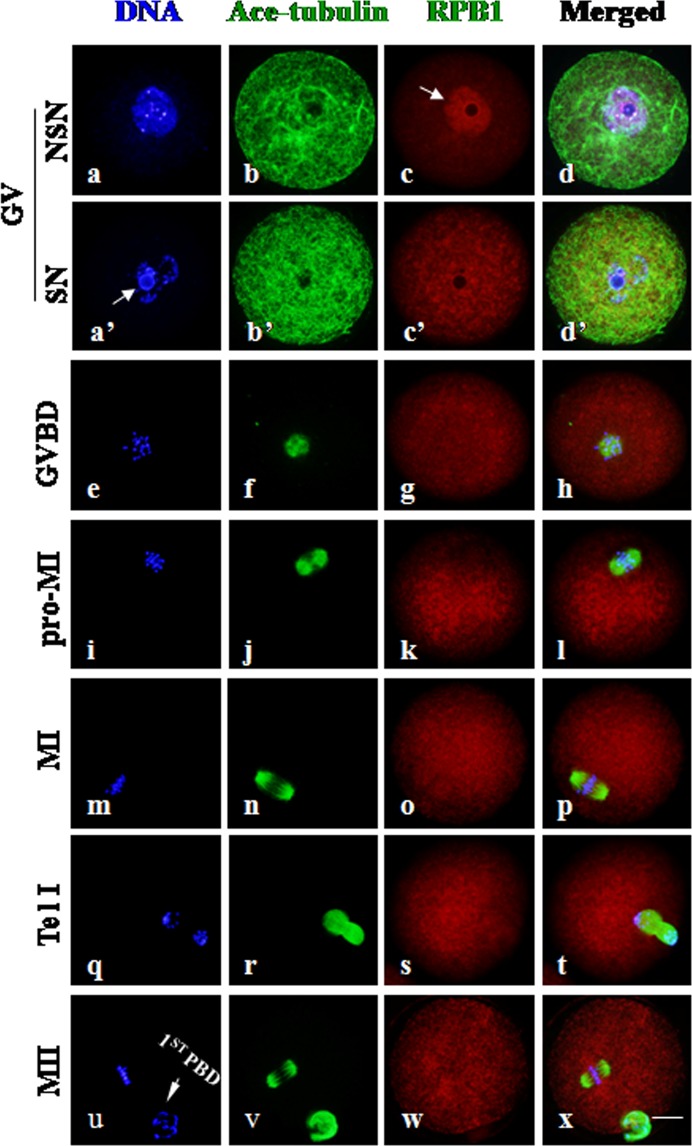
RPB1 was uniformly distributed throughout the cytoplasm after GVBD
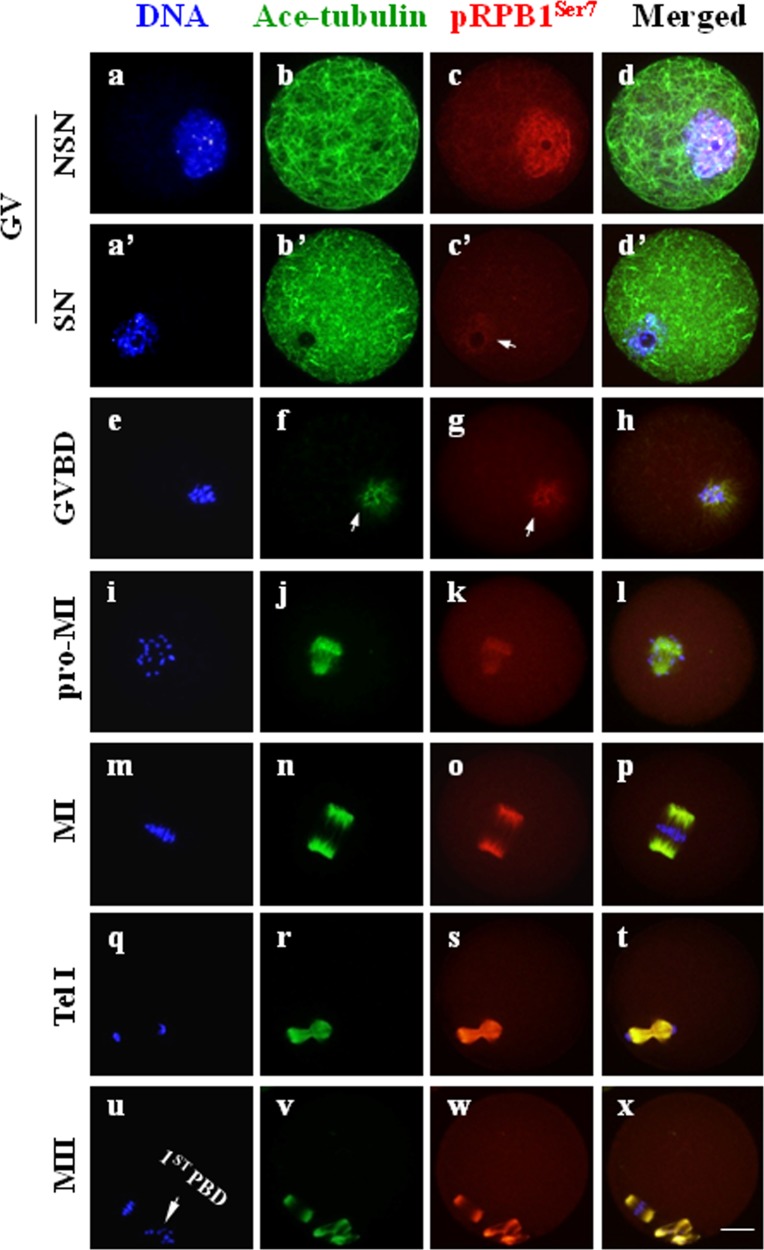
As shown in Fig. 2, the intracellular distribution of RPB1 was largely confined inside the enlarged nucleus when the nuclear envelope was intact at the GV stage, and no special accumulation of RPB1 was labeled in the cytoplasm. Notably, RPB1 accumulation was rarely observed in nuclei with a nucleolus surrounded by a rim of DAPI-positive chromatin (surrounded nucleolus, SN) (Fig.2, a’: arrow); it was mainly labeled in nuclei without such a chromatin rim around the nucleolus (non-surrounded nucleolus, NSN) (Fig.2, c: arrow). This is consistent with the fact that no transcriptional activity was detected in the SN-type oocytes [17]. Obviously, no fluorescence signal of RPB1 was detected in negative control oocytes (Supplementary Fig. 1, A: online only). With oocyte progression from the GVBD to MII stage, RPB1 was uniformly distributed throughout the cytoplasm, unequivocally, and no fluorescent signal was labeled on chromosomes or the spindle structure. It can be claimed that RPB1 protein was transferred from the nucleus to the cytoplasm and uniformly distributed in the oocyte upon resumption of meiosis.
