In C. elegans, the germ line is set apart from the soma early in embryogenesis. Several important themes have emerged in specifying and guiding the development of the nascent germ line. At early stages, the germline blastomeres are maintained in a transcriptionally silent state by the transcriptional repressor PIE-1. When this silencing is lifted, it is postulated that correct patterns of germline gene expression are controlled, at least in part, by MES-mediated regulation of chromatin state. Accompanying transcriptional regulation by PIE-1 and the MES proteins, RNA metabolism in germ cells is likely to be regulated by perinuclear RNA-rich cytoplasmic granules, termed P granules. This chapter discusses the molecular nature and possible roles of these various germline regulators, and describes a recently discovered mechanism to protect somatic cells from following a germline fate.
1. Overview
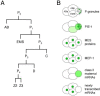
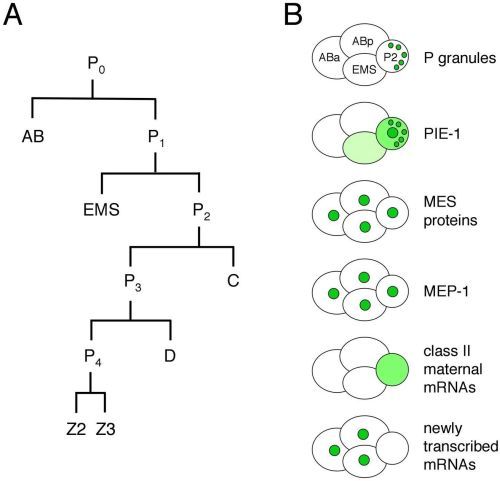
The germ line is set apart from the soma by the 16-24-cell stage of embryogenesis (see Asymmetric cell division and axis formation in the embryo). During early development of the embryo (Figure 1), through a series of asymmetric partitioning events and cell divisions, the germline blastomeres (P0, P1, P2, P3, P4) inherit two unique regulatory systems: cytoplasmic RNA-rich organelles called P granules, and transcriptional repression mediated by PIE-1. In addition, a chromatin-regulation system involving four MES proteins, although not partitioned specifically to the germline blastomeres, is crucial for their subsequent development. This chapter discusses the molecular nature and likely roles of P granules, PIE-1, and the MES proteins in germline specification, and the proposed role of MEP-1 in antagonizing germline fate in somatic cells.
2. pie-1 and transcriptional repression
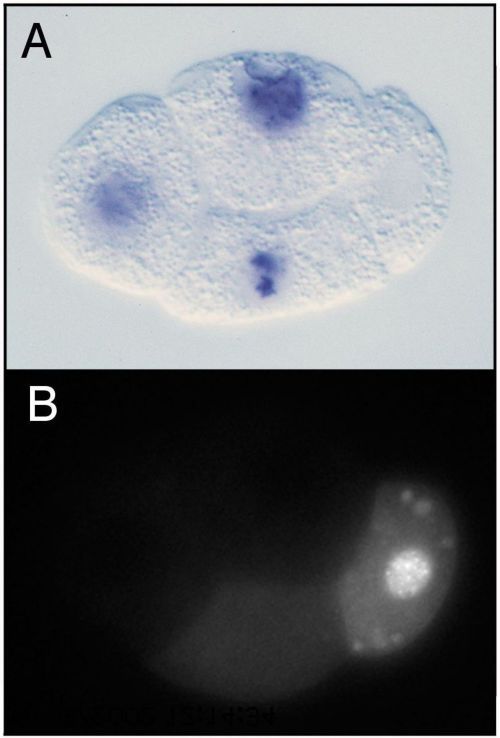
The pie-1 gene was identified in a screen for maternal-effect mutant embryos that produce too many pharyngeal and intestinal cells, hence the name pie for "pharynx and intestine excess" (Mello et al., 1992). The excess of specific somatic cell types in mutant embryos is due to transformation of the P2 germline blastomere into a somatic blastomere like its sister EMS (Mello et al., 1992). Thus, pie-1 is required for P2 to follow a germline fate instead of a somatic fate. PIE-1 is a CCCH-type zinc finger protein with a dynamic localization pattern in the early embryo (Mello et al., 1996; Figure 1). PIE-1 accumulates in the oocyte and 1-cell embryo and during each of the initial four divisions becomes enriched in the germline daughter (Figure 2B), by at least two mechanisms: pre-division enrichment in the cytoplasm destined for the germline daughter cell, and post-division degradation in the cytoplasm of the somatic daughter cell (Reese et al., 2000; DeRenzo et al., 2003; see Asymmetric cell division and axis formation in the embryo). The latter is accomplished by an elongin C/CUL-2 E3 ubiquitin ligase, which is targeted to pie-1 by a bridging protein (ZIF-1) that interacts with both the E3 ligase complex and PIE-1's first zinc finger (DeRenzo et al., 2003). The association of some PIE-1 with P granules (Figure 2B and see below) and centrosomes is not required for its segregation to the germline blastomeres (Reese et al., 2000). Thus, PIE-1 is maternally provided, becomes enriched in each germline blastomere, and is required to maintain germline fate.
