This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License. To view a copy of this license, visit https://creativecommons.org/licenses/by-nc-nd/4.0/
NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Gilmore MS, Clewell DB, Ike Y, et al., editors. Enterococci: From Commensals to Leading Causes of Drug Resistant Infection [Internet]. Boston: Massachusetts Eye and Ear Infirmary; 2014-.

Enterococci: From Commensals to Leading Causes of Drug Resistant Infection [Internet].
Show detailsBiofilm Formation by Enterococci: Concepts and Caveats
This chapter discusses biofilm formation in enterococci and its role in the biology of these organisms, especially in relation to opportunistic infections. As is the case for many microbes, the pace of enterococcal biofilm research has quickened in the past few years, and numerous genes and gene products affecting biofilm formation have been identified. At the same time, this research has not resulted in a comprehensive understanding of the critical steps in the process, particularly those steps that are involved in initiating the transition from planktonic growth to biofilm growth. Both the physical/chemical environment and the type of surface substratum on which the biofilm develops have a tremendous impact on the process, even with a single strain. A cursory scan of 15-20 years of publications on enterococcal biofilms indicates that the number of different experimental conditions employed to measure biofilm formation is comparable to the number of published papers on the topic. Clearly, these efforts have contributed to the discovery of different biofilm determinants and processes by different research groups, and in some cases, have provided contradictory reports and conclusions. Therefore, we begin this chapter with a brief summary of central principles, definitions, and questions to provide a framework for subsequent sections that contain more detailed descriptions of relevant research in this area. We conclude with a discussion of future research directions that may clarify and increase the level of understanding of this important microbial activity.
Biofilms are microbial communities resulting from the adherence of planktonic organisms to an abiotic surface, usually followed by growth. It is generally recognized that microbial species spend much of their time in the biofilm state, and that biofilm formation plays a critical role in infections. Thus, research on enterococcal biofilms is driven by its potential to yield new insights into the pathogenesis of opportunistic infections, their treatment, and their prevention. It is commonly hypothesized that the formation of biofilms represents a developmental process that involves a shift in physiology from planktonic growth, organization of the adherent bacteria into structured communities physically linked by an extracellular matrix, and where communication between members of the community coordinates gene expression and metabolic activity (O'Toole, Kaplan, & Kolter, 2000). While this model has been a useful paradigm to guide experimental investigations, it has not been completely validated, especially for non-sporulating bacteria, and strong arguments have been made for considering alternatives (Monds & O'Toole, 2009). The developmental model implies that there are critical functions specific for biofilm formation, and it can also be inferred that there may be conservation of these functions among different bacteria.
Examination of the published literature suggests that many, if not most, determinants of enterococcal biofilm formation identified to date are members of global networks important for adaptation to a variety of environments, and that factors such as the medium composition, physical/chemical conditions, and type of surface used for cultivation of biofilms may have a larger influence on the “biofilm functions” than biofilm growth itself. Investigators have also used different criteria to quantify biofilm formation. Conclusions about optimal biofilm formation can vary greatly depending on whether total biomass, enumeration of cell counts by microscopy or plate counting, or the ratio of biofilm biomass to bacterial growth in the planktonic phase is used as the primary criterion (Di Rosa, et al., 2006; Creti, Koch, Fabretti, Baldassarri, & Huebner, 2006; Kristich, Li, Cvitkovich, & Dunny, 2004; Hufnagel, Koch, Creti, Baldassarri, & Huebner, 2004; Sandoe, Witherden, Cove, Heritage, & Wilcox, 2003; Baldassarri, et al., 2001). Given these discrepancies, researchers interested in linking biofilm formation to pathogenesis need to carefully consider the ways in which their laboratory assays closely reflect conditions in an animal or human host, to compare results using different in vitro assays, and to examine the role of biofilm determinants of interest in the context of a relevant animal model. Pathogenesis and models of enterococcal infection discusses the use of various animal models to investigate enterococcal virulence in more detail. While it is conceivable that biofilm defects could result in the attenuation of virulence in any animal model, the rabbit cardiac catheterization model for experimental endocarditis (Frank, Barnes, Grindle, Manias, Schlievert, & Dunny, 2012; Singh K. V., Nallapareddy, Sillanpää, & Murray, 2010; Chuang, Schlievert, Wells, Manias, TRipp, & Dunny, 2009; Nallapareddy, Singh, & Murray, 2008; Nallapareddy S. R., et al., 2006) and the mouse urinary tract infection models (Sillanpää, et al., 2010; Guiton, Hung, Hancock, Caparon, & Hultgren, 2010; Singh, Nallapareddy, & Murray, 2007; Shankar, Lockatell, Baghdayan, Drachenberg, Gilmore, & Johnson, 2001) may be especially informative in examining the role of biofilm formation in the host on pathogenesis.
Epidemiology of Biofilm-Related Enterococcal Infections
The earliest reports of enterococci in association with infection-related biofilms were probably from the studies that identified Enterococcus faecalis in infected vascular ports from patients (Reed, Moody, Newman, Light, & Costerton, 1986) and in a urinary stone (Nickel, Reid, Bruce, & Costerton, 1986). Later, the expression of two E. faecalis-specific surface antigens, shown to be enriched when the bacteria were cultured in serum or brain heart infusion in vitro, or during growth on silastic discs in the rabbit peritoneum (but not in a chemically defined broth), were correlated to the potential to form biofilms during infection (Lambert, Shorrock, Aitchison, Dominique, Power, & Costerton, 1990). A number of studies in the early 1990s evaluated bacterial communities that were associated with indwelling catheters, and frequently isolated either E. faecalis alone or as part of a polymicrobial species on these devices (Reid, Denstedt, Kang, Lam, & Nause, 1992; Jansen, Goodman, & Ruiten, 1993; Stickler, King, Winters, & Morris, 1993; Jass, Phillips, Allan, Costerton, & Lappin-Scott, 1994; Keane, Bonner, Johnston, Zafar, & Gorman, 1994; Koivusalo, et al., 1996).
In an effort to examine approaches to combat biofilm-associated infections, numerous studies evaluated multiple antimicrobials, either alone or in combination, as agents used to coat catheter surfaces to prevent microbial adhesion. For instance, Farber et al (Farber & Wolff, 1993) reported that salicylic-acid–coated catheters showed diminished adherence of many Gram-negative bacteria, yeast, and E. faecalis. In another study, Raad et al (Raad, Darouiche, Hachem, Sacilowski, & Bodey, 1995) found that the inhibitory activities of catheters coated with minocycline and rifampin against Staphylococcus epidermidis, Staphylococcus aureus, and E. faecalis strains, for example, were significantly better than those of catheters coated with vancomycin (P < 0.05). In other studies, the efficacies of clinical dosing schedules of once-daily versus thrice-daily regimens of aminoglycosides in treating endocarditis were correlated to the killing of E. faecalis coated on glass beads, and concluded that no differences existed between the two regimens (Schwank & Blaser, 1996). Static in vitro biofilms perfused with the glycopeptide antibiotics vancomycin and teicoplanin, either alone or in combination, showed slower growth in the presence of either antibiotic, and the effect of the combination was a further 2-3 log reduction in growth rate and a 3-log reduction in viability of the biofilm (Foley & Gilbert, 1997). In other related studies, Bowers et al (Bower, Daeschel, & McGuire, 1998) explored the potential of peptide antimicrobials, such as lysozyme and nisin, to inhibit E. faecalis adhesion to treated catheters. The studies reported above established the fact that E. faecalis is a prominent bacterium encountered in biofilms, especially in catheter-related infections, and raised the concern that the therapeutic options to treat such infections might be somewhat limited.
Later studies attempted to correlate the ability of enterococcal isolates from clinical settings to biofilm formation in vitro, and also began to associate specific bacterial factors with this phenotype. One prospective study in Italy determined that 80% of E. faecalis clinical isolates and 48% of E. faecium isolates formed biofilms (Baldassarri, et al., 2001), while another study of 47 clinical isolates found the biofilm phenotype to be associated with 87% of E. faecalis, but only 16% of E. faecium strains (Duprè, Zanetti, Schito, Fadda, & Sechi, 2003). A later study in the same geographical region found that 96% of 52 E. faecalis isolates from orthopedic infections produced biofilms in vitro (Baldassarri, et al., 2006). A study in Spain found that a little over one-half of 152 E. faecalis clinical isolates were able to form biofilms in vitro with no biofilm phenotype exhibited by Enterococcus faecium, Enterococcus gallinarum, or Enterococcus avium strains (Toledo-Arana, et al., 2001). Studies in the UK analyzed 109 isolates from bloodstream infections and showed that, while all of the E. faecalis isolates (n=71) formed biofilms, less than one-half (16/38) of the E. faecium isolates did so (7). This study further found that E. faecalis isolates from catheter-related bloodstream infections (CRBSI) produced more biofilm, as compared to non-CRBSI isolates. Another study in Europe also found a significantly higher ability to form biofilms among E. faecalis isolates, but not from the four other enterococcal species studied (Dworniczek, Wojciech, Sobieszczańska, & Seniuk, 2005). A total of 163 E. faecalis isolates (51 from outside the US) were evaluated in a US study, which concluded that biofilm formation is common among E. faecalis clinical as well as fecal isolates. This study also found that the percentage and degree of biofilm formation are significantly greater (P = 0.001) among endocarditis isolates than among isolates from other sources. All 352 samples of E. faecalis isolated from urinary tract infections in one study in Japan were capable of forming biofilms and could be grouped into weak, medium, or strong biofilm formers (Seno, Kariyama, Mistuhata, Monden, & Kumon, 2005). A study of 171 clinical isolates of enterococci at a tertiary care hospital in South India concluded that about a quarter of the E. faecalis strains (n=44) were capable of forming biofilms in vitro, in contrast to none of the 25 E. faecium isolates in that cohort (Prakash, Rao, & Parija, 2005).
Enterococci play a role in endodontic failure and are often isolated from the root canal system. The results of one study showed that out of 100 root-filled teeth with apical periodontitis, 69% of the isolated bacteria were facultative and 50% of those were enterococci (Dahlén, Samuelsson, Molander, & Reit, 2000). E. faecalis is responsible for the vast majority of human enterococcal endodontic infection, and is usually the only Enterococcus species isolated from the obturated root canal (Love, 2001). The biofilm-forming ability of E. faecalis isolated from the oral cavity has also recently been evaluated (Duggan & Sedgley, 2007). This study revealed that unlike other clinical isolates, especially endocarditis strains, the E. faecalis from oral and endodontic sources were poor biofilm formers. In contrast, another recent study observed that surface conditioning of dentin with saliva and starvation can enhance the adherence of E. faecalis to dentin (George & Kishen, 2007). No significant difference in the prevalence of this species between subgingival biofilm (34.6%) and saliva (35.1%) samples was observed in another study (Souto & Colombo, 2008). E. faecalis was detected significantly more often in saliva and subgingival samples of periodontitis patients (40.5% and 47.8%, respectively) as compared to controls (14.6% and 17.1%, respectively; P <0.05), but the correlation with the biofilm phenotype is unclear. The consensus from the epidemiological studies above is that E. faecalis strains are better biofilm formers overall than E. faecium strains, and that the biofilm phenotype is an important contributory factor to enterococcal pathogenesis. These observations set the stage to explore the role of specific bacterial factors that mediate biofilm formation in E. faecalis and E. faecium in greater depth.
Factors that Affect Biofilm Formation
The past decade has seen the identification of a number of enterococcal genes that play a role in biofilm formation and maturation, especially in E. faecalis (Paganelli, Willems, & Leavis, 2012). These include surface adhesions, such as cell wall-associated proteins, autolysins, and glycolipids, which predominate early during the adhesion phase, and extend to polysaccharides, lipoteichoic acid, extracellular DNA, and proteases, which contribute to biofilm maturation. A brief description of each of these biofilm factors is presented below and summarized in Table 1.
Growth conditions
An indirect observation of the influence of growth medium on the ability of E. faecalis to form biofilms was probably made in 1990 by Lambert et al (Lambert, Shorrock, Aitchison, Dominique, Power, & Costerton, 1990), who observed that the expression of two E. faecalis surface antigens was enriched when the organisms were cultured in serum or brain heart infusion in vitro, or during growth on silastic discs in the rabbit peritoneum, but not in a chemically defined broth. Kristich et al (Kristich, Li, Cvitkovich, & Dunny, 2004) carefully evaluated different media with respect to biofilm formation by E. faecalis OG1RF and revealed a significant effect of growth medium on biofilm dynamics. While growth in TSB, M17, and M9YE medium caused biofilm accumulation to slow after 6-8 hours of growth, biofilm production in brain-heart infusion (BHI) or Todd-Hewitt yeast extract reached a plateau at 4 hours and subsequently declined. It must be noted that this study defined biofilm formation as the ratio of optical density in biofilm to optical density in planktonic mode, while most other studies below have relied on absolute optical densities of biofilms.