Fig. 2.
RPB1 subcellular distribution in mouse oocytes during meiosis. Immunocytochemical staining showed the sub-cellular localization of RPB1 in oocytes at different stages of meiotic maturation. Scale bar = 20 μm. DNA was visualized in blue, RPB1 was visualized in red, and Ace-tubulin was visualized in green. RPB1 was highly concentrated in the nucleus of NSN-type oocytes at the GV stage (c: arrow) but faintly labeled in SN oocytes (a’: arrow). RPB1 was evenly distributed throughout the cytoplasm after meiotic resumption, exhibiting no specific association with chromosomes and the spindle structure (e–x). The arrowheads in u indicate the first polar body (1st PBD) in MII oocytes.
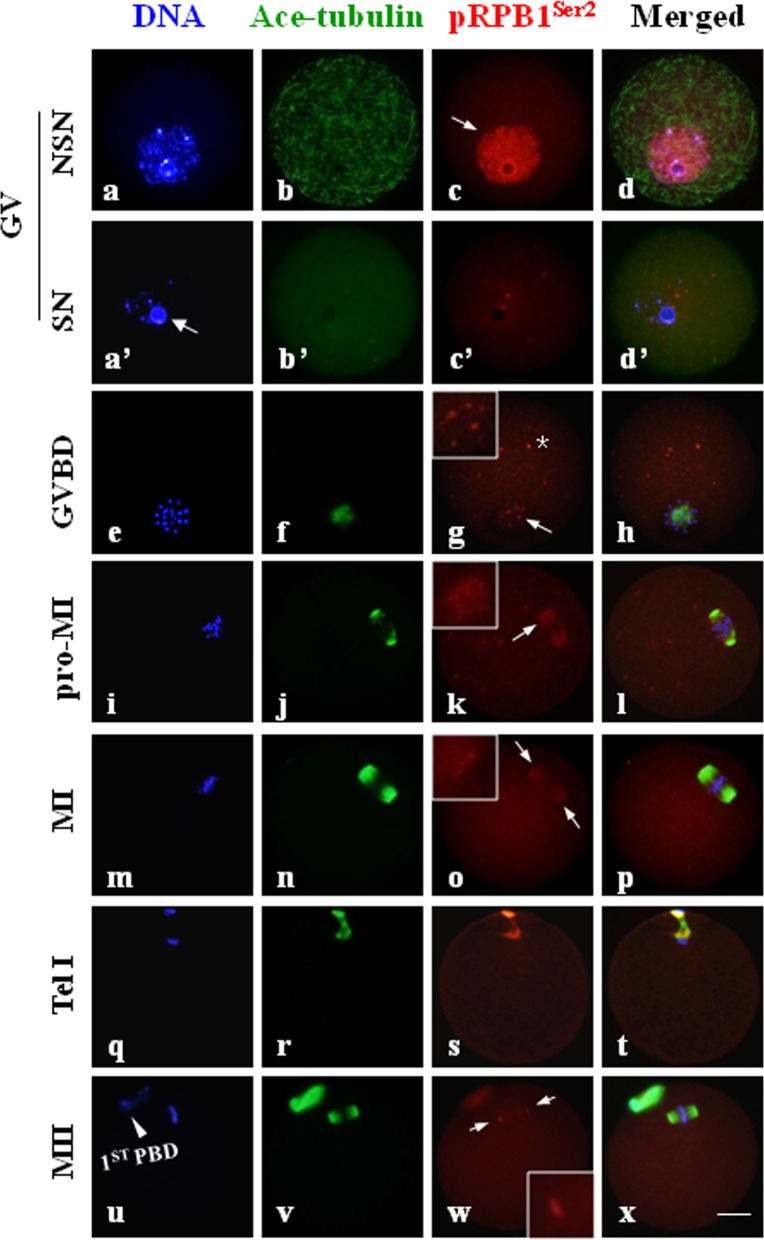
pRPB1Ser2 was concentrated in MTOCs and centromeres during meiotic division
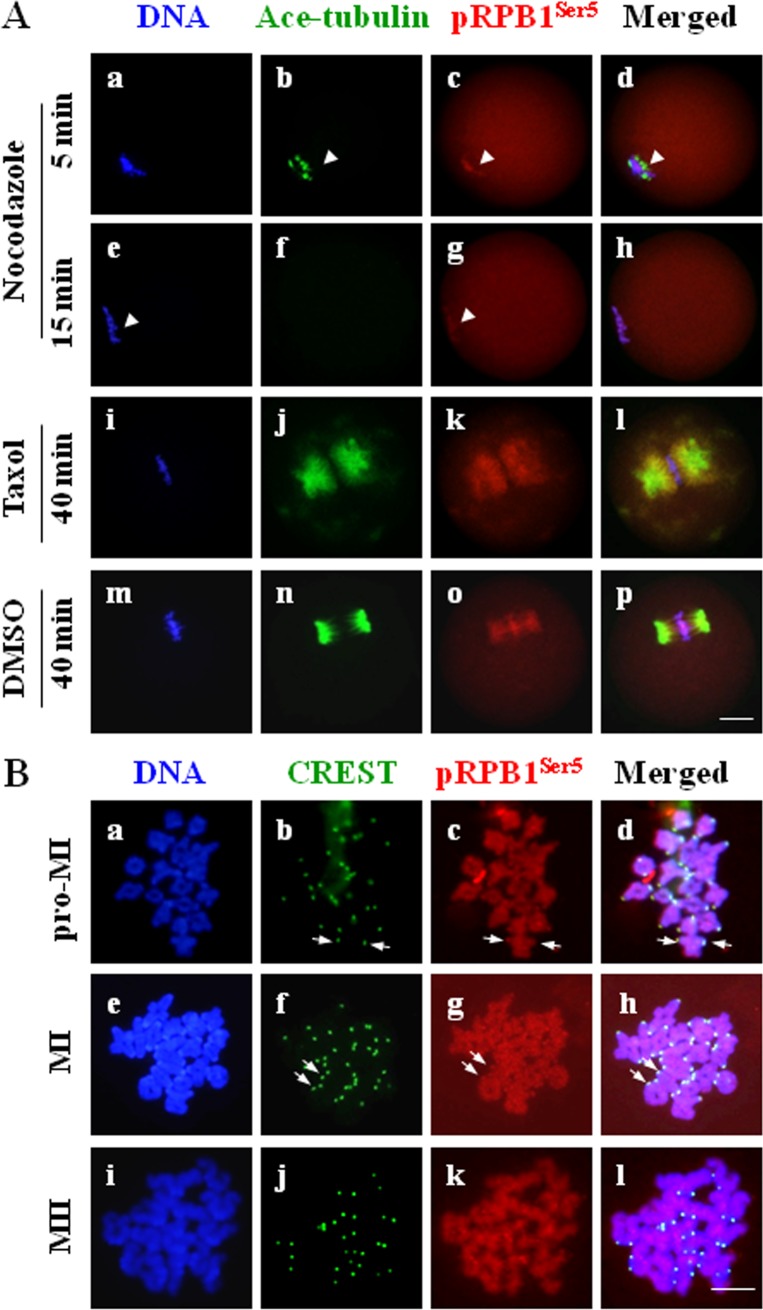
Immunofluorescence clearly demonstrated that the subcellular distribution of phosphorylated RPB1s was distinguishably different from that of unphosphorylated RPB1s and associated with cellular events responsible for chromosomes separation. As shown in Fig. 3, pRPB1Ser2 was evenly distributed within the nucleus in NSN-type GV oocytes (Fig. 3, c: arrow); in contrast, it was only faintly labeled in SN-type oocytes. Upon GVBD, pRPB1Ser2 emerged again as many small foci surrounding the condensing chromatin (Fig. 3, g: arrow). These foci gradually fused and clustered into two separate groups along with the meiotic progression from the GVBD to pro-MI stage (Fig. 