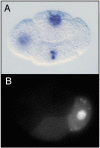
Within each germline blastomere, PIE-1 becomes concentrated in the nucleus (Figure 1 and Figure 2), where it functions as a trancriptional repressor. The initial clues for this repressor function came from three observations (Seydoux et al., 1996): 1) In early wild-type embryos, newly transcribed RNAs are detected in the somatic blastomeres but not in the germline blastomeres (Figure 2A). 2) In pie-1 mutant embryos, newly transcribed RNAs are additionally detected in the germline blastomeres. 3) Driving ectopic pie-1 expression in somatic cells represses embryonic transcription. The mechanism of trancriptional repression has been elegantly dissected (Seydoux and Dunn, 1997; Batchelder et al., 1999; Zhang et al., 2003). Unexpectedly, instead of inhibiting transcriptional initiation, pie-1 appears to function at the level of transcriptional elongation. The process of elongation requires phosphorylation by CDK9 (associated with cyclin T in the P-TEFb complex) of a repeated heptapeptide in the carboxy-terminal domain (CTD) of RNA polymerase II. In transfected mammalian cells, pie-1 inhibits CDK9, through a pie-1 sequence that is very similar to the repeated heptapepide sequence in the RNA pol CTD and that apparently competes for CDK9 (Zhang et al., 2003). Thus, the current view is that pie-1 protects germline fate by blocking transcriptional elongation and preventing expression in the germline blastomeres of genes that promote somatic differentiation.
The pie-1 story highlights two key features of early germline specification: segregation of maternally provided factors to the germline blastomeres, and protection of the germline blastomeres from somatic differentiation signals. pie-1 gene products are maternally provided to the 1-cell embryo along with numerous gene products that encode somatic differentiation cues (e.g., skn-1 mRNA). PIE-1 protein is specifically partitioned to the germline blastomeres (Figure 1 and Figure 2B), while various somatic differentiation factors are delivered to various subsets of blastomeres. SKN-1 illustrates the germline challenge. SKN-1 is a transcription factor that functions in EMS to turn on genes required for E and MS somatic cell fates, but SKN-1 is present in both EMS and its germline sister P2 (Bowerman et al., 1992; Bowerman et al., 1993; see E cell specification). One job of pie-1 is to prevent expression of SKN-1 target genes in P2 (and presumably prevent expression of other somatic differentiation genes in P3 and P4; Mello et al., 1992), and thereby protect germline fate. PIE-1 levels drop shortly after the primordial germ cell P4 divides into Z2 and Z3 at the ~100-cell stage (Mello et al., 1996). Lifting of pie-1 repression presumably sets the stage for turn-on of expression in the nascent germ line of genes appropriate for germline development.
3. The MES proteins and regulation of chromatin
The mes genes were discovered in screens for maternal-effect sterile or "grandchildless" mutants (Capowski et al., 1991). Such genes define factors that are maternally supplied and that function in development of the germ line in offspring. Indeed, mes/mes mothers produce healthy-looking hermaphrodite offspring whose germ line degenerates midway through larval development (Capowski et al., 1991; Paulsen et al., 1995). Interestingly, male offspring usually have a healthy germ line and are fertile (Garvin et al., 1998; see below). The emerging model is that the MES system helps specify the chromatin organization that the nascent germ line (i.e. Z2 and Z3) inherits and that participates in specifying patterns of gene expression once pie-1 repression is lifted.