The influence of additional sugar, such as glucose, in the growth medium used to cultivate enterococci for biofilm assays became apparent early on, when adhesion to polystyrene microtiter plates was adopted as the method of choice for evaluating biofilm formation in vitro (Toledo-Arana, et al., 2001). In these initial studies, trypticase soy broth (TSB) supplemented with either 0.25 or 0.5% (w/v) of glucose was used to cultivate the bacteria. Subsequent studies indicated that the addition of 1% glucose (w/v) to TSB enhanced biofilm production by some strains, as compared to growth without additional glucose (Baldassarri, et al., 2001; Pillai, Sakoulas, Eliopoulos, Moellering, Jr., Murray, & Inouye, 2004). Another study in agreement with this phenomenon noted optimal biofilm formation with 0.5% glucose added to TSB, as compared to 0.2% or no glucose added (Kristich, Li, Cvitkovich, & Dunny, 2004). The enhancement of biofilm formation by E. faecalis OG1RF was noted with 1% glucose added to TSB, but the same effect was not evident in a fsr mutant or a gelE mutant (Pillai, Sakoulas, Eliopoulos, Moellering, Jr., Murray, & Inouye, 2004). One possible explanation suggested for this observed effect was that a glucose-dependent transcriptional regulator controlled fsr, either directly or indirectly, and that fsr exerts catabolite control over biofilm formation through downstream proteases GelE and SprE. The bopABCD operon (biofilm on plastic) in E. faecalis, deemed to have a putative maltose metabolism function, is regulated by fsr and important for biofilm formation (Hufnagel, Koch, Creti, Baldassarri, & Huebner, 2004; Bourgogne, Hilsenbeck, Dunny, & Murray, 2006). It has been shown that maltose can influence a bopABC triple-deletion mutant to produce more biofilm than the wild type in a medium containing 1% glucose, whereas in the presence of 1% maltose, the mutant only produced about 4% of biofilm with respect to the wild type (Creti, Koch, Fabretti, Baldassarri, & Huebner, 2006). While these observations suggest that the availability of different sugars in the growth environment can drastically alter biofilm production in E. faecalis, it remains to be established if this process is related to catabolite repression or the total availability of fermentable carbon sources.
The addition of 10% human serum to the culture medium has been reported to enhance the adhesion of E. faecalis ATCC 29212 to glass and silicone surfaces (Gallardo-Moreno, González-Martín, Perez-Giraldo, Bruque, & Gómez-García, 2002). Although a specific mechanism for the observed effect has not yet been proposed, another study examining a salB mutant noted enhanced biofilms when grown in TSB supplemented with 0.25% glucose and 10% serum (Mohamed, Teng, Nallapareddy, & Murray, 2005). The same effect was not seen with the wild-type OG1RF strain. A recent study has also revealed the enhanced expression of the Ebp pilus in the presence of 0.1 M sodium bicarbonate added to TSB containing glucose (Bourgogne, Thomson, & Murray, 2010), although the molecular basis of this is unclear. Along these lines, the E. faecalis pathogenicity island encoded AraC-type transcriptional regulator (PerA) was suggested to be a repressor of biofilm formation (Coburn P. S., Baghdayan, Dolan, & Shankar, 2008). Comparison of wild type E. faecalis E99, a perA-insertion mutant, and the complemented strain showed that when grown in Todd-Hewitt broth (THB) containing 1% glucose, biofilm formation of the mutant in vitro was significantly greater (P < 0.0001) relative to the wild type, as well as the complement. However, a similar effect was not seen when these strains were grown in TSB that contained 1% glucose (Coburn P. S., Baghdayan, Dolan, & Shankar, 2008). Additional studies are needed to understand the mechanism behind these observations, but they highlight the importance of how the growth environment can significantly affect biofilm formation in E. faecalis.
Enterococcal Surface Adhesins
Aggregation substance
Aggregation substance (AS) is a surface adhesin that mediates cell-cell contact during pheromone responsive mating of donor and recipient E. faecalis cells, which is crucial for plasmid transfer (Clewell & Weaver, 1989). It has been suggested that AS may facilitate the translocation of E. faecalis across the intestinal epithelium, due to its involvement with adhesion to and invasion of intestinal epithelial cells (Wells, Moore, Hoag, Hirt, Dunny, & Erlandsen, 2000). AS has also been reported to mediate adherence to renal epithelial cells (Süßmuth, Muscholl-Silberhorn, Wirth, Susa, Marre, & Rozdzinski, 2000) and components of the extracellular matrix (Rozdzinski, Marre, Susa, Wirth, & Muschol-Silberhorn, 2001), as well as increase survival in polymorphonuclear leukocytes (Rakita, et al., 1999; Vanek, Simon, Jacques-Palaz, Mariscalco, Dunny, & Rakita, 1999) and macrophages (Süßmuth, Muscholl-Silberhorn, Wirth, Susa, Marre, & Rozdzinski, 2000). Deletion of the N-terminal region or the RGD domain of AS resulted in significantly reduced virulence of E. faecalis in the endocarditis model. Interestingly, while antibodies to the N-terminal region were not protective (McCormick, Hirt, Waters, Tripp, Dunny, & Schlievert, 2001), polyclonal antibodies to the full-length AS inecreased the severity of endocarditis (Schlievert, Chuang-Smith, Peterson, Cook, & Dunny, 2010). The same study also showed that Fab fragments of IgG from rabbits immunized against AS were able to reduce total vegetation size and microbial counts when passively administered to rabbits prior to challenge with E. faecalis-expressing AS. In vitro, the Fab fragments also prevented enterococcal aggregation. While AS did not appear to play a role in colonization in an ascending model of mouse urinary tract infection (Johnson, Clabots, Hirt, Waters, & Dunny, 2004), it has been reported to increase the severity of infective endocarditis by increasing E. faecalis vegetation (biofilm) weights on aortic heart valves (Chuang, Schlievert, Wells, Manias, TRipp, & Dunny, 2009). In recent work using an ex vivo porcine heart valve adherence model (Chuang-Smith, Wells, Henry-Stanley, & Dunny, 2010), it has been shown that AS significantly accelerated biofilm development on this biotic surface, relative to abiotic membranes, which implies that there is a significant role for AS in vivo. Reconciling all of these observations into a single model for the contribution of aggregation substance to the molecular pathogenesis of infection is an ongoing process, but it seems fairly certain to promote interbacterial association, which directly or indirectly contributes to increased adherence to human tissues, increasing the likelihood of formation of a quorum of E. faecalis cells and subsequent expression of factors that are dependent on quorum-sensing signals, such as the cytolysin or gelatinase.
Enterococcal Surface Protein, Esp
One of the first cell wall-associated proteins implicated in biofilm formation in vitro was the enterococcal surface protein Esp. First identified in E. faecalis as a large surface-anchored protein enriched among infection-derived isolates (Shankar V. , Baghdayan, Huycke, Lindahl, & Gilmore, 1999), it led to the discovery of a homolog in E. faecium (Willems, et al., 2001; Coque, Willems, Cantón, Del Campo, & Baquero, 2002; Eaton & Gasson, 2002). Subsequent studies localized the esp gene to pathogenicity islands in both species (Leavis, et al., 2004; Shankar, Baghdayan, & Gilmore, 2002). An early report suggested that there was a strong correlation of E. faecalis esp with the ability to form biofilms, with 93.5% of esp-positive isolates forming biofilms on polystyrene, while none of the esp-negative isolates did so (Toledo-Arana, et al., 2001). Biofilm formation was impaired in two esp insertion mutants, but not in a third strain tested, leading the authors to conclude that while esp was important for biofilm formation, it was probably one among many other factors in E. faecalis that mediated this phenotype.
Using a genetic approach, Tendolkar et al (Tendolkar, Baghdayan, Gilmore, & Shankar, 2004) showed increased biofilm formation by two E. faecalis strains OG1RF and FA2-2 (natively lacking the esp gene) when transformed with plasmid constructs expressing esp. However, a similar effect was not observed when esp from E. faecalis MMH594 was expressed in E. faecium or L. lactis, which led the authors to conclude that Esp may act in concert with other surface factors unique to E. faecalis. In a follow-up study, the same authors also showed that the non-repeat N-terminal region of mature Esp was sufficient for biofilm enhancement (67). Related studies in E. faecium established a high degree of sequence identity between Esp from E. faecalis and E. faecium, and further showed that primary adherence and biofilm formation correlated with levels of Esp expression at the cell surface (Leavis, et al., 2004; Van Wamel, Hendrickx, Bonten, Top, Posthuma, & Willems, 2007).
The multifactorial nature of biofilm formation by enterococci was borne out by one study that demonstrated biofilm formation by E. faecalis strain OG1RF lacking esp (Kristich, Li, Cvitkovich, & Dunny, 2004) and another that made the observation that 77 of 89 esp-negative clinical isolates of E. faecalis were also able to form biofilms categorized as weak, medium, or strong (69). However, all 74 esp-positive isolates in this study formed biofilms, strengthening the argument that, when present, esp significantly enhances biofilm formation. A number of other recent reports that examined esp-positive and esp-negative E. faecalis and E. faecium have made similar observations (Di Rosa, et al., 2006; Sandoe, Witherden, Cove, Heritage, & Wilcox, 2003; Duprè, Zanetti, Schito, Fadda, & Sechi, 2003; Dworniczek, et al., 2003; Raad, et al., 2005; Ramadhan & Hegedus, 2005). Murine intestinal colonization models failed to show a definitive role for esp in E. faecalis or E. faecium in gut colonization or persistence (Pultz, Shankar, Baghdayan, & Donskey, 2005; Heikens, et al., 2009). However, in a mouse ascending urinary tract infection model, esp-deficient E. faecalis were recovered in lower numbers from the bladder and urine of infected mice as compared to the wild type strain, which suggests that Esp may facilitate persistence and colonization at this site (Shankar, Lockatell, Baghdayan, Drachenberg, Gilmore, & Johnson, 2001).
Microbial Surface Components Recognizing Adhesive Matrix Molecules (MSCRAMMs)
Microbial surface components recognizing adhesive matrix molecules (MSCRAMMs) (MSCRAMMs) facilitate the colonization of host tissues and binding of bacterial cells to indwelling abiotic surfaces that become coated with host-derived extracellular matrix components. Both E. faecalis and E. faecium appear to have more than a dozen MSCRAMMs apiece, as deduced from genome sequences currently available (Sillanpää, Nallapareddy, Prakash, Qin, Höök, & Weinstock, 2008; Sillanpää, Xu, Nallapareddy, Murray, & Höök , 2004). Adhesion of collagen from E. faecalis (Ace) was the first of three MSCRAMMs that have been extensively characterized in enterococci, and have been shown to bind to collagen type I, collagen type IV, laminin, and dentin (Kowalski, Kasper, Hatton, Murray, Nallapareddy, & Gillespie, 2006; Nallapareddy S. R., Singh, Duh, Weinstock, & Murray, 2000; Nallapareddy, Qin, Weinstock, Höök, & Murray, 2000). An Ace homolog in E. faecium, Acm, predominantly interacts with collagen type I and interacts somewhat weakly with collagen type IV (Nallapareddy S. R., et al., 2006; Nallapareddy & Murray, 2006; Nallapareddy, Weinstock, & Murray, 2003). A second collagen adhesion of E. faecium, Scm, was shown to mediate binding to collagen type V and fibrinogen (Sillanpää, Nallapareddy, Prakash, Qin, Höök, & Weinstock, 2008). These three MSCRAMMs were ubiquitous among both clinical and non-clinical isolates tested; however, E. faecium isolates of non-clinical origin lacked binding to collagen type I and showed an insertion element disruption of the acm gene (Nallapareddy S. R., Singh, Okhuysen, & Murray, 2008). The structural features of these enterococcal MSCRAMMs have been described in a recent review (Hendrickx, Willems, Bonten, & van Schaik, 2009).