3, k: arrow), and they finally concentrated on two separate poles of the spindle formed at the MI stage (Fig. 3, o: arrow). Multiple distinct foci were also labeled in other areas of the cytoplasm (Fig. 3, g: asterisk). During anaphase I (AI)/telophase I (TI) transition, pRPB1Ser2 disappeared from the polar area and cytoplasm but was localized to the midbody derived from the MI spindle. By the MII stage, a barrel-shape spindle was formed, and pRPB1Ser2 was once again focused on the spindle poles (Fig. 3, w: arrow). Polar concentration of pRPB1Ser2 implies it might be associated with MTOCs in oocytes. No special accumulation of pRPB1Ser2 was observed in the negative control (Supplementary Fig. 1).
Fig. 3.
pRPB1Ser2 localization and its association with the spindle in mouse oocytes during meiotic maturation. Immunofluorescence analysis showed the unique subcellular distribution pattern of pRPB1Ser2 in oocytes during meiotic division. Scale bar = 20 μm. DNA was visualized in blue, pRPB1Ser2 was visualized in red, and Ace-tubulin was visualized in green. Yellow indicates the overlapping of pRPB1Ser2 and microtubules. A high concentration of pRPB1Ser2 was confined within the nucleus of NSN-type oocytes (c: arrow), and no aggregation was detected in SN oocytes (a’, b’, c’, d’). pRPB1Ser2 reappeared as many small foci around condensing chromatin after GVBD (e–k). Highly concentrated pRPB1Ser2 was localized on two opposite poles of the meiotic spindle at the MI and MII stages (m–p, u–x: arrow).
