This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License. To view a copy of this license, visit https://creativecommons.org/licenses/by-nc-nd/4.0/
NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Gilmore MS, Clewell DB, Ike Y, et al., editors. Enterococci: From Commensals to Leading Causes of Drug Resistant Infection [Internet]. Boston: Massachusetts Eye and Ear Infirmary; 2014-.

Enterococci: From Commensals to Leading Causes of Drug Resistant Infection [Internet].
Show detailsAbstract
Enterococci have evolved over eons as highly adapted members of the gastrointestinal consortia of a wide variety of hosts—humans and other mammals, birds, reptiles and insects—but for reasons that are not entirely clear, they emerged in the 1970s as some of the leading causes of multidrug-resistant, hospital-acquired infections. The taxonomy of enterococci has changed considerably over the past ten years, and the genus now includes over forty distinct species with various habitats, tropisms, and metabolic and phenotypic characteristics. These habitats include animal hosts, as well as plants, soil and water, and man-made products, including fermented foods and dairy products. Antibiotic-resistant strains of enterococci have emerged in many of these habitats, and strains with novel resistance mechanisms are isolated with alarming regularity. As a result, the relationship between the non-therapeutic use of antibiotics and the occurrence of enterococci in various non-human habitats is of substantial interest.
Introduction
The term "enterococcus" has its origin at the end of the 19th century when Thiercelin described a saprophytic coccus, of intestinal origin, capable of causing infection (Thiercelin & Jouhaud, 1899; Thiercelin, 1899). The same year, MacCallum and Hastings characterized a similar organism, now known to be Enterococcus faecalis, from a lethal case of endocarditis, thus providing a first detailed description of its pathogenic capabilities (MacCallum & Hastings, 1899). These early descriptions constitute the basis for the paradigm of enterococcal pathogenesis as that of a commensal opportunist. The emergence of multi-drug resistant enterococci over the past 50 years has substantially compounded this problem, and antibiotic resistant enterococci are now leading causes of nosocomial infection worldwide (Prabaker & Weinstein, 2011; Werner, et al., 2008).
Phylogenetically, the genus Enterococcus belongs to the low GC branch of Gram-positive bacteria (i.e. 37.5% of GC in the Enterococcus faecalis chromosome) (Paulsen, et al., 2003). Enterococci were classified as group D streptococci (Sherman J. M., The streptococci, 1937; Sherman J. M., The enterococci and related streptococci, 1938) until 1984, when Streptococcus faecalis and Streptococcus faecium were reclassified as Enterococcus faecalis and Enterococcus faecium, respectively (Schleifer & Kilpper-Balz, 1984).
The Enterococcus genus is placed in the Enterococcaceae family (Ludwig, Schleifer, & Whitman, 2009), along with genus Bavariicoccus, Catellicoccus, Melissococcus, Pilibacter, Tetragenococcus and Vagococcus, and consists of species that occur in human and animal gastro-intestinal (GI) tracts (Mundt, 1963; Mundt, 1986), as well as in the guts of insects (Martin & Mundt, 1972), traditional fermented food and dairy products (Mundt, 1986), and in various environments including plants ((Mundt, 1963), soil (Mundt, 1961) and water (Ator & Starzyk, 1976). Undoubtedly, an important key to their broad distribution in nature stems from being remarkably resilient organisms that are capable of enduring a broad range in pH and temperatures, as well as hypotonic and hypertonic conditions (Sherman J. M., The enterococci and related streptococci, 1938). Enterococci are ubiquitous in GI tracts even though they constitute a small proportion of the gut consortium, typically comprising less than 1% of the adult microflora (Finegold, Sutter, & Mathisen, 1983; Sghir, Gramet, Suau, Violaine, Pochart, & Dore, 2000). Little is known about the main mechanisms used by enterococci to colonize GI tracts of either healthy individuals or hospitalized patients. Nevertheless, exposure of hospitalized patients to antibiotics results in major modifications of the gut microbiota, which facilitate colonization of the GI tract by drug-resistant enterococci (Donskey, et al., 2000; Ubeda, et al., 2010).
In this chapter, we review the literature on the diversity of species that belong to the Enterococcus genus, discuss their origin and distribution in nature, and focus on the gut colonization by the enterococci and their transition from commensal to opportunistic pathogens.
The Enterococcus genus: definition and diversity
Historical perspective
In his report to the French Society of Biology in 1899, Thiercelin shared his observations of a saprophytic microbe that pullulates in the GI tract of humans along with the Bacterium coli (Thiercelin & Jouhaud, 1899). He described this microbe as a Gram-positive diplococcus and proposed the name "Enterocoque" to emphasize its morphology and its intestinal origin (Thiercelin & Jouhaud, 1899). Among the many initial observations in this paper was the observation that this microbe was pathogenic for mice, and susceptible to becoming pathogenic for humans (Thiercelin & Jouhaud, 1899). Microscopically, Thiercelin observed a halo surrounding some cells, similar to that observed for Pneumococcus, especially in the context of an intestinal disease, which suggested the presence of a capsule in situ (Hancock & Gilmore, 2002; Thiercelin & Jouhaud, 1899). Two months later, he described the morphological polymorphism of enterococcal cells (Thiercelin, 1899). He noted that since his previous report, this microbe had been found to be predominant in the GI tract of a patient with diarrhea, which then translocated from the intestine to the blood, which led to septicemia (Thiercelin, 1899). At nearly the same time, MacCallum and Hastings provided a detailed report of a case of acute endocarditis in a 37-year-old patient, caused by an organism they called Micrococcus zymogenes (163), now recognized as a strain of Enterococcus faecalis. This bacterium was continuously isolated from the patient’s blood until his death due to cardiac failure (MacCallum & Hastings, 1899). The authors described that this bacterium was "very hardy and tenacious of life" (MacCallum & Hastings, 1899), which are now well-recognized traits of the Enterococcus genus. MacCallum and Hastings thoroughly investigated the biochemical capabilities of the enterococci and showed that this particular isolate was hemolytic (MacCallum & Hastings, 1899). The accurate detail in the descriptions of the pathogenic traits of these opportunistic commensal bacteria is particularly striking.
In 1906 the name Streptococcus faecalis was used by Andrewes and Horder (Andrewes & Horder, 1906) to characterize an organism of fecal origin that clotted milk, similar to the observations of MacCallum and Hastings, and that also fermented mannitol and lactose, but not raffinose. The name faecalis was chosen to emphasize that this Streptococcus was characteristic of the human intestine, even though the organism was isolated from a patient with endocarditis in this study (Andrewes & Horder, 1906). Orla-Jensen subsequently reported the first description of Streptococcus faecium, which differed from the fermentation patterns described for S. faecalis (Orla-Jensen, 1919). About twenty years later, a third species, Streptococcus durans, was described by Sherman and Wing (Sherman & Wing, 1935) as being similar to S. faecium, but with more restricted fermentation capabilities. Sherman subsequently proposed a classification scheme that separated streptococci into four groups: pyogenic, viridans, lactic, and enterococcus (Sherman, 1937). The term "enterococcal group" included S. faecalis (hemolysis negative and proteolysis negative), S. faecalis var. liquefaciens (hemolysis negative and proteolysis positive), S. faecalis var. hemolyticus (hemolysis positive and proteolysis negative), S. faecalis var. zymogenes (hemolysis positive and proteolysis positive), and S. durans; and was used to describe streptococci that grew between 10 and 45° C, grew in broth at pH 9.6, grew in 6.5% NaCl, and survived heating to 60°C for 30 min (Sherman, 1937). Noteworthy, S. faecium, described by Orla-Jensen earlier, was considered by Sherman to be the same as S. faecalis (Sherman, 1937), and was only officially recognized as a separate species in the mid-1960s (Barnes, 1956; Breed, Murray, & Smith, 1957; Deibel, 1964).
In 1957, motile and yellow-pigmented enterococci were described and the name S. faecium var. casseliflavus (emphasizing the yellow coloration) was proposed (Graudal, 1957; Mundt, 1986). A decade later, Nowlan and Deibel (Nowlan & Deibel, 1967) added Streptococcus avium to the enterococcal group. As early as 1970, Kalina (Kalina, 1970) proposed the creation of the taxon Enterococcus, based on cellular arrangement and phenotypic characteristics of species in the enterococcal group. However, this genus was not formally accepted until 1984, when Schleifer and Kilpper-Balz (Schleifer & Kilpper-Balz, 1984) provided genetic evidence that S. faecalis and S. faecium were sufficiently distant from other members of the Streptococcus genus. Collins and collaborators (Collins, Jones, Farrow, Kilpper-Balz, & Schleifer, 1984) used a similar genetic approach to show that strains previously named Streptococcus avium, Streptococcus casseliflavus, Streptococcus durans and Streptococcus gallinarum, were distinct species but closely related to those of the genus Enterococcus. In 2002, an extensive review of the literature (Facklam, Carvalho, & Teixeira, 2002) reported the existence of 23 distinct Enterococcus species.
Considering the ubiquitous distribution in nature of the Enterococcus genus, it is noteworthy that the three Enterococcus species (E. faecalis, E. faecium and E. durans) that were recognized prior to 1950 (Figure 1), are capable of causing human infection. E. faecalis and E. faecium are considered to be the most abundant enterococci in human feces (Devriese, Pot, Van Damme, Kersters, & Haesebrouck, 1994; Finegold, Sutter, & Mathisen, 1983; Noble, 1978; Patel, et al., 1998), and account for most enterococcal disease (Malani, Kauffman, & Zervos, 2002). E. durans occurs occasionally among the fecal flora of healthy adults (Ruoff, 1990). Until the mid-1990s, E. faecalis accounted for 90–95% of clinical isolates, and E. faecium infection was rare (Huycke, Sahm, & Gilmore, 1998). Since then, the proportion of E. faecium isolates has increased, largely due to the spread of resistance to antibiotics, particularly vancomycin and ampicillin (Arias & Murray, 2012; Gilmore, Lebreton, & van Schaik, 2013; Huycke M. M., et al., 2002). Other species, including Enterococcus avium, gallinarum, casseliflavus, hirae, mundtii, and raffinosus, also have been isolated from human infection (Devriese, Pot, Van Damme, Kersters, & Haesebrouck, 1994; Hammerum A. M., 2012; Murray, 1990). Enterococcus gallinarum and Enterococcus casseliflavus infections are of special interest because of their intrinsic resistance to vancomycin, an antibiotic used to treat the aminoglycoside-resistant enterococcal infections that became problematic in the mid-1980s.
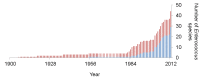
Figure 1.
Cumulative Enterococcus species identified. Red bars show the number of species associated with human disease.
Among the 30 species of Enterococcus described between 1992 and 2012 (Fig. 1, Table 1), only four (E. sanguinicola, E. gilvus, E. pallens and E. canintestini) have been associated with human infection (Carvalho, et al., 2008; Tan, et al., 2010; Tyrell, et al., 2002). Additionally, E. caccae has been isolated from human feces (Figure 2) (Carvalho, et al., 2006). The remaining species described since 1992 were isolated from non-human sources, including animals (De Graef, et al., 2003; de Vaux, Laguerre, Diviès, & Prévost, 1998; Devriese, Pot, & Collins, 1993; Koort, Coenye, Vandamme, Sukura, & Björkroth, 2004; Law-Brown & Meyers, 2003; Švec, et al., 2005), plants (Rahkila, Johansson, Säde, & Björkroth, 2011; Sedláček, et al., 2013; Sukontasing, Tanasupawat, Moonmangmee, Lee, & Suzuki, 2007; Švec, et al., 2011), water (Niemi, et al., 2012; Sedláček, et al., 2013; Sistek, et al., 2012; Svec, et al., 2001; Svec, et al., 2005; Svec, et al., 2006) and food (Fortina, Ricci, Mora, & Manachini, 2004; Koort, Coenye, Vandamme, Sukura, & Björkroth, 2004; Morandi, Cremonesi, Povolo, & Brasca, 2012; Tanasupawat, Sukontasing, & Lee, 2008) (Figure 2). Although much of the interest in enterococci historically has been driven by their association with infection and human carriage, it is clear that this is only the tip of the Enterococcus genus diversity iceberg, and comparison of these species to enterococci from other environments will reveal much about the basic features and origins of the genus.
Table 1.
Species part of the Enterococcus genus
Definition of the genus and criteria for identification
The core morphological and physiological features of all enterococci include being Gram-positive spherical or ovoid cells arranged in pairs or chains (Thiercelin, 1899). They are non-spore-forming facultative anaerobes and obligatory fermentative chemoorganotrophs. They typically have an optimum growth temperature of 35°C and a growth range from 10 to 45°C (Sherman, 1937). They typically grow in broth containing 6.5% NaCl, and hydrolyze esculin in the presence of 40% bile salts (Facklam, 1973). They are catalase reaction negative, and do not express complete cytochromes, although some species produce a catalase and appear catalase positive with weak effervescence. They are usually homofermentative, producing lactic acid as the end product of glucose fermentation, without production of gas (Klein, 2003; Murray, 1990). Some species are motile, such as E. gallinarum and E. casseliflavus (Graudal, 1957; Mundt, 1986). Pigmentation is a variable trait, with yellow-pigmented species including E. sulfureus, E. casseliflavus, and E. mundtii (Martinez-Murcia & Collins, 1991; Graudal, 1957; Mundt, 1986)), and pigmented species are commonly found among plants (Aarestrup, Butaye, & Witte, 2002). Most enterococci, apart from E. cecorum, E. columbae, E. pallens, and E. saccharolyticus, E. devriesei (variable among strains), E. canintestini, E. termitis and E. viikiensis are capable of hydrolyzing pyrrolidonyl β naphthylamide and producing leucine aminopeptidase (Facklam, Carvalho, & Teixeira, 2002; Naser, et al., 2005; Rahkila, Johansson, Säde, & Björkroth, 2011; Švec, et al., 2005; Svec, et al., 2006).
Phylogenetic analysis of catalase-negative Gram-positive cocci, based on the comparison of ~1400 bases of the 16s rRNA gene, shows that the Enterococcus are more closely related to the Vagococcus, Tetragenococcus, and Carnobacterium than they are to the Streptococcus and Lactococcus (Facklam, Carvalho, & Teixeira, 2002). The G + C content of DNA ranges from 37 to 45 mol% (237). Enterococcus faecalis has long been known to require a number of amino acids (including Val, Leu, Ile, Ser, Met, Glu, Arg, His and Trp) and vitamins (including biotin, nicotinic acid, pantothenate, pyridoxine, riboflavin, and sometimes folic acid) for maximal growth, with other species being similar in their fastidiousness (Facklam, Carvalho, & Teixeira, 2002). As a reflection of their highly evolved role as members of a consortium in an extremely competitive environment, enterococci have reduced genomes that range from 2.7 Mb to 3.6 Mb across species sequenced so far (Gaechter, Wunderlin, Schmidheini, & Solioz, 2012; Qin, et al., 2012; Palmer, et al., 2012; van Schaik & Willems, 2010).
Species of the Enterococcus genus identified by DNA-DNA reassociation, 16S rRNA gene sequencing, and/or whole-cell protein analysis to rigorously establish the novel species, and the dates of their initial description are shown in Table 1. DNA-DNA reassociation remains the gold standard for establishing bacterial species (Johnson & Ordal, 1968; Tindall, Rosselló-Móra, Busse, Ludwig, & Kämpfer, 2010), and this analysis has been carried out for most enterococci (Facklam, Carvalho, & Teixeira, 2002). Sequencing of 16S rDNA has become a practical and reliable additional tool (Vandamme, et al., 1996). Additionally, whole-cell protein analysis using standardized techniques correlate well with DNA hybridization, and give unique signatures for each Enterococcus species tested (Devriese, Pot, Van Damme, Kersters, & Haesebrouck, 1994; Merquoir, Netz, Camello, & Texeira, 1997). For practical reasons, 16S rDNA sequencing and whole-cell protein analysis are beginning to replace DNA-DNA reassociation, and are increasingly being used to describe new species. These methodological improvements are important developments, since many enterococcal species vary by only one phenotypic trait (Knudtson & Hartman, 1992; Moore, Zhowandai, Ferguson, McGee, Mott, & Stewart, 2006; Scheidegger, Fracalanzza, Texeira, & Caradarelli-Leite, 2009; Bodnar, Noskin, Suriano, Cooper, Reisberg, & Peterson, 1996) using conventional biochemical tests or commercial test systems (Devriese, Pot, & Collins, 1993; Facklam, Carvalho, & Teixeira, 2002). By itself, 16S rRNA sequence analysis does not appear to provide the resolution needed to distinguish Enterococcus species such as E. casseliflavus and E. gallinarum, which are 99.9% identical by this criterion (Patel, et al., 1998) (as opposed to 97% identity being the threshold usually used for species identification). As a consequence, additional complementary rapid molecular methods have been developed for routine identification of enterococcal isolates (Facklam, Carvalho, & Teixeira, 2002), including randomly amplified polymorphic DNA (RAPD) (Descheemaeker, Lammens, Pot, Vandamme, & Goossens, 1997; Monstein, Tiveljung, Kraft, Borch, & Jonasson, 2000; Quednau, Ahrné, Petersson, & Molin, 1998), sequencing of the domain V of the 23S rRNA gene (Poyart, Quesnes, & Trieu-Cuot, 2000), amplification of rRNA or tRNA intergenic spacers (Naimi, Beck, Monique, Lefèbvre, & Branlanti, 1999; Tyrell, Bethune, Willey, & Low, 1997), sequencing of the D-ala:D-ala ligase genes (ddl) (Ozawa, Courvalin, & Gaiimand, 2000) or the manganese-dependent superoxide dismutase (sodA) genes (Poyart, Quesnes, & Trieu-Cuot, 2000), sequencing and hybridization of the chaperonin 60 (cpn60) gene (Goh, et al., 2000), and amplification and probing of the Enterococcus protein A (efaA) gene (Singh, Coque, Weinstock, & Murray, 1998). Several additional molecular tests have been recently introduced to differentiate the Enterococcus species, including sequencing genes encoding the RNA polymerase α-subunit (rpoA), the phenylalanyl-tRNA synthase (pheS) or the elongation factor Tu (tufA) (Naser, et al., 2005; Naser, et al., 2005; Picard, et al., 2004), sequencing repetitive extragenic palindromic PCR (REP-PCR) or BOX-PCR (Nayak, Badgley, & Harwood, 2011), pulsed-field gel electrophoresis (PFGE) (Gordillo, Singh, & Murray, 1993; Tenover, Arbeit, & Goering, 1997), and multilocus sequence typing (MLST) (Homan, et al., 2002; Ruiz-Garbajosa, et al., 2006). MLST has proven to be useful in epidemiological studies of E. faecalis and E. faecium (Leavis, Bonten, & Willems, 2006), and shows similar accuracy to PFGE for the identification of organisms at the subspecies level (Nallapareddy, Duh, Singh, & Murray, 2002).
New species and reclassifications
The taxonomy of the enterococci has changed considerably, as the genus consisted of only 20 species at the end of the 20th century. Numerous new species have been described as the result of improvements in methods for differentiation, combined with growing interest in the enterococci (Figure 1) (Euzéby, 2013). Currently, the genus Enterococcus consists of 35 recognized species (Euzéby, 2013), with 8 more species (Enterococcus lactis, plantarum, quebecensis, rivorum, rotai, ureasiticus, ureilyticus, and viikiensis) likely to be added (Morandi, Cremonesi, Povolo, & Brasca, 2012; Niemi, et al., 2012; Rahkila, Johansson, Säde, & Björkroth, 2011; Sedláček, et al., 2013; Sistek, et al., 2012; Švec, et al., 2011). E. sanguinicola (Carvalho, et al., 2008) was recently described as likely to be a junior synonym of Enterococcus thailandicus (Shewmaker, et al., 2011), and probably will be removed. The list of Enterococcus species (Table 1) will doubtlessly continue to increase and be subject to reevaluation in the rapidly expanding genomic era, which offers new insights into distinctions between bacterial species (Konstantinidis, Ramette, & Tiedje, 2006). Since the diversity of the Enterococcus genus was extensively reviewed in 2002 (Facklam, Carvalho, & Teixeira, 2002), this review focuses on new enterococcal species that have been reported since then.
Enterococcus aquimarinus sp. nov. was isolated from sea water and was described by Švec et al. (Svec, et al., 2005) based on whole-cell protein analysis and 16S rDNA sequence. The 16S rDNA sequence shares 96.1–96.9% identity with its closest phylogenetic neighbors, E. saccharolyticus, E. sulfureus, and E. italicus (Svec, et al., 2005). Additional distinguishing features include pheS gene sequence polymorphism, (GTG)5-PCR fingerprinting (which identifies short sequence repeats (Lupski & Weinstock, 1992)), and biochemical characteristics. For example, E. aquimarinus is not able to produce acid from ribose, which distinguishes it from E. sulfurous, E. saccharolyticus, and the majority of other enterococcal species; but produces acid from L-Arabinose and other substrates which distinguishes it from E. italicus (Svec, et al., 2005). E. aquimarinus LMG 16607T was identified as the type strain (Svec, et al., 2005).