In vitro studies have shown that microspheres coated with the A domain of E. faecalis Ace can mediate binding to human intestinal and umbilical vein endothelial cells, a process that could be blocked by either soluble collagen or Ace-specific antibodies (Hall, et al., 2007). An ace-deletion mutant has recently been shown to be significantly attenuated, as compared to wild-type OG1RF in a mixed-infection rat endocarditis model (P < 0.0001), while no differences were observed in a peritonitis model (Singh K. V., Nallapareddy, Sillanpää, & Murray, 2010). In these experiments, Ace appeared to be important to the early colonization of heart valves, and rats actively immunized against rAce were less likely to develop E. faecalis endocarditis (P = 0.0001) and showed fewer counts in vegetations (P = 0.0146). A deletion mutant of ace has also been shown recently to be less virulent in an insect (Galleria mellonella) virulence model (Lebreton, et al., 2009) and attenuated in an experimental UTI model (87). Likewise, an acm deletion mutant has been shown to be less virulent in a rat endocarditis model (Nallapareddy, Singh, & Murray, 2008).
Two other MSCRAMMs in E. faecium have been recently reported (Hendrickx, et al., 2009). The surface adhesion SgrA binds to extracellular matrix molecules nidogen 1 and nidogen 2, while EcbA mediates binding to collagen type V. An E. faecium sgrA insertion mutant displayed reduced binding to both nidogens and fibrinogen. SgrA did not mediate binding of E. faecium to human intestinal epithelial, bladder, or kidney cells, but did contribute to E. faecium biofilm formation in vitro.
Pili
Expression of pili at the bacterial cell surface facilitates adhesion, which is the first step in the biofilm process. In Gram-positive organisms, the synthesis of pili is a complex process that involves the assembly of pilin subunits into a pilus polymer, which is eventually anchored to the cell wall through the action of housekeeping and dedicated sortase enzymes (Mandlik, Swierczynski, Das, & Ton-That, 2008). Genes for pilus biosynthesis are generally clustered as operons called pilin gene clusters (PGCs). The pilin protein subunits are known to contain MSCRAMM-like features, as well as conserved pilin motifs (HLYPK) and E box motifs (ETxAPExY) that facilitate assembly of the pilus. Gram-positive pili appear to play a major role in mediating cell-cell contact, colonization of host tissue, and biofilm formation (Telford, Barocchi, Margarit, Rappuoli, & Grandi, 2006).
Pilus-like structures in E. faecalis JH2 were reported long before the genetic basis for pilus biogenesis was worked out (Handley & Jacob, 1981). Subsequent to the sequencing of multiple enterococcal genomes, it became apparent that both E. faecalis and E. faecium may harbor PGCs that encode proteins bearing the classic motifs identified in pilin subunits (Nallapareddy S. R., et al., 2006; Tendolkar, Baghdayan, & Shankar, 2006; Hendrickx, Bonten, van Luit-Asbroek, Schapendonk, Kragten, & Willems, 2008). Two PGCs have been reported in E. faecalis, the ubiquitous endocarditis and biofilm-associated pili (ebp) operon (Nallapareddy S. R., et al., 2006; Cobo Molinos, Abriouel, Omar, López, & Galvez, 2008), and the much less frequently detected and plasmid encoded bee locus (Tendolkar, Baghdayan, & Shankar, 2006). In E. faecium, four PGCs have been described, all of which appear to be enriched among hospital-derived E. faecium isolates (Hendrickx, Bonten, van Luit-Asbroek, Schapendonk, Kragten, & Willems, 2008).
The detection of antibodies to Ebp subunits in the sera isolated from endocarditis patients suggests that this pilus plays an important role in this biofilm-type infection (Nallapareddy S. R., et al., 2006). Mutants defective in expression of one or more of the subunits or the dedicated sortase, SrtC, were found to be attenuated in attachment and biofilm formation in vitro. Further, recent data has also revealed a role for the Ebp pilus in mediating binding to human platelets (Nallapareddy, et al., 2011) and fibrinogen, with a lesser role in binding to collagen (Nallapareddy S. R., Singh, Sillanpää, Zhao, & Murray, 2011). The ebp locus was also found to contribute to virulence in a mouse ascending UTI model (Singh, Nallapareddy, & Murray, 2007), although it appeared that other factors may play a role in this model.
The roles of housekeeping sortase SrtA and Ebp pilus-specific sortase Bps (SrtC) have been evaluated for their relative contribution to biofilm formation in E. faecalis (Kemp, Singh, Nallapareddy, & Murray, 2007). A srtA deletion mutant of OG1RF showed little effect on biofilm formation (P = 0.037) as compared to the wild type, whereas both a bps deletion mutant and srtA-bps double deletion mutant showed significantly impaired biofilm formation (P < 0.001) relative to its parent strain. These data were interpreted as suggesting that Bps, and/or the protein(s) that it anchors, may compensate for some of the SrtA functions. Examining biofilm formation under static and hydrodynamic conditions others have determined that SrtA primarily functions during the initial attachment phase, which involves the initiation of biofilm formation (Guiton, et al., 2009).
Although it is ubiquitous among E. faecalis isolates, the expression of the Ebp pilus varies widely within a population of cells (30-72%), depending on growth conditions (Nallapareddy, et al., 2011). The ebp locus is under positive control of the EbpR regulator, which in turn is under the control of fsr (BourgogneA., Singh, Fox, Pflughoeft, Murray, & Garsin, 2007). A significant enhancement in the expression of the Ebp pilus was noted when cells were exposed to bicarbonate—although the precise mechanism for this effect remains to be deciphered (Bourgogne, Thomson, & Murray, 2010). An effect of RNA processing on Ebp pilus expression was also noted, whereby deletion of rnjB encoding RNase J2 decreased transcript levels for the ebp operon (Gao, Pinkston, Nallapareddy, van Hoof, Murray, & Harvey, 2010).
The bee locus was identified as a cluster of five genes encoded on a large conjugative plasmid in an E. faecalis strain E99 (Tendolkar, Baghdayan, & Shankar, 2006; Coburn P. S., et al., 2010). The structural subunits of the Bee pilus are Bee-1, Bee-2, and Bee-3, which bear the cell wall sorting and pilin motifs characteristic of pilin subunits. Unlike in the ebp operon, the bee structural genes are followed by two sortase genes in tandem. While pilus expression at the cell surface has been confirmed by electron microscopy in strain E99, as well as in E. faecalis OG1RF and JH2-2 transconjugants, the role of the Bee pilus in infection models remain to be established, although a pilot study concluded that the Bee pilus did not contribute to virulence or persistence in a mouse model of UTI (unpublished). However, in in vitro biofilm assays, the Bee pilus has been shown to confer a high biofilm phenotype upon E. faecalis (Tendolkar, Baghdayan, & Shankar, 2006).
E. faecium clinical isolates, including those from endocarditis, harbor genetic loci that encode two pilus-like structures, PilA and PilB (Hendrickx, Bonten, van Luit-Asbroek, Schapendonk, Kragten, & Willems, 2008; Hendrickx, Schapendonk, van Luit-Asbroek, Bonten, van Schaik, & Willems, 2010). The pilA locus is encoded on a large conjugative plasmid (Kim, et al., 2010) and appears to be a minor pilus, while the pilB gene was found to be part of the ebpABC(fm) cluster (Sillanpää, et al., 2010) that encodes the major pilus PilB. The differential expression of pilA and pilB was noted, with the former being expressed only on solid growth media, while the latter was also expressed in broth. Loss of PilB was evident in a ebpABC deletion mutant with concomitant reduction in in vitro biofilm formation and attenuated virulence of the mutant in a murine UTI model, as compared to wild type (Sillanpää, et al., 2010).
Polysaccharides
An E. faecalis polysaccharide antigen locus (epa) has been described (Teng, Jacques-Palaz, Weinstock, & Murray, 2002) as being involved in synthesis of cell wall-associated polysaccharides, which contributes to biofilm formation, among other virulence properties. Biofilm formation was specifically attenuated by mutations in epaB and epaE. A more recent characterization has defined the epa locus to be a 26-kb region comprising genes epaA to epaR (Teng, Singh, Bourgogne, Zeng, & Murray, 2009). Similar to that observed with epaB and epaE mutants, disruptions in epaA, epaM, and epaN resulted in the alteration of Epa polysaccharide content and decreased biofilm formation.
A putative sugar binding transcriptional regulator, BopD, which shows sequence similarity with a number of proteins involved in maltose metabolism, has also been shown to influence biofilm formation in E. faecalis (Hufnagel, Koch, Creti, Baldassarri, & Huebner, 2004). By comparing a transposon insertion mutant and a deletion mutant, a sugar-specific effect on biofilm production was observed (Creti, Koch, Fabretti, Baldassarri, & Huebner, 2006). When grown in medium containing 1% glucose, the transposon mutant produced less biofilm and the non-polar deletion mutant produced more biofilm than the wild type. However, in the presence of 1% maltose in the medium, the transposon mutant produced more biofilm than the wild type and biofilm formation by the deletion mutant was abrogated. These data suggested that BopD was likely to be a maltose-sensitive negative regulatory protein that may repress both bopABC and the divergently transcribed malT operon. The lower expression of bopD in the transposon mutant could thus lead to the derepression of bopA (a maltose phosphorylase) and of the sugar transport gene malT, which may also lead to the increased utilization of maltose and enhanced biofilm formation when bacteria are grown in maltose.
Secreted factors, autolysin and eDNA
The zinc metalloprotease GelE is ubiquitous among E. faecalis strains, and is a significant factor in promoting the formation of biofilm (Hancock & Perego, 2004). The gelE determinant is adjacent and upstream of sprE, the determinant that encodes a serine protease. Both genes are cotranscribed and are under the control of the fsr quorum sensing system (Qin, Singh, Weinstock, & Murray, 2001). A detailed description of the organization and regulation of this operon is presented later in this chapter. The influence of gelatinase and the fsr locus in relation to biofilm development have been described (Mohamed, Huang, Nallapareddy, Teng, & Murray, 2004; Hancock & Perego, 2004; Thomas, Thurlow, Boyle, & Hancock, 2008; Carniol & Gilmore, 2004), and both GelE and SprE act in concert with autolysin (AtlA) processing to regulate cell death and eDNA release by a process akin to fratricide (Guiton, et al., 2009; Thomas, Thurlow, Boyle, & Hancock, 2008; Thomas, Hiromasa, Harms, Thurlow, Tomich, & Hancock, 2009; Thomas & Hancock, 2009). Additional confirmation of the importance of GelE in biofilm formation was put forth by Kristich et al (Kristich, Li, Cvitkovich, & Dunny, 2004), who showed that conditioned media from OG1RF (gelatinase positive) promoted biofilm formation by E. faecalis JH2, a poor biofilm former, as did GelE expressed under a nisin-inducible promoter in JH2.
In biofilm-associated infection models, both GelE and SprE were found to be important for virulence in experimental rat endocarditis (Singh K. V., Nallapareddy, Nannini, & Murray, 2005). A double gelE/sprE mutant were found to be more attenuated for virulence then a fsr mutant affecting GelE production in this model; however, the opposite effect was seen in the rabbit endophthalmitis (Mylonakis, et al., 2002) and C. elegans (Sifri, et al., 2002) models. It has also been shown in the rabbit endocarditis model that GelE contributes to increased bacterial counts at disseminated sites of infection (Thurlow, Thomas, Narayanan, Olson, Fleming, & Hancock, 2010).
Other enterococcal biofilm mediators
A number of different studies have attempted to identify additional factors that mediate biofilm formation. Inactivation of the biofilm-associated glycolipid synthesis A (bgsA) gene that encodes a putative glycosyltransferase in E. faecalis had a significant impact on initial adherence and biofilm formation on plastic, adhesion to colonic epithelial cells, and virulence in a mouse bacteremia model (Theilacker, et al., 2009). No changes were observed in levels of surface protein expression, autolysis, or sensitivity to antimicrobial peptides. In similar studies, a dltA mutant of E. faecalis 12030 lacking D-alanine esters in lipoteichoic acid (LTA), due to the disruption within the dltABCD operon, showed reduced in vitro biofilm formation and binding to eukaryotic cells (Fabretti, et al., 2006). However, a dltA mutant in E. faecalis OG1RF failed to show the same effect. Two secreted E. faecalis proteins, SalA and SalB, homologous to the E. faecium SagA protein, have been implicated in in vitro biofilm formation on polystyrene (Mohamed, Teng, Nallapareddy, & Murray, 2005; Breton, Mazé, Hartke, Lemarinier, Auffray, & Rincé, 2002). The effect was more pronounced in the salB mutant, which exhibited marked morphological changes, as well as a 54% reduction in biofilm production. A putative E. faecalis surface-exposed antigenic protein EF3314 that showed a delayed effect on biofilm after culturing in medium containing glucose diminished adherence of the deletion mutant to HeLa cells and less killing in a C. elegans model (Creti, et al., 2009).