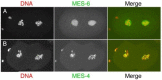
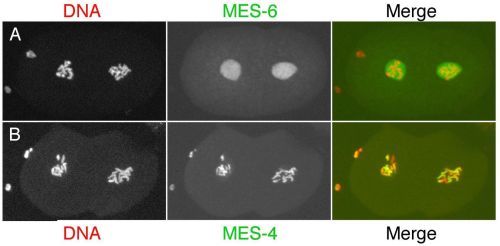
Although the four mes genes have similar mutant phenotypes, they appear to function in two distinct complexes. MES-2, MES-3, and MES-6 are in a complex that resembles the Enhancer of zeste [E(z)] complex in insects and vertebrates (Xu et al., 2001a). Like E(z), MES-2 has a SET domain, which functions in methylating histone tails (Holdeman et al., 1998; Bender et al., 2004). Both E(z) and MES-2, in their respective complexes, methylate Lys27 of histone H3 (Bender et al., 2004; Cao and Zhang, 2004). Trimethylated H3-Lys27 (abbreviated H3K27me3) is a "repressive" chromatin mark - it causes chromatin to be organized into a silenced state (Cao and Zhang, 2004). MES-4 is not in the MES-2/3/6 complex (Xu et al., 2001a) and likely functions with a unique set of partners that have not yet been identified. MES-4 also has a SET domain (Fong et al., 2002) and so, like MES-2, is predicted to methylate histone tails.
The distributions of the MES proteins figure prominently in modeling their probable functions. MES-4 associates with chromatin and shows a striking pattern in the germ line and in early embryos (Figure 3B): it binds in a banded pattern along the lengths of the 5 autosomes and is excluded from the X chromosomes (except the leftmost tip; Fong et al., 2002; Carroll and Strome, unpublished). That distribution is controlled by the MES-2/3/6 complex, as MES-4 binds along the length of the X chromosomes in mes-2, mes-3, or mes-6 mutants (Fong et al., 2002; Carroll and Strome, unpublished). MES-2, MES-3, and MES-6 are concentrated in the nucleoplasm but not significantly on chromatin (Figure 3A; Holdeman et al., 1998; Korf et al., 1998; Xu et al., 2001b). Consequently, the binding specificity of the MES-2/3/6 complex is inferred largely from the distribution of the chromatin mark (H3K27me) that is made by the complex. H3K27me3 is concentrated on the X chromosomes and in addition lightly distributed in a banded pattern along the autosomes (Bender et al., 2004). An appealing scenario is that MES-4 and MES-2/3/6 bind to mutually exclusive chromatin domains and differentially modify histone tails in those domains: the MES-2/3/6 complex creates domains of repressed chromatin, while MES-4 may create domains of active or potentially activatable chromatin. Another possibility for MES-4 is that its main role is to exclude MES-2/3/6 or other repressors from autosomal regions and thereby focus repressor action on the X chromosomes (Fong et al., 2002).
Based on the above and additional information, the working model is that the MES system operates in the maternal germ line and during early embryogenesis (Xu et al., 2001b) to epigenetically mark chromatin domains (or individual genes) and in that way regulate patterns of gene expression in Z2 and Z3 and their descendants. Genes to be kept in a silent state when pie-1 repression is lifted and transcription commences in Z2 and Z3 are marked by MES-2/3/6 with H3K27me3. As discussed in Germline chromatin and Germline genomics, both X chromosomes in hermaphrodites and the single X in males are maintained in a silent state throughout most of germline development. The concentration of the repressive H3K27me3 mark on the Xs is consistent with MES-2/3/6 participating in silencing. As further support, the X chromosomes acquire marks of active chromatin in hermaphrodite offspring of mes-2, mes-3, and mes-6 mothers (Fong et al., 2002). The prediction is that X-linked genes are desilenced in those animals. The two models for MES-4 function, marking genes to be expressed or alternatively participating indirectly in X-chromosome silencing, make different predictions about gene expression patterns in the nascent germ line of mes-4 mutants. Those predictions are currently being tested.
The nascent germ line in males does not absolutely require the MES system (Garvin et al., 1998). This suggests that an alternative system mediates X-chromosome silencing in males (see Germline chromatin) and challenges the view that the MES system serves an important role in regulating expression of autosomal genes in the germ line. Interestingly, in the soma of males, mes-2, mes-3, and mes-6 are known to participate in repressing gene expression from the autosomal Hox gene cluster (Ross and Zarkower, 2003).
4. P granules and regulation of RNA
RNA-rich cytoplasmic "germ granules" are a common feature of germ cells across species and have been invoked for decades as germline "determinants" (reviewed in Saffman and Lasko, 1999). At least some of the proteins associated with the worm versions of these granules, called P granules, are indeed required for germline development (e.g., Kawasaki et al., 1998; Kuznicki et al., 2000). The questions that have dominated P-granule investigations are: how are P granules segregated to the germline blastomeres during early embryogenesis, what are they made of, and what roles do they serve during germline development?