Enterococcus caccae sp. nov. (Carvalho, et al., 2006) was recovered from human stool, and was distinguished from other species by DNA–DNA hybridization, whole-cell protein analysis, and sequencing of the 16S rDNA (Carvalho, et al., 2006). Comparative 16S rDNA sequence analysis showed that E. caccae shared 99.6% identity with E. moraviensis (Carvalho, et al., 2006). However, the level of DNA–DNA relatedness between this novel taxon and any of the currently recognized species of Enterococcus was surprisingly low (32% for E. moraviensis), and the differences in the protein pattern supported the novelty of this species (Carvalho, et al., 2006). E. caccae can be differentiated from E. moraviensis by its ability to grow at 45 °C and a positive Voges-Proskauer test, as well as its inability to produce acid from arabinose, lactose and mannitol (Carvalho, et al., 2006). The type strain of the E. caccae species is 2215-02T (Carvalho, et al., 2006).
Enterococcus camelliae sp. nov., isolated from fermented tea leaves in Thailand, was proposed as a new species based on sequence analysis of its 16S rDNA and rpoA genes, and global DNA–DNA hybridization analysis (Sukontasing, Tanasupawat, Moonmangmee, Lee, & Suzuki, 2007). On the basis of the 16S rRNA and the rpoA sequence comparison, E. camelliae is closely related to Enterococcus italicus (99.2% and 93.8% sequence identity, respectively) (Sukontasing, Tanasupawat, Moonmangmee, Lee, & Suzuki, 2007). However, it shared only 33.8% genome identity based on DNA-DNA hybridization, and possessed phenotypic characteristics, including its inability to produce acid from D-galactose and lactose, which distinguishes it from E. italicus and other enterococci (Sukontasing, Tanasupawat, Moonmangmee, Lee, & Suzuki, 2007). Its type strain is E. camelliae FP15-1T (Sukontasing, Tanasupawat, Moonmangmee, Lee, & Suzuki, 2007).
The taxonomic position of isolates obtained from healthy dog faeces and originally classified as Enterococcus dispar was reinvestigated by Naser and collaborators (Naser, et al., 2005), who determined them to be a different species that they named Enterococcus canintestini sp. nov. The 16S rDNA gene sequence showed the highest similarities (between 98–99%) with E. dispar, E. canis and E. asini; however, E. canintestini shared only 33.8% genome identity, based on DNA-DNA hybridization with E. dispar, and showed a substantially different whole-cell protein profile (Naser, et al., 2005). Multilocus sequence analysis (MLSA) of the genes that encode the alpha subunit of ATP synthase (atpA), rpoA, and pheS was also performed (Naser, et al., 2005). Additionally, biochemical characteristics, including the inability to produce acid from raffinose and D-melibiose, distinguished E. canintestini from E. dispar and other enterococci (Naser, et al., 2005). The type strain is E. canintestini LMG 13590T (Naser, et al., 2005).
Enterococcus canis sp. nov. was proposed by De Graef and collaborators (De Graef, et al., 2003) for strains isolated from rectal swabs and chronic otitis in dogs. While the 16S rDNA sequence shared 98.4% to 99% similarity with members of the E. faecium species group, the level of DNA-DNA reassociation with these species was very low (7–13%) (De Graef, et al., 2003). E. canis shares a number of phenotypic characteristics with the E. faecium species group, but can be distinguished by its inability to hydrolyse arginine or to grow on selective media that contain 0.04% sodium azide (55). The type strain is E. canis LMG 12316T (De Graef, et al., 2003).
The species Enterococcus devriesei sp. nov. was proposed by Švec et al. (Švec, et al., 2005), after having reinvestigated the taxonomic position of two isolates of bovine origin that were originally assigned to the species Enterococcus raffinosus based on biochemical reactions. These strains, as well as two additional strains obtained from a charcoal-broiled river lamprey and from the air of a poultry slaughter by-product processing factory, showed key differences from E. raffinosus and all other enterococcal species (Švec, et al., 2005). The 16S rDNA sequence indicated a phylogenetic position in the Enterococcus avium species group, and showed Enterococcus pseudoavium as the closest phylogenetic relative (99.8–99.9% identity) (Švec, et al., 2005). However, whole cell protein electrophoresis, (GTG)5-PCR fingerprinting, ribotyping, and DNA-DNA hybridization (with reassociation values between 25% and 34% with the closest phylogenetic neighbors) confirmed that these strains were a novel species (Švec, et al., 2005). A combination of phenotypic traits, including the ability to produce acid from lactose (but not from melibiose or 2-ketogluconate) is helpful for distinguishing E. devriesei from species of the E. avium group (Švec, et al., 2005). However, there is variability within the phenotypic traits of this species (Švec, et al., 2005). The type strain is E. devriesei LMG 14595T (Švec, et al., 2005).
Enterococcus hermanniensis sp. nov. was proposed by Koort (Koort, Coenye, Vandamme, Sukura, & Björkroth, 2004) for strains isolated from modified-atmosphere-packaged broiler meat, and was also detected on canine tonsils. E. hermanniensis isolates were placed in the Enterococcus avium group, with E. pallens as their closest phylogenetic neighbor, based on 16S rDNA sequence (99% sequence similarity) (Koort, Coenye, Vandamme, Sukura, & Björkroth, 2004). Whole-cell protein analysis as well as a low level of DNA-DNA hybridization (12-30%), however, distinguishes this species from E. pallens and other known enterococci (Koort, Coenye, Vandamme, Sukura, & Björkroth, 2004). Interestingly, E. hermanniensis strains fails to produce acid from most common substrates, further distinguishing this species from other enterococci (Koort, Coenye, Vandamme, Sukura, & Björkroth, 2004). The type strain is E. hermanniensis LMG 12317T (Koort, Coenye, Vandamme, Sukura, & Björkroth, 2004).
Fortina et al. (Fortina, Ricci, Mora, & Manachini, 2004) investigated the taxonomic position of atypical Enterococcus strains isolated from artisanal Italian cheeses, and proposed the name Enterococcus italicus sp. nov. for strains that were found to be unique. The closest relatives are Enterococcus sulfureus and Enterococcus saccharolyticus, which possess 96% and 97% similarity based on 16S rDNA sequence, respectively (94). E. italicus strains can be easily differentiated from the other Enterococcus species by DNA–DNA hybridization and phenotypic characteristics (e.g., lack of growth in 6.5% NaCl, and lack of acid production from L-arabinose, melezitose, melibiose, raffinose or ribose). The type strain is E. italicus DSM 15952T (Fortina, Ricci, Mora, & Manachini, 2004).
Enterococcus lactis sp. nov. was proposed by Morandi et al. (Morandi, Cremonesi, Povolo, & Brasca, 2012) for other atypical Enterococcus strains isolated from raw milk Italian cheeses. On the basis of 16S rDNA sequence analysis, the strains were found to be closely related to E. hirae, E. durans, and E. faecium, with a sequence similarity greater that 98.8% (Morandi, Cremonesi, Povolo, & Brasca, 2012). Since no DNA-DNA hybridization or whole-cell protein analysis have been conducted, the authors based the description of this new species on the analysis of the cellular fatty acid composition between E. lactis and its phylogenetic neighbors E. faecium and E. durans (Morandi, Cremonesi, Povolo, & Brasca, 2012). The authors indicated that E. lactis strains can be differentiated from other Enterococcus species based on the pheS gene sequence, RAPD-PCR, 16S–23S rRNA gene intergenic region analysis (Morandi, Cremonesi, Povolo, & Brasca, 2012). The proposed type strain is E. lactis DSM 23655T (Morandi, Cremonesi, Povolo, & Brasca, 2012).
Enterococcus phoeniculicola sp. nov. was proposed by Law-Brown and Meyers (Law-Brown & Meyers, 2003) to identify a single strain isolated from the uropygial gland of wild red-billed woodhoopoes (Phoeniculus purpureus). Examination of the 16S rDNA sequence showed that its closest relatives are E. faecium and E. avium (97.3% and 97.5% respectively). However, neither DNA-DNA hybridization nor whole-cell protein analyses were conducted. E. phoeniculicola may be differentiated from E. faecium and E. avium by its inability to form acid from lactose, D-mannitol, D(+)-melezitose or D-sorbitol, as well as its inability to grow in the presence of 40% bile or 6.5% NaCl (153). Its type strain is E. phoeniculicola DSM 14726T (Law-Brown & Meyers, 2003).
Enterococcus plantarum sp. nov. was proposed after investigation of the taxonomic position of eight strains isolated during characterization of enterococcal populations on plants (Švec, et al., 2011). Rep-PCR fingerprinting indicated that the isolates constituted a single cluster that was distinct from known enterococcal species (Švec, et al., 2011). In addition, the 16S rDNA sequence showed that these isolates clustered with the E. faecalis species group (Švec, et al., 2011). E. plantarum was confirmed to be a novel species by whole-cell protein fingerprinting, and can be differentiated from its closest phylogenetic neighbors by pheS and rpoA gene sequence analysis (Švec, et al., 2011). The authors described the colonies as "yellowish" on agar, but did not describe this species as pigmented (Švec, et al., 2011). The type strain is E. plantarum LMG 26214T (Švec, et al., 2011).
Enterococcus quebecensis sp. nov. was recently proposed by Sistek and collaborators (Sistek, et al., 2012) to identify a single isolate that originated from water sampled in the Province of Québec, Canada. Sequence analysis of genes 16S RNA, rpoA, pheS, tufA, and atpD, as well as the results of AFLP DNA fingerprinting and DNA–DNA hybridization, confirmed the status of this strain as distinct from other enterococcal species (Sistek, et al., 2012). Based on the 16S rDNA phylogeny, E. quebecensis is closely related to E. moraviensis, and clusters in the E. faecalis group. E. quebecensis strains are negative for the Voges-Proskauer test, meaning that they do not produce acetoin as a result of fermentation (Sistek, et al., 2012). The type strain is E. quebecensis DSM 23327T LMG 26306T (Sistek, et al., 2012).
Enterococcus rivorum sp. nov. was proposed by Niemi and collaborators (Niemi, et al., 2012) after taxonomic investigation of strains that originated from pristine waters in Finland. The authors showed by genotypic analysis that E. rivorum strains are closely related to the E. faecalis group (Niemi, et al., 2012). However, DNA-DNA hybridization confirms that they are a new species (Niemi, et al., 2012). Growth at 45 °C, or in broth containing 6.5% NaCl, is weak or absent; and production of the D antigen is variable (200). Slow growth occurs at 10 °C (Niemi, et al., 2012). The type strain is E. rivorum is LMG 25899T = CCM 7986T (Niemi, et al., 2012).
Very recently, Sedláček et al. (Sedláček, et al., 2013), proposed the name Enterococcus rotai sp. nov. for urease-producing, yellow-pigmented enterococci isolated from environmental sources, including drinking water, plants, and mosquitoes, and placed them in the Enterococcus faecalis species group. These strains show resistance to clindamycin, chloramphenicol, and oxacillin, with most also being resistant to ofloxacin—but they are sensitive to imipenem, and most strains are sensitive to gentamicin as well (Sedláček, et al., 2013). The type strain is E. rotai LMG 26678T (Sedláček, et al., 2013).
Two isolates from drinking water in the region of Silesia in Czech Republic were shown to be distinct from other enterococcal species by DNA-DNA hybridization, and were named Enterococcus silesiacus sp. nov. (Svec, et al., 2006). The 16S rRNA gene sequence analysis classified these strains in the E. faecalis species group. The type strain is E. silesiacus LMG 23085T (Svec, et al., 2006). During the same study, Švec and collaborators (Svec, et al., 2006) identified an isolate from the gut of a termite, and proposed the name Enterococcus termitis sp. nov. for this species. As for E. silesiacus, E. termitis is part of the E. faecalis species group based on 16S rDNA analysis (Svec, et al., 2006). E. termitis strains are pyruvate and Voges-Proskauer test negative, which is unusual among the enterococci (Svec, et al., 2006). The type strain is E. termitis LMG 8895T (Svec, et al., 2006).
In 2008, a single strain was isolated from fermented sausage in Thailand, named Enterococcus thailandicus sp. nov., was examined and proposed as a new species (Tanasupawat, Sukontasing, & Lee, 2008). By DNA-DNA hybridization, this strain shared less than 70% identity with known species, but was found to be related to E. hirae, E. durans, and E. faecium, based on 16S rDNA and rpoA gene sequence analysis (Tanasupawat, Sukontasing, & Lee, 2008). The type strain is E. thailandicus FP48-3T (Tanasupawat, Sukontasing, & Lee, 2008).
Enterococcus ureasiticus sp. nov. (which refers to the presence of urease activity) was proposed based on the study of two enterococcal strains isolated from water samples that differed from other Enterococcus species by more than 30%, based on DNA-DNA hybridization (Sistek, et al., 2012). 16S rDNA sequence analysis classified them in the E. faecalis species group, but this new species harbored divergent rpoA, pheS, tufA, and atpD sequences, as well as AFLP-DNA fingerprinting patterns (Sistek, et al., 2012). The type strain is E. ureasiticus FP48-3T (Sistek, et al., 2012).
Enterococcus ureilyticus sp. nov. was proposed as a name for a novel Enterococcus species identified in a study of strains that inhabit both water and plants (Sedláček, et al., 2013). These urease-producing, yellow-pigmented strains were placed in the E. faecalis species group, based on 16S rDNA sequence (Sedláček, et al., 2013). Analysis of pheS and rpoA genes sequences, as well as as whole-cell protein electrophoresis and DNA-DNA hybridization, indicated significant divergence from other enterococci (Sedláček, et al., 2013). Strains of this species showed resistance to chloramphenicol, oxacillin, and tetracycline, as well as sensitivity to imipenem (Sedláček, et al., 2013). The type strain is E. ureilyticus LMG 26676T (Sedláček, et al., 2013).
Finally, Rahkila et al. (Rahkila, Johansson, Säde, & Björkroth, 2011) proposed the species Enterococcus viikkiensis, with strain LMG 26075T being the type strain. This strain and four others were isolated from air from a broiler processing plant. Based on 16S rDNA sequence analysis, they belong to the E. avium group, with E. devriesei being their closest neighbor (Rahkila, Johansson, Säde, & Björkroth, 2011). Notably, these strains are also negative for pyruvate utilization (Rahkila, Johansson, Säde, & Björkroth, 2011).
The identification of enterococcal species is dynamic and is continually being revised. The species Enterococcus seriolicida, originally proposed by Kusuda (Kusuda, Kawai, Salati, Banner, & Fryer, 1991), has been reclassified as Lactococcus garvieae, based on DNA relatedness (Elliott, Collins, Pigott, & Facklam, 1991; Teixeira, et al., 1996)). Additionally, based on biochemical and genetic evidence, Enterococcus solitarius (Collins, Facklam, Farrow, & Williamson, 1989), was moved to the Tetragenococcus genus, and was named Tetragenococcus solitarius (Ennahar & Cai, 2005). Within the Enterococcus genus, E. saccharominimus (Vancanneyt, et al., 2004) was shown to be a synonym of Enterococcus italicus (Fortina, Ricci, Mora, & Manachini, 2004; Naser, Vancanneyt, Hoste, Snauwaert, Vandemuelebroecke, & Swings, 2006), and E. casseliflavus was shown to be the senior subjective synonym of Enterococcus flavescens (Naser, Vancanneyt, Hoste, Snauwaert, Vandemuelebroecke, & Swings, 2006; Teixeira, et al., 1997). In 2001, Enterococcus porcinus, which was isolated and associated with enteric disorders in animals (Teixeira, et al., 2001), was shown to be the same species as Enterococcus villorum (Vancanneyt, et al., 2001), as no phenotypic or genotypic differences could be found between these species (De Graef, et al., 2003). Finally, Enterococcus sanguinicola sp. nov. was proposed by Carvalho et al. (Carvalho, et al., 2008) in 2008, after taxonomic analysis of two isolates recovered from the blood of patients in the United States and in Sweden (Carvalho, et al., 2008), and the proposed type strain was E. sanguinicola SS-1729T (Carvalho, et al., 2008). One of the isolates showed a vancomycin-resistant phenotype, due to the presence of the vanA operon. However, the authors noted that this species has not been fully validated (Carvalho, et al., 2008), and in fact, a recent study reported that E. sanguinicola should be reclassified as E. thailandicus, based on DNA-DNA reassociation and sequence of the 16S rDNA and rpoB genes (Shewmaker, et al., 2011).
Phenotypic characteristics of the Enterococcus genus
The phenotypic tests routinely used to identify members of the Enterococcus genus and species have been previously reviewed (Facklam, Carvalho, & Teixeira, 2002). Briefly, Facklam and collaborators (Facklam, Carvalho, & Teixeira, 2002) divided enterococcal species into 5 groups based on acid formation in mannitol and sorbose broth, as well as hydrolysis of arginine. This clustering was applied to the 22 recently identified Enterococcus species, based on phenotypic tests performed along species descriptions (Table 2). It is important to note that this phenotypic classification should be distinguished from the 16S rRNA sequence relationship, so although these tests are potentially useful for diagnostics, it does not appear to reflect the evolutionary relationships between Enterococcus species.
Table 2.
Phenotypic characteristics of recently identified Enterococcus species
Group I consists of enterococcal species that form acid in both carbohydrate broths, but do not hydrolyze arginine (Facklam, Carvalho, & Teixeira, 2002). The new species E. phoeniculicoa, E. devriesei, and E. canis (De Graef, et al., 2003; Law-Brown & Meyers, 2003; Švec, et al., 2005) belong to Group I, along with E. avium, E. malodoratus, E. raffinosus, E. pseudoavium, E. saccharolyticus, E. pallens, and E. gilvus (Table 2). Group II includes E. faecalis and E. faecium, and consists of species that form acid in mannitol broth and hydrolyze arginine but fail to form acid in sorbose broth (Facklam, Carvalho, & Teixeira, 2002). New species E. canintestini, E. lactis, E. thailandicus, and E. sanguinicola belong to this group (40, 184, 196, 264) (Carvalho, et al., 2008; Morandi, Cremonesi, Povolo, & Brasca, 2012; Naser, et al., 2005; Tanasupawat, Sukontasing, & Lee, 2008). Group III includes species that are not able to form acid in either mannitol or sorbitol broth, but that hydrolyze arginine (Facklam, Carvalho, & Teixeira, 2002). This group now includes E. villorum, E. durans, E. dispar, E. hirae, E. silesiacus, and E. rotai (Sedláček, et al., 2013; Svec, et al., 2006), as well as variants of E. faecalis and E. faecium (Table 2) (Facklam, Carvalho, & Teixeira, 2002). E. asini, E. sulfureus, and E. cecorum previously formed Group IV, to which we may now add E. aquamarinus, E. plantarum, E. caccae, and E. termitis (Carvalho, et al., 2006; Facklam, Carvalho, & Teixeira, 2002; Švec, et al., 2005; Svec, et al., 2006; Švec, et al., 2011). These species are negative for acid formation in mannitol and sorbose broth and do not hydrolyze arginine (Table 2). Finally, Group V consists of the species that form acid on mannitol, but not sorbose broth, and fail to hydrolyse arginine (Facklam, Carvalho, & Teixeira, 2002). This group includes E. columbae, E. rivorum, E. hermaniensis, E. camelliae, and E. viikiensis, as well as variants of E. casseliflavus, E. gallinarum, and E. faecalis that fail to hydrolyse arginine (Table 2) (Facklam, Carvalho, & Teixeira, 2002; Koort, Coenye, Vandamme, Sukura, & Björkroth, 2004; Niemi, et al., 2012; Rahkila, Johansson, Säde, & Björkroth, 2011; Sukontasing, Tanasupawat, Moonmangmee, Lee, & Suzuki, 2007). The recently described E. ureilyticus (Sedláček, et al., 2013) does not appear to fit into any existing group, and we place it as the sole member of Group VI, based on its ability to formate acid from sorbose but not in mannitol broth, as well as its inability to hydrolyze arginine. Because of lack of available information on the fermentation capabilities of E. quebencensis, E. italicus, and E. ureasiticus, they are unable to be classified by this scheme (Table 2) (Fortina, Ricci, Mora, & Manachini, 2004; Sistek, et al., 2012). Zhang and collaborators have identified a gene cluster in the E. faecium strain E980 that is responsible for the metabolism of the α-galactoside sugar raffinose (Zhang, Vrijenhoek, Bonten, Willems, & van Schaik, 2011). While (Table 2) E. faecium is placed in Group II in the presented table, which is the group unable to use raffinose, these authors showed that raffinose utilization is actually a trait carried by megaplasmids, which indicates that these mobile elements can have important roles in shaping the competitive fitness of the enterococci in the environment; for example, by expanding their metabolic repertoire (Zhang, Vrijenhoek, Bonten, Willems, & van Schaik, 2011).