Two independent genome-wide approaches have recently identified a number of new and novel genetic determinants in the core genome of E. faecalis OG1RF that influence biofilm formation (Ballering, Kristich, Grindle, Oromendia, Beattie, & Dunny, 2009; Kristich, Nguyen, Le, Barnes, Grindle, & Dunny, 2008). The first approach employed a modified mini-mariner transposable element (EfaMarTn) that produced mostly single insertions distributed throughout the genome (Kristich, Nguyen, Le, Barnes, Grindle, & Dunny, 2008). Mutants were screened for impaired biofilm formation in vitro on polystyrene, and led to the identification of not only a number of the previously identified loci, but also additional genes, such as those that encode heat-shock proteins (GrpE, DnaK, and DnaJ), as well as putative regulators (EF0676 and EF0983) that belong to the ArgR family. In the second study, a recombinase-in vivo expression technology (RIVET) approach was used to identify chromosomally-encoded biofilm determinants (Ballering, Kristich, Grindle, Oromendia, Beattie, & Dunny, 2009). This study identified 68 different promoters that were active at different stages of biofilm growth, with at least 17 conserved hypothetical genes, and 7 of them were predicted to encode DNA-binding proteins not previously associated with biofilms. Using the same approach, a more recent study has highlighted the way in which the RIVET screen may provide information on temporal activation of genes during infection (Frank, Barnes, Grindle, Manias, Schlievert, & Dunny, 2012). In this study, deletions in the in vivo-activated eep gene (encoding a membrane metalloprotease, Eep) affected the cellular organization of in vitro biofilms, and the deletion mutant was severely attenuated in a rabbit endocarditis model. Microscopic analysis of in vitro biofilms revealed numerous small aggregates not present in wild-type biofilms. Another independent study that evolved from the results of the RIVET screen has shown that in the presence of uric acid, which is freely available in both urine and blood, E. faecalis may shift from a planktonic to biofilm mode of growth (Srivastava, Mallard, Barke, Hancock, & Self, 2011). Enhanced biofilm production was seen to be a selenium-dependent process, driven through the production of peroxide, and mutations in the selD (selenophosphate synthetase) and xdh (xanthine dehydrogenase) genes that reside within the selenium-dependent molybdenum hydroxylase (SDMH) operon both abolished biofilm formation.
Regulation of Biofilm Development
The recent increase of literature on the topic of enterococcal biofilms has coincided with the emergence of accessible genomic information, starting with the published sequence of the first enterococcal genome in 2003 (Paulsen, et al., 2003). Since that time, various enterococcal factors have been shown to contribute to in vitro biofilms (Paganelli, Willems, & Leavis, 2012), while only a handful of factors have also been shown to play a key role in pathogenesis (Frank, Barnes, Grindle, Manias, Schlievert, & Dunny, 2012; Nallapareddy S. R., et al., 2006; Singh, Nallapareddy, & Murray, 2007; Shankar, Lockatell, Baghdayan, Drachenberg, Gilmore, & Johnson, 2001; Singh K. V., Nallapareddy, Nannini, & Murray, 2005; Thurlow, Thomas, Narayanan, Olson, Fleming, & Hancock, 2010). Pili and Esp are known to be important adhesins for attachment to biotic and/or abiotic surfaces, but the subsequent steps that lead to organization of the community structure of the biofilm are still poorly defined. Here we focus on what is known about such mechanisms, as well as raise the specter of additional signaling pathways that may facilitate such development. Because E. faecalis causes the majority of enterococcal infections, much of our understanding of the way in which cell signaling influences biofilm development comes from studies on this species.
Signal Transduction in Enterococcal Biofilms
In microbial species, the best characterized signal transduction system involves a two-component system comprised of a histidine kinase and its cognate response regulator (Hoch & Silhavy, 1995). As the histidine kinase perceives a sensory signal, it undergoes autophosphorylation and subsequently transfers a phosphoryl group to the response regulator. The phosphorylated form of the response regulator allows it to regulate gene expression. An analysis of the E. faecalis V583 genome sequence revealed that it possesses 17 paired two-component systems and an additional orphan response regulator (Hancock & Perego, 2002). Systematic inactivation and phenotypic characterization of the 18 response regulator mutants in V583 showed that mutant RR15 was attenuated in its biofilm-forming ability (Hancock & Perego, 2004). The response regulator disrupted in this mutant was previously characterized by Qin et al. (Qin, Singh, Weinstock, & Murray, 2001) as being part of the quorum-sensing signal transduction system Fsr, which responds to the accumulation of GBAP, an 11 amino acid peptide lactone (Nakayama, et al., 2001). The Fsr quorum system of E. faecalis partially resembles the staphylococcal Agr system (Novick & Geisinger, 2008), where a histidine kinase (FsrC) responds to the accumulation of a quorum peptide (GBAP) to phosphorylate the cognate response regulator (FsrA). Del Papa and Perego (Del Papa & Perego, 2011) recently showed that the phosphorylated form of FsrA is required in order to recognize its target DNA sequence to promote transcription. The production of GBAP arises from the processing of a 53 amino acid precursor propeptide encoded by fsrD (Nakayama, et al., 2006). Processing of the propeptide and export by the membrane-localized FsrB drives the external accumulation of GBAP. Several genes have been shown to be directly regulated by the Fsr system (Bourgogne, Hilsenbeck, Dunny, & Murray, 2006; Qin, Singh, Weinstock, & Murray, 2001) and include autoregulation of the fsrBDC operon, as well as the co-transcribed gelEsprE genes, which encode two extracellular proteases, gelatinase (GelE) and serine protease (SprE).
Several independent laboratories have established a role for the Fsr system and proteases in biofilm formation (Kristich, Li, Cvitkovich, & Dunny, 2004; Pillai, Sakoulas, Eliopoulos, Moellering, Jr., Murray, & Inouye, 2004; Mohamed, Huang, Nallapareddy, Teng, & Murray, 2004; Hancock & Perego, 2004). Thomas et al. (Thomas, Thurlow, Boyle, & Hancock, 2008) went on to show that the production of GelE, a zinc metalloprotease, resulted in the release of extracellular DNA (eDNA), presumably through cell lytic processes. Consistent with observations from other bacterial pathogens (Montanaro, et al., 2011), eDNA proved to be an important early biofilm matrix component, as treatment of developing E. faecalis biofilms with DNase I significantly reduced biofilm accumulation at 6 and 12 hours, but not at 24 hours. A role for eDNA as a matrix component in E. faecalis OG1RF was also reported by Guiton et al. (Guiton, et al., 2009), who showed that the major autolysin (Atn), AtlA, also contributed to eDNA release. A role for Atn in biofilm development was initially reported by Mohamed et al. (Mohamed, Huang, Nallapareddy, Teng, & Murray, 2004), and was subsequently confirmed by a mariner-based transposon screen to identify biofilm defective mutants (Kristich, Nguyen, Le, Barnes, Grindle, & Dunny, 2008). From a pool of ~15,000 transposon mutants, 25 were biofilm-defective, and three mapped to transposon insertions in the atn gene (Kristich, Nguyen, Le, Barnes, Grindle, & Dunny, 2008). This same screen further highlighted the importance of the Ebp pilus in biofilm formation, as nearly one-third of the biofilm-defective mutants mapped to this locus (EbpR, EbpABC or SrtC).
Thomas et al. presented a mechanism for how a quorum response mediated through Fsr could give rise to a lytic process that governs biofilm development (Thomas, Thurlow, Boyle, & Hancock, 2008; Thomas, Hiromasa, Harms, Thurlow, Tomich, & Hancock, 2009). These studies implicated GelE as being a pro-lytic effector of lysis, as the deletion of gelE rendered the cells less susceptible to autolysis. In contrast, the deletion of sprE resulted in a more rapid rate of lysis, which suggests that it possessed anti-lytic properties. The observation that these two proteases governed the lytic properties of the cell in opposing ways suggested that they might target the same downstream effector. The use of cell wall zymography identified AtlA as being differentially targeted by GelE and SprE. The targeting of AtlA by GelE results in multiple AtlA active forms, whereas SprE processes AtlA to a discrete ~ 62 kDa active form that has a high affinity for cell walls and renders this AtlA form immune to further processing by GelE. Mutation of atlA also gives rise to a cellular chaining phenotype (Qin, Xu, Singh, Weinstock, & Murray, 1998), and this phenotype is also partially observed in a gelE mutant (Waters, Antiporta, Murray, & Dunny, 2003), which provides further evidence for an association of these proteins.
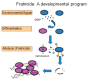
Fratricide
The observation that cell-cell communication through quorum signaling resulted in the co-transcription of genes whose products trigger opposing roles in cell death is reminiscent of competence development in Streptococcus pneumoniae (Claverys & Håvarstein, 2007). In the pneumococcal model, termed fratricide (Gilmore & Haas, 2005), the competence-stimulating peptide serves as a quorum molecule to activate the early competence regulon, which includes cell wall lytic enzymes and bacteriocins, as well as immunity proteins that prevent self-induced killing. In the E. faecalis model of fratricide (Figure 1) and biofilm development, cells that respond to GBAP enhance expression of both GelE (effector) and SprE (immunity), whereas quorum non-responders fail to express these proteins and would therefore be lysis-susceptible. Consistent with a requirement for a bimodal population of quorum responders and non-responders, roughly 10-15% of the cells failed to respond to the quorum signal in stationary phase cultures, as noted by FACS analysis with a gelE-GFP reporter fusion (Thomas, Hiromasa, Harms, Thurlow, Tomich, & Hancock, 2009). Non-responders would then be more susceptible to activation of their AtlA by GelE diffusing from a neighboring cell in the absence of SprE. This was demonstrated through co-culture experiments, in which a GFP-tagged prey population (GelE-SprE-) was mixed with various predator populations. When GelE+ strains served as the predator, lysis of the prey occurred, and this lytic activity was enhanced in the absence of SprE activity. A dependence on functional AtlA is also required in this model, as mutation of atlA in both the predator and prey populations is necessary to abolish lysis, suggesting that soluble forms of AtlA derived from the predator can also mediate prey lysis. The susceptibility of the prey or non-responder population to lysis would release eDNA to serve as a biofilm matrix component. The rates of diffusion away from producing cells, as well as an affinity for AtlA by GelE and SprE, would likely govern the extent of non-responder death. As death in E. faecalis biofilms occurs in only a small minority of cells, it suggests that this process is highly regulated to favor survival.
Cell-cell communication
In addition to the aforementioned Fsr quorum signaling in cell-cell communication, one of the hallmarks of E. faecalis biology is the use of peptide pheromones secreted by recipient cells to induce the conjugative apparatus of the donor cell to mediate the transfer of pheromone responsive plasmids (reviewed in Extrachromosomal and Mobile Elements in Enteroocci). A recent study by Frank et al. (Frank, Barnes, Grindle, Manias, Schlievert, & Dunny, 2012) identified the regulated intramembrane protease Eep as being important to the endocarditis model of infection, as an eep mutant was attenuated by 3-logs compared to the wild-type OG1RF strain. Eep was originally identified by Clewell’s group (An, Sulavik, & Clewell, 1999) as an enhancer of enterococcal pheromones. As peptide pheromones are known to be derived from the signal sequence of putative lipoproteins (An & Clewell, 2002; Antiporta & Dunny, 2002), Eep is required to further process the signal peptide into the active pheromone (Chandler & Dunny, 2008). The fact that an eep mutant is highly attenuated in the endocarditis model, and that this model provides an ideal environment to study in vivo biofilms, raises the intriguing prospect that peptide pheromones might be playing an important signaling role in biofilm development (Frank, Barnes, Grindle, Manias, Schlievert, & Dunny, 2012). This potential peptide pool arising from the numerous lipoprotein signal peptides could provide a treasure trove of signaling molecules by which biofilm development could be modulated. Consistent with this prediction, in vitro imaging of eep mutant biofilms compared to the isogenic parent OG1RF revealed altered biofilm architecture in the mutant with small cellular aggregates. Whether peptide pheromones play roles in biofilm development beyond the mating response awaits further investigation.
A large number of bacterial species, including enterococci, possess the capacity to produce autoinducer-2 (AI-2) (Schauder, Shokat, Surette, & Bassler, 2001; Federle, 2009). Unlike other cell-cell signaling pathways reserved for intraspecies communication, AI-2 signaling has evolved to permit crosstalk among bacteria from varying genera that might occupy the same environmental niche (Kaper & Sperandio, 2005). The production of AI-2 is driven by the enzyme LuxS, which is centered on the metabolism of S-adenosyl methionine as it converts ribose-homocysteine into homocysteine and 4.5-dihydro-2,3-pentanedione, which spontaneously cyclizes in the presence of water to produce furanone, the precursor of AI-2 (Schauder, Shokat, Surette, & Bassler, 2001). AI-2 signaling is known to be important in biofilm formation in firmicutes closely related to enterococci, such as S. pneumoniae (Vidal, Ludewick, Kunkel, Zähner, & Klugman, 2011). A recent study has linked AI-2 signaling with fratricide and competence development in pneumococci (Trappetti, Potter, Paton, Oggioni, & Paton, 2011). The known link between fratricide and enterococcal biofilms raises the question of whether AI-2 signaling might also play a role in this process, but this examination will await further study. What is clear is that the study of cell-cell communication in enterococcal biofilm development portends an exciting and potentially fruitful area of research.