P granules are maternally loaded into the oocyte and progressively partitioned to P1. P2, P3, and finally P4 (Figure 1) by a combination of mechanisms (Hird et al., 1996; Cheeks et al., 2004; see Asymmetric cell division and axis formation in the embryo): 1) posterior movement with bulk cytoplasmic flow, 2) hitching a ride on the nucleus to the future germline side of the cell, and 3) destabilization of granules left behind in the somatic daughter's cytoplasm. These mechanisms accomplish segregation of the majority of detectable granules to P4. Once delivered to P4, P granules become localized primarily around the outside of the nuclear envelope, and that is their primary residence throughout the rest of germline development. When considering what roles P granules are likely to serve, two possibilities seem likely: segregating P granules to each P blastomere serves to deliver maternal proteins and RNAs to the nascent germ line, and once there, nucleus-associated P granules regulate new transcripts as they exit the nucleus (see below).
P granules contain RNA and numerous proteins, all of which are predicted to bind RNA and several of which have been implicated in translational control (see RNA-binding proteins and RNA regulation in the germline). As shown in Table 1, protein components include members of the PGL (RGG motif) family, members of the VASA-related GLH (DEAD box helicase motif) family, the predicted RNA helicase CGH-1, a cap-binding translation initiation factor IFE-1, Sm proteins (best known for their role in RNA splicing), PIE-1 (Figure 2B and see above) and other CCCH finger proteins (MEX-1, POS-1, OMA-1, and OMA-2), the KH-domain proteins MEX-3 and GLD-1, and the GLD-2/GLD-3 poly(A) polymerase. The only components known to be in granules at all stages are the PGL and GLH proteins (Kawasaki et al., 1998; Kawasaki et al., 2004; Gruidl et al., 1996; Kuznicki et al., 2000), leading to the hypothesis that they are important for assembly/stability of P granules. Indeed, molecular epistasis studies suggest a P-granule assembly pathway in which GLH-1 accumulation in granules is necessary for efficient recruitment or retention of the PGL proteins (Kawasaki et al., 1998; Kawasaki et al., 2004; Kuznicki et al., 2000). Interestingly, no one protein has been identified whose loss results in a total absence of P granules.
Table 1
Proteins present in P granules.
The list of RNA components of P granules includes nos-2, pos-1, mex-1, mex-3, skn-1, and gld-1 (Subramaniam and Seydoux, 1999; Schisa et al., 2001). Basically, all of the six developmentally regulated maternal mRNAs tested showed an accumulation in P granules, while structural RNAs (5S, 5.8S, 18S, and 26S) and abundant housekeeping mRNAs (actin and β-tubulin) did not (Schisa et al., 2001). Most of these studies were done in adult germ lines, where P granules are perinuclear and in fact associated with clusters of nuclear pores (Pitt et al., 2000). Even P granules dissociated from the nuclear envelope contain nuclear-pore proteins and pore-like structures (Pitt et al., 2000). Thus, a reasonable scenario is that many or most developmentally regulated maternal mRNAs pass through and are transiently retained in P granules as they exit from nuclei. An additional scenario is that P granules participate in delivering and then stabilizing certain maternal mRNAs in the germline cells of embryos. Indeed, the P-granule-associated mRNAs listed above all belong to the class of messengers that are selectively retained in the germline blastomeres and degraded in the somatic blastomeres (Seydoux and Fire, 1994; see Translational control of maternal RNAs).
It is likely that P granules serve diverse functions at different times of development. Most relevant to a discussion of germline specification is what roles maternally contributed P granules serve in the nascent germ line. Mutants lacking multiple PGL proteins offer some insights. The offspring from homozygous pgl-1; pgl-3 double mutant mothers show severe defects in germline proliferation and gametogenesis and develop into sterile adults (Kawasaki et al., 2004; see Germline proliferation and its control). pgl-1 single mutants show similar defects, but only at elevated temperature (Kawasaki et al., 1998), suggesting that PGL-1 is required to maintain P-granule integrity and function at elevated temperature, but that PGL-3 can serve this role at low temperature. A similar situation appears to hold for GLH-1 and GLH-4 (Kuznicki et al., 2000). We still do not understand the molecular causes of the germline proliferation or gametogenesis defects observed in pgl and glh mutants. Important directions for future analysis are figuring out the roles of various P-granule proteins, when and how various RNA components of P granules are regulated by their granule association, and how such regulation impacts germline events.