Phylogeny of the Enterococcus genus
As a reflection of specialization for life in the gut of a host, over millions of years of co-evolution, genome analysis have highlighted the selective advantage for enterococci of their acquisition of many nutrients required for growth and survival from their habitats (usually either from the diet of the host, or through cross-feeding relationships with other microbes in the gut consortium), as opposed to carrying the additional genetic material necessary for their biosynthesis from simpler precursors (Gilmore, Lebreton, & van Schaik, 2013). Figure 2 shows the phylogenetic relationship among the different species of Enterococcus based on single-nucleotide polyphormism (SNP) analysis of the 16S rDNA sequences (Fig. 2). This tree can be divided into at least 7 groups, based on deep divides within the genus (Fig. 2). Some patterns emerge with respect to the habitats in which these species were identified and/or reported (Figure 3). Group I members are species that are farthest from other enterococci, which likely means that their speciation from the rest of the genus occurred comparatively early (Fig. 3). This is translated phenotypically by their unusual negative result in the pyruvate utilization test. E. columbae was exclusively isolated from pigeon faecal flora (Devriese, Ceyssens, Rodrigues, & Collins, 1990), and E. cecorum also occurs mainly in the gut of birds and less frequently in the GI tract of mammals (some mammals like cattle or swine are fed with poultry) (Aarestrup, Butaye, & Witte, 2002). These strains have not yet been isolated from insects that are at the base of the food chain for many birds (Fig. 3), leading to speculation that speciation may have occurred after the evolutionary split among mammals, reptiles, birds, and insects. For strains of Group II, there appears to be a tropism for mammals, as none of these five strains have been isolated from insects, lizards, or birds (Fig. 3). The presence of the strains in the environment may be the consequence of their occurence in the GI tract of humans and animals (Fig. 3), whereas their presence in food products reflects intentional use and not a natural habitat.
E. faecalis in Group III is found ubiquitously along the food chain, in addition to having emerged as an important opportunistic hospital pathogen (Fig. 3). This would suggest that since at least the early Devonian period, ~412 MYA (the time of the last common ancestor of mammals, reptiles, birds, and insects), this species has been a member of gut microbiomes (Gilmore, Lebreton, & van Schaik, 2013). E. faecalis is closely related to the recently described E. rivorum, which has been isolated from natural water (Niemi, et al., 2012). It would be very interesting to know the extent to which the genome of these two species is conserved. Other species found in Group III have a strong tropism (and almost specificity) to environments like water or plants. In several cases, an attempt was made to exclude contamination from human or animal feces, but it still remains unknown whether these species live freely in the environment (and if so, how their metabolic requirements are satisfied), or whether they are associated with simple eukaryotes like amoeba, as suggested by recent reports (Huws, Morley, Jones, Brown, & Smith, 2008).
Species found in Groups IV, V, and VI show some tropism for humans and animals (especially mammals) (Fig. 3). Enterococcal species that belong to Group V appear to be specific to humans and mammals (Fig. 3). In contrast, species of Groups IV and VI are more ubiquitous, and can be found along the food chain from insects to mammals. Their occurrence in environments such as water or on plants may stem from faecal contamination (Casanovas-Massana & Blanch, 2013; Staley, Reckhow, Lukasik, & Harwood, 2012). It will be of substantial interest to determine precisely how the nutritional requirements of the various species relate to their host or environmental specializations.
Enterococcal species: Natural and man-made habitats
In the 1960’s and 1970’s Mundt and collaborators explored a variety of environments and found enterococci in the GI tracts and feces of mammals (71.3%), reptiles (85.7%), and birds (31.8%) (Mundt, 1963), as well as insects (53%) (Martin & Mundt, 1972). Culture-positive rates underestimate the presence of microbes (Ward, Weller, & Bateson, 1990), which suggests that the actual rate of colonization may be closer to 100%. This widespread pattern of colonization suggests that since at least the early Devonian period, ~412 MYA (the time of the last common ancestor of mammals, reptiles, birds and insects (Selden, 2007), enterococci have been members of gut microbiomes, which likely place them among the earliest members of GI tract consortia (Gilmore, Lebreton, & van Schaik, 2013).
Enterococci in humans: gut colonization and infection
The human body (especially the GI tract, but also the skin, the upper respiratory tract, the oral cavity, and the vagina) is colonized by microbial communities that, together, constitute the “normal microbiota.” Each colonized body site represents an ecosystem that is defined by unique physicochemical and histological characteristics that constitute a competitive environment and select for adapted microbes (Tannock, 1988). The human colon contains about 1012 diverse bacteria per gram of contents, with the highest counts in the colon (Sartor, 2008; Vollaard & Clasener, 1994), which contribute to tissue development and immune system homeostasis (Berg, 1996) (Figure 4).
Infections in humans. Enterococcal infection and its treatment are comprehensively covered elsewhere in this volume. Here, we briefly highlight select features of infection that inform the dynamic between colonization and infection by diverse species of enterococci. Enterococci began to emerge as leading causes of multidrug-resistant hospital-acquired infections in the 1970s and 1980s (Huycke, Sahm, & Gilmore, 1998; Jett, Huycke, & Gilmore, 1994), and they now rank among leading causes of hospital-acquired infections of the bloodstream, urinary tract, surgical wounds, and other sites (Hidron, et al., 2008). They are also associated with obligate anaerobes in mixed infections that result in intra-abdominal abscesses (Onderdonk, Bartlett, Louie, & Sullivan-Seigler, 1976). The most recent data available on enterococcal infection from all infection sites and all classes of hospitals in the US, which covers the period 01/01/10–06/30/12, identified 9,309 bloodstream isolates, 54,709 urinary tract isolates, and 20,032 wound isolates (a total of 84,050 isolates) (TSN® Database, Eurofins, Inc., personal communication Daniel F. Sahm, Ph.D.). Of the total, 17,360 are vancomycin-resistant (20.6%) and 64,015 (76%) are E. faecalis. Although 24% of isolates are E. faecium, they represent 14,998 of the 20,038 (75%) of the vancomycin-resistant isolates. Similar trends have been reported for the European Union (56, 294). E. durans, E. avium, E. gallinarum, E. casseliflavus, E. hirae, E. muntdii, E. dispar, and E. raffinosus only occasionally cause infection in humans (Devriese, Pot, Van Damme, Kersters, & Haesebrouck, 1994; Tannock & Cook, 2002). In 2002, Tyrrell and collaborators reported the isolation of E. gilvus and E. pallens from the bile of a patient with cholecystitis and the peritoneal dialysate of another patient with peritonitis, respectively (Tyrell, et al., 2002). This has been the only description of these species causing human disease thus far. Recently, Ahmed and coworkers (Ahmed, Baig, Gascoyne-Binzi, & Sandoe, 2011) reported a case of human aortic valve infective endocarditis caused by E. cecorum, while a study conducted between 2000 and 2008 identified this species as responsible for 0.2% (2 out of 1887) of the bacteremia cases diagnosed at a medical center in Taiwan (Tan, Liu, Li, Huang, Sun, & Qu, 2013). These authors also reported a unique case of bacteremia caused by E. canintestini (Tan, Liu, Li, Huang, Sun, & Qu, 2013). While we lack studies that characterize the prevalence of the recently described species in the genus, we can generally assume, based on their recent discovery, that these are not often found as etiological agents of infections in humans.
Gut colonization. Much of what is known about enterococcal colonization of the gut stems from studies of the human GI tract and feces. Enterococci are primarily localized to the human small and large intestine, where enterococci are prominent members of jejunal, ileal, cecal, and recto-sigmoidal consortia (Hayashi, Takahashi, Nishi, Sakamoto, & Benno, 2005). They are also found in human feces, although they constitute a minority population (up to 1%) within the gut microflora (Eckburg, et al., 2005; Sghir, Gramet, Suau, Violaine, Pochart, & Dore, 2000). Enterococci are common in the oral cavity (Smyth, Matthews, Halpenny, Brandis, & Colman, 1987), but occur more rarely in the stomach (Bik, et al., 2005; Monstein, Tiveljung, Kraft, Borch, & Jonasson, 2000). E. faecalis and E. faecium are most common in human feces, and E. durans and E. avium are occasionally detected (Finegold, Sutter, & Mathisen, 1983; Tannock & Cook, 2002). E. caccae was isolated from human feces, but its prevalence is low (Carvalho, et al., 2006). One study quantified E. avium (11%), E. durans (33%), E. faecalis (78%), E. faecium (100%), E. gallinarum (33%), and E. hirae (11%) in human fecal specimens (Layton, Walters, Lam, & Boehm, 2010), while E. casseliflavus and E. saccharolyticus were absent from the nine samples tested (Layton, Walters, Lam, & Boehm, 2010). Other members of the genus Enterococcus identified in clinical samples are rarely part of the normal human intestinal microflora, but are present in the gut of other animals or on plants (Fig. 3).
Some members of the microbiota, including indigenous commensal enterococci, can act as opportunistic pathogens and translocate across the mucosal barrier to cause systemic infection in immune-compromised hosts (Berg, 1996; Donskey, 2004). More commonly, however, infection results from the colonization, overgrowth, and translocation of hospital-adapted antibiotic-resistant strains with enhanced pathogenicity. Studies using animal models have shed light on the mechanisms of microflora-mediated colonization resistance to enterococci (Brandl, et al., 2008; Kinnebrew, Ubeda, Zenewicz, Smith, Flavell, & Pamer, 2010). Components of bacterial cells, which include lipoteichoic acid, lipopolysaccharide, and flagellin, are recognized by Toll-like and Nod receptors expressed by intestinal epithelial and Paneth cells. By using a mouse model, Brandl and co-workers (Brandl, et al., 2008) showed that antibiotic reduction of the population of Gram-negative bacteria in the intestine resulted in decreased production of RegIIIγ by Paneth cells, which resulted in intestinal overgrowth of vancomycin-resistant enterococci (VRE) (Brandl, et al., 2008). RegIIIγ is a secreted C-type lectin that selectively kills Gram-positive bacteria, including VRE. RegIIIγ expression was restored in antibiotic treated animals by oral administration of the TLR4 agonist LPS, or systemic administration of the TLR5 agonist flagellin, which reduced VRE counts in the gut of mice (Figure 4B) (Brandl, et al., 2008; Kinnebrew, Ubeda, Zenewicz, Smith, Flavell, & Pamer, 2010). The same mechanisms may also take place in humans, as antibiotic intake by hospitalized patients is associated with an Enterococcus outgrowth (Ruiz-Garbajosa, et al., 2012; Ubeda, et al., 2010) that often precedes VRE bloodstream invasion (Ubeda, et al., 2010). Despite increasing knowledge about the interplay between Gram-negative commensals, the host’s immune system, and Enterococcus, the understanding of synergies and antagonisms between enterococci and other microbes is far from complete. A recent study by Ubeda and coworkers (Ubeda, et al., 2013) observed that the reintroduction of a diverse intestinal microbiota to the GI tracts of mice densely colonized with VRE resulted in a substantial reduction in VRE numbers. They further showed that obligate anaerobic commensal bacteria belonging to the Barnesiella genus specifically promoted the clearance of intestinal VRE (Ubeda, et al., 2013). These authors further showed that, in patients undergoing allogeneic hematopoietic stem cell transplantation, intestinal colonization with Barnesiella confers resistance to intestinal domination and bloodstream infection with VRE (Ubeda, et al., 2013), which may provide a novel approach for the prevention of infection and the spread of highly antibiotic-resistant bacteria.
Enterococci in animals: gut colonization and infection
Animal GI tracts likely represent the greatest reservoir for enterococci (Gilmore, Lebreton, & van Schaik, 2013). As in humans, enterococci also can infect animals (Aarestrup, Butaye, & Witte, 2002). Ostrolenk and Hunter (Ostrolenk & Hunter, 1946) recovered low levels of enterococci from 49 of 51 fecal samples from various animals, including human, cat, mouse, guinea pig, rabbit, dog, rat, chicken, fly, and monkey. Similarly, Haenel and Mueller-Buethow (Haenel & Muller-Beuthow, 1957) obtained enterococci from human, rat, chicken, and dog, but not in the samples isolated from rabbit, guinea pig, or horse. In 1963, Mundt and colleagues (Mundt, 1963) conducted a thorough analysis of the occurrence of enterococci in animals and natural environments. Enterococci were obtained from the feces of 71% of 216 mammals, 86% of 70 reptiles, and 32% of 22 birds sampled in Great Smoky Mountains National Park (Mundt, 1963). Enterococci occurred only sporadically among the primarily herbivorous mammals, whereas enterococci appear to naturally colonize rodents and larger animals that have varied diets (Mundt, 1963). Enterococci were obtained from most specimens of bats and from the carnivorous mammals, such as fox, bear, raccoon, skunk, and boar (Mundt, 1963). Most probably, microbiota composition in relation to some animals’ particular diets determine whether enterococci are able to survive and thrive in their GI tracts (Mieth, 1960).
As new species are described, their associations with hosts are discovered. E. columbae is specific to pigeons so far (Devriese, Ceyssens, Rodrigues, & Collins, 1990), and E. asini has only been found in donkeys (de Vaux, Laguerre, Diviès, & Prévost, 1998). E. asini has not been reported in additional studies since its description in 1998, and as a result, is likely to be rare. E. columbae is difficult to isolate, as this species does not grow on selective media that are commonly used for isolation of enterococci and also requires CO2 for growth, so the prevalence of this species may therefore be underestimated (Devriese, Ceyssens, Rodrigues, & Collins, 1990).
The most commonly encountered enterococcal species in the gut of mammals are E. faecalis, E. faecium, E. hirae, and E. durans (Devriese & De Pelsmaecker, 1987). Other species are found only occasionally (Devriese & De Pelsmaecker, 1987), or in particular age groups (such as E. cecorum in older poultry) (Aarestrup, Butaye, & Witte, 2002). In fact, an age-dependent succession of enterococcal species colonization appears to occur in chickens (Devriese, Hommez, Wijfels, & Haesebrouck, 1991; Kaukas, Hinton, & Linton, 1987). Chickens are initially colonized by E. faecalis, but this population is then displaced, mainly by E. faecium, and it was proposed that the use of tylosin (to which E. faecium is commonly resistant) as a growth promoter is the reason of this replacement (Kaukas, Hinton, & Linton, 1987). These species then appear to be replaced by E. cecorum in the mature chicken. It is of interest that E. avium and E. gallinarum, originally described from chickens, were rarely found, suggesting that these species may not belong to the normal intestinal flora of poultry (Devriese, Hommez, Wijfels, & Haesebrouck, 1991). E. avium was originally described from human feces (Guthof, 1955), but is common in chicken feces (Nowlan & Deibel, 1967). The extent to which the flora of chickens now reflects that native to poultry, or reflects the consequences of intensive production and modern poultry husbandry practices (namely, the frequent use of antibiotics including aminoglycosides), is controversial, but has undoubtedly impacted the representation of various species of enterococci (Aarestrup, Butaye, & Witte, 2002; Devriese, Pot, Van Damme, Kersters, & Haesebrouck, 1994). Kobayashi and collaborators reported that aminoglycoside modifying enzymes are less prevalent in E. avium strains, as compared to E. faecium and E. faecalis strains isolated from poultry (Kobayashi, Alam, Nishimoto, Urasawa, Uehara, & Watanabe, 2001).
Age-dependent enterococcal colonization also has been described in cattle (Devriese, Laurier, De Herdt, & Haesebrouck, 1992). In preruminant calves, the enterococcal flora mainly consists of E. faecalis, E. faecium, and E. avium. This flora is gradually replaced by E. cecorum (Devriese, Laurier, De Herdt, & Haesebrouck, 1992). Enterococci occur on the tonsils of preruminating calves; mainly E. faecalis, as well as E. raffinosus, to a lesser extent (Devriese, Laurier, De Herdt, & Haesebrouck, 1992).
The enterococcal species most frequently isolated from the intestines of swine are E. faecalis and E. faecium; however, the latter species occurs in low numbers (Devriese & De Pelsmaecker, 1987; Devriese, Pot, Van Damme, Kersters, & Haesebrouck, 1994). Other enterococcal species found among swine flora include E. hirae and E. cecorum (Devriese & De Pelsmaecker, 1987; Devriese & Haesebrouck, 1991; Devriese, Pot, Van Damme, Kersters, & Haesebrouck, 1994).
A study on the flora of cats and dogs found that E. faecalis was the most frequently encountered enterococcal species in the gut, as well as on the tonsils of these animals, with E. faecium and E. hirae occurring infrequently (Devriese, Cruz Colque, De Herdt, & Haesebrouck, 1992). Other species, including E. avium, E. raffinosus, E. durans, E. cecorum, and E. gallinarum were also occasionally isolated (Devriese, Cruz Colque, De Herdt, & Haesebrouck, 1992). As noted above, E. canis and E. canintestini have more recently been isolated from healthy dogs (De Graef, et al., 2003; Naser, et al., 2005). E. faecalis, E. faecium, E. hirae, E. gallinarum, E. casseliflavus, and E. mundtii have also been found in horses (Devriese & De Pelsmaecker, 1987; Thal, et al., 1995).
Recent studies in Portugal identified enterococci in a broad range of environments, including wild animals. A total of 144 enterococci (120 E. faecium, 14 E. hirae, 8 E. faecalis, 2 E. gallinarum) were recovered in echinoderms collected from Azorean waters (Marinho, et al., 2013). Resistance to erythromycin, ampicillin, tetracycline, and ciprofloxacin in these enterococci was found to be common among them (Marinho, et al., 2013). VRE (E. faecium, E. gallinarum and E. casseliflavus) were recovered in 8 of the 365 analysed fecal samples from Iberian wolf and lynx (Gonçalves, et al., 2013). Similarly, vancomycin-resistant E. faecalis, E. faecium, and E. durans were isolated in 7 of 118 fecal samples (5.9%) of natural gilthead seabream recovered off the coast of Portugal (Barros, et al., 2012).
In other studies, Radhouani and collaborators (Radhouani, Poeta, Gonçalves, Pacheco, Sargo, & Igrejas, 2012) examined 31 enterococcal isolates from 42 common buzzard fecal samples, and found E. faecium as the most commonly encountered species (48.4%) recovered. E. faecium, E. durans, and E. gallinarum isolates were detected in fecal samples of other wild birds (Silva, et al., 2011). Enterococci were also isolated from wild boars, partridges and fish (Almeida, et al., 2011), red foxes (Radhouani, et al., 2011), and wild rabbits (Silva, et al., 2010). In addition, enterococci have been found in wild geese (Han, et al., 2011) and from cattle reared with traditional practices in Ethiopia (Bekele & Ashenafi, 2010). In this last study, a total of 298 enterococcal isolates were obtained, which consisted of E. faecium (49.6%), E. durans (26.9%), E. hirea (11.9%), and E. faecalis (11.5%).
A large variety of insects, including beetles, flies, bees, termites, and worms have also been found to harbor enterococci. E. faecalis and E. faecium are predominant but other species occur at lower prevalence. Martin and Mundt (Martin & Mundt, 1972) observed the association of E. faecalis, E. faecium, and E. casseliflavus with a broad range of insect orders. E. faecalis (32%), E. faecium (22.4%), and E. casseliflavus (43.5%) were associated with wild insects from 37 different taxa (Martin & Mundt, 1972). In a survey of laboratory-reared and wild Drosophila, Cox and Gilmore (Cox & Gilmore, 2007) found several species of enterococci, including E. faecalis, E. faecium, E. gallinaraum, and E. durans, and localized them to the digestive tract. Differences between human and Drosophila GI tract anatomy and physiology undoubtedly play a role in determining the composition and physical location of their respective microbial populations.
Anatomically, the GI tract of Drosophila (Bodenstein, Cooper, Ferris, Miller, Poulson, & Sonnenblick, 1950) has an overall organization that has parallels to that of humans. With the exception of the Drosophila crop, the two systems possess a single alimentary canal that begins at the esophagus, connects to a ventriculus (stomach), extends to the intestine, proceeds to the rectum, and terminates at the anus (Figure 5). In terms of digestive function, however, they are quite different. Drosophila possesses an acidic crop, but maintains an alkaline ventriculus and a neutral to acidic hindgut (Clark, 1999; Dow & Harvey, 1988). Another major difference is the absence of obligate anaerobic phylotypes in Drosophila (Cox & Gilmore, 2007), while the majority of human intestinal microbial phylotypes are obligate or microaerophilic anaerobes (Eckburg, et al., 2005).
Enterococci have been detected on insects in hospitals, and concerns have been raised about this possible reservoir for human contamination (Fotedar, Banerjee, Singh, Shriniwas, & Verma, 1992; Pai, Chen, & Peng, 2004). The importance of insects to the transmission of resistant or infectious enterococci has not been established. Ahmed and collaborators (Ahmed, Baig, Gascoyne-Binzi, & Sandoe, 2011) showed that house flies and cockroaches in the confined swine production environment may act as vectors and/or reservoirs of antibiotic resistant and potentially virulent enterococci. Macovei et al. (Macovei, Miles, & Zurek, 2008) showed that houseflies captured in fast-food restaurants commonly carry antibiotic-resistant germs. Among newly characterized Enterococcus species, E. termitis was isolated from the gut of a termite, while some E. rotai strains were isolated from mosquitoes (Sedláček, et al., 2013; Svec, et al., 2006).