Future Directions
Since the publication of the first edition of this book, many more databases and experimental tools have become available for studies of enterococcal biofilms. These have great potential to address many current gaps in our understanding, including the following areas.
Genetic basis for biofilm formation
While a great deal of progress has been made in the identification of biofilm determinants, a comprehensive list of these determinants has not been generated. We still do not know whether unique biofilm functions actually exist, what the relative contributions of the conserved core genome versus mobile elements to biofilm-formation are, or the true impact of biofilm growth on expression and transfer of antibiotic resistance. Several researchers have suggested that new paradigms for antimicrobial development are needed to overcome the rapid increases in resistance to currently available antimicrobials. Strategies such as preventing disease production without directly killing or inhibiting growth, or targeting the genetic transfer machinery, should be considered in this regard (Rasko & Sperandio, 2010; Baquero, Coque, & de la Cruz, 2011). Gene products involved in biofilm growth and biofilm-associated antibiotic resistance certainly represent potential targets for alternative approaches to drug development.
Biofilms and pathogenesis
The “gold standard” for implicating a particular bacterial determinant in pathogenesis is the demonstration of attenuation of virulence in an animal model upon disruption of the determinant. It will be essential to continue to examine the effects of mutations that affect biofilm formation in vitro in relevant animal models, such as experimental endocarditis and urinary tract infections. Comparative studies using these models, in conjunction with ex vivo tissue, tissue culture cell, insect, or nematode systems, may be informative in determining the relative contributions of attachment and surface growth functions with evasion of host immunity. For example, it has been shown that AS expression is associated with resistance to phagocytic killing (Süßmuth, Muscholl-Silberhorn, Wirth, Susa, Marre, & Rozdzinski, 2000; Rakita, et al., 1999), as well as with enhancing attachment and biofilm development on heart valves in vitro (Chuang-Smith, Wells, Henry-Stanley, & Dunny, 2010). Thus, it is likely that the AS-mediated enhancement of virulence in experimental endocarditis results in multiple functions of the protein, which are likely encoded in separate domains (Chuang, Schlievert, Wells, Manias, TRipp, & Dunny, 2009).
Sensing and signaling
A previous section of this chapter described the important role of fsr-mediated quorum sensing in regulation of proteases controlling fratricidal cell lysis and DNA release E. faecalis biofilm development. This is the only well-documented example of an important effect of cell-cell signaling in enterococcal systems that fits well with the developmental model for microbial biofilms. However, recent work has shown that biofilm growth affects the dynamics of peptide pheromone signaling as well as its control of plasmid transfer (Cook, Chatterjee, Barnes, Yarwood, Hu, & Dunny, 2011). Given the genetic potential for E. faecalis to produce a plethora of extracellular peptide signals, it would not be surprising to find new cell-cell communication systems that affect biofilm development. In addition, the mechanism(s) by which the initial attachment of a planktonic cell to a surface triggers the physiological transition from planktonic to biofilm growth is not known. The answer to this question may represent the “holy grail” for all biofilm researchers, and finding it will require analysis of events that occur very early in biofilm development. Comparatively little analysis of the initiation of the process has been done, and the tiny amount of cell-associated microbial biomass present at this stage likely precludes standard “-omics” approaches. Success in addressing this topic will likely come from the creative use of genetic and microscopic tools. The most attractive drug or vaccine targets may be those required to complete the initial transition to biofilm growth, so their identification and characterization is critical.
The extracellular matrix
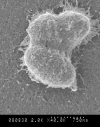
Based on work with other bacteria, it is likely that polysaccharides, DNA, and protein comprise a substantial portion of the enterococcal biofilm matrix, and the importance of eDNA in the structure and adhesive function of the enterococcal matrix is well documented, as noted above. High-resolution scanning electron microscopy (Figure 2) and transmission electron microscopy suggest an important structural role of anionic polysaccharides in the enterococcal matrix (Erlandsen, Kristich, Dunny, & Wells, 2004), and genetic evidence is also supportive of this finding (Teng, Jacques-Palaz, Weinstock, & Murray, 2002; Teng, Singh, Bourgogne, Zeng, & Murray, 2009; Singh, Lewis, & Murray, 2009). However, a detailed biochemical characterization of the extracellular matrix of an enterococcal biofilm has not been presented, and studies of the way in which the matrix changes temporally or in different growth conditions have not been reported. Likewise, while emerging information from Staphylococci point to mechanisms of biofilm disassembly that involve the production of extracellular proteases, deoxyribonucleases, and surfactants, little such information is known in enterococci (Boles & Horswill, 2011). It will be essential to obtain this information in the future to complement continuing genetic analyses of biofilm formation and dispersal.
Biofilm and the evolution of the adaptable enterococcal life style
Perhaps the most remarkable aspect of enterococci is their ability to survive and proliferate in a remarkable diversity of host-associated and environmental niches that are lethal to many phylogenetically-related pathogens. How does biofilm-forming ability impact the fitness of enterococci in these diverse environments, and how many genetic determinants important to one niche play a role in others? Since much of the evolution of the core genomes of these organisms occurred outside a mammalian host, further study of biofilms and their effects on enterococcal ecology in diverse environments, and on strain and gene transmission between niches should be a high priority. Given the availability of many new tools for genetic, physiological, and evolutionary studies, there is unlimited potential for this area of research. There is a tremendous amount to be learned about the role of biofilm formation in the intestinal versus extra-intestinal ecology within the human host, both in healthy individuals and in hospital patients. In the long run, the insights gained from these studies may prove as useful in prevention and control of enterococcal diseases as those obtained in more focused studies of pathogenesis.
Acknowledgements
The studies from the authors’ laboratories described in this chapter were supported, in part, by grants GM49530 and AI58134 (G.M.D.), AI77782 (L.E.H.) and AI59673 and DE20928 (N.S.) from the National Institutes of Health.
References
- An FY, Clewell DB. Identification of the cAD1 Sex Pheromone Precursor in Enterococcus faecalis. J Bacteriol. 2002;184(7):1880–1887. [PMC free article: PMC134937] [PubMed: 11889094]
- An FY, Sulavik MC, Clewell DB. Identification and Characterization of a Determinant (eep) on the Enterococcus faecalis Chromosome That Is Involved in Production of the Peptide Sex Pheromone cAD1. J Bacteriol. 1999;181(19):5915–5921. [PMC free article: PMC103617] [PubMed: 10498702]
- Antiporta MH, Dunny GM. ccfA, the Genetic Determinant for the cCF10 Peptide Pheromone in Enterococcus faecalis OG1RF. J Bacteriol. 2002;184(4):1155–1162. [PMC free article: PMC134800] [PubMed: 11807076]
- Baldassarri L, Cecchini R, Bertuccini L, Ammendolia MG, Iosi F, Arciola CR, et al. Enterococcus spp. produces slime and survives in rat peritoneal macrophages. Med Microbiol Immunol (Berl). 2001;190(3):113–120. [PubMed: 11827199]
- Baldassarri L, Creti R, Recchia S, Pataracchia M, Alfarone G, Orefici G, et al. Virulence factors in enterococcal infections of orthopedic devices. Int J Artif Organs. 2006;29(4):402–406. [PubMed: 16705609]
- Ballering KS, Kristich CJ, Grindle SM, Oromendia A, Beattie DT, Dunny GM. Functional genomics of Enterococcus faecalis: multiple novel genetic determinants for biofilm formation in the core genome. J Bacteriol. 2009;191(8):2806–2814. [PMC free article: PMC2668403] [PubMed: 19218379]
- Baquero F, Coque TM, de la Cruz F. Ecology and evolution as targets: the need for novel eco-evo drugs and strategies to fight antibiotic resistance. Antimicrob Agents Chemother. 2011;55(8):3649–3660. [PMC free article: PMC3147629] [PubMed: 21576439]
- Boles BR, Horswill AR. Staphylococcal biofilm disassembly. Trends Microbiol. 2011;19(9):449–455. [PMC free article: PMC3164736] [PubMed: 21784640]
- Bourgogne A, Hilsenbeck SG, Dunny GM, Murray BE. Comparison of OG1RF and an isogenic fsrB deletion mutant by transcriptional analysis: the Fsr system of Enterococcus faecalis is more than the activator of gelatinase and serine protease. J Bacteriol. 2006;188(8):2875–2884. [PMC free article: PMC1446981] [PubMed: 16585749]
- Bourgogne A, Thomson LC, Murray BE. Bicarbonate enhances expression of the endocarditis and biofilm associated pilus locus, ebpR-ebpABC, in Enterococcus faecalis. BMC Microbiol. 2010;10:17. [PMC free article: PMC2824692] [PubMed: 20092636]
- Bourgogne A, Singh KV, Fox A, Pflughoeft KJ, Murray BE, Garsin DA. EbpR is important for biofilm formation by activating expression of the endocarditis and biofilm-associated pilus operon (ebpABC) of Enterococcus faecalis OG1RF. J Bacteriol. 2007;189(17):6490–6493. [PMC free article: PMC1951926] [PubMed: 17586623]
- Bower CK, Daeschel MA, McGuire J. Protein antimicrobial barriers to bacterial adhesion. J Dairy Sci. 1998;81(10):2771–2778. [PubMed: 9812283]
- Breton YL, Mazé A, Hartke A, Lemarinier S, Auffray Y, Rincé A. Isolation and characterization of bile salts-sensitive mutants of Enterococcus faecalis. Curr Microbiol. 2002;45(6):434–439. [PubMed: 12402085]
