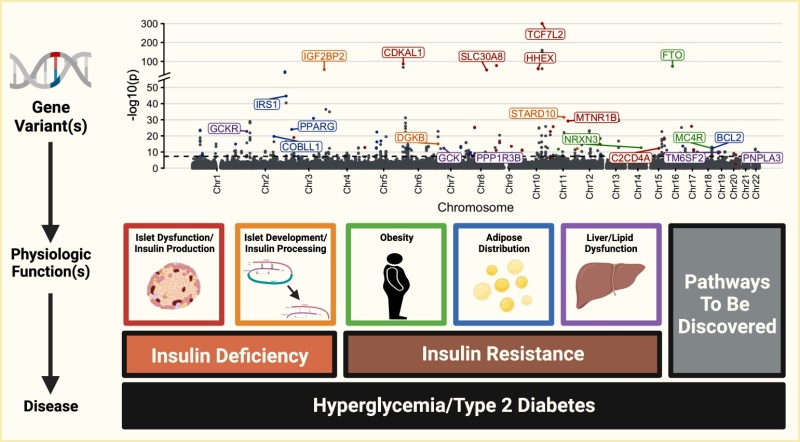
From: Genetics of Type 2 Diabetes

Diabetes in America is in the public domain of the United States. You may use the work without restriction in the United States.
NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.