Comparison of enterococci isolated from human and animals
In 1993, Bates and collaborators (Bates, Jordens, & Selkon, 1993) described the first vancomycin-resistant enterococci isolated from a non-human reservoir. These authors detected a vancomycin-resistant strain of E. faecium (VREF) in farm animals in the UK (Bates, Jordens, & Selkon, 1993). To understand the relationship between enterococci, and more particularly, between antibiotic-resistant strains of agricultural and human origin, E. faecium isolates were compared using PFGE, and similar PFGE profiles were found for VRE isolated from stool samples from both humans and animals (Hammerum, et al., 2004; van den Bogaard & Stobberingh, 2000). Besides PFGE, amplified fragment length polymorphism (AFLP) analysis and MLST were used to compare these E. faecium isolates (Willems R. J., et al., 2000). VREF strains were largely found to be host-specific, and strains isolated from hospitalized patients were genetically different from the prevailing VREF strains present in the fecal flora of non-hospitalized humans (Willems R. J., et al., 2000). Further, MLST showed that outbreak isolates from hospitalized patients clustered in a subgroup, which was originally termed a clonal complex 17 (CC17), while E. faecium isolated from mainly domesticated animals belonged to other types (Willems R. J., et al., 2005; Willems R. J., et al., 2012). Although there is some evidence of host adaptation to E. faecium strains, typical hospital-associated clones have been identified in farm animals (Hammerum, 2012), and dogs are frequently colonized by strains similar to those identified in clinical isolates (Damborg, Sørensen, & Guardabassi, 2008; de Regt, et al., 2012). This indicates that sharing a habitat is an important risk factor for the spread of these organisms.
Similarly to E. faecium, E. faecalis isolates from hospitalized patients tend to derive from select STs and CCs (McBride, Fischetti, LeBlanc, Moellering Jr., & Gilmore, 2007; Ruiz-Garbajosa, et al., 2006), but a diversity of STs can be detected in E. faecalis recovered from animals, meat, fecal samples from humans in the community, and patients with bloodstream infections (Kawalec, et al., 2007; Kuch, et al., 2012; McBride, Fischetti, LeBlanc, Moellering Jr., & Gilmore, 2007; Nallapareddy, Wenxiang, Weinstock, & Murray, 2005; Ruiz-Garbajosa, et al., 2006). E. faecalis human isolates have been found to be highly related to pig isolates, based on their antimicrobial resistance pattern, virulence gene profile, and MLST/ PFGE types (Larsen, et al., 2011). Similarly, the presence of the pathogenicity island (PAI) of E. faecalis can be found in isolates from pigs (Shankar, Baghdayan, Willems, Hammerum, & Jensen, 2006).
Enterococci in the environment (plants, water and soil)
Enterococci are widely used as indicators of fecal contamination from animal or human origin in the environment (see Enterococci as indicators of environmental fecal contamination). However, the extent to which enterococci in the environment (including in water sources) reflects human or animal waste contamination continues to be debated (Harwood, Whitlock, & Withington, 2000). Comparing the genomes of enterococci obtained from environmental sources to those derived from humans and animals will shed considerable light on this question. Among the extra-enteric niches where enterococci are routinely isolated include soil and sediments, aquatic and terrestrial plants, and ambient waters (Fig. 3). These are heterothermic habitats, in which temperatures are variable—in contrast to the gastrointestinal tract of warm-blooded animals, where the temperature is relatively constant. In addition, a variety of environmental stressors such as UV sunlight, salinity, starvation, and predation also occur (Byappanahalli, Nevers, Korajkic, Staley, & Harwood, 2012). The transition from the animal gastrointestinal tract, an environment rich in many nutrients with key nutrients restricted, to oligotrophic environments that are generally nutrient-poor, exposes enterococci to nutrient starvation. One of the first reports on the survival of enterococci under nutrient-starvation conditions indicated that E. faecalis survived for extended periods in sterilized sewage (presumably due to the availability of organic nutrients), but declined rapidly in sterile lake water and in water with a phosphate buffer, indicating that oligotrophic conditions (exemplified by the sterile lake water and phosphate buffer) were deleterious to the survival of enterococci (Sinclair & Alexander, 1984). Indeed, enterococci are auxotrophic bacteria. It is important to note that although the authors observed survival of E. faecalis in sterilized sewage, they did not observe growth (Sinclair & Alexander, 1984).
In and on aquatic and terrestrial vegetation. While various enterococcal species can be found on plants (Fig. 3), it is unclear whether or not these strains are contaminants from fecal consortia that are simply surviving, or whether these environments actually constitute a habitat. Ostrolenk and Hunter (Ostrolenk & Hunter, 1946) considered their presence in these locations more an indication of survival, whereas Sherman (Sherman, 1937) considered as evidence against accidental occurrence and survival, given the fact that, of the many cultures isolated from plants, none of the hemolytic types of enterococci had been obtained. Mundt (Mundt, 1961; Mundt, 1963) recovered enterococci from the surface of a variety of plant species, and proposed that enterococci were transient populations, most likely introduced by insects and wind, since their occurrence was seasonal (Mundt, 1961; Mundt, 1963). In another study, however, he demonstrated what appeared to be the ability of E. faecalis to grow on plants (Mundt, Coggin, Jr., & Johnson, 1962). Occurrence in seed or soil, movement from the seed to the emergent parts, and ability to reproduce on the growing plant are criteria for bacterial plant epiphytes (Burri, 1903; Dueggeli, 1904; Thaysen & Galloway, 1930; Voznyakovskaya & Khudyakov, 1960). In his work (Mundt, Coggin, Jr., & Johnson, 1962), Mundt suggested that S. faecalis is a potential epiphyte on plants, since it was found consistently on most plant parts, it established a cycle on plants in which transmission occurs via the seed, and it reproduced on the growing plant. Although no details are provided, the author concluded that these findings indicate that these bacteria are capable of adaptation to an environment substantially different from that of the intestinal tract in its quantity of nutrients, temperature, and moisture (Mundt, Coggin, Jr., & Johnson, 1962). However, it is important to note that the possibility that the enterococcal cells were commensals of macro or micro-invertebrates, or that they may have benefitted from a more complex bacterial population, were not addressed. Huws and collaborators (Huws, Morley, Jones, Brown, & Smith, 2008) investigated the interactions of E. faecalis, among other bacteria, with the amoeba Acanthamoeba polyphaga. They found evidence of protozoal predation of E. faecalis. Interestingly, the extracellular numbers of E. faecalis JH2-2 were significantly higher when cultured with amoebae, as compared to growth in the absence of amoebae (Huws, Morley, Jones, Brown, & Smith, 2008). The authors stated that, since not all bacteria are digested by protozoa, outgrowth of select populations occurs, as was observed for the tested enterococcal strain (Huws, Morley, Jones, Brown, & Smith, 2008).
The occurrence of enterococci has been investigated in association with various vegetation, including algae (Whitman, Shively, Pawlik, Nevers, & Byappanahalli, 2003), beach wrack (Anderson, Turner, & Lewis, 1997; Grant, et al., 2001; Inamura, Thompson, Boehm, & Jay, 2011), submerged vegetation (Badgley, Nayak, & Harwood, 2010; Badgley, Thomas, & Harwood, 2010), flowering plants (Mundt, 1963), and forage crops (Müller, Ulrich, Ott, & Müller, 2001; Byappanahalli, Nevers, Korajkic, Staley, & Harwood, 2012). Whitman and collaborators (Whitman, Shively, Pawlik, Nevers, & Byappanahalli, 2003) showed that the Cladophora mats, a green alga found in both fresh and marine waters, were a significant source of E. coli and enterococci. In addition to these fecal bacteria, enteric pathogens such as Shigella, Campylobacter, and Salmonella also were isolated (Whitman, Shively, Pawlik, Nevers, & Byappanahalli, 2003). The high densities of enterococci in fresh Cladophora have been attributed to in situ growth (Byappanahalli, Shively, Nevers, Sadowsky, & Whitman, 2003). Indeed, these authors stated that algal leachate readily supported the in-vitro multiplication of enterococci (100-fold in undiluted algal leachate at 35 °C over 24 h), and that such growth was directly related to the concentration of algal leachate. This suggests that leachates contain the necessary growth-promoting substances to sustain these bacteria (Byappanahalli, Shively, Nevers, Sadowsky, & Whitman, 2003). However, other adherent bacteria present on these algae may contribute nutrients that enterococci require. Enterococci can persist and survive on submerged vegetations (Badgley, Nayak, & Harwood, 2010; Badgley, Thomas, & Harwood, 2010) or dried algae (Byappanahalli, Shively, Nevers, Sadowsky, & Whitman, 2003), and enterococci have also been isolated from plankton and macro-invertebrates (Maugeri, Carbone, Fera, Irrera, & Gugliandolo, 2004; Signoretto, et al., 2004).
Several recently characterized Enterococcus species were identified on plants. E. plantarum was isolated from plant samples obtained from a meadow in the Czech Republic, not influenced by farm animals and exploited only for hay harvesting; and E. rotai and E. ureilyticus were isolated from plants, as well as water samples (Švec, et al., 2011).
In water. Large quantities of human and animal wastes are distributed into the environment through sewage or non-sewage systems. Human wastes empty into waterways, usually through a sewage treatment plant, while animal wastes are often used composted (but otherwise untreated) as fertilizers on fields. Enterococci are found in both fresh (Byappanahalli, Nevers, Korajkic, Staley, & Harwood, 2012; Fujioka, Tenno, & Kansako, 198) and marine water (Boehm, et al., 2002; Korajkic, Brownell, & Harwood, 2011). They can survive and persist in fresh (Anderson, Whitlock, & Harwood, 2005) as well as marine water (Litopoulo-Tzanetaki & Tzanetakis, 1992), but it is not clear from the literature whether enterococci can also multiply in these environments. Growth in the absence of essential nutrients obviously will not occur, and studies that report growth in water in the presence of algae (Byappanahalli, Shively, Nevers, Sadowsky, & Whitman, 2003) or sediments (Yamahara, Walters, & Boehm, 2009) rarely precisely define their position in more complex food webs.
Shifts in Enterococcus species composition generally occur during domestic wastewater treatment. While E. faecalis was predominant, different species including E. pseudoavium, E. casseliflavus, E. faecium, E. mundtii, E. gallinarum, E. dispar, E. hirae, E. durans, E. flavescens, Enterococcus haemoperoxidus, and Enterococcus moraviensis also can be recovered (Kühn, et al., 2003). This may have implications for the identification of fecal pollution based on the presence of specific bacterial types associated with domestic wastewater (Graves & Weaver, 2010). In water, the species considered as fecal contaminants are mainly E. faecium and E. faecalis, and the origins of other species are less clear (Aarestrup, Butaye, & Witte, 2002). Data reported by Signoretto et al. (Signoretto, et al., 2004) suggested that the adhesion to plankton of viable but non-culturable cells contribute to the prolonged survival of enterococci in marine waters. Numerous recently-characterized Enterococcus species have been isolated from water samples. E. rotai and E. ureilyticus were isolated from drinking water, E. acquimarinus was isolated from sea water, and E. rivorum was isolated from a pristine brook (Niemi, et al., 2012; Svec, et al., 2005). Similarly, E. moraviensis, E. silesiacus, E. ureasiticus, and E. quebecensis have been isolated from surface water (Sistek, et al., 2012; Svec, et al., 2001; Svec, et al., 2006). Whether they represent flora of microscopic eukaryotes or other larger organisms remains to be definitively determined.
In soil and sediments. Enterococci occur in temperate and tropical soils (Byappanahalli, Nevers, Korajkic, Staley, & Harwood, 2012; Mundt, 1961), fresh or marine water sediments (Ferguson, Moore, Getrich, & Zhowandai, 2005; Obiri-Danso & Jones, 2000), and can be recovered from sand (Halliday & Gast, 2011; Yamahara, Layton, Santoro, & Boehm, 2007). The prolonged survival of enterococci has been observed in freshwater and estuarine sediments (Anderson, Whitlock, & Harwood, 2005). van Donsel et al. (Van Donsel, Geldreich, & Clarke, 1967) found that the rates of survival of S. faecalis were higher than those of fecal coliforms during spring and winter, but that fecal coliforms survived longer than S. faecalis during the summer months. Interestingly, many of the early investigations of the survival and persistence of enterococci in soil environments focused on watersheds impacted by anthropogenic activities, particularly in cattle grazing and field lot operations (Byappanahalli, Nevers, Korajkic, Staley, & Harwood, 2012). All samples taken from urban sewage, and from farmland using pig manure, as well as crops grown on this land, readily yielded Enterococcus species (Kühn, et al., 2003). In crops to which animal fertilizer had not been applied, the incidence of Enterococcus species was reduced to 33% (Kühn, et al., 2003). Enterococcus species can be recovered from sand from freshwater and marine beaches (E. faecium, E. casseliflavus, E. durans), and from marine sediments (E. faecalis, E. faecium, E. hirae, E. casseliflavus, and E. mundtii) (Ferguson, Moore, Getrich, & Zhowandai, 2005). As for other environments, whether enterococci grow as free living organisms in sediments, sands, or soils remains highly speculative. While the growth requirements of many bacteria, including E. coli, are relatively simple (Andrews, 1991), enterococci require complex nutrients.
Enterococci in food
It is likely that because of their natural occurrence as contaminants of raw meat and dairy products (Franz, Holzapfel, & Stiles, 1999), thermotolerance, low toxicity, and their ability to acidify an environment, enterococci have become an important ingredient in fermented foods. Enterococci are often used for the preparation of traditional cheeses manufactured in Mediterranean countries, including Greece, Italy, Spain and Portugal, from raw or pasteurized goat, sheep, or bovine milk (Foulquié Moreno, Sarantinopoulos, Tsakalidou, & De Vuyst, 2006). Levels of enterococci in different cheeses range from 104 to 106 CFU/g, with E. faecium and E. faecalis being the most common (Foulquié Moreno, Sarantinopoulos, Tsakalidou, & De Vuyst, 2006). Enterococci contribute to the ripening and development of aroma in these products because of their proteolytic and esterolytic properties, as well as the production of diacetyl and other important volatile compounds (Foulquié Moreno, Sarantinopoulos, Tsakalidou, & De Vuyst, 2006). Enterococci play an important role in improving flavor development and overall quality of other traditional fermented foods as well, including vegetables and sausages (Franz, Holzapfel, & Stiles, 1999; Giraffa, 2002), including salami and chorizo (Foulquié Moreno, Sarantinopoulos, Tsakalidou, & De Vuyst, 2006). In addition to desired fermentation properties, both E. faecalis and E. faecium have been implicated in the spoilage of cured meat products, including canned hams and chub-packed luncheon meats (Magnus, Ingledew, & McCurdy, 1986; Magnus, McCurdy, & Ingledew, 1988). Finally, enterococci commonly occur in large numbers in vegetables (Ben Omar, et al., 2004; Fernandez-Diaz, 1983; Franz, Holzapfel, & Stiles, 1999; Giraffa, 2002; Mundt, 1963); and occur in Spanish-style green olive fermentations (de Castro, Montaño, Casado, Sánchez, & Rejano, 2002; Fernandez-Diaz, 1983; Floriano, Ruiz-Barba, & Jiménez-Díaz, 1998), in which E. faecalis and E. faecium are frequent contaminants.
While enterococci are used as an indicator of fecal contamination in the environment, their value as hygiene indicators in the industrial processing of foods has been questioned (Birollo, Reinheimer, & Vinderola, 2001). The resistance of enterococci to pasteurization and their adaptability to different substrates and growth conditions (extreme pH and salinity) explains their occurrence in food products manufactured from raw materials or in nominally–heat-treated food products. The contribution of enterococci to the organoleptic properties of fermented food products (Coppola, Villani, Coppola, Coppola, Parente, & Parente, 1990; Giraffa & Carminati, 1997; Pisano, Fadda, Deplano, Corda, Casula, & Cosentino, 1994; Litopoulo-Tzanetaki & Tzanetakis, 1992; Manolopoulou, Sarantinopoulos, Zoidou, Aktypis, Moschopoulou, & Kandarakis, 2003), and their ability to produce bacteriocins (enterocins) (Cleveland, Montville, Nes, & Chikindas, 2002; De Vuyst & Vandamme, 1994; Ennahar & Cai, 2005; Klaenhammer, 1993; Moll, Konings, & Driessen, 1999; Nes, Diep, Håvarstein, Brurberg, Eijinsk, & Holo, 1996), are important characteristics in food technology. Despite their use in traditionally fermented products, the continuing use of enterococci in food processes is somewhat controversial, because of their association with human infection (De Vuyst, Foulquié Moreno, & Revets, 2003; Vancanneyt, et al., 2002).
E. faecium and E. faecalis are generally the most frequently encountered species of Enterococcus in food products (Lopez-Diaz, Santos, Gonzalez, Moreno, & Garcia, 1995; Suzzi, et al., 2000). Martin and collaborators (Martín, Corominas, Garriga, & Aymerich, 2009) investigated the species distribution of enterococci in traditional fermented sausages by species-specific multiplx PCR, and by atpA and 16S rDNA gene sequencing. They identified E. faecalis (31.4%) and E. faecium (30.7%), and more rarely E. sanguinicola (14.9%), E. devriesei (9.7%), E. malodoratus (7.2%), E. casseliflavus (3.4%), E. gallinarum (1.3%), E. gilvus (1.0%), E. hermanniensis (0.2%), and E. durans (0.2%) (Martín, Corominas, Garriga, & Aymerich, 2009). The species E. camelliae, E. italicus, E. thailandicus, and E. lactis were originally isolated from tea leaves, an Italian cheese, fermented sausages, and raw milk cheese, respectively (Fortina, Ricci, Mora, & Manachini, 2004; Morandi, Cremonesi, Povolo, & Brasca, 2012; Sukontasing, Tanasupawat, Moonmangmee, Lee, & Suzuki, 2007; Tanasupawat, Sukontasing, & Lee, 2008). The natural habitat of these Enterococcus species is very likely not fermented food, but elsewhere in nature in ecologies yet to be defined.
Enterococci are permitted in some countries as probiotics (Franz, Holzapfel, & Stiles, 1999; Franz, Stiles, Schleifer, & Holzapfel, 2003). One E. faecium strain (SF68) used as a probiotic, has been suggested to be clinically effective in the treatment of diarrhea in children (Bellomo, Mangiagle, Nicastro, & Frigerio, 1980) and to prevent diarrhea caused by antibiotic treatments (Wunderlich, et al., 1989). In a comprehensive review on the various enterococcal strains used as probiotics, Franz and collaborators (Franz, Huch, Abriouel, Holzapfel, & Gálvez, 2011) concluded that data in support of the use of enterococci as probiotics is controversial. While it is possible to argue for the probiotic benefit of some strains, the emergence of enterococci in human disease and as multidrug-resistant germs raises concern.
Drug resistant enterococci in non-human reservoirs
Besides the treatment of human infection, antimicrobial agents are used in food preservation, to treat and promote the growth of animals and pets, in plant production, and for other non-health-related purposes (Dibner & Richards, 2005; Turnidge, 2004). The contribution of each of these applications to resistance among human pathogens is still debated.
In animals. Enterococcal infections of animals are rarely treated with antimicrobial agents; however, enterococci are exposed to antimicrobial selection as members of the GI tract during the course of treatment of other infections, or when antibiotics are used as growth promoters or prophylaxis (Turnidge, 2004). The virtues of antibiotics as growth promoters have been debated since the 1960s (Swann, 1969). Studies conducted in the 1950s confirmed that the use of tetracycline for treatment or growth promotion selected for tetracycline resistance among group D streptococci (enterococci) in chickens (Barnes, 1958; Elliott & Barnes, 1959). More recently, epidemiological studies and feeding experiments have shown that the use of different antimicrobial growth promoters, including glycopeptides (avoparcin), macrolides (tylosin) and oligosaccharides (avilamycin), select for resistance among enterococci in poultry and pigs (Aarestrup, Bager, Jensen, Madsen, Meyling, & Wegener, 1998; Aarestrup, Bager, Jensen, Madsen, Meyling, & Wegener, 1998; Aarestrup F. M., 2000; Bager, Madsen, Christensen, & Aarestrup, 1997; Christie, Davidson, Novick, & Dunny, 1983; Hinton, Kaukas, Lim, & Linton, 1986). Most interest has focused on the use of avoparcin and the selection of VRE in animal husbandry (Aarestrup F. M., 1995; Bager, Madsen, Christensen, & Aarestrup, 1997). A multitude of global studies describe the occurrence of VRE in many different non-human reservoirs, including pet cats and dogs, horses, birds, wood frogs, ostriches, pigs, pork, broilers, poultry meat, environmental samples, and sewage, as well as from stool samples from farmers and non-hospitalized humans in the community, mainly in Europe (Hammerum A. M., 2012). The linkage of VRE in non-human reservoirs to the use of avoparcin as a growth promoter is debated. Avoparcin was used in animal husbandry in the EU, Australia, and several other locations, but was not allowed for growth promotion in the USA and Canada. As a likely result, there is a low prevalence of VRE in the US and Canada, but nevertheless, there is a high level of occurrence of VRE in US hospitals (Hammerum A. M., 2012). Feeding animals low doses of antimicrobials, in certain conditions, increases productivity by improving feed conversion and decreasing morbidity and mortality caused by infection (Butaye, Baele, Devriese, & Haesebrouck, 2002). The glycopeptide avoparcin was first introduced for growth promotion in 1975 (Hammerum, Lester, & Heuer, 2010), and was mainly used for broilers and pigs, but also for turkeys, veal calves, and other animals (Bates J. , 1997; Hammerum, Lester, & Heuer, 2010; Hammerum A. M., 2012). At that time, it was used extensively in most parts of Europe and the rest of the world, with the notable exceptions of Canada and the USA (McDonald, Kuehnert, Tenover, & Jarvis, 1997). As avoparcin confers cross-resistance to vancomycin, its use selected for outgrowth of VRE, and as a consequence, VRE was common in the intestinal flora of farm animals in Europe during the 1990s (Bager, Madsen, Christensen, & Aarestrup, 1997). In contrast, until 2008, VRE had never been isolated from farm animals in the US. Because of this connection, the use of avoparcin as a growth promoter was banned and the prevalence of VRE in farm animals in Europe rapidly declined, but VRE did not disappear (Aarestrup F. M., 2000). To explain why VRE persist among farm animals, it has been suggested that the use of the macrolide tylosin in pigs co-selects for vancomycin resistance among enterococci, since both the resistance determinants are often on the same plasmid (Aarestrup F. M., 2000). Besides tylosin, Johnsen and collaborators (Johnsen, et al., 200) hypothesized that plasmid addiction systems may contribute to retention of the resistance.