- Carniol K, Gilmore MS. Signal transduction, quorum-sensing, and extracellular protease activity in Enterococcus faecalis biofilm formation. J Bacteriol. 2004;186(24):8161–8163. [PMC free article: PMC532445] [PubMed: 15576763]
- Chandler JR, Dunny GM. Characterization of the sequence specificity determinants required for processing and control of sex pheromone by the intramembrane protease Eep and the plasmid-encoded protein PrgY. J Bacteriol. 2008;190(4):1172–1183. [PMC free article: PMC2238190] [PubMed: 18083822]
- Chuang O. N., Schlievert P. M., Wells C. L., Manias D. A.TRipp, T. J., & Dunny, G. MMultiple functional domains of Enterococcus faecalis aggregation substance Asc10 contribute to endocarditis virulence Infect Immun 200977(1):539–548. [PMC free article: PMC2612263] [PubMed: 18955479]
- Chuang-Smith ON, Wells CL, Henry-Stanley MJ, Dunny GM. Acceleration of Enterococcus faecalis biofilm formation by aggregation substance expression in an ex vivo model of cardiac valve colonization. PLoS ONE. 2010;5(12):e15798. [PMC free article: PMC3012704] [PubMed: 21209892]
- Claverys J-P, Håvarstein LS. Cannibalism and fratricide: mechanisms and raisons d'etre. Nat Rev Microbiol. 2007;5:219–229. [PubMed: 17277796]
- Clewell DB, Weaver KE. Sex pheromones and plasmid transfer in Enterococcus faecalis. Plasmid. 1989;21(3):175–184. [PubMed: 2550976]
- Cobo Molinos A, Abriouel H, Omar NB, López RL, Galvez A. Detection of ebp (endocarditis- and biofilm-associated pilus) genes in enterococcal isolates from clinical and non-clinical origin. Int J Food Microbiol. 2008;126(1-2):123–126. [PubMed: 18571263]
- Coburn PS, Baghdayan AS, Craig N, Burroughs A, Tendolkar P, Miller K, et al. A novel conjugative plasmid from Enterococcus faecalis E99 enhances resistance to ultraviolet radiation. Plasmid. 2010;64(1):18–25. [PMC free article: PMC2891438] [PubMed: 20307569]
- Coburn PS, Baghdayan AS, Dolan GT, Shankar N. An AraC-type transcriptional regulator encoded on the Enterococcus faecalis pathogenicity island contributes to pathogenesis and intracellular macrophage survival. Infect Immun. 2008;76(12):5668–5676. [PMC free article: PMC2583563] [PubMed: 18824537]
- Cook L, Chatterjee A, Barnes A, Yarwood J, Hu WS, Dunny G. Biofilm growth alters regulation of conjugation by a bacterial pheromone. Mol Microbiol. 2011;81(6):1499–1510. [PMC free article: PMC3187857] [PubMed: 21843206]
- Coque TM, Willems R, Cantón R, Del Campo R, Baquero F. High occurrence of esp among ampicillin-resistant and vancomycin-susceptible Enterococcus faecium clones from hospitalized patients. J Antimicrob Chemother. 2002;50(6):1035–1038. [PubMed: 12461029]
- Creti R, Fabretti F, Koch S, Huebner J, Garsin DA, Baldassarri L, et al. Surface protein EF3314 contributes to virulence properties of Enterococcus faecalis. Int J Artif Organs. 2009;32(9):611–620. [PMC free article: PMC4351668] [PubMed: 19856273]
- Creti R, Koch S, Fabretti F, Baldassarri L, Huebner J. Enterococcal colonization of the gastro-intestinal tract: role of biofilm and environmental oligosaccharides. BMC Microbiol. 2006;6:60. [PMC free article: PMC1534043] [PubMed: 16834772]
- Dahlén G, Samuelsson W, Molander A, Reit C. Identification and antimicrobial susceptibility of enterococci isolated from the root canal. Oral Microbiol Immunol. 2000;15(5):309–312. [PubMed: 11154422]
- Del Papa MF, Perego M. Enterococcus faecalis virulence regulator FsrA binding to target promoters. J Bacteriol. 2011;193(7):1527–1532. [PMC free article: PMC3067650] [PubMed: 21257771]
- Di Rosa R, Creti R, Venditti M, D'Amelio R, Arciola CR, Montanaro L, et al. Relationship between biofilm formation, the enterococcal surface protein (Esp) and gelatinase in clinical isolates of Enterococcus faecalis and Enterococcus faecium. FEMS Microbiol Lett. 2006;256(1):145–150. [PubMed: 16487332]
- Duggan JM, Sedgley CM. Biofilm formation of oral and endodontic Enterococcus faecalis. J Endod. 2007;33(7):815–818. [PubMed: 17804318]
- Duprè I, Zanetti S, Schito AM, Fadda G, Sechi LA. Incidence of virulence determinants in clinical Enterococcus faecium and Enterococcus faecalis isolates collected in Sardinia (Italy). J Med Microbiol. 2003;52(Pt 6):491–498. [PubMed: 12748268]
- Dworniczek E, Kuzko K, Mróz E, Wojciech Ł, Adamski R, Sobieszczańska B, et al. Virulence factors and in vitro adherence of Enterococcus strains to urinary catheters. Folia Microbiol (Praha). 2003;48(5):671–678. [PubMed: 14976727]
- Dworniczek E, Wojciech Ł, Sobieszczańska B, Seniuk A. Virulence of Enterococcus isolates collected in Lower Silesia (Poland). Scand J Infect Dis. 2005;37(9):630–636. [PubMed: 16126561]
- Eaton TJ, Gasson MJ. A variant enterococcal surface protein Esp(fm) in Enterococcus faecium; distribution among food, commensal, medical, and environmental isolates. FEMS Microbiol Lett. 2002;216(2):269–275. [PubMed: 12435513]
- Erlandsen SL, Kristich CJ, Dunny GM, Wells CL. High-resolution visualization of the microbial glycocalyx with low-voltage scanning electron microscopy: dependence on cationic dyes. J Histochem Cytochem. 2004;52(11):1427–1435. [PMC free article: PMC3957825] [PubMed: 15505337]
- Fabretti F, Theilacker C, Baldassarri L, Kaczynski Z, Kropec A, Holst O, et al. Alanine Esters of Enterococcal Lipoteichoic Acid Play a Role in Biofilm Formation and Resistance to Antimicrobial Peptides. Infect Immun. 2006;74(7):4164–4171. [PMC free article: PMC1489678] [PubMed: 16790791]
- Farber BF, Wolff AG. Salicylic acid prevents the adherence of bacteria and yeast to silastic catheters. J Biomed Mater Res. 1993;27(5):599–602. [PubMed: 8390997]
- Federle MJ. Autoinducer-2-based chemical communication in bacteria: complexities of interspecies signaling. Contrib Microbiol. 2009;16:18–32. [PMC free article: PMC3042238] [PubMed: 19494577]
- Foley I, Gilbert P. In-vitro studies of the activity of glycopeptide combinations against Enterococcus faecalis biofilms. J Antimicrob Chemother. 1997;40(5):667–672. [PubMed: 9421314]
- Frank KL, Barnes AM, Grindle SM, Manias DA, Schlievert PM, Dunny GM. Use of recombinase-based in vivo expression technology to characterize Enterococcus faecalis gene expression during infection identifies in vivo-expressed antisense RNAs and implicates the protease Eep in pathogenesis. Infect Immun. 2012;80(2):539–549. [PMC free article: PMC3264308] [PubMed: 22144481]
- Gallardo-Moreno AM, González-Martín ML, Perez-Giraldo C, Bruque JM, Gómez-García AC. Serum as a factor influencing adhesion of Enterococcus faecalis to glass and silicone. Appl Environ Microbiol. 2002;68(11):5784–5787. [PMC free article: PMC129886] [PubMed: 12406782]
- Gao P, Pinkston KL, Nallapareddy SR, van Hoof A, Murray BE, Harvey BR. Enterococcus faecalis rnjB is required for pilin gene expression and biofilm formation. J Bacteriol. 2010;192(20):5489–5498. [PMC free article: PMC2950486] [PubMed: 20729365]
- George S, Kishen A. Effect of tissue fluids on hydrophobicity and adherence of Enterococcus faecalis to dentin. J Endod. 2007;33(12):1421–1425. [PubMed: 18037050]
- Gilmore MS, Haas W. The selective advantage of microbial fratricide. Proc Natl Acad Sci U S A. 2005;102(24):8401–8402. [PMC free article: PMC1150870] [PubMed: 15939890]
- Guiton PS, Hung CS, Hancock LE, Caparon MG, Hultgren SJ. Enterococcal biofilm formation and virulence in an optimized murine model of foreign body-associated urinary tract infections. Infect Immun. 2010;78(10):4166–4175. [PMC free article: PMC2950371] [PubMed: 20696830]
- Guiton PS, Hung CS, Kline KA, Roth R, Kau AL, Hayes E, et al. Contribution of autolysin and Sortase a during Enterococcus faecalis DNA-dependent biofilm development. Infect Immun. 2009;77(9):3626–3638. [PMC free article: PMC2738007] [PubMed: 19528211]
- Hall AE, Gorovits EL, Syribeys PJ, Domanski PJ, Ames BR, Chang CY, et al. Monoclonal antibodies recognizing the Enterococcus faecalis collagen-binding MSCRAMM Ace: conditional expression and binding analysis. Microb Pathog. 2007;43(2-3):55–66. [PubMed: 17521860]
- Hancock LE, Perego M. Systematic inactivation and phenotypic characterization of two-component signal transduction systems of Enterococcus faecalis V583. J Bacteriol. 2004;186(23):7951–7958. [PMC free article: PMC529088] [PubMed: 15547267]
- Hancock LE, Perego M. The Enterococcus faecalis fsr two-component system controls biofilm development through production of gelatinase. J Bacteriol. 2004;186(17):5629–5639. [PMC free article: PMC516840] [PubMed: 15317767]
- Hancock L, Perego M. Two-Component Signal Transduction in Enterococcus faecalis. J Bacteriol. 2002;184(21):5819–5825. [PMC free article: PMC135378] [PubMed: 12374813]
- Handley PS, Jacob AE. Some structural and physiological properties of fimbriae of Streptococcus faecalis. Microbiology. 1981;127(2):289–293. [PubMed: 6123542]
- Heikens E, Leendertse M, Wijnands LM, van Luit-Asbroek M, Bonten MJ, van der Poll T, et al. Enterococcal surface protein Esp is not essential for cell adhesion and intestinal colonization of Enterococcus faecium in mice. BMC Microbiol. 2009;9:19. [PMC free article: PMC2639590] [PubMed: 19178704]
- Hendrickx AP, Bonten MJ, van Luit-Asbroek M, Schapendonk CM, Kragten AH, Willems RJ. Expression of two distinct types of pili by a hospital-acquired Enterococcus faecium isolate. Microbiology. 2008;154(Pt 10):3212–3223. [PubMed: 18832326]
- Hendrickx AP, Schapendonk CM, van Luit-Asbroek M, Bonten MJ, van Schaik W, Willems RJ. Differential PilA pilus assembly by a hospital-acquired and a community-derived Enterococcus faecium isolate. Microbiology. 2010;156(Pt 9):2649–2659. [PubMed: 20542929]
- Hendrickx AP, van Luit-Asbroek M, Schapendonk CM, van Wamel WJ, Braat JC, Wijnands LM, et al. SgrA, a nidogen-binding LPXTG surface adhesin implicated in biofilm formation, and EcbA, a collagen binding MSCRAMM, are two novel adhesins of hospital-acquired Enterococcus faecium. Infect Immun. 2009;77(11):5097–5106. [PMC free article: PMC2772516] [PubMed: 19737906]
- Hendrickx AP, Willems RJ, Bonten MJ, van Schaik W. LPxTG surface proteins of enterococci. Trends Microbiol. 2009;17(9):423–430. [PubMed: 19726195]
- Hoch JA, Silhavy TJ. (1995). Two-component signal transduction. Washington, D. C.: ASM Press.
