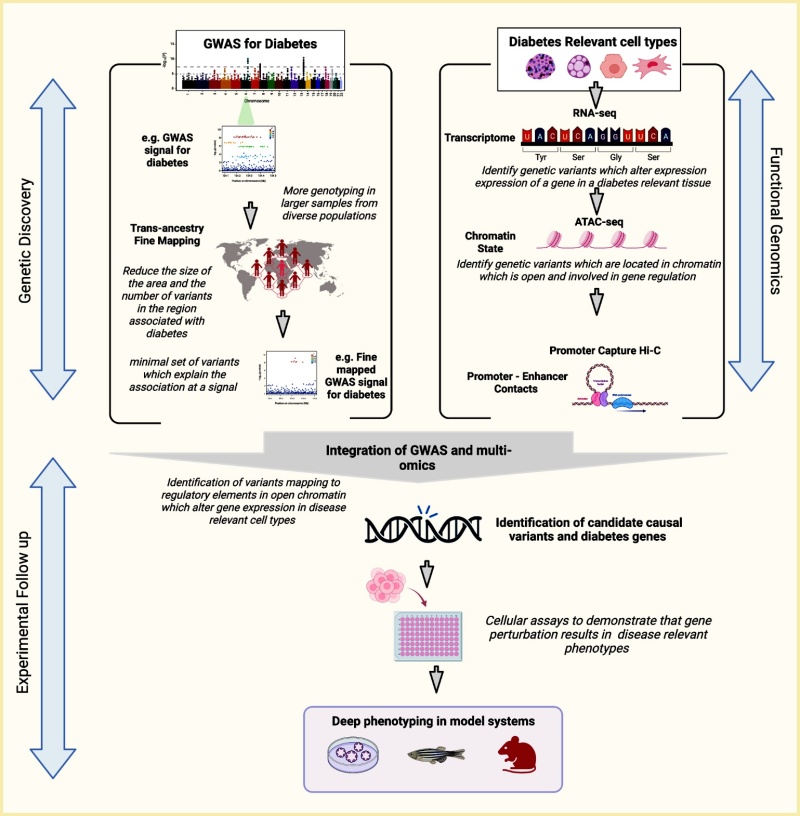
Moving From Variant to Function Using Epigenomics.
Any given GWAS locus is likely to contain multiple potential causal variants that are indistinguishable due to LD. Fine-mapping, particularly in populations that have different LD structures, can be used to reduce the number of potential variants. Variants in a DNA sequence with a presumed functional effect, such as a protein-coding sequence or regulatory annotation in a disease-relevant tissue, are more likely to be the causal variant. The potential for a noncoding variant to be functional can be assessed by integrating DNA sequence data with other types of -omics data, such as chromatin state and gene expression in diabetes-relevant cell types (e.g., islets, adipocytes, hepatocytes, and myocytes). Genes that are influenced by altered regulation can be identified through transcriptomics (cis-eQTLs) and through chromatin capture experiments in disease-relevant cell types. Candidate causal variants and diabetes genes can then be prioritized for validation and functional follow-up in cell and animal models. ATAC-seq, assay for transposase-accessible chromatin using sequencing; eQTL, expression quantitative trait loci; GWAS, genome-wide association study; LD, linkage disequilibrium.
SOURCE: Original figure constructed by A. L. Gloyn, using BioRender.com