High-level aminoglycoside resistance has been observed among enterococci isolated from a variety of animals. Highly gentamicin-resistant strains were isolated from bovine mastitis (Jayaro & Oliver, 1992) and pigs (Aarestrup, Bager, Jensen, Madsen, Meyling, & Wegener, 1998; Aarestrup F. M., 2000). These gentamicin-resistant isolates include E. faecalis, E. faecium, E. gallinarum, and E. casseliflavus (Aarestrup, Butaye, & Witte, 2002). Resistance to kanamycin has been observed in 30% of E. faecalis and E. faecium isolates from bovine mastitis (Jayaro & Oliver, 1992), 62% of 62 isolates from chickens, and 36% of 72 isolates from pigs (Rollins, Lee, & LeBlanc, 1985). Streptomycin resistance has been observed among enterococcal isolates from bovine mastitis (80%) (Jayaro & Oliver, 1992), chickens (64%), beef cattle (6%), and pigs (57%) (Rollins, Lee, & LeBlanc, 1985). Thal and collaborators (Thal, et al., 1995) also isolated streptomycin-resistant enterococci from horses and birds. Chloramphenicol use has been limited in food animals for the last several decades. There are few reports on the occurrence of resistance to chloramphenicol among enterococci isolated from animals. Saikia et al. (Saikia, Dutta, Devriese, & Kalita, 1995) found that one-third of E. faecalis from ducks in India were resistant to chloramphenicol. A low frequency (1–7%) of chloramphenicol resistance occurs in E. faecium and E. faecalis isolates from broilers and pigs in Denmark (Aarestrup F. M., 2000).
The first finding of quinupristin-dalfopristin–resistant E. faecium in non-hospitalized humans was reported in the United Kingdom (UK) by Woodford and collaborators (Woodford, Adebiyi, Palepou, & Cookson, 1998) in the late 1990s. Interestingly, quinupristin-dalfopristin and other streptogramins used for human therapy were not available at that time in the UK for use in humans. Similarly to vancomycin and avoparcin, the finding of quinupristin-dalfopristin–resistant E. faecium isolates outside hospitals was assumed to be associated with the use of virginiamycin in animals (McDonald, et al., 2001). The genetic determinant that encodes streptogramin A resistance has been detected in E. faecium isolates from poultry, pigs, pork, and sewage worldwide. Enterococci from animals are also frequently resistant to penicillin or ampicillin. Frequencies of resistance comparable to those observed among isolates from humans have also been reported (Aarestrup F. M., 2000; Yoshimura, Ishimaru, Endoh, & Kojima, 2000).
In food. Antibiotic-resistant enterococci occur in meat products, dairy products, and even within strains used as probiotics (Giraffa, 2002). The contamination of meat with antibiotic resistant enterococci during slaughter of animals to which antibiotics were administered has been reported (Bager, Madsen, Christensen, & Aarestrup, 1997; Del Grosso, et al., 2000; Klare, Badstübner, Konstabel, Böhme, Claus, & Witte, 1999; Kruse, Johansen, Rørvik, & Schaller, 1999). In 2002, Giraffa and collaborators (Giraffa, 2002) highlighted the role played by food reservoirs of VRE in the dissemination of antibiotic resistance traits in the environment. VRE and tetracycline-resistant enterococci have been found among food animals in several countries in Europe (Aarestrup, Butaye, & Witte, 2002). Recently, Ronconi and collaborators (Ronconi, Merino, & Fernández, 2002) assessed the presence of highly aminoglycosides and glycopeptide-resistant enterococci in uncooked foods, and more particularly, in lettuce. The most frequently detected species was E. faecium (32.61%), followed by E. faecalis (21.74%), E. gallinarum (13.04%), E. casseliflavus and E. mundtii (7.60%), E. hirae, (6.52%), E. durans (4.35%), E. raffinossus and E. saccharolyticus (2.17%), and E. avium and E. malodoratus (1.10%) (Ronconi, Merino, & Fernández, 2002). Susceptibility profiles were found to be similar to those of strains occurring in hospitals (Ronconi, Merino, & Fernández, 2002).
High-level aminoglycoside resistance has been observed among enterococci isolated from food of animal origin (Aarestrup, Butaye, & Witte, 2002). In a US study, gentamicin resistance was found in 7 of 18 enterococci isolates from chicken meat (Thal, et al., 1995). Streptomycin resistance occurred at a similar rate (Thal, et al., 1995). Streptomycin resistance was found at a lower rate (1% of enterococcal isolates) from food in Germany; and has been detected among E. faecalis (10%) and E. faecium isolates from broilers (2-3%) in Denmark (Aarestrup F. M., 2000). The use of chloramphenicol has been banned in food animal production for more than 30 years, and is largely restricted in human medicine to limited use for systemic treatment or for the treatment of eye infections. As a consequence, a low frequency (1-7%) of chloramphenical resistance, encoded by the gene catpIP501, has been observed among E. faecium and E. faecalis isolates from broilers (Aarestrup F. M., 2000). However, Pavia et al. (Pavia, Nobile, Salpietro, & Angelillo, 2000) found more than 40% of enterococci from raw meat in Italy to be resistant to chloramphenicol, whereas Klein et al. (Klein, Pack, & Reuter, 1998) found 1% of vancomycin susceptible and 29% of vancomycin-resistant enterococci from raw meat in Germany to also be chloramphenicol-resistant.
In the environment. Sewage is an important reservoir of resistant enterococci. As early as the 1970s, tetracycline-resistant enterococci were isolated from sewage (van Embden, Engel, & van Klingeren, 1977). Glycopeptide resistance in E. faecium was first detected in 1986 in clinical isolates from hospitals in France (Leclercq, Derlot, Duval, & Courvalin, 1988) and the United Kingdom (Uttley, et al., 1989). The first indication of a reservoir outside hospitals was the detection of VRE in wastewater treatment plants of small towns in Germany that had no hospital (Klare, Heier, Claus, & Witte, 1993); and in sewage in the United Kingdom (Bates, Jordens, & Selkon, 1993). High-level aminoglycoside resistance has been observed worldwide among enterococci isolated in the environment (Aarestrup, Butaye, & Witte, 2002). More specifically, high-level resistance to kanamycin has been observed among 34% of 248 enterococcal isolates from sewage and water in the US (Rice, Messer, Johnson, & Reasoner, 1995). Streptomycin resistance was found in 5% of sewage isolates in the US. Highly gentamicin-resistant enterococci have been isolated from sewage and water in the US (Rice, Messer, Johnson, & Reasoner, 1995) and Germany (Klare, Heier, Claus, & Witte, 1993) as well.
Conclusions
It is clear that the few Enterococcus strains and species regularly associated with human infection represent just the tip of the iceberg, as microbes of the genus Enterococcus are mainly ancient and highly evolved members of GI tract consortia of various hosts. More information is sorely needed on the natural habitats of many of the species, including new species that have recently come to our attention, due to of their rare occurrence in clinical specimens. The extent to which enterococcal species naturally inhabit and proliferate in and on plants, soil, and water (as opposed to contaminating them through animal excretion) remains controversial. Genes mirror the requirements of life. As our understanding of enterococcal genomics grows, bacterial genomics will become an important tool for providing new insights into the nature, biology, and habitats of the enterococci.
References
- Aarestrup F. M. Occurrence of glycopeptide resistance among Enterococcus faecium isolates from conventional and ecological poultry farms. Microbial Drug Resistance. 1995;1(3):255–257. [PubMed: 9158784]
- Aarestrup F. M. Characterization of glycopeptide-resistant Enterococcus faecium (GRE) from broilers and pigs in Denmark: genetic evidence that persistence of GRE in pig herds is associated with coselection by resistance to macrolides. Journal of Clinical Microbiology. 2000;38(7):2774–2777. [PMC free article: PMC87029] [PubMed: 10878086]
- Aarestrup F. M., Carstensen B. Effect of tylosin used as a growth promoter on the occurrence of macrolide-resistant enterococci and staphylococci in pigs. Microbial Drug Resistance. 1998;4(4):307–312. [PubMed: 9988049]
- Aarestrup F. M., Ahrens P., Madsen M., Pallesen L. V., Poulsen R. L., Westh H. Glycopeptide susceptibility among Danish Enterococcus faecium and Enterococcus faecalis isolates of animal and human origin and PCR identification of genes within the VanA cluster. Antimicrobial Agents and Chemotherapy. 1996;40(8):1938–1940. [PMC free article: PMC163445] [PubMed: 8843309]
- Aarestrup F. M., Bager F., Jensen N. E., Madsen M., Meyling A., Wegener H. C. Resistance to antimicrobial agents used for animal therapy in pathogenic-, zoonotic- and indicator bacteria isolated from different food animals in Denmark: a baseline study for the Danish Integrated Antimicrobial Resistance Monitoring Programme (DANMAP). APMIS. 1998;106(8):745–770. [PubMed: 9744762]
- Aarestrup F. M., Bager F., Jensen N. E., Madsen M., Meyling A., Wegener H. C. Surveillance of antimicrobial resistance in bacteria isolated from food animals to antimicrobial growth promoters and related therapeutic agents in Denmark. APMIS. 1998;106(6):606–622. [PubMed: 9725794]
- Aarestrup, F. M., Butaye, P., & Witte, W. (2002). Non-human reservoirs of enterococci. In M. S. Gilmore, D. B. Clewell, P. Courvalin, G. M. Dunny, B. E. Murray, & L. B. Rice (Eds.), The Enterococci: Pathogenesis, Molecular Biology, and Antibiotic Resistance (pp. 55-100). Washington, District of Columbia: ASM Press.
- Aarestrup F. M., Wegener H. C., Rosdahl V. T., Jensen N. E. Staphylococcal and other bacterial species associated with intramammary infections in Danish dairy herds. Acta Veterinaria Scandinavica. 1995;36(4):475–487. [PMC free article: PMC8095438] [PubMed: 8669375]
- Ahmed F. Z., Baig M. W., Gascoyne-Binzi D., Sandoe J. A. Enterococcus cecorum aortic valve endocarditis. Diagnostic Microbiology and Infectious Disease. 2011;70(4):525–527. [PubMed: 21767709]
- Almeida T., Brandão A., Muñoz-Atienza E., Gonçalves A., Torres C., Igrejas G., et al. Identification of bacteriocin genes in enterococci isolated from game animals and saltwater fish. Journal of Food Protection. 2011;74(8):1252–1260. [PubMed: 21819651]
- Anderson K. L., Whitlock J. E., Harwood V. J. Persistence and differential survival of fecal indicator bacteria in subtropical waters and sediments. Applied and Environmental Microbiology. 2005;71(6):3041–3048. [PMC free article: PMC1151827] [PubMed: 15933000]
- Anderson S. A., Turner S. J., Lewis G. D. Enterococci in the New Zealand environment: implications for water quality monitoring. Water Science and Technology. 1997;35(11-12):325–331.
- Andrewes F. W., Horder T. J. A study of streptococci pathogenic for man. The Lancet. 1906;168(4335):852–855.
- Andrews, J. H. (1991). Comparative Ecology of Microorganisms and Macroorganisms. New York: Springer.
- Arias C. A., Murray B. E. The rise of the Enterococcus: beyond vancomycin resistance. Nature Reviews Microbiology. 2012;10(4):266–278. [PMC free article: PMC3621121] [PubMed: 22421879]
- Ator L. L., Starzyk M. J. Distribution of group D streptococci in rivers and streams. Microbios. 1976;16(64):91–104. [PubMed: 829267]
- Badgley B. D., Nayak B. S., Harwood V. J. The importance of sediment and submerged aquatic vegetation as potential habitats for persistent strains of enterococci in a subtropical watershed. Water Research. 2010;44(20):5857–5866. [PubMed: 20678788]
- Badgley, B. D., Thomas, F. I., & Harwood, V. J. (2010). The effects of submerged aquatic vegetation on the persistence of environmental populations of Enterococcus spp. (1271-1281, Ed.) Environmental Microbiology, 12(5). [PubMed: 20192964]
- Bager F., Madsen M., Christensen J., Aarestrup F. M. Avoparcin used as a growth promoter is associated with the occurrence of vancomycin-resistant Enterococcus faecium on Danish poultry and pig farms. Preventive Veterinary Medicine. 1997;31(1-2):95–112. [PubMed: 9234429]
- Barnes E. M. Tetrazolium reduction as a means of differentiating Streptococcus faecalis from Streptococcus faecium. Journal of General Microbiology. 1956;14(1):57–68. [PubMed: 13306890]
- Barnes E. M. The effect of antibiotic supplements on the faecal streptococci (Lancefield group D) of poultry. British Veterinary Journal. 1958;114:333–334.
- Barros J., Andrade M., Radhouani H., López M., Igrejas J., Poeta P., et al. Detection of vanA-containing Enterococcus species in faecal microbiota of gilthead seabream (Sparus aurata). Microbes and Environments. 2012;27(4):509–511. [PMC free article: PMC4103562] [PubMed: 22641152]
- Bates J. Epidemiology of vancomycin-resistant enterococci in the community and the relevance of farm animals to human infection. Journal of Hospital Infection. 1997;37(2):89–101. [PubMed: 9364258]
- Bates J., Jordens Z., Selkon J. B. Evidence for an animal origin of vancomycin-resistant enterococci. The Lancet. 1993;342(8869):490–491. [PubMed: 8102440]
- Bekele B., Ashenafi M. Distribution of drug resistance among enterococci and Salmonella from poultry and cattle in Ethiopia. Tropical Animal Health and Production. 2010;42(5):857–864. [PubMed: 19921457]
- Bellomo G., Mangiagle A., Nicastro L., Frigerio G. A controlled double-blind study of SF68 strain as a new biological preparation for the treatment of diarrhoea in pediatrics. Current Therapeutic Research. 1980;28:927–936.
- Ben Omar N., Castro A., Lucas R., Abriouel H., Yousif N. M., Franz C., et al. Functional and safety aspects of Enterococci isolated from different Spanish foods. Systematic and Applied Microbiology. 2004;27(1):118–130. [PubMed: 15053328]
- Berg R. D. The indigenous gastrointestinal microflora. Trends in Microbiology. 1996;4(11):430–435. [PubMed: 8950812]
- Bik E. M., Eckburg P. B., Gill S. R., Nelson K. E., Purdom E. A., Francois F., et al. Molecular analysis of the bacterial microbiota in the human stomach. Proceedings of the National Academy of Sciences of the United States of America. 2005;103(3):732–737. [PMC free article: PMC1334644] [PubMed: 16407106]
- Birollo G. A., Reinheimer J. A., Vinderola C. G. Enterococci vs non-lactic acid microflora as hygiene indicators for sweetened yoghurt. Food Microbiology. 2001;18(6):597–604.
- Bodenstein, D., Cooper, K. W., Ferris, G. F., Miller, A., Poulson, D. F., & Sonnenblick, B. P. (1950). Biology of Drosophila. (M. Demerec, Ed.) New York: John Wiley and Sons.
- Bodnar U. R., Noskin G. A., Suriano T., Cooper I., Reisberg B. E., Peterson L. R. The use of molecular typing techniques in the epidemiologic investigation of resistant enterococci. Infection Control and Hospital Epidemiology. 1996;34(9):2129–2132. [PMC free article: PMC229203] [PubMed: 8862571]
- Boehm A. B., Grant S. B., Kim J. H., Mowbray S. L., McGee C. D., Clark C. D., et al. Decadal and shorter period variability of surf zone water quality at Huntington Beach, California. Environmental Science & Technology. 2002;18:3885–3892. [PubMed: 12269739]
- Brandl K., Plitas G., Mihu C. N., Ubeda C., Jia T., Fleisher M., et al. Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature. 2008;455(7214):804–807. [PMC free article: PMC2663337] [PubMed: 18724361]
- Breed, R. S., Murray, E. G., & Smith, N. R. (1957). Bergey's manual of determinative bacteriology, 7th ed. Baltimore: The Williams and Wilkins Company.
- Bridge P. D., Sneath P. H. Streptococcus gallinarum sp. nov. and Streptococcus oralis sp. nov. International Journal of Systematic Bacteriology. 1982;32(4):410–415.
- Burri, R. (1903). Die Bakterienvegetation auf der Oberfliache normal entwickelter Pflanzen. Zentr. Bakteriol. Parasitenk., Abt. II, 10, 756-763.
- Butaye P., Baele M., Devriese L. A., Haesebrouck F. Comparison of susceptibility to antimicrobials of the enterococcal species isolated from pigeons (Columba livia). Microbial Drug Resistance. 2002;8(3):215–218. [PubMed: 12363011]
- Byappanahalli M. N., Nevers M. B., Korajkic A., Staley Z. R., Harwood V. J. Enterococci in the environment. Microbiology and Molecular Biology Reviews. 2012;76(4):685–706. [PMC free article: PMC3510518] [PubMed: 23204362]
- Byappanahalli M. N., Shively D. A., Nevers M. B., Sadowsky M. J., Whitman R. L. Growth and survival of Escherichia coli and enterococci populations in the macro-alga Cladophora (Chlorophyta). FEMS Microbiology Ecology. 2003;46(2):203–211. [PubMed: 19719574]
- Carvalho M. G., Shewmaker P. L., Steigerwalt A. G., Morey R. E., Sampson A. J., Joyce K., et al. Enterococcus caccae sp. nov. isolated from human stools. International Journal of Systematic and Evolutionary Microbiology. 2006;56(7):1505–1508. [PubMed: 16825620]
- Carvalho M. G., Steigerwalt A. G., Morey R. E., Shewmaker P. L., Falsen E., Facklam R. R., et al. Designation of the provisional new enterococcus species CDC PNS-E2 as Enterococcus sanguinicola sp. nov., isolated from human blood, and identification of a strain previously named Enterococcus CDC PNS-E1 as Enterococcus italicus... Journal of Clinical Microbiology. 2008;46(10):3473–3476. [PMC free article: PMC2566103] [PubMed: 18667594]
- Casanovas-Massana A., Blanch A. R. Determination of fecal contamination origin inreclaimed water open-air ponds using biochemical fingerprinting of enterococciand fecal coliforms. Environmental Science and Pollution Research. 2013;20(5):3003–3010. [PubMed: 23054766]
- Christie P. J., Davidson J. N., Novick R. P., Dunny G. M. Effects of tylosin feeding on the antibiotic resistance of selected gram-positive bacteria in pigs. American Journal of Veterinary Research. 1983;44(1):126–128. [PubMed: 6824216]
- Clark T. M. Evolution and adaptive significance of larval midgut alkalinization in the insect superorder. Journal of Chemical Ecology. 1999;25(8):1945–1960.
- Cleveland J., Montville T. J., Nes I. F., Chikindas M. L. Bacteriocins: safe, natural antimicrobials for food preservation. International Journal of Food Microbiology. 2002;71:1–20. [PubMed: 11764886]
- Collins M. D., Ash C., Farrow J. A., Wallbanks S., Williams A. M. 16S Ribosomal ribonucleic acid sequence analyses of lactococci and related taxa. Description of Vagococcus fluvalis gen. nov, sp. nov. Journal of Applied Bacteriology. 1989;67(4):453–460. [PubMed: 2479630]
- Collins M. D., Facklam R. R., Farrow J. A., Williamson R. Enterococcus raffinosus sp. nov., Enterococcus solitarius sp. nov. and Enterococcus pseudoavium sp. nov. FEMS Microbiology Letters. 1989;48(3):283–288. [PubMed: 2721919]
- Collins M. D., Farrow J. A., Jones D. Enterococcus mundtii sp. nov. International Journal of Systematic and Evolutionary Microbiology. 1986;36(1):8–12.
- Collins M. D., Jones D., Farrow J. A., Kilpper-Balz R., Schleifer K. H. Enterococcus avium nom. rev, comb. nov; E. casseliflavus nom. rev; E. durans nom. rev, comb. nov; E. gallinarum comb. nov; and E. malodoratus sp. nov. International Journal of Systematic and Evolutionary Biology. 1984;34:220–223.