5. mep-1 and avoiding germline specification
Germline blastomeres are thought to be protected from following a somatic fate at least in part by pie-1-mediated transcriptional repression, as discussed above. Recent studies suggest that the converse, somatic cells being protected from following a germline fate, may also occur. Members of the synMuv B class of chromatin regulators appear to play a crucial role, which was discovered through analysis of the mep-1 gene. In mep-1 mutant embryos, somatic cells display germline traits (e.g., activate pgl-1 and other P-granule genes) as well as somatic traits; mutants arrest as young larvae (Unhavaithaya et al., 2002; Belfiore et al., 2002). Surprisingly, loss of mes gene function suppresses the mep-1 "ectopic P granules" and larval-arrest phenotypes; many mep-1; mes double mutants develop into viable albeit sterile adults (Unhavaithaya et al., 2002). MEP-1 is in a complex with LET-418 and HDA-1, two subunits of the nucleosome remodeling and histone deacetylase (NuRD) complex (see Germline chromatin). Intriguingly, PIE-1 appears to regulate the MEP-1/LET-418/HDA-1 complex. In transfected vertebrate cells, PIE-1 interacts directly with MEP-1 and, perhaps through that interaction, inhibits the histone deacetylase activity of HDA-1. Furthermore, ectopic expression of pie-1 in C. elegans somatic cells mimics the mep-1 mutant phenotype and causes larval arrest and ectopic expression of at least one P-granule protein (Unhavaithaya et al., 2002).
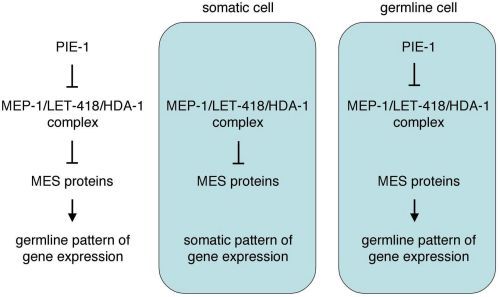
A model that may explain the genetic interactions of MEP-1, PIE-1, and the MES proteins is shown in Figure 4 (Unhavaithaya et al., 2002; Shin and Mello, 2003). The MES proteins establish and maintain a particular "germline state" of chromatin that is required for germline patterns of gene expression. All cells of the early embryo inherit that germline chromatin state from the oocyte and sperm (Figure 1). In somatic cells, the MEP-1 complex remodels that chromatin organization, to allow somatic patterns of gene expression and appropriate somatic differentiation. In the germline blastomeres, PIE-1 antagonizes the MEP-1 complex, to protect the germline state of chromatin. According to this model, loss of mep-1 would cause the germline chromatin state to persist in somatic cells, leading to defective somatic development. Concomitant loss of the MES's would eliminate the germline chromatin state and make MEP-1 dispensable in somatic cells, restoring somatic development. Loss of pie-1 would allow MEP-1 to remodel the germline chromatin state to a somatic state in the germline cells, leading to defective germline development. Tests of this model will reveal whether MEP-1 in the soma selectively represses germline fate or more generally regulates gene expression patterns, and whether PIE-1 inhibits the MEP-1 complex in C. elegans embryos. Another area for investigation is the essential role of mep-1 during later germline development (Belfiore et al., 2002).
Figure 4
A model to explain the genetic interactions between MEP-1, PIE-1, and the MES proteins.
6. Summary and future directions
Germline specification appears to involve: establishment of a particular chromatin state in the parental germ line and transmission of that state to the early embryo (via the MES system); preservation of that state specifically in the germline blastomeres (via PIE-1 inhibition of MEP-1); transcriptional repression in the germline blastomeres (via PIE-1); and regulation of RNA metabolism in the primordial germ cells (via PIE-1 and P granules). An exciting frontier for investigation is figuring out the epigenetic components of this regulation: what "chromatin states" are established by the MES proteins, which genes or chromosomal domains are regulated in this way, and whether and how the MEP-1/LET-418/HDA-1 complex "remodels" chromatin. PIE-1 is clearly a major germline player, with nuclear functions that include blocking transcriptional elongation and perhaps inhibiting MEP-1/LET-418/HDA-1, and cytoplasmic functions that include regulation of mRNA stability and expression (Tenenhaus et al., 2001). One mRNA target of PIE-1 is nos-2 RNA, whose function promotes incorporation of Z2 and Z3 into the somatic gonad (Subramaniam and Seydoux, 1999; see Translational control of maternal RNAs). Elucidating PIE-1's various functions and their relationships to one another is a high priority. Finally, P granules, although known to exist for over 20 years, remain relatively mysterious. Learning which mRNAs they regulate and whether regulation is at the level of delivery, stability, processing, translation, or degradation will significantly advance our understanding of these unique germline organelles.