- Hufnagel M, Koch S, Creti R, Baldassarri L, Huebner J. A putative sugar-binding transcriptional regulator in a novel gene locus in Enterococcus faecalis contributes to production of biofilm and prolonged bacteremia in mice. J Infect Dis. 2004;189(3):420–430. [PubMed: 14745699]
- Jansen B, Goodman LP, Ruiten D. Bacterial adherence to hydrophilic polymer-coated polyurethane stents. Gastrointest Endosc. 1993;39(5):670–673. [PubMed: 8224690]
- Jass J, Phillips LE, Allan EJ, Costerton JW, Lappin-Scott HM. Growth and adhesion of Enterococcus faecium L-forms. FEMS Microbiol Lett. 1994;115(2-3):157–162. [PubMed: 8138130]
- Johnson JR, Clabots C, Hirt H, Waters C, Dunny G. Enterococcal aggregation substance and binding substance are not major contributors to urinary tract colonization by Enterococcus faecalis in a mouse model of ascending unobstructed urinary tract infection. Infect Immun. 2004;72(4):2445–2448. [PMC free article: PMC375174] [PubMed: 15039379]
- Kaper JB, Sperandio V. Bacterial Cell-to-Cell Signaling in the Gastrointestinal Tract. Infect Immun. 2005;73(6):3197–3209. [PMC free article: PMC1111840] [PubMed: 15908344]
- Keane PF, Bonner MC, Johnston SR, Zafar A, Gorman SP. Characterization of biofilm and encrustation on ureteric stents in vivo. Br J Urol. 1994;73(6):687–691. [PubMed: 8032837]
- Kemp KD, Singh KV, Nallapareddy SR, Murray BE. Relative contributions of Enterococcus faecalis OG1RF sortase-encoding genes, srtA and bps (srtC), to biofilm formation and a murine model of urinary tract infection. Infect Immun. 2007;75(11):5399–5404. [PMC free article: PMC2168291] [PubMed: 17785477]
- Kim DS, Singh KV, Nallapareddy SR, Qin X, Panesso D, Arias CA, et al. The fms21 (pilA)-fms20 locus encoding one of four distinct pili of Enterococcus faecium is harboured on a large transferable plasmid associated with gut colonization and virulence. J Med Microbiol. 2010;59(Pt 4):505–507. [PMC free article: PMC2850606] [PubMed: 20075113]
- Koivusalo A, Mäkisalo H, Talja M, Siitonen A, Vuopio-Varkila J, Ruutu M, et al. Bacterial adherence and biofilm formation on latex and silicone T-tubes in relation to bacterial contamination of bile. Scand J Gastroenterol. 1996;31(4):398–403. [PubMed: 8726310]
- Kowalski WJ, Kasper EL, Hatton JF, Murray BE, Nallapareddy SR, Gillespie MJ. Enterococcus faecalis adhesin, Ace, mediates attachment to particulate dentin. J Endod. 2006;32(7):634–637. [PubMed: 16793469]
- Kristich CJ, Li Y-H, Cvitkovich DG, Dunny GM. Esp-independent biofilm formation by Enterococcus faecalis. J Bacteriol. 2004;186(1):154–163. [PMC free article: PMC365672] [PubMed: 14679235]
- Kristich CJ, Nguyen VT, Le T, Barnes AM, Grindle S, Dunny GM. Development and use of an efficient system for random mariner transposon mutagenesis to identify novel genetic determinants of biofilm formation in the core Enterococcus faecalis genome. Appl Environ Microbiol. 2008;74(11):3377–3386. [PMC free article: PMC2423031] [PubMed: 18408066]
- Lambert PA, Shorrock PJ, Aitchison EJ, Dominique PA, Power ME, Costerton JW. Effect of in vivo growth conditions upon expression of surface protein antigens in Enterococcus faecalis. FEMS Immunol Med Microbiol. 1990;2(1):51–54. [PubMed: 2114887]
- Leavis H, Top J, Shankar N, Borgen K, Bonten M, van Embden J, et al. A novel putative enterococcal pathogenicity island linked to the esp virulence gene of Enterococcus faecium and associated with epidemicity. J Bacteriol. 2004;186(3):672–682. [PMC free article: PMC321477] [PubMed: 14729692]
- Lebreton F, Riboulet-Bisson E, Serror P, Sanguinetti M, Posteraro B, Torelli R, et al. ace, Which encodes an adhesin in Enterococcus faecalis, is regulated by Ers and is involved in virulence. Infect Immun. 2009;77(7):2832–2839. [PMC free article: PMC2708572] [PubMed: 19433548]
- Love RM. Enterococcus faecalis--a mechanism for its role in endodontic failure. Int Endod J. 2001;34(5):399–405. [PubMed: 11482724]
- Mandlik A, Swierczynski A, Das A, Ton-That H. Pili in Gram-positive bacteria: assembly, involvement in colonization and biofilm development. Trends Microbiol. 2008;16(1):33–40. [PMC free article: PMC2841691] [PubMed: 18083568]
- McCormick J, Hirt H, Waters CM, Tripp TJ, Dunny GM, Schlievert PM. Antibodies to a surface-exposed, N-terminal domain of aggregation substance are not protective in the rabbit model of Enterococcus faecalis infective endocarditis. Infect Immun. 2001;69(5):3305–3314. [PMC free article: PMC98289] [PubMed: 11292753]
- Mohamed JA, Huang W, Nallapareddy SR, Teng F, Murray BE. Influence of origin of isolates, especially endocarditis isolates, and various genes on biofilm formation by Enterococcus faecalis. Infect Immun. 2004;72(6):3658–3663. [PMC free article: PMC415661] [PubMed: 15155680]
- Mohamed JA, Teng F, Nallapareddy SR, Murray BE. Pleiotrophic effects of 2 Enterococcus faecalis sagA-like genes, salA and salB, which encode proteins that are antigenic during human infection, on biofilm formation and binding to collagen type I and fibronectin. J Infect Dis. 2005;193:231–240. [PubMed: 16362887]
- Monds RD, O'Toole GA. The developmental model of microbial biofilms: ten years of a paradigm up for review. Trends Microbiol. 2009;17(2):73–87. [PubMed: 19162483]
- Montanaro L, Poggi A, Visai L, Ravaioli S, Campoccia D, Speziale P, et al. Extracellular DNA in biofilms. Int J Artif Organs. 2011;34(9):824–831. [PubMed: 22094562]
- Mylonakis E, Engelbert M, Qin X, Sifri CD, Murray BE, Ausubel FM, et al. The Enterococcus faecalis fsrB gene, a key component of the Fsr quorum-sensing system, is associated with virulence in the rabbit endophthalmitis model. Infect Immun. 2002;70(8):4678–4681. [PMC free article: PMC128160] [PubMed: 12117982]
- Nakayama J, Cao Y, Horii T, Sakuda S, Akkermans AD, de Vos WM, et al. Gelatinase biosynthesis-activating pheromone: a peptide lactone that mediates a quorum sensing in Enterococcus faecalis. Mol Microbiol. 2001;41(1):145–154. [PubMed: 11454207]
- Nakayama J, Chen S, Oyama N, Nishiguchi K, Azab EA, Tanaka E, et al. Revised model for Enterococcus faecalis fsr quorum-sensing system: the small open reading frame fsrD encodes the gelatinase biosynthesis-activating pheromone propeptide corresponding to staphylococcal agrD. J Bacteriol. 2006;188(23):8321–8326. [PMC free article: PMC1698201] [PubMed: 16980448]
- Nallapareddy SR, Murray BE. Ligand-signaled upregulation of Enterococcus faecalis ace transcription, a mechanism for modulating host-E. faecalis interaction. Infect Immun. 2006;74(9):4982–4989. [PMC free article: PMC1594855] [PubMed: 16926389]
- Nallapareddy SR, Qin X, Weinstock GM, Höök M, Murray BE. Enterococcus faecalis adhesin, Ace, mediates attachment to extracellular matrix proteins collagen type IV and laminin as well as collagen type I. Infect Immun. 2000;68(9):5218–5224. [PMC free article: PMC101781] [PubMed: 10948147]
- Nallapareddy SR, Sillanpää J, Mitchell J, Singh KV, Chowdhury SA, Weinstock GM, et al. Conservation of Ebp-type pilus genes among Enterococci and demonstration of their role in adherence of Enterococcus faecalis to human platelets. Infect Immun. 2011;79(7):2911–2920. [PMC free article: PMC3191956] [PubMed: 21502588]
- Nallapareddy SR, Singh KV, Murray BE. Contribution of the collagen adhesin Acm to pathogenesis of Enterococcus faecium in experimental endocarditis. Infect Immun. 2008;76(9):4120–4128. [PMC free article: PMC2519397] [PubMed: 18591236]
- Nallapareddy SR, Singh KV, Duh RW, Weinstock GM, Murray BE. Diversity of ace, a gene encoding a microbial surface component recognizing adhesive matrix molecules, from different strains of Enterococcus faecalis and evidence for production of Ace during human infections. Infect Immun. 2000;68(9):5210–5217. [PMC free article: PMC101780] [PubMed: 10948146]
- Nallapareddy SR, Singh KV, Okhuysen PC, Murray BE. A functional collagen adhesin gene, acm, in clinical isolates of Enterococcus faecium correlates with the recent success of this emerging nosocomial pathogen. Infect Immun. 2008;76(9):4110–4119. [PMC free article: PMC2519430] [PubMed: 18591238]
- Nallapareddy SR, Singh KV, Sillanpää J, Garsin DA, Höök M, Erlandsen SL, et al. Endocarditis and biofilm-associated pili of Enterococcus faecalis. J Clin Invest. 2006;116(10):2799–2807. [PMC free article: PMC1578622] [PubMed: 17016560]
- Nallapareddy SR, Singh KV, Sillanpää J, Zhao M, Murray BE. Relative contributions of Ebp pili and the collagen adhesin Ace to host extracellular matrix protein adherence and experimental urinary tract infection by Enterococcus faecalis OG1RF. Infect Immun. 2011;79(7):2901–2910. [PMC free article: PMC3191960] [PubMed: 21505082]
- Nallapareddy SR, Weinstock GM, Murray BE. Clinical isolates of Enterococcus faecium exhibit strain-specific collagen binding mediated by Acm, a new member of the MSCRAMM family. Mol Microbiol. 2003;47(6):1733–1747. [PubMed: 12622825]
- Nickel JC, Reid G, Bruce AW, Costerton JW. Ultrastructural microbiology of infected urinary stone. Urology. 1986;28(6):512–515. [PubMed: 3097904]
- Novick RP, Geisinger E. Quorum sensing in staphylococci. Annu Rev Genet. 2008;42:541–564. [PubMed: 18713030]
- O'Toole G, Kaplan HB, Kolter R. Biofilm formation as microbial development. Annu Rev Microbiol. 2000;54:49–79. [PubMed: 11018124]
- Paganelli FL, Willems RJ, Leavis HL. Optimizing future treatment of enterococcal infections: attacking the biofilm? Trends Microbiol. 2012;20(1):40–49. [PubMed: 22169461]
- Paulsen IT, Banerjei L, Myers GS, Nelson KE, Seshadri R, Read TD, et al. Role of Mobile DNA in the Evolution of Vancomycin-Resistant Enterococcus faecalis. Science. 2003;299(5615):2071–2074. [PubMed: 12663927]
- Pillai SK, Sakoulas G, Eliopoulos GM, Moellering RC Jr, Murray BE, Inouye RT. Effects of glucose on fsr-mediated biofilm formation in Enterococcus faecalis. J Infect Dis. 2004;190(5):967–970. [PubMed: 15295702]
- Prakash VP, Rao SR, Parija SC. Emergence of unusual species of enterococci causing infections, South India. BMC Infect Dis. 2005;5:14. [PMC free article: PMC555955] [PubMed: 15774018]
- Pultz NJ, Shankar N, Baghdayan AS, Donskey CJ. Enterococcal surface protein Esp does not facilitate intestinal colonization or translocation of Enterococcus faecalis in clindamycin-treated mice. FEMS Microbiol Lett. 2005;242:217–219. [PubMed: 15621440]
- Qin X, Singh KV, Weinstock GM, Murray BE. Characterization of Fsr, a regulator controlling expression of gelatinase and serine protease in Enterococcus faecalis OG1RF. J Bacteriol. 2001;183(11):3372–3382. [PMC free article: PMC99635] [PubMed: 11344145]
- Qin X, Xu Y, Singh KV, Weinstock GM, Murray BE. Effect of disruption of a gene encoding an autolysin of Enterococcus faecalis OG1RF. Antimicrob Agents Chemother. 1998;42(11):2883–2888. [PMC free article: PMC105960] [PubMed: 9797220]
- Raad II, Hanna HA, Boktour M, Chaiban G, Hachem RY, Dvorak T, et al. Vancomycin-resistant Enterococcus faecium: catheter colonization, esp gene, and decreased susceptibility to antibiotics in biofilm. Antimicrob Agents Chemother. 2005;49(12):5046–5050. [PMC free article: PMC1315928] [PubMed: 16304171]
- Raad I, Darouiche R, Hachem R, Sacilowski M, Bodey GP. Antibiotics and prevention of microbial colonization of catheters. Antimicrob Agents Chemother. 1995;39(11):2397–2400. [PMC free article: PMC162954] [PubMed: 8585715]
- Rakita RM, Vanek NN, Jacques-Palaz K, Mee M, Mariscalco MM, Dunny GM, et al. Enterococcus faecalis bearing aggregation substance is resistant to killing by human neutrophils despite phagocytosis and neutrophil activation. Infect Immun. 1999;67(11):6067–6075. [PMC free article: PMC96994] [PubMed: 10531268]
- Ramadhan AA, Hegedus E. Biofilm formation and esp gene carriage in enterococci. J Clin Pathol. 2005;58(7):685–686. [PMC free article: PMC1770729] [PubMed: 15976332]
- Rasko DA, Sperandio V. Anti-virulence strategies to combat bacteria-mediated disease. Nat Rev Drug Discov. 2010;9:117–128. [PubMed: 20081869]
- Reed WP, Moody MR, Newman KA, Light PD, Costerton JW. Bacterial colonization of Hemasite access devices. Surgery. 1986;99(3):308–317. [PubMed: 3082026]