- Collins M. D., Rodrigues U. M., Pigott N. E., Facklam R. R. Enterococcus dispar sp. nov, a new Enterococcus species from human sources. Letters in Applied Microbiology. 1991;12(3):95–98. [PubMed: 1370024]
- Coppola S., Villani F., Coppola R., Coppola R., Parente E., Parente E. Comparison of different starter systems for water-buffalo Mozzarella cheese manufacture. Lait. 1990;70(5-6):411–423.
- Cox C. R., Gilmore M. S. Native microbial colonization of Drosophila melanogaster and its use as a model of Enterococcus faecalis pathogenesis. Infection and Immunity. 2007;75(4):1565–1576. [PMC free article: PMC1865669] [PubMed: 17220307]
- Damborg P., Sørensen A. H., Guardabassi L. Monitoring of antimicrobial resistance in healthy dogs: first report of canine ampicillin- resistant Enterococcus faecium clonal complex 17. Veterinary Microbiology. 2008;132(1-2):190–196. [PubMed: 18524501]
- de Castro A., Montaño A., Casado F., Sánchez A., Rejano L. Utilization of Enterococcus casseliflavus and Lactobacillus pentosus as starter cultures for Spanish-style green olive fermentation. Food Microbiology. 2002;19(6):637–644.
- De Graef E. M., Devriese L. A., Vancanneyt M., Baele M., Collins M. D., Lefebvre K., et al. International Journal of Systematic and Evolutionary Microbiology. 2003;53(4):1069–1074. Description of Enterococcus canis sp. nov. from dogs and reclassification of Enterococcus porcinus Teixeira et al. 2001 as a junior synonym of Enterococcus villorum Vancanneyt et al 2001. [PubMed: 12892128]
- de Kraker M. E., Jarlier V., Monen J. C., Heuer O. E., van de Sande N., Grundmann H. The changing epidemiology of bacteraemias in Europe: trends from the European Antimicrobial Resistance Surveillance System. Clinical Microbiology and Infection. 2013;19(9):860–868. [PubMed: 23039210]
- de Regt M. J., van Schaik W., van Luit-Asbroek M., Dekker H. A., van Duijkeren E., Koning C. J., et al. Hospital and community ampicillin-resistant Enterococcus faecium are evolutionarily closely linked but have diversified through niche adaptation. PLoS One. 2012;7:e30319. [PMC free article: PMC3281830] [PubMed: 22363425]
- de Vaux A., Laguerre G., Diviès C., Prévost H. Enterococcus asini sp. nov. isolated from the caecum of donkeys (Equus asinus). International Journal of Systematic and Evolutionary Microbiology. 1998;48(2):383–387. [PubMed: 9731276]
- De Vuyst, L., & Vandamme, E. J. (1994). Bacteriocins of lactic acid bacteria: microbiology, genetics, and applications. London: Blackie Academic and Professional.
- De Vuyst L., Foulquié Moreno M. R., Revets H. Screening for enterocins and detection of hemolysin and vancomycin resistance in enterococci of different origins. International Journal of Food Microbiology. 2003;84(3):299–318. [PubMed: 12810293]
- Deibel R. H. The group D streptococci. Bacteriological Reviews. 1964;28(3):330–366. [PMC free article: PMC441228] [PubMed: 14220658]
- Del Grosso M., Caprioli A., Chinzari P., Fontana M. C., Pezzotti G., Manfrin A., et al. Detection and characterization of vancomycin-resistant enterococci in farm animals and raw meat products in Italy. Microbial Drug Resistance. 2000;6(4):313–318. [PubMed: 11272260]
- Descheemaeker P., Lammens C., Pot B., Vandamme P., Goossens H. Evaluation of arbitrarily primed PCR analysis and pulsed- field gel electrophoresis of large genomic DNA fragments for identification of enterococci important in human medicine. International Journal of Systematic and Evolutionary Microbiology. 1997;47(2):555–561. [PubMed: 9103648]
- Devriese L. A., De Pelsmaecker K. The anal region as a main carrier site of Staphylococcus intermedius and Streptococcus canis in dogs. Veterinary Record. 1987;121:302–303. [PubMed: 3424574]
- Devriese L. A., Haesebrouck F. Enterococcus hirae in different animal species. Veterinary Record. 1991;129:391–392. [PubMed: 1746125]
- Devriese L. A., Ceyssens K., Rodrigues U. M., Collins M. D. Enterococcus columbae, a species from pigeon intestines. FEMS Microbiology Letters. 1990;59(3):247–251. [PubMed: 2272492]
- Devriese L. A., Cruz Colque J. I., De Herdt P., Haesebrouck F. Identification and composition of the tonsillar and anal enterococcal and streptococcal flora of dogs and cats. Journal of Applied Bacteriology. 1992;73(5):421–425. [PubMed: 1447058]
- Devriese L. A., Cruz Colque J. I., Haesebrouck F., Desmidt M., Uyttebroek E., Ducatelle R. Enterococcus hirae in septicaemia of psittacine birds. Veterinary Record. 1992;130:558–559. [PubMed: 1496756]
- Devriese L. A., Dutta G. N., Farrow J. A., Van De Kerckhove A., Phillips B. A. Streptococcus cecorum, a new species isolated from chickens. International Journal of Systematic and Evolutionary Microbiology. 1983;33(4):772–776.
- Devriese L. A., Hommez J., Wijfels R., Haesebrouck F. Composition of the enterococcal and streptococcal intestinal flora of poultry. Journal of Applied Bacteriology. 1991;71(1):46–50. [PubMed: 1910033]
- Devriese L. A., Laurier L., De Herdt P., Haesebrouck F. Enterococcal and streptococcal species isolated from faeces of calves, young cattle and dairy cows. Journal of Applied Bacteriology. 1992;72(1):29–31. [PubMed: 1541597]
- Devriese L. A., Pot B., Collins M. D. Phenotypic identification of the genus Enterococcus and differentiation of phylogenetically distinct enterococcal species and species groups. Journal of Applied Bacteriology. 1993;75(5):399–408. [PubMed: 8300442]
- Devriese L. A., Pot B., Van Damme L., Kersters K., Haesebrouck F. Identification of Enterococcus species isolated from foods of animal origin. International Journal of Food Microbiology. 1994;26(2):187–197. [PubMed: 7577357]
- Dibner J. J., Richards J. D. Antibiotic growth promoters in agriculture: history and mode of action. Poultry Science. 2005;84(4):634–643. [PubMed: 15844822]
- Domig K. J., Mayer H. K., Kneifel W. Methods used for the isolation, enumeration, characterisation and identification of Enterococcus spp. 2. Pheno- and genotypic criteria. International Journal of Food Microbiology. 2003;88(2-3):165–188. [PubMed: 14596988]
- Donskey C. J. The role of the intestinal tract as a reservoir and source for transmission of nosocomial pathogens. Clinical Infectious Diseases. 2004;39(2):219–226. [PubMed: 15307031]
- Donskey C. J., Chowdhry T. K., Hecker M. T., Hoyen C. K., Hanrahan J. A., Hujer A. M., et al. Effect of antibiotic therapy on the density of vancomycin-resistant enterococci in the stool of colonized patients. The New England Journal of Medicine. 2000;343(26):1925–1932. [PMC free article: PMC4370337] [PubMed: 11136263]
- Dow J. A., Harvey W. R. Role of midgut electrogenic K+ pump potential difference in regulating lumen K+ and pH in larval lepidoptera. The Journal of Experimental Biology. 1988;140:455–463. [PubMed: 2849625]
- Dueggeli, M. (1904). Die Bakterienflora gesunder Samen und daraus gezogener Keimpflanzchen. Zentr. Bakteriol. Parasitenk., Abt. II, 13, 56-63.
- Eckburg P. B., Bik E. M., Bernstein C. N., Purdom E., Dethlefsen L., Sargent M., et al. Diversity of the human intestinal microbial flora. Science. 2005;308(5728):1635–1638. [PMC free article: PMC1395357] [PubMed: 15831718]
- Elliott J. A., Collins M. D., Pigott N. E., Facklam R. R. Differentiation of Lactococcus lactis and Lactococcus garvieae from humans by comparison of whole cell protein patterns. Journal of Clinical Microbiology. 1991;29(12):2731–2734. [PMC free article: PMC270423] [PubMed: 1757541]
- Elliott S. D., Barnes E. M. Changes in serological type and antibiotic resistance on Lancefield group D streptococci in chickens receiving dietary chlortetracycline. Journal of General Microbiology. 1959;20(2):426–433. [PubMed: 13654735]
- Ennahar S., Cai Y. Biochemical and genetic evidence for the transfer of Enterococcus solitarius Collins et al. 1989 to the genus Tetragenococcus as Tetragenococcus solitarius comb. nov. Int J Syst Evol Microbiol. 2005;55(Pt 2):589–592. [PubMed: 15774629]
- Ennahar S., Asou Y., Zendo T., Sonomoto K., Ishizaki A. Biochemical and genetic evidence for production of enterocins A and B by Enterococcus faecium WHE 81. International Journal of Food Microbiology. 2001;70:291–301. [PubMed: 11764194]
- Euzéby, J. P. (2013, September 24). List of Prokaryotic names with Standing in Nomenclature. (A. C. Parte, Editor) Retrieved December 7, 2013, from List of Prokaryotic names with Standing in Nomenclature: http://www
.bacterio.cict .fr/e/enterococcus.html. - Facklam R. R. Comparison of several laboratory media for presumptive identification of enterococci and group D streptococci. Journal of Applied Microbiology. 1973;26(2):138–145. [PMC free article: PMC379740] [PubMed: 4490481]
- Facklam, R. R., Carvalho, M. G., & Teixeira, L. M. (2002). Enterococcus. In M. S. Gilmore, D. B. Clewell, P. Courvalin, G. M. Dunny, B. E. Murray, & L. B. Rice, The Enterococci: Pathogenesis, Molecular Biology, and Antibiotic Resistance (pp. 1-54). Washington, DC: ASM Press.
- Farrow J. A., Collins M. D. Enterococcus hirae, a new species that includes amino acid assay strain NCDO 1258 and strains causing growth depression in young chickens. International Journal of Systematic and Evolutionary Microbiology. 1985;35:73–75.
- Farrow J. A., Kruze J., Phillips B. A., Bramley A. J., Collins M. D. Taxonomic studies on Streptococcus bovis and Streptococcus equinus: description of Streptococcus alactolyticus sp. nov. and Streptococcus saccharolyticus sp nov. Systematic and Applied Microbiology. 1984;5:467–482.
- Ferguson D. M., Moore D. F., Getrich M. A., Zhowandai M. H. Enumeration and speciation of enterococci found in marine and intertidal sediments and coastal water in southern California. Journal of Applied Microbiology. 2005;99(3):598–608. [PubMed: 16108802]
- Fernandez-Diaz, M. J. (1983). Olives. In H. J. Rehm, & G. Reed, Biotechnology. Food and Feed Production by Microorganisms, vol. 5. (pp. 379-397). Hoboken: Wiley-Blackwell.
- Finegold, S. M., Sutter, V. L., & Mathisen, G. E. (1983). Normal indigenous intestinal flora. In D. J. Hentges, Human intestinal microflora in health and disease (pp. 3-29). Waltham: Academic Press.
- Floriano B., Ruiz-Barba J. L., Jiménez-Díaz R. Purification and genetic characterization of enterocin I from Enterococcus faecium 6T1a, a novel antilisterial plasmid-encoded bacteriocin which does not belong to the pediocin family of bacteriocins. Applied and Environmental Microbiology. 1998;64(12):4883–4890. [PMC free article: PMC90938] [PubMed: 9835578]
- Fortina M. G., Ricci G., Mora D., Manachini P. L. Molecular analysis of artisanal Italian cheeses reveals Enterococcus italicus sp. nov. International Journal of Systematic and Applied Microbiology. 2004;54(5):1717–1721. [PubMed: 15388734]
- Fotedar R, Banerjee U, Singh S, Shriniwas, Verma A. K. The housefly (Musca domestica) as a carrier of pathogenic microorganisms in a hospital environment. Journal of Hospital Infection. 1992;20(3):209–215. [PubMed: 1348776]
- Foulquié Moreno M. R., Sarantinopoulos P., Tsakalidou E., De Vuyst L. The role and application of enterococci in food and health. International Journal of Food Microbiology. 2006;106(1):1–24. [PubMed: 16216368]
- Franz C. M., Holzapfel W. H., Stiles M. E. Enterococci at the crossroads of food safety? International Journal of Food Microbiology. 1999;47(1-2):1–24. [PubMed: 10357269]
- Franz C. M., Huch M., Abriouel H., Holzapfel W., Gálvez A. Enterococci as probiotics and their implications in food safety. International Journal of Food Microbiology. 2011;151(2):125–140. [PubMed: 21962867]
- Franz C. M., Stiles M. E., Schleifer K. L., Holzapfel W. H. Enterococci in foods--a conundrum for food safety. International Journal of Food Microbiology. 2003;88(2-3):105–122. [PubMed: 14596984]
- Fujioka, R. S., Tenno, K., & Kansako, S. (198). Naturally occurring fecal coliforms and fecal streptococci in Hawaii’s freshwater streams. Toxicity Assessment, 3(5), 613-630.
- Gaechter T., Wunderlin C., Schmidheini T., Solioz M. Genome sequence of Enterococcus hirae (Streptococcus faecalis) ATCC 9790, a model organism for the study of ion transport, bioenergetics, and copper homeostasis. Journal of Bacteriology. 2012;194(18):5126–5127. [PMC free article: PMC3430321] [PubMed: 22933757]
- Gilmore M. S., Lebreton F., van Schaik W. Genomic transition of enterococci from gut commensals to leading causes of multidrug-resistant hospital infection in the antibiotic era. Current Opinion in Microbiology. 2013;16(1):10–6. [PMC free article: PMC3649759] [PubMed: 23395351]
- Giraffa G. Enterococci from foods. FEMS Microbiology Reviews. 2002;26(2):163–171. [PubMed: 12069881]
- Giraffa G., Carminati D. Control of Listeria monocytogenes in the rind of Taleggio, a surface-smear cheese, by a bacteriocin from Enterococcus faecium 7C5. Sciences des Ailments. 1997;17:383–391.
- Goh S. H., Facklam R. R., Chang M., Hill J. E., Tyrrell G. J., Burns E. C., et al. Identification of Enterococcus species and phenotypically similar Lactococcus and Vagococcus species by reverse checkerboard hybridization to chaperonin 60 gene sequences. Journal of Clinical Microbiology. 2000;38(11):3953–3959. [PMC free article: PMC87524] [PubMed: 11060051]
- Gonçalves A., Igrejas G., Radhouani H., Correia S., Pacheco R., Santos T., et al. Antimicrobial resistance in faecal enterococci and Escherichia coli isolates recovered from Iberian wolf. Letters in Applied Microbiology. 2013;56(4):268–274. [PubMed: 23336611]
- Gordillo M. E., Singh K. V., Murray B. E. Comparison of ribotyping and pulsed-field gel electrophoresis for subspecies differentiation of strains of Enterococcus faecalis. Journal of Clinical Microbiology. 1993;31(6):150–1574. [PMC free article: PMC265579] [PubMed: 8314998]
- Grant S. B., Sanders B. F., Boehm A. B., Redman J. A., Kim J. H., Mrse R. D., et al. Generation of enterococci bacteria in a costal saltwater marsh and its impact on surf zone water quality. Environmental Science & Technology. 2001;15(35):2407–2416. [PubMed: 11432541]
- Graudal H. The classification of motile streptococci within the enterococcus group. Acta Pathologica Microbiota Scandinavica. 1957;41:403–410. [PubMed: 13497684]
- Graves A. K., Weaver R. W. Characterization of enterococci populations collected from a subsurface flow constructed wetland. Journal of Applied Microbiology. 2010;108(4):1226–1234. [PubMed: 19778344]
- Guthof O. A new serologic group of alphahemolytic Streptococci (serologic group Q). Zentralbl Bakteriol Orig. 1955;164:60–3. [PubMed: 13371820]
- Haenel H., Muller-Beuthow W. Vitamin metabolism of coli cultures in vitamin-free and vitamin containing culture solutions. Experientia. 1957;13:149. [PubMed: 13447902]
- Halliday E., Gast R. J. Bacteria in beach sands: an emerging challenge in protecting coastal water quality and bather health. Environmental Science & Technology. 2011;45(2):370–379. [PMC free article: PMC3109870] [PubMed: 21162561]
- Hammerum A. M. Enterococci of animal origin and their significance for public health. Clinical Microbiology and Infection. 2012;18(7):619–625. [PubMed: 22487203]
- Hammerum A. M., Lester C. H., Heuer O. E. Antimicrobial-resistant enterococci in animals and meat: a human health hazard? Foodborne Pathogens and Disease. 2010;7(10):1137–1146. [PubMed: 20578915]
- Hammerum A. M., Lester C. H., Neimann J., Porsbo L. J., Olsen K. E., Jensen L. B., et al. A vancomycin-resistant Enterococcus faecium isolate from a Danish healthy volunteer, detected 7 years after the ban of avoparcin, is possibly related to pig isolates. Journal of Antimicrobial Chemotherapy. 2004;53(3):547–549. [PubMed: 14762053]
- Han D., Unno T., Jang J., Lim K., Lee S. N., Ko G., et al. The occurrence of virulence traits among high-level aminoglycosides resistant Enterococcus isolates obtained from feces of humans, animals, and birds in South Korea. International Journal of Food Microbiology. 2011;144(3):387–392. [PubMed: 21078531]
- Hancock L. E., Gilmore M. S. The capsular polysaccharide of Enterococcus faecalis and its relationship to other polysaccharides in the cell wall. Proceedings of the National Academy of Sciences. 2002;5(99):1574–1579. [PMC free article: PMC122232] [PubMed: 11830672]
- Hartenstein, V. (1993). Atlas of Drosophila development. Cold Spring Harbor: Cold Spring Harbor Laboratory Press.
- Harwood V. J., Whitlock J., Withington V. Classification of antibiotic resistance patterns of indicator bacteria by discriminant analysis: use in predicting the source of fecal contamination in subtropical waters. Applied and Environmental Microbiology. 2000;66(9):3698–3704. [PMC free article: PMC92209] [PubMed: 10966379]
- Hayashi H., Takahashi R., Nishi T., Sakamoto M., Benno Y. Molecular analysis of jejunal, ileal, caecal and recto-sigmoidal human colonic microbiota using 16S rRNA gene libraries and terminal restriction fragment length polymorphism. Journal of Medical Microbiology. 2005;54(11):1093–1101. [PubMed: 16192442]
- Hidron A. I., Edwards J. R., Patel J., Horan T. C., Sievert D. M., Pollock D. A., et al. National Healthcare Safety Network Team and Participating National Healthcare Safety Network Facilities: NHSN Annual Update-Antimicrobial-Resistant Pathogens Associated With Healthcare- Associated Infections: Annual Summary of Data Reported... Infection Control and Hospital Epidemiology. 2008;29(11):996–1011. [PubMed: 18947320]
- Hinton M., Kaukas A., Lim S. S., Linton A. H. Preliminary observations on the influence of antibiotics on the ecology of Escherichia coli and the enterococci in the faecal flora of healthy young chickens. Journal of Antimicrobial Chemotherapy. 1986;18 Supplement C:165–173. [PubMed: 3542935]
- Homan W. L., Tribe D., Poznanski S., Li M., Hogg G., Spalburg E., et al. Multilocus sequence typing scheme for Enterococcus faecium. Journal of Clinical Microbiology. 2002;40(6):1963–1971. [PMC free article: PMC130786] [PubMed: 12037049]
- Huws S. A., Morley R. J., Jones M. V., Brown M. R., Smith A. W. Interactions of some common pathogenic bacteria with Acanthamoeba polyphaga. FEMS Microbiology Letters. 2008;282(2):258–265. [PubMed: 18399997]
- Huycke, M. M., Gilmore, M. S., Clewell, D. B., Courvalin, P., Dunny, G. M., Murray, B. E., et al. (2002). The Enterococci: Pathogenesis, Molecular Biology, and Antibiotic Resistance. Washington, D. C.: ASM Press.