7. References
- Amiri A. , Keiper B.D. , Kawasaki I. , Fan Y. , Kohara Y. , Rhoads R.E. , Strome S. An isoform of eIF4E is a component of germ granules and is required for spermatogenesis in C. elegans. Development. (2001);128:3899–3912. [PMC free article: PMC2430591] [PubMed: 11641215]
- Barbee S.A. , Lublin A.L. , Evans T.C. A novel function for the Sm proteins in germ granule localization during C. elegans embryogenesis. Curr. Biol. (2002);12:1502–1506. [PubMed: 12225665]
- Batchelder C. , Dunn M.A. , Choy B. , Suh Y. , Cassie C. , Shim E.Y. , Shin T.H. , Mello C. , Seydoux G. , Blackwell T.K. Transcriptional repression by the Caenorhabditis elegans germ-line protein PIE-1. Genes Dev. (1999);13:202–212. [PMC free article: PMC316391] [PubMed: 9925644]
- Belfiore M. , Mathies L.D. , Pugnale P. , Moulder G. , Barstead R. , Kimble J. , Puoti A. The MEP-1 zinc-finger protein acts with MOG DEAH box proteins to control gene expression via the fem-3 3' untranslated region in Caenorhabditis elegans. RNA. (2002);8:725–739. [PMC free article: PMC1370292] [PubMed: 12088146]
- Bender L.B. , Cao R. , Zhang Y. , Strome S. The MES-2/MES-3/MES-6 complex and regulation of histone H3 methylation in C. elegans. Curr. Biol. (2004);14:1639–1643. [PubMed: 15380065]
- Bowerman B. , Draper B.W. , Mello C.C. , Priess J.R. The maternal gene skn-1 encodes a protein that is distributed unequally in early C. elegans embryos. Cell. (1993);74:443–452. [PubMed: 8348611]
- Bowerman B. , Eaton B.A. , Priess J.R. skn-1, a maternally expressed gene required to specify the fate of ventral blastomeres in the early C. elegans embryo. Cell. (1992);68:1061–1075. [PubMed: 1547503]
- Cao R. , Zhang Y. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr. Opin. Genet. Dev. (2004);14:155–164. [PubMed: 15196462]
- Capowski E.E. , Martin P. , Garvin C. , Strome S. Identification of grandchildless loci whose products are required for normal germ-line development in the nematode Caenorhabditis elegans. Genetics. (1991);129:1061–1072. [PMC free article: PMC1204771] [PubMed: 1783292]
- Cheeks R.J. , Canman J.C. , Gabriel W.N. , Meyer N. , Strome S. , Goldstein B. C. elegans PAR proteins function by mobilizing and stabilizing asymmetrically localized protein complexes. Curr. Biol. (2004);14:851–862. [PubMed: 15186741]
- DeRenzo C. , Reese K.J. , Seydoux G. Exclusion of germ plasm proteins from somatic lineages by cullin-dependent degradation. Nature. (2003);424:685–689. [PMC free article: PMC1892537] [PubMed: 12894212]
- Draper B.W. , Mello C.C. , Bowerman B. , Hardin J. , Priess J.R. MEX-3 is a KH domain protein that regulates blastomere identity in early C. elegans embryos. Cell. (1996);87:205–216. [PubMed: 8861905]
- Eckmann C.R. , Kraemer B. , Wickens M. , Kimble J. GLD-3, a bicaudal-C homolog that inhibits FBF to control germline sex determination in C. elegans. Dev. Cell. (2002);3:697–710. [PubMed: 12431376]
- Fong Y. , Bender L. , Wang W. , Strome S. Regulation of the different chromatin states of autosomes and X chromosomes in the germ line of C. elegans. Science. (2002);296:2235–2238. [PMC free article: PMC2435369] [PubMed: 12077420]
- Garvin C. , Holdeman R. , Strome S. The phenotype of mes-2, mes-3, mes-4 and mes-6, maternal-effect genes required for survival of the germline in Caenorhabditis elegans, is sensitive to chromosome dosage. Genetics. (1998);148:167–185. [PMC free article: PMC1459798] [PubMed: 9475730]
- Gruidl M.E. , Smith P.A. , Kuznicki K.A. , McCrone J.S. , Kirchner J. , Roussell D.L. , Strome S. , Bennett K.L. Multiple potential germ-line helicases are components of the germ-line-specific P granules of Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. (1996);93:13837–13842. [PMC free article: PMC19442] [PubMed: 8943022]
- Guedes S. , Priess J.R. The C. elegans MEX-1 protein is present in germline blastomeres and is a P granule component. Development. (1997);124:731–739. [PubMed: 9043088]