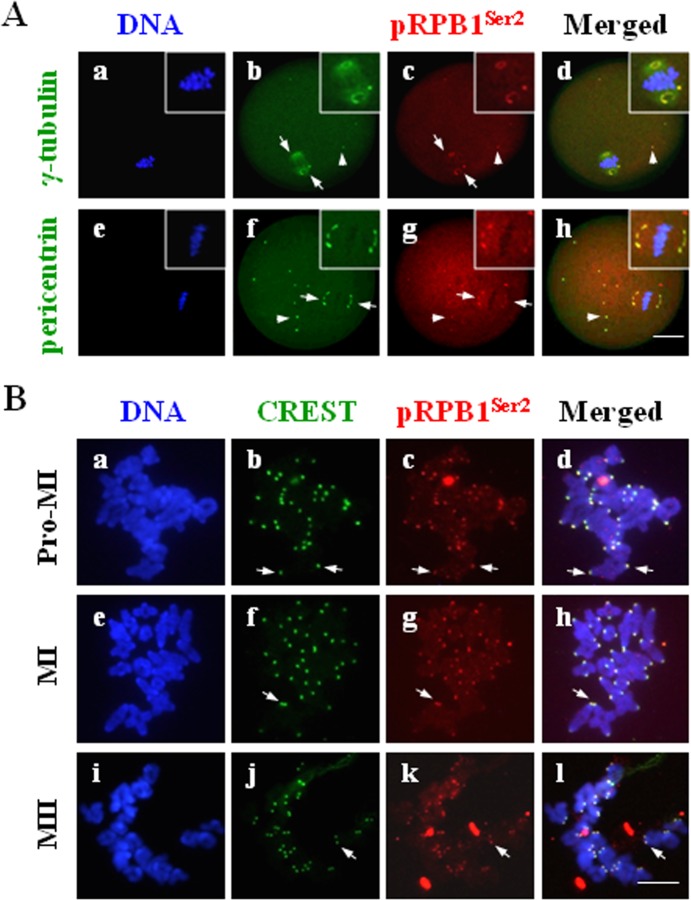
Further immunocytochemistry was carried out to reveal the spatial relationship between pRPB1Ser2 and key components of the MTOC, γ-tubulin and pericentrin, in oocytes. As shown in Fig. 4A, pRPB1Ser2 was essentially co-localized with both γ-tubulin and pericentrin on spindle poles (Fig. 4A: arrow), as well as in several cytoplasmic MTOCs (Fig. 4A: arrowhead) in MI oocytes. Actually, the tight colocalization of pRPB1Ser2 with pericentrin and γ-tubulin was consistently observed from GVBD to MII (data not shown). The data suggest that pRPB1Ser2 is a newly proved MTOC-associated protein and that it may play a role in meiotic spindle formation through regulation of MTOC organization and maintenance.
Fig. 4.
A: Colocalization of pRPB1Ser2 with MTOC proteins in mouse oocytes. Immunofluorescence analysis revealed colocalization of pRPB1Ser2 with the MTOC core components, γ-tubulin or pericentrin. DNA was visualized in blue, pRPB1Ser2 was visualized in red, and γ-tubulin or pericentrin was visualized in green. Scale bar = 20 μm. pRPB1Ser2 was specially co-localized with γ-tubulin or pericentrin in the spindle polar area, as well as in some cytoplasmic MTOCs (b–d, f–h: arrows). B: pRPB1Ser2 concentration on chromosome centromeres in mouse oocytes. pRPB1Ser2 was visualized in red, CREST was visualized in green, and DNA was visualized in blue. Scale bar = 10 μm. pRPB1Ser2 was persistently aggregated on the chromosome centromere area in oocytes from the pro-MI to MII stage (b–c, f–h, k–m: arrows).
To determine the precise localization of pRPB1Ser2 on chromosomes, oocyte chromosome spreads were prepared and immunolabeled with pRPB1Ser2 antibody and centromere autoantibody (CREST). As shown in Fig. 4B, pRPB1Ser2 was especially concentrated as bright dots on chromosome centromeres (Fig. 4B: arrow). It is noteworthy that pRPB1Ser2 was persistently detected in the centromere area from the pro-MI to MI stage and at the MII stage. No pRPB1Ser2 was labeled on the chromosome arm outside the centromere region. The centromeric localization of pRPB1Ser2 implies that CTD phosphorylation might be involved in the regulation of microtubules attaching to kinetochores in the centromere area.
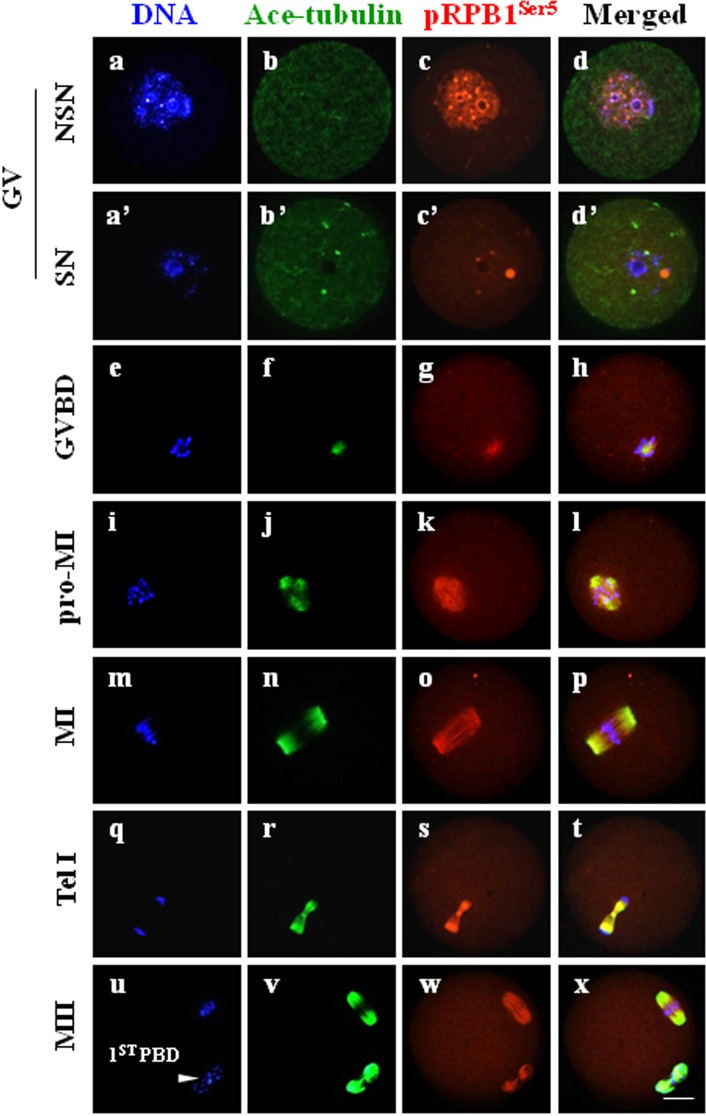
pRPB1Ser5 was localized on spindles and chromosomes after GVBD
To determine the distribution pattern of pRPB1Ser5 during mouse oocyte meiotic division, immunocytochemistry was carried out on wholly fixed oocytes and chromosome spreads. The overall subcellular distribution of pRPB1Ser5 was different from that of pRPB1Ser2 (Fig. 5). In NSN-type GV oocytes, pRPB1Ser5 was mainly present as fine particles uniformly distributed in the germinal vesicle (Fig. 5, c); at the same time, it was also noted as a number of bright dots dispersed randomly within the nucleus. In SN oocytes, pRPB1Ser5 accumulation almost disappeared, and only a few bright dots of different sizes were detected in the region surrounding the nucleolus (Fig. 5, c’). After GVBD, these pRPB1Ser5 foci were totally disassembled, reaggregated into filamentous bundles around condensing chromatin and completely co-localized with microtubules, especially at the MI and MII stages (Fig. 5, e–x). pRPB1Ser5 was organized into bipolar structure that was precisely overlapped with the spindle organized from microtubules (Fig. 5, n, o, p). No special organization of pRPB1Ser5 was found in oocytes without primary antibody incubation (Supplementary Fig. 1).