- Huycke M. M., Gilmore M. S., Jett B. D., Booth J. L. Transfer of pheromone-inducible plasmids between Enterococcus faecalis in the Syrian hamster gastrointestinal tract. The Journal of Infectious Diseases. 1992;166(5):1188–1191. [PubMed: 1402034]
- Huycke M. M., Sahm D. F., Gilmore M. S. Multiple-drug resistant enterococci: the nature of the problem and an agenda for the future. Emerging Infectious Diseases. 1998;4(2):239–249. [PMC free article: PMC2640141] [PubMed: 9621194]
- Inamura G. J., Thompson R. S., Boehm A. B., Jay J. A. Wrack promotes the persistence of fecal indicator bacteria in marine sands and seawater. FEMS Microbiology Ecology. 2011;77(1):40–9. [PubMed: 21385189]
- Jayaro B. M., Oliver S. P. Aminoglycoside-resistant Streptococcus and Enterococcus species isolated from bovine mammary secretions. Journal of Dairy Science. 1992;75(4):991–997. [PubMed: 1578037]
- Jett B. D., Huycke M. M., Gilmore M. S. Virulence of enterococci. Clinical Microbiological Reviews. 1994;7(4):462–478. [PMC free article: PMC358337] [PubMed: 7834601]
- Johnsen P. J., Østerhus J. I., Sletvold H., Sørum M., Kruse H., Nielsen K., et al. (200). Persistence of animal and human glycopeptide-resistant enterococci on two Norwegian poultry farms formerly exposed to avoparcin is associated with a widespread plasmid-mediated vanA element within a polyclonal Enterococcus faecium population. Applied and Environmental Microbiology. 71(1):159–168. [PMC free article: PMC544227] [PubMed: 15640183]
- Johnson J. L., Ordal E. J. Deoxyribonucleic acid homology in bacterial taxonomy: effect of incubation temperature on reaction specificity. Journal of Bacteriology. 1968;95(3):893–900. [PMC free article: PMC252107] [PubMed: 4966832]
- Kalina A. P. The position of enterococci in the system of microorganisms. Zhurnal Mikrobiologii, Epidemiologii, I Immunobiologii. 1970;47:20–1. [PubMed: 4992950]
- Kaukas A., Hinton M., Linton A. H. The effect of ampicillin and tylosin on the faecal enterococci of healthy young chickens. Journal of Applied Bacteriology. 1987;62(5):441–447. [PubMed: 3112089]
- Kawalec M., Pietras Z., Daniłowicz E., Jakubczak A., Gniadkowski M., Hryniewicz W., et al. Clonal structure of Enterococcus faecalis isolated from Polish hospitals: characterization of epidemic clones. Journal of Clinical Microbiology. 2007;45(1):147–153. [PMC free article: PMC1828945] [PubMed: 17093020]
- Kinnebrew M. A., Ubeda C., Zenewicz L. A., Smith N., Flavell R. A., Pamer E. G. Bacterial flagellin stimulates Toll-like receptor 5-dependent defense against vancomycin-resistant Enterococcus infection. The Journal of Infectious Diseases. 2010;201(4):534–543. [PMC free article: PMC2811237] [PubMed: 20064069]
- Klaenhammer T. R. Genetics of bacteriocins produced by lactic acid bacteria. FEMS Microbiology Reviews. 1993;12(1-3):39–85. [PubMed: 8398217]
- Klare I., Badstübner D., Konstabel C., Böhme G., Claus H., Witte W. Decreased incidence of VanA-type vancomycin-resistant enterococci isolated from poultry meat and from fecal samples of humans in the community after discontinuation of avoparcin usage in animal husbandry. Microbial Drug Resistance. 1999;5(1):45–52. [PubMed: 10332721]
- Klare I., Heier H., Claus H., Witte W. Environmental strains of Enterococcus faecium with inducible high-level resistance to glycopeptides. FEMS Microbiology Letters. 1993;106(1):23–29. [PubMed: 8440464]
- Klein G. Taxonomy., ecology and antibiotic resistance of enterococci from food and the gastro-intestinal tract. International Journal of Food Microbiology. 2003;88(2-3):123–131. [PubMed: 14596985]
- Klein G., Pack A., Reuter G. Antibiotic resistance patterns of enterococci and occurrence of vancomycin-resistant enterococci in raw minced beef and pork in Germany. Applied and Environmental Microbiology. 1998;64(5):1825–1830. [PMC free article: PMC106237] [PubMed: 9572958]
- Knudtson L. M., Hartman P. A. Routine procedures for isolation and identification of enterococci and fecal streptococci. Applied and Environmental Microbiology. 1992;58(9):3027–3031. [PMC free article: PMC183043] [PubMed: 1444416]
- Kobayashi N., Alam M., Nishimoto Y., Urasawa S., Uehara N., Watanabe N. Distribution of aminoglycoside resistance genes in recent clinical isolates of Enterococcus faecalis, Enterococcus faecium and Enterococcus avium. Epidemiology & Infection. 2001;126(2):197–204. [PMC free article: PMC2869683] [PubMed: 11349969]
- Konstantinidis K. T., Ramette A., Tiedje J. M. The bacterial species definition in the genomic era. Philosophical Transactions of the Royal Society B: Biological Sciences. 2006;361(1475):1929–1940. [PMC free article: PMC1764935] [PubMed: 17062412]
- Koort J., Coenye T., Vandamme P., Sukura A., Björkroth J. Enterococcus hermanniensis sp. nov. from modified-atmosphere-packaged broiler meat and canine tonsils. International Journal of Systematic and Evolutionary Microbiology. 2004;54(Pt 5):1823–1827. [PubMed: 15388750]
- Korajkic A., Brownell M. J., Harwood V. J. Investigation of human sewage pollution and pathogen analysis at Florida Gulf Coast beaches. Journal of Applied Microbiology. 2011;110(1):174–183. [PubMed: 21029275]
- Kruse H., Johansen B. K., Rørvik L. M., Schaller G. The use of avoparcin as a growth promoter and the occurrence of vancomycin-resistant Enterococcus species in Norwegian poultry and swine production. Microbial Drug Resistance. 1999;5(2):135–139. [PubMed: 10432274]
- Kuch A., Willems R. J., Werner G., Coque T. M., Hammerum A. M., Sundsfjord A., et al. Insight into antimicrobial susceptibility and population structure of contemporary human Enterococcus faecalis isolates from Europe. Journal of Antimicrobial Chemotherapy. 2012;67(3):551–558. [PubMed: 22207599]
- Kühn I., Iversen A., Burman L. G., Olsson-Liljequist B., Franklin A., Finn M., et al. 2003 Comparison of enterococcal populations in animals, humans, and the environment - a European study. International Journal of Food Microbiology. 2003;88(2-3):133–145. [PubMed: 14596986]
- Kusuda R., Kawai K., Salati F., Banner C. R., Fryer J. L. Enterococcus seriolicida sp. nov. a fish pathogen. International Journal of Systematic and Evolutionary Biology. 1991;41(3):406–409. [PubMed: 1883715]
- Larsen J., Schønheyder H. C., Singh K. V., Lester C. H., Olsen S. S., Porsbo L. J., et al. Porcine and human community reservoirs of Enterococcus faecalis, Denmark. Emerging Infectious Diseases. 2011;17(12):2395–2397. [PMC free article: PMC3311169] [PubMed: 22172303]
- Law-Brown J., Meyers P. R. Enterococcus phoeniculicola sp. nov. a novel member of the enterococci isolated from the uropygial gland of the Red-billed Woodhoopoe, Phoeniculus purpureus. International Journal of Systematic and Evolutionary Microbiology. 2003;53(Pt 3):683–685. [PubMed: 12807187]
- Layton B. A., Walters S. P., Lam L. H., Boehm A. B. Enterococcus species distribution among human and animal hosts using multiplex PCR. Journal of Applied Microbiology. 2010;109(2):539–547. [PubMed: 20132375]
- Leavis H. L., Bonten M. J., Willems R. J. Identification of high-risk enterococcal clonal complexes: global dispersion and antibiotic resistance. Current Opinion in Microbiology. 2006;9(5):454–460. [PubMed: 16880002]
- Leclercq R., Derlot E., Duval J., Courvalin P. Plasmid-mediated resistance to vancomycin and teicoplanin in Enterococcus faecium. The New England Journal of Medicine. 1988;319(3):157–161. [PubMed: 2968517]
- Lessard E. J., Sieburth J. M. Survival of natural sewage populations of enteric bacteria in diffusion and batch chambers in the marine environment. Applied and Environmental Microbiology. 1983;45(3):950–959. [PMC free article: PMC242396] [PubMed: 6405693]
- Litopoulo-Tzanetaki E., Tzanetakis N. Microbiological study of whitebrined cheese made from raw goat milk. Food Microbiology. 1992;9:13–19.
- Lopez-Diaz T. M., Santos J. A., Gonzalez C. J., Moreno B., Garcia M. L. Bacteriological quality of traditional Spanish blue cheese. Milchwissenschaft (Milk Science International). 1995;50:503–505.
- Ludwig, W., Schleifer, K., & Whitman, W. B. (2009). Family IV. Enterococcaceae fam. nov. In P. Vos, G. Garrity, D. Jones, N. R. Krieg, W. Ludwig, F. A. Rainey, et al., Bergey’s Manual of Systematic Bacteriology., 2nd edn, vol. 3 (The Firmicutes) (p. 594). New York: Springer.
- Lupski J. R., Weinstock G. M. Short, interspersed repetitive DNA sequences in prokaryotic genomes. Journal of Bacteriology. 1992;174(14):4525–4529. [PMC free article: PMC206247] [PubMed: 1624445]
- MacCallum W. G., Hastings T. W. A case of acute endocarditis caused by Micrococcus zymogenes (Nov. Spec.), with a description of the microorganism. The Journal of Experimental Medicine. 1899;4(5-6):521–534. [PMC free article: PMC2118017] [PubMed: 19866921]
- Macovei L., Miles B., Zurek L. Potential of houseflies to contaminate ready-to-eat food with antibiotic-resistant enterococci. Journal of Food Protection. 2008;71(2):435–439. [PubMed: 18326202]
- Magnus C. A., Ingledew W. M., McCurdy A. R. Thermal resistance of streptococci isolated from pasteurized ham. Canadian Institute of Food Science and Technology Journal. 1986;19(2):62–67.
- Magnus C. A., McCurdy A. R., Ingledew W. M. Further studies on the thermal resistance of Streptococcus faecium and Streptococcus faecalis in pasteurized ham. Canadian Institute of Food Science and Technology Journal. 1988;21:209–212.
- Malani, P. N., Kauffman, C. A., & Zervos, M. J. (2002). Enterococcal disease, epidemiology, and treatment. In M. S. Gilmore, D. B. Clewell, P. Courvalin, G. M. Dunny, B. E. Murray, & L. B. Rice, The Enterococci: Pathogenesis., Molecular Biology., and Antibiotic Resistance (pp. 385-408). Washington, D. C.: ASM Press.
- Manolopoulou E., Sarantinopoulos P., Zoidou E., Aktypis A., Moschopoulou E., Kandarakis I. G. Evolution of microbial populations during traditional Feta cheese manufacture and ripening. International Journal of Food Microbiology. 2003;82(2):153–161. [PubMed: 12568755]
- Marinho C., Silva N., Pombo S., Santos T., Monteiro R., Gonçalves A., et al. Echinoderms from Azores islands: An unexpected source of antibiotic resistant Enterococcus spp. and Escherichia coli isolates. Marine Pollution Bulletin. 2013;69(1-2):122–127. [PubMed: 23419753]
- Martín B., Corominas L., Garriga M., Aymerich T. Identification and tracing of Enterococcus spp. by RAPD-PCR in traditional fermented sausages and meat environment. Journal of Applied Microbiology. 2009;106(1):66–77. [PubMed: 19120623]
- Martin J. D., Mundt J. O. Enterococci in insects. Journal of Applied Microbiology. 1972;24(4):575–580. [PMC free article: PMC380616] [PubMed: 4628796]
- Martinez-Murcia A. J., Collins M. D. Enterococcus sulfureus, a new yellow-pigmented Enterococcus species. FEMS Microbiology Letters. 1991;80(1):69–74. [PubMed: 1855650]
- Maugeri T. L., Carbone M., Fera M. T., Irrera G. P., Gugliandolo C. Distribution of potentially pathogenic bacteria as free living and plankton associated in a marine coastal zone. Journal of Applied Microbiology. 2004;5:354–361. [PubMed: 15239702]
- McBride S. M., Fischetti V. A., LeBlanc D. J., Moellering R. C. Jr, Gilmore M. S. Genetic diversity among Enterococcus faecalis. PLOS One. 2007;2(7):e582. [PMC free article: PMC1899230] [PubMed: 17611618]
- McDonald L. C., Kuehnert M. J., Tenover F. C., Jarvis W. R. Vancomycin-resistant enterococci outside the health-care setting: prevalence, sources, and public health implications. Emerging Infectious Diseases. 1997;3(3):311–317. [PMC free article: PMC2627647] [PubMed: 9284375]
- McDonald L. C., Rossiter S., Mackinson C., Wang Y. Y., Johnson S., Sullivan M., et al. Quinupristin-dalfopristin-resistant Enterococcus faecium on chicken and in human stool specimens. The New England Journal of Medicine. 2001;345(16):1155–1160. [PubMed: 11642231]
- Merquoir V. L., Netz D. J., Camello T. C., Texeira L. M. Characterization of enterococci isolated from nosocomial and community infections in Brazil. Advances in Experimental Medicine and Biology. 1997;418:281–283. [PubMed: 9331653]
- Mieth H. Untersuchungen iiber das Vorkommen von Enterokokken bei Tieren und Menschen. I. Mitteilung: ihr Vorkommen im Darm von gesunden Schlachtschweinen. Zentr. Bakteriol. Parasitenk. Abt. I. Orig. 1960;179:456–482.
- Mieth H. Untersuchungen uber das Vorkommen von Enterokokken bei Tieren und Menschen. III. Mitteilung:die Enterokokkenflora in den Faeces von Rindern. Zentr. Bakteriol. Parasitenk. Abt. I. Orig. 1962;185:47–52.
- Moll G. N., Konings W. N., Driessen J. M. Bacteriocins: mechanism of membrane insertion and pore formation. Antonie van Leeuwenhoek. 1999;76(1-4):185–198. [PubMed: 10532378]
- Monstein H. J., Quednau M., Samuelsson A., Ahrné S., Isaksson B., Jonasson J. Division of the genus Enterococcus into species groups using PCR-based molecular typing methods. Microbiology. 1998;144(Pt 5):1171–1179. [PubMed: 9611791]
- Monstein H. J., Tiveljung A., Kraft C. H., Borch K., Jonasson J. Profiling of bacterial flora in gastric biopsies from patients with Helicobacter pylori-associated gastritis and histologically normal control individuals by temperature gradient gel electrophoresis and 16S rDNA sequence analysis. Journal of Medical Microbiology. 2000;49(9):817–822. [PubMed: 10966230]
- Moore D. F., Zhowandai M. H., Ferguson D. M., McGee C., Mott J. B., Stewart J. C. Comparison of 16S rRNA sequencing with conventional and commercial phenotypic techniques for identification of enterococci from the marine environment. Journal of Applied Microbiology. 2006;100(6):1272–1281. [PubMed: 16696674]
- Morandi S., Cremonesi P., Povolo M., Brasca M. Enterococcus lactis sp. nov., from Italian raw milk cheeses. International Journal of Systematic and Evolutionary Microbiology. 2012;62(Pt 8):1992–1996. [PubMed: 22003038]
- Müller T., Ulrich A., Ott E.-M., Müller M. Identification of plant-associated enterococci. Journal of Applied Microbiology. 2001;91(2):268–278. [PubMed: 11473591]
- Mundt J. O. Occurrence of Enterococci: Bud., Blossom., and Soil Studies. Journal of Applied Microbiology. 1961;9(6):541–544. [PMC free article: PMC1057784] [PubMed: 16349612]
- Mundt J. O. Occurrence of enterococci in animals in a wild environment. Journal of Applied Microbiology. 1963;11(2):136–140. [PMC free article: PMC1057957] [PubMed: 13936610]
- Mundt J. O. Occurrence of enterococci on plants in a wild environment. Journal of Applied Microbiology. 1963;11(2):141–144. [PMC free article: PMC1057958] [PubMed: 13936611]
- Mundt, J. O. (1986). Enterococci. In J. G. Holt, P. H. Sneath, & N. R. Krieg, Bergey’s Manual of Systematic Bacteriology, vol. 2. (pp. 1063-1065). Baltimore: Williams & Wilkins.
- Mundt J. O., Coggin J. H. Jr, Johnson L. F. Growth of Streptococcus faecalis var. liquefaciens on plants. Journal of Applied Microbiology. 1962;10(6):552–555. [PMC free article: PMC1057913] [PubMed: 13936612]
- Murray B. E. The life and times of the Enterococcus. Clinical Microbiology Reviews. 1990;3(1):46–65. [PMC free article: PMC358140] [PubMed: 2404568]
- Naimi A., Beck G., Monique M., Lefèbvre G., Branlanti C. Determination of the nucleotide sequence of the 23S ribosomal RNA and flanking spacers of an Enterococcus faecium strain, reveals insertion- deletion events in the ribosomal spacer 1 of enterococci. Systematic and Applied Microbiology. 1999;22(1):9–21. [PubMed: 10188274]
- Nallapareddy S. R., Duh R. W., Singh K. V., Murray B. E. Molecular typing of selected Enterococcus faecalis isolates: pilot study using multilocus sequence typing and pulsed-field gel electrophoresis. Journal of Clinical Microbiology. 2002;40(3):868–876. [PMC free article: PMC120268] [PubMed: 11880407]
- Nallapareddy S. R., Wenxiang H., Weinstock G. M., Murray B. E. Molecular characterization of a widespread, pathogenic, and antibiotic resistance-receptive Enterococcus faecalis lineage and dissemination of its putative pathogenicity island. Journal of Bacteriology. 2005;187(16):5709–5718. [PMC free article: PMC1196071] [PubMed: 16077117]
- Naser S. M., Thompson F. L., Hoste B., Gevers D., Dawyndt P., Vancanneyt M., et al. Application of multilocus sequence analysis (MLSA) for rapid identification of Enterococcus species based on rpoA and pheS genes. Microbiology. 2005;151(Pt 7):2141–2150. [PubMed: 16000705]
- Naser S. M., Vancanneyt M., De Graef E., Devriese L. A., Snauwaert C., Lefebrve K., et al. Enterococcus canintestini sp. nov., from faecal samples of healthy dogs. International Journal of Systematic and Evolutionary Microbiology. 2005;55(Pt 5):2177–2182. [PubMed: 16166728]
- Naser S. M., Vancanneyt M., Hoste B., Snauwaert C., Vandemuelebroecke K., Swings J. International Journal of Systematic and Evolutionary Microbiology. 2006;56(Pt 2):413–416. Reclassification of Enterococcus flavescens Pompei et al. 1992 as a later synonym of Enterococcus casseliflavus (ex Vaughan et al. 1979) Collins et al. 1984 and Enterococcus saccharominimus Vancanneyt et al. 2004 as a later synonym. [PubMed: 16449449]
- Nayak B. S., Badgley B., Harwood V. J. Comparison of genotypic and phylogenetic relationships of environmental Enterococcus isolates by BOX-PCR typing and 16SrRNA gene sequencing. Applied and Environmental Microbiology. 2011;77(14):5050–5055. [PMC free article: PMC3147396] [PubMed: 21622792]
- Nes I. F., Diep D. B., Håvarstein L. S., Brurberg M. B., Eijinsk V., Holo H. Biosynthesis of bacteriocins in lactic acid bacteria. Antonie van Leeuwenhoek. 1996;70(2-4):113–128. [PubMed: 8879403]
- Niemi R. M., Ollinkangas T., Paulin L., Švec P., Vandamme P., Karkman A., et al. Enterococcus rivorum sp. nov. from water of pristine brooks. International Journal of Systematic and Evolutionary Microbiology. 2012;62(Pt 9):2169–2173. [PubMed: 22058322]
- Nissen-Meyer J., Nes I. F. Ribosomally synthesized antimicrobial peptides: their function, structure, biogenesis, and mechanism of action. Archives of Microbiology. 1997;167(2-3):67–77. [PubMed: 9133328]
- Noble C. J. Carriage of group D streptococci in the human bowel. Journal of Clinical Pathology. 1978;31(12):1182–1186. [PMC free article: PMC1145528] [PubMed: 107199]
- Nowlan S. S., Deibel R. H. Group Q streptococci. I. Ecology., serology, physiology, and relationship to established enterococci. Journal of Bacteriology. 1967;94(2):291–296. [PMC free article: PMC315038] [PubMed: 4962699]
- Obiri-Danso K., Jones K. Intertidal sediments as reservoirs for hippurate negative campylobacters, salmonellae and faecal indicators in three EU recognised bathing waters in north west England. Water Research. 2000;34:519–527.
- Onderdonk A. B., Bartlett J. G., Louie T., Sullivan-Seigler N. Microbial synergy in experimental intra-abdominal abscess. Infection and Immunity. 1976;13(1):22–26. [PMC free article: PMC420571] [PubMed: 814099]
- Orla-Jensen S. The lactic acid bacteria. Memoirs of the Academy of the Royal Society of Denmark. Section of Sciences Series. 1919;85:81–197.