- Reid G, Denstedt JD, Kang YS, Lam D, Nause C. Microbial adhesion and biofilm formation on ureteral stents in vitro and in vivo. J Urol. 1992;148(5):1592–1594. [PubMed: 1433574]
- Rozdzinski E, Marre R, Susa M, Wirth R, Muschol-Silberhorn A. Aggregation substance-mediated adherence of Enterococcus faecalis to immobilized extracellular matrix proteins. Microb Pathog. 2001;30(4):211–220. [PubMed: 11312614]
- Sandoe JA, Witherden IR, Cove JH, Heritage J, Wilcox MH. Correlation between enterococcal biofilm formation in vitro and medical-device-related infection potential in vivo. J Med Microbiol. 2003;52(Pt 7):547–550. [PubMed: 12808074]
- Schauder S, Shokat K, Surette MG, Bassler BL. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol Microbiol. 2001;41(2):463–476. [PubMed: 11489131]
- Schlievert PM, Chuang-Smith ON, Peterson ML, Cook LC, Dunny GM. Enterococcus faecalis endocarditis severity in rabbits is reduced by IgG Fabs interfering with aggregation substance. PLoS ONE. 2010;5(10):e13194. [PMC free article: PMC2949389] [PubMed: 20957231]
- Schwank S, Blaser J. Once-versus thrice-daily netilmicin combined with amoxicillin, penicillin, or vancomycin against Enterococcus faecalis in a pharmacodynamic in vitro model. Antimicrob Agents Chemother. 1996;40(10):2258–2261. [PMC free article: PMC163514] [PubMed: 8891125]
- Seno Y, Kariyama R, Mistuhata R, Monden K, Kumon H. Clinical implications of biofilm formation by Enterococcus faecalis in the urinary tract. Acta Med Okayama. 2005;59(3):79–87. [PubMed: 16049560]
- Shankar N, Baghdayan AS, Gilmore MS. Modulation of virulence within a pathogenicity island in vancomycin-resistant Enterococcus faecalis. Nature. 2002;417(6890):746–750. [PubMed: 12066186]
- Shankar N, Lockatell CV, Baghdayan AS, Drachenberg C, Gilmore MS, Johnson DE. Role of Enterococcus faecalis surface protein Esp in the pathogenesis of ascending urinary tract infection. Infect Immun. 2001;69(7):4366–4372. [PMC free article: PMC98508] [PubMed: 11401975]
- Shankar V, Baghdayan AS, Huycke MM, Lindahl G, Gilmore MS. Infection-derived Enterococcus faecalis strains are enriched in esp, a gene encoding a novel surface protein. Infect Immun. 1999;67(1):193–200. [PMC free article: PMC96296] [PubMed: 9864215]
- Sifri CD, Mylonakis E, Singh KV, Qin X, Garsin DA, Murray BE, et al. Virulence effect of Enterococcus faecalis protease genes and the quorum-sensing locus fsr in Caenorhabditis elegans and mice. Infect Immun. 2002;70(10):5647–5650. [PMC free article: PMC128331] [PubMed: 12228293]
- Sillanpää J, Nallapareddy SR, Prakash VP, Qin X, Höök M, Weinstock GM. Identification and phenotypic characterization of a second collagen adhesin, Scm, and genome-based identification and analysis of 13 other predicted MSCRAMMs, including four distinct pilus loci, in Enterococcus faecium. Microbiology. 2008;154(Pt 10):3199–3211. [PMC free article: PMC2677164] [PubMed: 18832325]
- Sillanpää J, Nallapareddy SR, Singh KV, Prakash VP, Fothergill T, Ton-That H, et al. Characterization of the ebp(fm) pilus-encoding operon of Enterococcus faecium and its role in biofilm formation and virulence in a murine model of urinary tract infection. Virulence. 2010;1(4):236–246. [PMC free article: PMC2910428] [PubMed: 20676385]
- Sillanpää J, Xu Y, Nallapareddy SR, Murray BE, Höök M. A family of putative MSCRAMMs from Enterococcus faecalis. Microbiology. 2004;150(Pt 7):2069–2078. [PubMed: 15256550]
- Singh KV, Lewis RJ, Murray BE. Importance of the epa locus of Enterococcus faecalis OG1RF in a mouse model of ascending urinary tract infection. J Infect Dis. 2009;200(3):417–420. [PMC free article: PMC3615707] [PubMed: 19545208]
- Singh KV, Nallapareddy SR, Murray BE. Importance of the ebp (endocarditis- and biofilm-associated pilus) locus in the pathogenesis of Enterococcus faecalis ascending urinary tract infection. J Infect Dis. 2007;195(11):1671–1677. [PMC free article: PMC2680192] [PubMed: 17471437]
- Singh KV, Nallapareddy SR, Nannini EC, Murray BE. Fsr-independent production of protease(s) may explain the lack of attenuation of an Enterococcus faecalis fsr mutant versus a gelE-sprE mutant in induction of endocarditis. Infect Immun. 2005;73(8):4888–4894. [PMC free article: PMC1201275] [PubMed: 16041002]
- Singh KV, Nallapareddy SR, Sillanpää J, Murray BE. Importance of the collagen adhesin Ace in pathogenesis and protection against Enterococcus faecalis experimental endocarditis. PLoS Pathog. 2010;6(1):e1000716. [PMC free article: PMC2798748] [PubMed: 20072611]
- Souto R, Colombo AP. Prevalence of Enterococcus faecalis in subgingival biofilm and saliva of subjects with chronic periodontal infection. Arch Oral Biol. 2008;53(2):155–160. [PubMed: 17897617]
- Srivastava M, Mallard C, Barke T, Hancock LE, Self WT. A selenium-dependent xanthine dehydrogenase triggers biofilm proliferation in Enterococcus faecalis through oxidant production. J Bacteriol. 2011;193(7):1643–1652. [PMC free article: PMC3067645] [PubMed: 21257770]
- Stickler DJ, King JB, Winters C, Morris SL. Blockage of urethral catheters by bacterial biofilms. J Infect. 1993;27(2):133–134. [PubMed: 8228293]
- Süßmuth SD, Muscholl-Silberhorn A, Wirth R, Susa M, Marre R, Rozdzinski E. Aggregation substance promotes adherence, phagocytosis, and intracellular survival of Enterococcus faecalis within human macrophages and suppresses respiratory burst. Infect Immun. 2000;68(9):4900–4906. [PMC free article: PMC101694] [PubMed: 10948103]
- Telford JL, Barocchi MA, Margarit I, Rappuoli R, Grandi G. Pili in Gram-positive pathogens. Nat Rev Microbiol. 2006;4:509–519. [PubMed: 16778837]
- Tendolkar PM, Baghdayan AS, Shankar N. The N-terminal domain of enterococcal surface protein, Esp, is sufficient for Esp-mediated biofilm enhancement in Enterococcus faecalis. J Bacteriol. 2005;187(17):6213–6222. [PMC free article: PMC1196143] [PubMed: 16109963]
- Tendolkar PM, Baghdayan AS, Shankar N. Putative surface proteins encoded within a novel transferable locus confer a high-biofilm phenotype to Enterococcus faecalis. J Bacteriol. 2006;188(6):2063–2072. [PMC free article: PMC1428127] [PubMed: 16513736]
- Tendolkar PM, Baghdayan AS, Gilmore MS, Shankar N. Enterococcal surface protein, Esp, enhances biofilm formation by Enterococcus faecalis. Infect Immun. 2004;72(10):6032–6039. [PMC free article: PMC517584] [PubMed: 15385507]
- Teng F, Jacques-Palaz KD, Weinstock GM, Murray BE. Evidence that the enterococcal polysaccharide antigen gene (epa) cluster is widespread in Enterococcus faecalis and influences resistance to phagocytic killing of E. faecalis. Infect Immun. 2002;70(4):2010–2015. [PMC free article: PMC127866] [PubMed: 11895965]
- Teng F, Singh KV, Bourgogne A, Zeng J, Murray BE. Further characterization of the epa gene cluster and Epa polysaccharides of Enterococcus faecalis. Infect Immun. 2009;77(9):3759–3767. [PMC free article: PMC2737988] [PubMed: 19581393]
- Theilacker C, Sanchez-Carballo P, Toma I, Fabretti F, Sava I, Kropec A, et al. Glycolipids are involved in biofilm accumulation and prolonged bacteraemia in Enterococcus faecalis. Mol Microbiol. 2009;71(4):1055–1069. [PubMed: 19170884]
- Thomas VC, Hancock LE. Suicide and fratricide in bacterial biofilms. Int J Artif Organs. 2009;32(9):537–544. [PubMed: 19851979]
- Thomas VC, Hiromasa Y, Harms N, Thurlow L, Tomich J, Hancock LE. A fratricidal mechanism is responsible for eDNA release and contributes to biofilm development of Enterococcus faecalis. Mol Microbiol. 2009;72(4):1022–1036. [PMC free article: PMC2779696] [PubMed: 19400795]
- Thomas VC, Thurlow LR, Boyle D, Hancock LE. Regulation of autolysis-dependent extracellular DNA release by Enterococcus faecalis extracellular proteases influences biofilm development. J Bacteriol. 2008;190(16):5690–5698. [PMC free article: PMC2519388] [PubMed: 18556793]
- Thurlow LR, Thomas VC, Narayanan S, Olson S, Fleming SD, Hancock LE. Gelatinase contributes to the pathogenesis of endocarditis caused by Enterococcus faecalis. Infect Immun. 2010;78(11):4936–4943. [PMC free article: PMC2976315] [PubMed: 20713628]
- Toledo-Arana A, Valle J, Solano C, Arrizubieta MJ, Cucarella C, Lamata M, et al. The enterococcal surface protein, Esp, is involved in Enterococcus faecalis biofilm formation. Appl Environ Microbiol. 2001;67(10):4538–4545. [PMC free article: PMC93200] [PubMed: 11571153]
- Trappetti C, Potter AJ, Paton AW, Oggioni MR, Paton JC. LuxS mediates iron-dependent biofilm formation, competence, and fratricide in Streptococcus pneumoniae. Infect Immun. 2011;79(11):4550–4558. [PMC free article: PMC3257940] [PubMed: 21875962]
- van Merode AE, van der Mei HC, Busscher HJ, Krom BP. Influence of culture heterogeneity in cell surface charge on adhesion and biofilm formation by Enterococcus faecalis. J Bacteriol. 2006;188(7):2421–2426. [PMC free article: PMC1428413] [PubMed: 16547028]
- Van Wamel WJ, Hendrickx AP, Bonten MJ, Top J, Posthuma G, Willems RJ. Growth condition-dependent Esp expression by Enterococcus faecium affects initial adherence and biofilm formation. Infect Immun. 2007;75(2):924–931. [PMC free article: PMC1828491] [PubMed: 17118984]
- Vanek NN, Simon SI, Jacques-Palaz K, Mariscalco MM, Dunny GM, Rakita RM. Enterococcus faecalis aggregation substance promotes opsonin-independent binding to human neutrophils via a complement receptor type 3-mediated mechanism. FEMS Immunol Med Microbiol. 1999;26(1):49–60. [PubMed: 10518042]
- Vidal JE, Ludewick HP, Kunkel RM, Zähner D, Klugman KP. The LuxS-dependent quorum-sensing system regulates early biofilm formation by Streptococcus pneumoniae strain D39. Infect Immun. 2011;79(10):4050–4060. [PMC free article: PMC3187250] [PubMed: 21825061]
- Waters CM, Antiporta MH, Murray BE, Dunny GM. Role of the Enterococcus faecalis GelE protease in determination of cellular chain length, supernatant pheromone levels, and degradation of fibrin and misfolded surface proteins. J Bacteriol. 2003;185(12):3613–3623. [PMC free article: PMC156229] [PubMed: 12775699]
- Wells CL, Moore EA, Hoag JA, Hirt H, Dunny GM, Erlandsen SL. Inducible expression of Enterococcus faecalis aggregation substance surface protein facilitates bacterial internalization by cultured enterocytes. Infect Immun. 2000;68(12):7190–7194. [PMC free article: PMC97839] [PubMed: 11083854]
- Willems RJ, Homan W, Top J, van Santen-Verheuvel M, Tribe D, Manzioros X, et al. Variant esp gene as a marker of a distinct genetic lineage of vancomycin-resistant Enterococcus faecium spreading in hospitals. Lancet. 2001;357(9259):853–855. [PubMed: 11265956]
- Review Streptococcus pyogenes Biofilm.[Streptococcus pyogenes: Basic ...]Review Streptococcus pyogenes Biofilm.Young C, Holder RC, Dubois L, Reid SD. Streptococcus pyogenes: Basic Biology to Clinical Manifestations. 2016
- Review [Pathogenicity of Enterococci].[Nihon Saikingaku Zasshi. 2017]Review [Pathogenicity of Enterococci].Ike Y. Nihon Saikingaku Zasshi. 2017; 72(2):189-211.
- Macromolecular crowding: chemistry and physics meet biology (Ascona, Switzerland, 10-14 June 2012).[Phys Biol. 2013]Macromolecular crowding: chemistry and physics meet biology (Ascona, Switzerland, 10-14 June 2012).Foffi G, Pastore A, Piazza F, Temussi PA. Phys Biol. 2013 Aug; 10(4):040301. Epub 2013 Aug 2.
- Review Transcriptional and Post Transcriptional Control of Enterococcal Gene Regulation.[Enterococci: From Commensals t...]Review Transcriptional and Post Transcriptional Control of Enterococcal Gene Regulation.DebRoy S, Gao P, Garsin DA, Harvey BR, Kos V, Nes IF, Solheim M. Enterococci: From Commensals to Leading Causes of Drug Resistant Infection. 2014
- Review Enterococcal Bacteriocins and Antimicrobial Proteins that Contribute to Niche Control.[Enterococci: From Commensals t...]Review Enterococcal Bacteriocins and Antimicrobial Proteins that Contribute to Niche Control.Ness IF, Diep DB, Ike Y. Enterococci: From Commensals to Leading Causes of Drug Resistant Infection. 2014
- Enterococcal Biofilm Structure and Role in Colonization and Disease - Enterococc...Enterococcal Biofilm Structure and Role in Colonization and Disease - Enterococci
Your browsing activity is empty.
Activity recording is turned off.
See more...
 3.
3.