Fig. 5.
pRPB1Ser5 subcellular distribution and its relationship with the spindle in oocytes during meiotic division. Immunocytochemical analysis showed that pRPB1Ser5 was precisely colocalized with microtubules on the spindle in oocytes during meiotic maturation. DNA was visualized in blue, pRPB1Ser5 was visualized in red, and Ace-tubulin was visualized in green. Yellow indicates the overlapping of pRPB1Ser5 and microtubules. Scale bar = 20 μm. At the GV stage, pRPB1Ser5 was labeled in the nucleus of NSN-type oocytes (a–d) and almost undetectable in SN oocytes, and only a few bright dots were detected around the nucleolus (a’ –d’). After GVBD, pRPB1Ser5 was aggregated into filamentous bundles and completely co-localized with microtubules, and it was shaped into a bipolar structure that was precisely overlapped by the meiotic spindle at the MI and MII stages (m–p, u–x; arrowhead: 1st polar body).
The morphological relationship between pRPB1Ser5 and microtubules was further confirmed using spindle-perturbing drugs, nocodazole and taxol. Firstly, the MI oocytes were treated for 5, 10 and 15 min with nocodazole, a microtubule-depolymerizing agent. As microtubules gradually depolymerized with the increase in drug processing time, pRPB1Ser5 fibers also disassembled, but they always maintained colocalization with microtubules (Fig. 6A, b and c: arrow head). Upon complete depolymerization of spindle microtubules, the filamentous pRPB1Ser5 was also totally dismantled. In addition, despite the morphological change of pRPB1Ser5 along with the spindle, the pRPB1Ser5 localization on chromosomes was persisted during the entire process of nocodazole treatment (Fig. 6A, g: arrowhead). Next, the MI oocytes were treated for 45 min with taxol, a microtubule-stabilizing drug. As shown in Fig. 6A, taxol suppressed microtubule dynamics in oocytes, thus causing excessive polymerization of microtubules and formation of astral microtubules in the spindle architecture and numerous MTOCs in the cytoplasm. Interestingly, pRPB1Ser5 exhibited the exactly same change pattern and was completely co-localized with microtubules in the spindle and cytoplasmic MTOCs (Fig. 6A, j–l). These data clearly indicates that the accumulation and distribution of pRPB1Ser5 were associated with the structural integrity of the spindle in oocytes.
Fig. 6.
A: Changes of pRPB1Ser5 distribution in MI oocytes treated with spindle-disturbing agents. pRPB1Ser5 always remained colocalized with microtubules as spindle integrity and microtubule stability were altered with nocodazole (10 μM) and taxol (10 μM). DNA was visualized in blue, pRPB1Ser5 was visualized in red, and Ace-tubulin was visualized in green. Scale bar = 20 μm. B: Immunolocalization of pRPB1Ser5 on chromosomes. Chromosome spreads were made at the pro-MI, MI and MII stages and processed for immunostaining using antibodies to pRPB1Ser2 and CREST. pRPB1Ser5 was visualized in red, CREST was visualized in green and DNA was visualized in blue. Scale bar = 10 μm. pRPB1Ser5 was uniformly distributed along the whole chromosome structure, with a relatively high concentration in the centromere area.
To determine the specific local positioning of pRPB1Ser5 on the chromosome structure in oocytes, chromosomes spreads were immunolabeled with pRPB1Ser5 antibody and CREST serum. As shown in Fig. 6B, highly concentrated pRPB1Ser5 was persistently observed on chromosome centromeres at the pro-MI, MI and MII stages (Fig. 6B, b–d, f–h: arrow). A little weaker pRPB1Ser5 signal was also distributed throughout individual chromosomes. This evidence suggests pRPB1Ser5 may be involved in cellular events associated with kinetochores, as well as the maintenance of chromosome configuration during oocyte meiosis.
pRPB1Ser7 was co-localized with spindle microtubules during meiotic division
As shown in Fig. 7, pRPB1Ser7 was highly detected and evenly distributed within the nucleus in NSN-type oocytes at the GV stage (Fig. 7, c), but rarely observed in SN-type cells (Fig. 7, c’: arrow). Upon GVBD, pRPB1Ser7 reappeared as filamentous aggregates in the vicinity around condensing chromatin and co-localized with the emerging microtubules (Fig. 7, f–h: arrow). Along with oocyte progression to the MII stage, pRPB1Ser7 had the exactly same distribution pattern as that of microtubules. At two meiotic metaphases, pRPB1Ser7 fibers were shaped into a bipolar architecture that was precisely overlapped with the microtubule-organized spindle (Fig. 7, n–p, v–x). No pRPB1Ser7 was detected on chromosomes from the GVBD to MII stage (data not shown). In the negative control, no fluorescence signal of pRPB1Ser7 was detected (Supplementary Fig. 1, B).