- Hird S.N. , Paulsen J.E. , Strome S. Segregation of germ granules in living Caenorhabditis elegans embryos: cell-type-specific mechanisms for cytoplasmic localisation. Development. (1996);122:1303–1312. [PubMed: 8620857]
- Holdeman R. , Nehrt S. , Strome S. MES-2, a maternal protein essential for viability of the germline in Caenorhabditis elegans, is homologous to a Drosophila Polycomb group protein. Development. (1998);125:2457–2467. [PubMed: 9609829]
- Jones A.R. , Francis R. , Schedl T. GLD-1, a cytoplasmic protein essential for oocyte differentiation, shows stage- and sex-specific expression during Caenorhabditis elegans germline development. Dev. Biol. (1996);180:165–183. [PubMed: 8948583]
- Kawasaki I. , Amiri A. , Fan Y. , Meyer N. , Dunkelbarger S. , Motohashi T. , Karashima T. , Bossinger O. , Strome S. The PGL family proteins associate with germ granules and function redundantly in Caenorhabditis elegans germline development. Genetics. (2004);167:645–661. [PMC free article: PMC1470885] [PubMed: 15238518]
- Kawasaki I. , Shim Y.H. , Kirchner J. , Kaminker J. , Wood W.B. , Strome S. PGL-1, a predicted RNA-binding component of germ granules, is essential for fertility in C. elegans. Cell. (1998);94:635–645. [PubMed: 9741628]
- Korf I. , Fan Y. , Strome S. The Polycomb group in Caenorhabditis elegans and maternal control of germline development. Development. (1998);125:2469–2478. [PubMed: 9609830]
- Kuznicki K.A. , Smith P.A. , Leung-Chiu W.M. , Estevez A.O. , Scott H.C. , Bennett K.L. Combinatorial RNA interference indicates GLH-4 can compensate for GLH-1; these two P granule components are critical for fertility in C. elegans. Development. (2000);127:2907–2916. [PubMed: 10851135]
- Lin R. A gain-of-function mutation in oma-1, a C. elegans gene required for oocyte maturation, results in delayed degradation of maternal proteins and embryonic lethality. Dev. Biol. (2003);258:226–239. [PubMed: 12781695]
- Mello C.C. , Draper B.W. , Krause M. , Weintraub H. , Priess J.R. The pie-1 and mex-1 genes and maternal control of blastomere identity in early C. elegans embryos. Cell. (1992);70:163–176. [PubMed: 1623520]
- Mello C.C. , Schubert C. , Draper B. , Zhang W. , Lobel R. , Priess J.R. The PIE-1 protein and germline specification in C. elegans embryos. Nature. (1996);382:710–712. [PubMed: 8751440]
- Navarro R.E. , Shim E.Y. , Kohara Y. , Singson A. , Blackwell T.K. cgh-1, a conserved predicted RNA helicase required for gametogenesis and protection from physiological germline apoptosis in C. elegans. Development. (2001);128:3221–3232. [PubMed: 11546739]
- Ogura K. , Kishimoto N. , Mitani S. , K. Gengyo-Ando , Kohara Y. Translational control of maternal glp-1 mRNA by POS-1 and its interacting protein SPN-4 in Caenorhabditis elegans. Development. (2003);130:2495–2503. [PubMed: 12702662]
- Paulsen J.E. , Capowski E.E. , Strome S. Phenotypic and molecular analysis of mes-3, a maternal-effect gene required for proliferation and viability of the germ line in C. elegans. Genetics. (1995);141:1383–1398. [PMC free article: PMC1206874] [PubMed: 8601481]
- Pitt J.N. , Schisa J.A. , Priess J.R. P granules in the germ cells of Caenorhabditis elegans adults are associated with clusters of nuclear pores and contain RNA. Dev. Biol. (2000);219:315–233. [PubMed: 10694425]
- Reese K.J. , Dunn M.A. , Waddle J.A. , Seydoux G. Asymmetric segregation of PIE-1 in C. elegans is mediated by two complementary mechanisms that act through separate PIE-1 protein domains. Mol. Cell. (2000);6:445–455. [PubMed: 10983990]
- Ross J.M. , Zarkower D. Polycomb group regulation of Hox gene expression in C. elegans. Dev. Cell. (2003);4:891–901. [PubMed: 12791273]
- Saffman E.E. , Lasko P. Germline development in vertebrates and invertebrates. Cell. Mol. Life Sci. (1999);55:1141–1163. [PubMed: 10442094]