Fig. 7.
pRPB1Ser7 subcellular localization in mouse oocytes. Immunocytochemical analysis revealed the dynamic subcellular distribution of pRPB1Ser7 in oocytes during meiotic maturation. DNA was visualized in blue, pRPB1Ser7 was visualized in red, and Ace-tubulin was visualized in green. Yellow suggests the overlapping of pRPB1Ser7 and microtubulin. Scale bar = 20 μm. pRPB1Ser7 was uniformly distributed in the nucleus of NSN-type oocytes (a–d) and rarely detected in SN oocytes (a’–d’). After oocytes resumed meiosis, chromatin was condensed into individual chromosomes, and pRPB1Ser5 reappeared as filamentous threads co-localized with microtubules from the GVBD to MII stage and were typically assembled into a spindle-like structure at the MI and MII stages, which was completely overlapped with the meiotic spindle organized from microtubules (m–p, u–x).
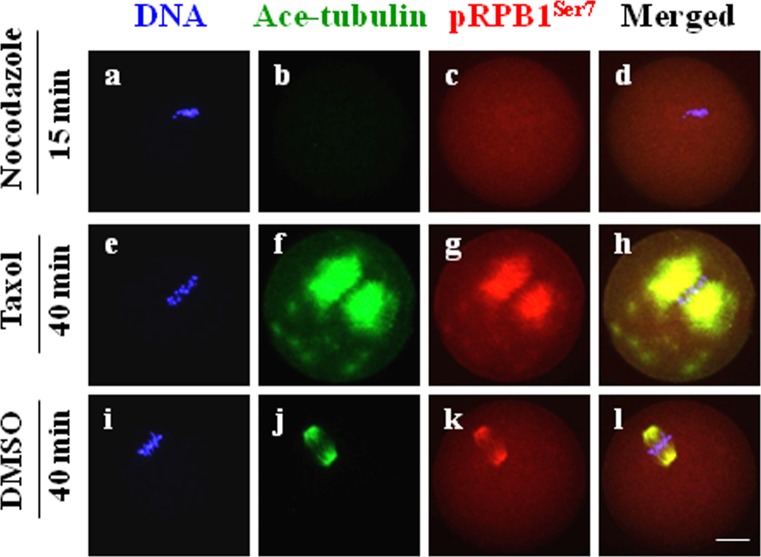
The correlation between pRPB1Ser7 and microtubules was further verified by using microtubule-targeting drugs. As shown in Fig. 8, pRPB1Ser7 disappeared along with the disassembly of microtubules in MI oocytes treated with nocodazole, while in taxol-treated oocytes, pRPB1Ser7 underwent the same change pattern as microtubules, which were stabilized in the spindle and cytoplasmic MTOCs. This evidence suggests that pRPB1Ser7 is associated with the microtubule polymerization state and might play a role in spindle formation during both meiotic divisions.
Fig. 8.
Pharmacological verification of the morphological relationship between pRPB1Ser7 and microtubules in MI oocytes. Changes in pRPB1Ser7 distribution were essentially the same as those of microtubules in MI oocytes treated with nocodazole (10 μM) and taxol (10 μM). DNA was visualized in blue, pRPB1Ser7 was visualized in red and Ace-tubulin was visualized in green. Scale bar = 20 μm.
Discussion
It is well known that general genomic transcription becomes silenced when the cell cycle enters mitosis or meiosis [1, 2], but the fate of the transcription machine during the subsequent cell division is still not fully clear. We investigated the status of CTD phosphorylation in mouse oocytes during meiotic maturation and confirmed that CTD phosphorylation definitely occurred in oocytes after meiotic resumption, which may be involved in spindle formation and kinetochore function, which are essential for ensuring accurate chromosome separation.
There were two different types of chromatin configuration in GV-stage oocytes: NSN or SN nucleolus chromatin configurations [17]. In NSN-type oocytes, the transcription process is active and characterized by a DNA-binding functional transcription complex with a hyperphosphorylated CTD, while in SN-type oocytes, transcription is arrested, the CTD was dephosphorylated and RNAP II and related factors are detached from the nucleus [14]. However, the phosphorylation pattern of specific individual amino residues in the CTD sequence during the nuclear transition from the NSN to SN type has not been determined. In the present study, immunofluorescence analysis revealed that CTD phosphorylation at Ser2, Ser5 and Ser7 (denoted as pRPB1Ser2, pRPB1Ser5 and pRPB1Ser7, respectively) was highly detected and uniformly distributed in the nucleus in NSN oocytes but only faintly detected in SN oocytes. This is logically consistent with the fact that transcription activity was totally shut down in SN-type oocytes. Immunoblotting analysis detected high levels of pRPB1Ser2 and pRPB1Ser5 not only at the GV stage but also at stages up to MII. Interestingly, the protein level of pRPB1Ser7 was significantly increased after meiotic resumption. Therefore, CTD phosphorylation definitely occurs in oocytes after meiotic resumption when transcription is silenced. RPB1 with a phosphorylated CTD was not degraded but was transferred from the nucleus to cytoplasm, and it might be involved in regulation of the meiotic cell cycle.