- Schisa J.A. , Pitt J.N. , Priess J.R. Analysis of RNA associated with P granules in germ cells of C. elegans adults. Development. (2001);128:1287–1298. [PubMed: 11262230]
- Seydoux G. , Dunn M.A. Transcriptionally repressed germ cells lack a subpopulation of phosphorylated RNA polymerase II in early embryos of Caenorhabditis elegans and Drosophila melanogaster. Development. (1997);124:2191–2201. [PubMed: 9187145]
- Seydoux G. , Fire A. Soma-germline asymmetry in the distributions of embryonic RNAs in Caenorhabditis elegans. Development. (1994);120:2823–2834. [PubMed: 7607073]
- Seydoux G. , Mello C.C. , Pettitt J. , Wood W.B. , Priess J.R. , Fire A. Repression of gene expression in the embryonic germ lineage of C. elegans. Nature. (1996);382:713–716. [PubMed: 8751441]
- Shimada M. , Kawahara H. , Doi H. Novel family of CCCH-type zinc-finger proteins, MOE-1, -2 and -3, participates in C. elegans oocyte maturation. Genes Cells. (2002);7:933–947. [PubMed: 12296824]
- Shin T.H. , Mello C.C. Chromatin regulation during C. elegans germline development. Curr. Opin. Genet. Dev. (2003);13:455–462. [PubMed: 14550409]
- Subramaniam K. , Seydoux G. nos-1 and nos-2, two genes related to Drosophila nanos, regulate primordial germ cell development and survival in Caenorhabditis elegans. Development. (1999);126:4861–4871. [PubMed: 10518502]
- Tabara H. , Hill R.J. , Mello C.C. , Priess J.R. , Kohara Y. pos-1 encodes a cytoplasmic zinc-finger protein essential for germline specification in C. elegans. Development. (1999);126:1–11. [PubMed: 9834181]
- Tenenhaus C. , Subramaniam K. , Dunn M.A. , Seydoux G. PIE-1 is a bifunctional protein that regulates maternal and zygotic gene expression in the embryonic germ line of Caenorhabditis elegans. Genes Dev. (2001);15:1031–1040. [PMC free article: PMC312670] [PubMed: 11316796]
- Unhavaithaya Y. , Shin T.H. , Miliaras N. , Lee J. , Oyama T. , Mello C.C. MEP-1 and a homolog of the NURD complex component Mi-2 act together to maintain germline-soma distinctions in C. elegans. Cell. (2002);111:991–1002. [PubMed: 12507426]
- Wang L. , Eckmann C.R. , Kadyk L.C. , Wickens M. , Kimble J. A regulatory cytoplasmic poly(A) polymerase in Caenorhabditis elegans. Nature. (2002);419:312–316. [PubMed: 12239571]
- Xu L. , Fong Y. , Strome S. The Caenorhabditis elegans maternal-effect sterile proteins, MES-2, MES-3, and MES-6, are associated in a complex in embryos. Proc. Natl. Acad. Sci. U.S.A. (2001a);98:5061–5066. [PMC free article: PMC33163] [PubMed: 11320248]
- Xu L. , Paulsen J. , Yoo Y. , Goodwin E.B. , Strome S. Caenorhabditis elegans MES-3 is a target of GLD-1 and functions epigenetically in germline development. Genetics. (2001b);159:1007–1017. [PMC free article: PMC1461849] [PubMed: 11729149]
- Zhang F. , Barboric M. , Blackwell T.K. , Peterlin B.M. A model of repression: CTD analogs and PIE-1 inhibit transcriptional elongation by P-TEFb. Genes Dev. (2003);17:748–758. [PMC free article: PMC196018] [PubMed: 12651893]
Footnotes
- *
Edited by Judith Kimble. Last revised July 8, 2005. Published July 28, 2005. This chapter should be cited as: Strome, S. Specification of the germ line (July 28, 2005), WormBook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.9.1, http://www
.wormbook.org [PMC free article: PMC4781390] [PubMed: 18050414].
- §
To whom correspondence should be addressed. E-mail: ude.anaidni.oib@emortss
Publication Details
Author Information and Affiliations
Authors
Susan Strome,§.Affiliations
Publication History
Created: July 28, 2005.
Copyright
All WormBook content, except where otherwise noted, is licensed under a Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Publisher
WormBook, Pasadena (CA)
NLM Citation
Strome S. Specification of the germ line. 2005 Jul 28. In: WormBook: The Online Review of C. elegans Biology [Internet]. Pasadena (CA): WormBook; 2005-2018.