With the progression of the cell cycle from GVBD to MII in mouse oocytes, pRPB1Ser2, pRPB1Ser5 and pRPB1Ser7 exhibited unique distribution patterns that were not associated with transcription but related to cellular events responsible for chromosome division. pRPB1Ser2 was specially co-localized with key MTOC components, γ-tubulin and pericentrin, on the spindle poles. Both pRPB1Ser5 and pRPB1Ser7 were co-localized with microtubules on the spindle, exhibiting the same change pattern as microtubules in response to microtubule-targeting drugs. This morphological evidence implies that CTD phosphorylation may play roles in MTOC organization and functional maturation and may be required for meiotic spindle formation and maintenance in mouse oocytes.
The spindle apparatus works as the direct power driving chromosome segregation, and any structural abnormality in the spindle can undermine the accuracy of chromosome separation, causing oocyte aneuploidy [18]. In the somatic cell, mitotic spindle formation is controlled by a functional complex, the centrosome [19]. The mammalian oocyte does not contain the typical centrosome structure, which is ascribed to the degeneration of centrioles at the early stage of oogenesis. The meiotic spindle formation is acentrosomal and depends on a unique MTOC [18, 20, 21]. However, the molecular composition and regulatory mechanisms of MTOCs in oocyte meiosis are still not fully understood [21, 22]. Pericentrin and γ-tubulin are the core components of MTOCs, and γ-tubulin forms a conical structure known as the γ-tubulin ring complex (γ-TuRC), which is anchored to the MTOC and catalyzes the assembly of α/β tubulin dimers into microtubules [23, 24]. Pericentrin functions as a scaffold for anchoring γ-tubulin and other protein to the MTOC platform [16, 22]. There are some other proteins, especially some protein kinases, including aurora kinase A (AURKA) [25], polo like kinase 1(Plk1) [26] and protein kinase C-delta (PKCδ) [27], that join the MTOC structure after meiotic resumption, promoting the functional maturation of MTOCs. Similar to these regulatory proteins, pRPB1Ser2 was co-localized with pericentrin and γ-tubulin on spindle poles after GVBD, strongly implying that it is a newly found MTOC-associated protein, but its exact role in MTOC organization requires further morphological and functional studies. The functional effectiveness of the spindle depends on its structural integrity, which is tightly associated with microtubule stability [28]. In the present study, filamentous aggregates of both pRPB1Ser5 and pRPB1Ser7 were precisely co-localized with microtubules, indicating that these two proteins might be involved in the regulation of microtubule stability, and this speculation was further supported by the fact that both pRPB1Ser5 and pRPB1Ser7 responded to spindle-disturbing agents. The multiple types of CTD phosphorylation may regulate different aspects of meiotic spindle formation in mouse oocytes.
Besides the localization on MTOCs, pRPB1Ser2 and pRPB1Ser5 were also persistently concentrated on centromeres of chromosomes, suggesting that these proteins might be related to the regulation of spindle microtubule attachment on chromosomes. The kinetochore, a complex structure assembled on each centromere of the chromatid, acts as the attachment site for spindle microtubules [29]. An unattached kinetochore is actually a platform recruiting spindle assembly checkpoint (SAC) proteins [30], and the SAC is a supervisory system guaranteeing the fidelity of chromosome segregation during the process of cell division by initiating a signaling pathway, which prevents separation of duplicated chromosomes until each chromosome is properly attached to the spindle apparatus [31, 32]. Currently, the molecular mechanism of SAC functional regulation is not yet fully understood, especially the recruitment and disassociation of SAC proteins in kinetochore area [33]. Whether or not the centromere-localized pRPB1Ser2 and pRPB1Ser5 are involved in SAC regulation requires further investigation.
In summary, RNAP II CTD phosphorylation occurs in oocytes during meiotic division, which is transcription independent, and might be involved in molecular mechanisms ensuring accurate chromosome separation in multiple aspects, such as MTOC organization and positioning, spindle formation and maintenance, and SAC functional regulation.
Supplementary
Acknowledgments
The authors thank Dr Shu Wang for critical reading of the manuscript. This work was supported by the National Natural Science Foundation of China (31471108 and 31271253), Doctoral Fund of the Ministry of Education of China (20121107120019) and Natural Science Foundation of Beijing, China (7132030).
References
- 1.Moore GP, Lintern-Moore S, Peters H, Faber M. RNA synthesis in the mouse oocyte. J Cell Biol 1974; 60: 416–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aoki F, Worrad DM, Schultz RM. Regulation of transcriptional activity during the first and second cell cycles in the preimplantation mouse embryo. Dev Biol 1997; 181: 296–307. [DOI] [PubMed] [Google Scholar]
- 3.Zurita M, Merino C. The transcriptional complexity of the TFIIH complex. Trends Genet 2003; 19: 578–584. [DOI] [PubMed] [Google Scholar]
- 4.Li X, Urwyler O, Suter B. Drosophila Xpd regulates Cdk7 localization, mitotic kinase activity, spindle dynamics, and chromosome segregation. PLoS Genet 2010; 6: e1000876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ito S, Tan LJ, Andoh D, Narita T, Seki M, Hirano Y, Narita K, Kuraoka I, Hiraoka Y, Tanaka K. MMXD, a TFIIH-independent XPD-MMS19 protein complex involved in chromosome segregation. Mol Cell 2010; 39: 632–640. [DOI] [PubMed] [Google Scholar]
- 6.Weber A, Chung HJ, Springer E, Heitzmann D, Warth R. The TFIIH subunit p89 (XPB) localizes to the centrosome during mitosis. Cell Oncol 2010; 32: 121–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shin H, Kwon S, Song H, Lim HJ. The transcription factor Egr3 is a putative component of the microtubule organizing center in mouse oocytes. PLoS ONE 2014; 9: e94708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corden JL. RNA polymerase II C-terminal domain: Tethering transcription to transcript and template. Chem Rev 2013; 113: 8423–8455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev 2006; 20: 2922–2936. [DOI] [PubMed] [Google Scholar]
- 10.Czudnochowski N, Bösken CA, Geyer M. Serine-7 but not serine-5 phosphorylation primes RNA polymerase II CTD for P-TEFb recognition. Nat Commun 2012; 3: 842. [DOI] [PubMed] [Google Scholar]
- 11.Gu B, Eick D, Bensaude O. CTD serine-2 plays a critical role in splicing and termination factor recruitment to RNA polymerase II in vivo. Nucleic Acids Res 2013; 41: 1591–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hintermair C, Heidemann M, Koch F, Descostes N, Gut M, Gut I, Fenouil R, Ferrier P, Flatley A, Kremmer E, Chapman RD, Andrau JC, Eick D. Threonine-4 of mammalian RNA polymerase II CTD is targeted by Polo-like kinase 3 and required for transcriptional elongation. EMBO J 2012; 31: 2784–2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heidemann M, Eick D. Tyrosine-1 and threonine-4 phosphorylation marks complete the RNA polymerase II CTD phospho-code. RNA Biol 2012; 9: 1144–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abe K, Inoue A, Suzuki MG, Aoki F. Global gene silencing is caused by the dissociation of RNA polymerase II from DNA in mouse oocytes. J Reprod Dev 2010; 56: 502–507. [DOI] [PubMed] [Google Scholar]
- 15.Bellier S, Dubois MF, Nishida E, Almouzni G, Bensaude O. Phosphorylation of the RNA polymerase II largest subunit during Xenopus laevis oocyte maturation. Mol Cell Biol 1997; 17: 1434–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma W, Baumann C, Viveiros MM. NEDD1 is crucial for meiotic spindle stability and accurate chromosome segregation in mammalian oocytes. Dev Biol 2010; 339: 439–450. [DOI] [PubMed] [Google Scholar]
- 17.Debey P, Szöllösi MS, Szöllösi D, Vautier D, Girousse A, Besombes D. Competent mouse oocytes isolated from antral follicles exhibit different chromatin organization and follow different maturation dynamics. Mol Reprod Dev 1993; 36: 59–74. [DOI] [PubMed] [Google Scholar]
- 18.Dumont J, Desai A. Acentrosomal spindle assembly and chromosome segregation during oocyte meiosis. Trends Cell Biol 2012; 22: 241–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hinchcliffe EH. Centrosomes and the art of mitotic spindle maintenance. Int Rev Cell Mol Biol 2014; 313: 179–217. [DOI] [PubMed] [Google Scholar]
- 20.Schuh M, Ellenberg J. Self-organization of MTOCs replaces centrosome function during acentrosomal spindle assembly in live mouse oocytes. Cell 2007; 130: 484–498. [DOI] [PubMed] [Google Scholar]
- 21.Luksza M, Queguigner I, Verlhac MH, Brunet S. Rebuilding MTOCs upon centriole loss during mouse oogenesis. Dev Biol 2013; 382: 48–56. [DOI] [PubMed] [Google Scholar]
- 22.Ma W, Viveiros MM. Depletion of pericentrin in mouse oocytes disrupts microtubule organizing center function and meiotic spindle organization. Mol Reprod Dev 2014; 81: 1019–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schatten H, Sun QY. Centrosome dynamics during mammalian oocyte maturation with a focus on meiotic spindle formation. Mol Reprod Dev 2011; 78: 757–768. [DOI] [PubMed] [Google Scholar]
- 24.Colombié N, Głuszek AA, Meireles AM, Ohkura H. Meiosis-specific stable binding of augmin to acentrosomal spindle poles promotes biased microtubule assembly in oocytes. PLoS Genet 2013; 9: e1003562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Solc P, Baran V, Mayer A, Bohmova T, Panenkova-Havlova G, Saskova A, Schultz RM, Motlik J. Aurora kinase A drives MTOC biogenesis but does not trigger resumption of meiosis in mouse oocytes matured in vivo. Biol Reprod 2012; 87: 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiong B, Sun SC, Lin SL, Li M, Xu BZ, OuYang YC, Hou Y, Chen DY, Sun QY. Involvement of Polo-like kinase 1 in MEK1/2-regulated spindle formation during mouse oocyte meiosis. Cell Cycle 2008; 7: 1804–1809. [DOI] [PubMed] [Google Scholar]
- 27.Ma W, Koch JA, Viveiros MM. Protein kinase C delta (PKCdelta) interacts with microtubule organizing center (MTOC)-associated proteins and participates in meiotic spindle organization. Dev Biol 2008; 320: 414–425. [DOI] [PubMed] [Google Scholar]
- 28.Selesniemi K, Lee HJ, Muhlhauser A, Tilly JL. Prevention of maternal aging-associated oocyte aneuploidy and meiotic spindle defects in mice by dietary and genetic strategies. Proc Natl Acad Sci USA 2011; 108: 12319–12324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fukagawa T, Earnshaw WC. The centromere: chromatin foundation for the kinetochore machinery. Dev Cell 2014; 30: 496–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sacristan C, Kops GJ. Joined at the hip: kinetochores, microtubules, and spindle assembly checkpoint signaling. Trends Cell Biol 2015; 25: 21–28. [DOI] [PubMed] [Google Scholar]
- 31.Jones KT, Lane SI. Molecular causes of aneuploidy in mammalian eggs. Development 2013; 140: 3719–3730. [DOI] [PubMed] [Google Scholar]
- 32.Foley EA, Kapoor TM. Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nat Rev Mol Cell Biol 2013; 14: 25–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y, Jin F, Higgins R, McKnight K. The current view for the silencing of the spindle assembly checkpoint. Cell Cycle 2014; 13: 1694–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.