- Ostrolenk M., Hunter A. C. The Distribution of Enteric Streptococci. Journal of Bacteriology. 1946;51(6):735–741. [PMC free article: PMC518119] [PubMed: 16561127]
- Ozawa Y., Courvalin P., Gaiimand M. Identification of enterococci at the species level by sequencing of the genes for D-alanine:D-alanine ligases. Systematic and Applied Microbiology. 2000;23(2):230–237. [PubMed: 10930075]
- Pai H. H., Chen W. C., Peng C. F. Cockroaches as potential vectors of nosocomial infections. Infection Control and Hospital Epidemiology. 2004;25(11):979–984. [PubMed: 15566034]
- Palmer K., Godfrey P., Griggs A., Kos V. N., Zucker J., Desjardins C., et al. Comparative genomics of enterococci: variation in Enterococcus faecalis, clade structure in E. faecium, and defining characteristics of E. gallinarum and E. casseliflavus. mBio. 2012;3(1):e00318–e00311. [PMC free article: PMC3374389] [PubMed: 22354958]
- Pangallo D., Drahovska H., Harichova J., Karelova E., Chovanova K., Aradska J., et al. Evaluation of different PCR-based approaches for the identification and typing of environmental enterococci. Antonie Van Leeuwenhoek. 2008;93(1-2):193–203. [PubMed: 17929191]
- Patel R., Piper K. E., Rouse M. S., Steckelberg J. M., Uhl J. R., Kohner P., et al. Determination of 16S rRNA sequences of enterococci and application to species identification of nonmotile Enterococcus gallinarum isolates. Journal of Clinical Microbiology. 1998;36(11):3399–3407. [PMC free article: PMC105342] [PubMed: 9774606]
- Paulsen I. T., Banerjei L., Myers G. S., Nelson K. E., Seshadri R., Read T. D., et al. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science. 2003;299(5615):2071–2074. [PubMed: 12663927]
- Pavia M., Nobile C. G., Salpietro L., Angelillo I. F. Vancomycin resistance and antibiotic susceptibility of enterococci in raw meat. Journal of Food Protection. 2000;63(7):912–915. [PubMed: 10914659]
- Pette J. W. De vorming van zwavelwaterstof in goudse kaas, veroorzaakt door melkzuurbacterien. Netherlands Milk and Dairy Journal. 1955;10:291–302.
- Picard F. J., Danbing K., Boudreau D. K., Boissinot M., Huletsky A., Richard D., et al. Use of tuf sequences for genus-specific PCR detection and phylogenetic analysis of 28 streptococcal species. Journal of Clinical Microbiology. 2004;42(8):3686–3695. [PMC free article: PMC497632] [PubMed: 15297518]
- Pisano M. B., Fadda M. E., Deplano M., Corda A., Casula M., Cosentino S. Technological characterization of lactococci and enterococci for the manufacture of Fiore Sardo sheep cheese. Scienza e Tecnica Lattiero-Casearia. 1994;45:443–456.
- Poyart C., Quesnes G., Trieu-Cuot P. Sequencing the gene encoding manganese-dependent superoxide dismutase for rapid species identification of enterococci. Journal of Clinical Microbiology. 2000;38(1):415–418. [PMC free article: PMC88737] [PubMed: 10618129]
- Prabaker K., Weinstein R. A. Trends in antimicrobial resistance in intensive care units in the United States. Current Opinion in Critical Care. 2011;17(5):472–479. [PubMed: 21900766]
- Qin X., Galloway-Peña J. R., Sillanpaa J., Roh J. H., Nallapareddy S. R., Chowdhury S., et al. Complete genome sequence of Enterococcus faecium strain TX16 and comparative genomic analysis of Enterococcus faecium genomes. BMC Microbiology. 2012;12:135. [PMC free article: PMC3433357] [PubMed: 22769602]
- Quednau M., Ahrné S., Petersson A. C., Molin G. Identification of clinically important species of Enterococcus within 1 day with randomly amplified polymorphic DNA (RAPD). Current Microbiology. 1998;36(6):332–336. [PubMed: 9608744]
- Radhouani H., Igrejas G., Carvalho C., Pinto L., Gonçalves A., Lopez M., et al. Clonal lineages, antibiotic resistance and virulence factors in vancomycin-resistant enterococci isolated from fecal samples of red foxes (Vulpes vulpes). Journal of Wildlife Diseases. 2011;47(3):769–773. [PubMed: 21719850]
- Radhouani H., Poeta P., Gonçalves A., Pacheco R., Sargo R., Igrejas G. Wild birds as biological indicators of environmental pollution: antimicrobial resistance patterns of Escherichia coli and enterococci isolated from common buzzards (Buteo buteo). Journal of Medical Microbiology. 2012;61(Pt 6):837–843. [PubMed: 22403140]
- Rahkila R., Johansson P., Säde E., Björkroth J. Identification of enterococci from broiler products and a broiler processing plant and description of Enterococcus viikkiensis sp. nov. Applied and Environmental Microbiology. 2011;77(4):1193–1203. [PMC free article: PMC3067211] [PubMed: 21183650]
- Reuter G. Culture media for enterococci and group D-streptococci. International Journal of Food Microbiology. 1992;17(2):101–111. [PubMed: 1486020]
- Rice E. W., Messer J. W., Johnson C. H., Reasoner D. J. Occurrence of high-level aminoglycoside resistance in environmental isolates of enterococci. Applied and Environmental Microbiology. 1995;61(1):374–376. [PMC free article: PMC167293] [PubMed: 7887619]
- Riley M. A., Wertz J. E. Bacteriocin diversity: ecological and evolutionary perspectives. Biochimie. 2002;84(5-6):357–364. [PubMed: 12423779]
- Rodrigues U., Collins M. D. Phylogenetic analysis of Streptococcus saccharolyticus based on 16S rRNA sequencing. FEMS Microbiology Letters. 1990;59(1-2):231–234. [PubMed: 1703505]
- Rollins L. D., Lee L. N., LeBlanc D. J. Evidence for a disseminated erythromycin resistance determinant mediated by Tn917-like sequences among group D streptococci isolated from pigs, chickens, and humans. Antimicrobial Agents and Chemotherapy. 1985;27(4):439–444. [PMC free article: PMC180070] [PubMed: 2988428]
- Ronconi M. C., Merino L. A., Fernández G. Detection of Enterococcus with high-level aminoglycoside and glycopeptide resistance in Lactuca sativa (lettuce). Enfermedades Infecciosas y Microbiología Clínica. 2002;20(8):380–383. [PubMed: 12372233]
- Ruiz-Garbajosa P., Bonten M. J., Robinson D. A., Top J., Nallapareddy S. R., Torres C., et al. Multilocus sequence typing scheme for Enterococcus faecalis reveals hospital-adapted genetic complexes in a background of high rates of recombination. Journal of Clinical Microbiology. 2006;44(6):2220–2228. [PMC free article: PMC1489431] [PubMed: 16757624]
- Ruiz-Garbajosa P., Cantón R., Pintado V., Coque T. M., Willems R., Baquero F., et al. Genetic and phenotypic differences among Enterococcus faecalis clones from intestinal colonisation and invasive disease. Clinical Microbiology and Infection. 2006;12(12):1193–1198. [PubMed: 17121625]
- Ruiz-Garbajosa P., de Regt M., Bonten M., Baquero F., Coque T. M., Cantón R., et al. High-density fecal Enterococcus faecium colonization in hospitalized patients is associated with the presence of the polyclonal subcluster CC17. European Journal of Clinical Microbiology & Infectious Diseases. 2012;31(4):519–522. [PubMed: 21789603]
- Ruoff K. L. Recent taxonomic changes in the genus Enterococcus. European Journal of Clinical Microbiology & Infectious Diseases. 1990;9(2):75–79. [PubMed: 2108030]
- Saikia P. K., Dutta G. N., Devriese L., Kalita C. C. Characterization and antimicrobial susceptibility of Enterococcus species from the intestines of ducks in Assam. Research in Veterinary Science. 1995;58(3):288–289. [PubMed: 7659859]
- Sartor R. B. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134(2):577–594. [PubMed: 18242222]
- Scheidegger E. M., Fracalanzza S. A., Texeira L. M., Caradarelli-Leite P. RFLP analysis of a PCR-amplified fragment of the 16S rRNA gene as a tool to identify Enterococcus strains. Memórias do Instituto Oswaldo Cruz. 2009;104(7):1003–1008. [PubMed: 20027468]
- Schleifer K. H., Kilpper-Balz R. Transfer of Streptococcus faecalis and Streptococcus faecium to the genus Enterococcus nom. rev. as Enterococcus faecalis comb. nov. and Enterococcus faecium comb. nov. International Journal of Systematic and Evolutionary Microbiology. 1984;34(1):31–34.
- Sedláček I., Holochová P., Mašlaňová I., Kosina M., Spröer C., Bryndová H., et al. Enterococcus ureilyticus sp. nov. and Enterococcus rotai sp. nov. two novel urease producing enterococci from the environment. International Journal of Systematic and Evolutionary Microbiology. 2013;63(Pt 2):502–510. [PubMed: 22523160]
- Selden P. A. Terrestrialization of animals 1.3.6. In D. Briggs, & P. R. Crowther. Paleobiology. 2007;II:71–74. (pp. Malden: Blackwell Science, Ltd.
- Sghir A., Gramet G., Suau A., Violaine R., Pochart P., Dore J. Quantification of bacterial groups within human fecal flora by oligonucleotide probe hybridization. Applied and Environmental Microbiology. 2000;66(5):2263–2266. [PMC free article: PMC101487] [PubMed: 10788414]
- Shankar N., Baghdayan A. S., Willems R., Hammerum A. M., Jensen L. B. Presence of pathogenicity island genes in Enterococcus faecalis isolates from pigs in Denmark. Journal of Clinical Microbiology. 2006;44(11):4200–4203. [PMC free article: PMC1698369] [PubMed: 16957034]
- Sherman J. M. The streptococci. Bacteriological Reviews. 1937;1:3–97. [PMC free article: PMC440821] [PubMed: 16350049]
- Sherman J. M. The enterococci and related streptococci. Journal of Bacteriology. 1938;35(2):81–93. [PMC free article: PMC374429] [PubMed: 16560094]
- Sherman J. M., Wing H. U. An unnoted hemolytic Streptococcus associated with milk products. Journal of Dairy Science. 1935;18(10):657–660.
- Shewmaker P. L., Steigerwalt A. J., Nicholson A. G., Carvalho G., Facklam R. R., Whitney A. M., et al. Reevaluation of the taxonomic status of recently described species of Enterococcus: evidence that E. thailandicus is a senior subjective synonym of "E. sanguinicola" and confirmation of E. caccae as a species distinct from E. silesiacus. Journal of Clinical Microbiology. 2011;49(7):2676–2679. [PMC free article: PMC3147853] [PubMed: 21543565]
- Signoretto C., Burlacchini G., Lleò M. M., Pruzzo C., Zampini M., Pane L., et al. Adhesion of Enterococcus faecalis in the nonculturable state to plankton is the main mechanism responsible for persistence of this bacterium in both lake and seawater. Applied and Environmental Microbiology. 2004;70(11):6892–6896. [PMC free article: PMC525140] [PubMed: 15528559]
- Silva N., Igrejas G., Figueireido N., Gonçalves A., Radhouani H., Rodrigues J., et al. Molecular characterization of antimicrobial resistance in enterococci and Escherichia coli isolates from European wild rabbit (Oryctolagus cuniculus). Science of the Total Environment. 2010;408(20):4871–4876. [PubMed: 20624632]
- Silva N., Igrejas G., Rodrigues T., Gonçalves A., Felgar A. C., Pacheco R., et al. Molecular characterization of vancomycin-resistant enterococci and extended-spectrum β-lactamase-containing Escherichia coli isolates in wild birds from the Azores Archipelago. Avian Pathology. 2011;40(5):473–479. [PubMed: 21834624]
- Sinclair J. L., Alexander M. Role of resistance to starvation in bacterial survival in sewage and lake water. Applied and Environmental Microbiology. 1984;8:410–415. [PMC free article: PMC241527] [PubMed: 6435525]
- Singh K. V., Coque T. M., Weinstock G. M., Murray B. E. In vivo testing of an Enterococcus faecalis efaA mutant and use of efaA homologs for species identification. FEMS Immunology and Medical Microbiology. 1998;21(4):323–331. [PubMed: 9753005]
- Sistek V., Maheux A. F., Boissinot M., Bernard K. A., Cantin P., Cleenwerck I., et al. Enterococcus ureasiticus sp. nov. and Enterococcus quebecensis sp. nov. isolated from water. International Journal of Systematic and Evolutionary Microbiology. 2012;62(Pt 6):1314–1320. [PubMed: 21788227]
- Smyth C. J., Matthews H., Halpenny M. K., Brandis H., Colman G. Biotyping, serotyping and phage typing of Streptococcus faecalis isolated from dental plaque in the human mouth. Journal of Medical Microbiology. 1987;23(1):45–54. [PubMed: 3102745]
- Staley C., Reckhow K. H., Lukasik J., Harwood V. J. Assessment of sources of human pathogens and fecal contamination in a Florida freshwater lake. Water Research. 2012;46(17):5799–5812. [PubMed: 22939220]
- Sukontasing S., Tanasupawat S., Moonmangmee S., Lee J. S., Suzuki K. Enterococcus camelliae sp. nov. isolated from fermented tea leaves in Thailand. International Journal of Systematic and Evolutionary Microbiology. 2007;57(Pt 9):2151–2154. [PubMed: 17766890]
- Suzzi G., Caruso M., Gardini F., Lombardi A., Vannini L., Guerzoni M. E., et al. A survey of the enterococci isolated from an artisanal Italian goat’s cheese (Semicotto Caprino). Journal of Applied Microbiology. 2000;89(2):267–274. [PubMed: 10971758]
- Svec P., Devriese L. A., Sedlacek I., Baele M., Vancanneyt M., Haesebrouck F., et al. Enterococcus haemoperoxidus sp. nov. and Enterococcus moraviensis sp. nov. isolated from water. International Journal of Systematic and Evolutionary Microbiology. 2001;51:1567–1574. [PubMed: 11491359]
- Svec P., Vancanneyt M., Devriese L. A., Naser S. M., Snauwaert C., Lefebvre K., et al. Enterococcus aquamarinus sp. nov. isolated from sea water. International Journal of Systematic and Evolutionary Microbiology. 2005;55:2183–2187. [PubMed: 16166729]
- Švec P., Vancanneyt M., Koort J., Naser S. M., Hoste B., Vihavainen E., et al. Enterococcus devriesei sp. nov. associated with animal sources. International Journal of Systematic and Evolutionary Microbiology. 2005;55(6):2479–2484. [PubMed: 16280513]
- Svec P., Vancanneyt M., Sedlácek I., Naser S. M., Snauwaert C., Lefebvre K., et al. Enterococcus silesiacus sp. nov. and Enterococcus termitis sp. nov. International Journal of Systematic and Evolutionary Microbiology. 2006;56(Pt 3):577–581. [PubMed: 16514030]
- Švec P., Vandamme P., Bryndová H., Holochova P., Kosina M., Maslanova I., et al. Enterococcus plantarum sp. nov. isolated from plants. International Journal of Systematic and Evolutionary Microbiology. 2011;62(Pt 7):1499–1505. [PubMed: 21856982]
- Swann, M. (1969). Joint Committee on the use of Antibiotics in Animal Husbandry and Veterinary Medicine. London: H.M.S.O.
- Tan C. K., Lai C. C., Wang J. Y., Lin S. H., Liao C. H., Huang Y. T., et al. Bacteremia caused by non-faecalis and non- faecium enterococcus species at a Medical center in Taiwan, 2000 to 2008. Journal of Infection. 2010;61(1):34–43. [PubMed: 20433865]
- Tan, R., Liu, J., Li, M., Huang, J., Sun, J., & Qu, H. (2013). Epidemiology and antimicrobial resistance among commonly encountered bacteria associated with infections and colonization in intensive care units in a university-affiliated hospital in Shanghai. Journal of Microbiology, Immunology and Infection, S1684-1182(12), 00241-1. [PubMed: 23357606]
- Tanasupawat S., Sukontasing S., Lee J. S. Enterococcus thailandicus sp. nov. isolated from fermented sausage ('mum') in Thailand. International Journal of Systematic and Evolutionary Microbiology. 2008;58(Pt 7):1630–1634. [PubMed: 18599707]
- Tannock G. W. The normal microflora: new concepts in health promotion. Science Journal Of Microbiology. 1988;5(1):4–8. [PubMed: 3079214]
- Tannock, G. W., & Cook, G. (2002). In M. S. Gilmore, D. B. Clewell, P. Courvalin, G. M. Dunny, B. E. Murray, & L. B. Rice, The Enterococci: Pathogenesis, Molecular Biology, and Antibiotic Resistance. Washington, D. C.: ASM Press.
- Teixeira L. M., Carvalho M. G., Espinola M. M., Steigerwalt A. G., Douglas M. P., Brenner D. J., et al. Enterococcus porcinus sp. nov. and Enterococcus ratti sp. nov. associated with enteric disorders in animals. International Journal of Systematic and Evolutionary Microbiology. 2001;51(Pt 5):1737–1743. [PubMed: 11594604]
- Teixeira L. M., Carvalho M. G., Merquior V. L., Steigerwalt A. G., Teixeira M. G., Brenner D. J., et al. Recent approaches on the taxonomy of the enterococci and some related microorganisms. Advances in Experimental Medicine and Biology. 1997;418:397–400. [PubMed: 9331679]
- Teixeira L. M., Merquior V. L., Vianni M. C., Carvalho M. G., Fracalanzza S. E., Steigerwalt A. G., et al. Phenotypic and genotypic characterization of atypical Lactococcus garvieae strains isolated from water buffalos with subclinical mastitis and confirmation of L. garvieae as a senior subjective synonym of Enterococcus seriolicida. International Journal of Systematic and Evolutionary Microbiology. 1996;46(3):664–668. [PubMed: 8782673]
- Tenover F. C., Arbeit R. D., Goering R. V. How to select and interpret molecular strain typing methods for epidemiological studies of bacterial infections: a review for healthcare epidemiologists. Molecular Typing Working Group of the Society for Healthcare Epidemiology of America. Infection Control and Hospital Epidemiology. 1997;18(6):426–439. [PubMed: 9181401]
- Thal L. A., Chow J. W., Mahayni R., Bonilla H., Perri M. B., Donabedian S. A., et al. Characterization of antimicrobial resistance in enterococci of animal origin. Antimicrobial Agents and Chemotherapy. 1995;39(9):2112–2115. [PMC free article: PMC162890] [PubMed: 8540725]
- Thaysen, A. C., & Galloway, L. D. (1930). The microbiology of starch and sugars. London: Oxford University Press.
- Thiercelin M. E. Morphologie et modes de reproduction de l'enterocoque. Comptes Rendus des Seances de la Societe de Biologie et des ses Filiales. 1899;11:551–553.
- Thiercelin M. E., Jouhaud L. Sur un diplococque saprophyte de l'intestin susceptible de devenir pathogene. CR Soc. Biol. 1899;5:269–271.
- Tindall B. J., Rosselló-Móra R., Busse H. J., Ludwig W., Kämpfer P. Notes on the characterization of prokaryote strains for taxonomic purposes. International Journal of Systematic and Evolutionary Microbiology. 2010;60(Pt 1):249–266. [PubMed: 19700448]
- Tsiodras S., Gold H. S., Coakley E. P., Wennersten C., Moellering R. C. Jr, Eliopoulos G. M. Diversity of domain V of 23S rRNA gene sequence in different Enterococcus species. Journal of Clinical Microbiology. 2000;38(11):3991–3993. [PMC free article: PMC87530] [PubMed: 11060057]
- Turnidge J. Antibiotic use in animals--prejudices, perceptions and realities. Journal of Antimicrobial Chemotherapy. 2004;53(1):26–27. [PubMed: 14657093]
- Tyrell G. J., Bethune R. N., Willey B., Low D. E. Species identification of enterococci via intergenic ribosomal PCR. Journal of Clinical Microbiology. 1997;35(5):1054–1060. [PMC free article: PMC232702] [PubMed: 9114380]
- Tyrell G. J., Turnbull L., Teixeira L. M., Lefebvre J., Carvalho M. G., Facklam R. R., et al. Enterococcus gilvus sp. nov. and Enterococcus pallens sp. nov. isolated from human clinical specimens. Journal of Clinical Microbiology. 2002;40(4):1140–1145. [PMC free article: PMC140378] [PubMed: 11923322]
- Ubeda C., Bucci V., Caballero S., Djukovic A., Toussaint N. C., Equinda M., et al. Intestinal microbiota containing Barnesiella cures vancomycin-resistant Enterococcus faecium colonization. Infection and Immunity. 2013;81(3):965–973. [PMC free article: PMC3584866] [PubMed: 23319552]
- Ubeda C., Taur Y., Jeng R. R., Equinda M. J., Son T., Samstein M., et al. Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. The Journal of Clinical Investigation. 2010;120(12):4332–4341. [PMC free article: PMC2993598] [PubMed: 21099116]
- Uttley A. H., George R. C., Naidoo J., Johnson P., Collins C. H., Morrison D., et al. High-level vancomycin-resistant enterococci causing hospital infections. Epidemiology & Infection. 1989;103(1):173–181. [PMC free article: PMC2249484] [PubMed: 2506070]
- van den Bogaard A. E., Stobberingh E. E. Epidemiology of resistance to antibiotics. Links between animals and humans. International Journal of Antimicrobial Agents. 2000;14(4):327–335. [PubMed: 10794955]
- Van Donsel D. J., Geldreich E. E., Clarke N. A. Seasonal variations insurvival of indicator bacteria in soil and their contribution to stormwater pollution. Journal of Applied Microbiology. 1967;15(6):1362–1370. [PMC free article: PMC547201] [PubMed: 16349746]
- van Embden J. D., Engel H. W., van Klingeren B. Drug resistance in group D streptococci of clinical and nonclinical origin: prevalence, transferability, and plasmid properties. Antimicrobial Agents and Chemotherapy. 1977;11(6):925–932. [PMC free article: PMC352104] [PubMed: 879758]
- van Schaik W., Willems R. J. Genome-based insights into the evolution of enterococci. Clinical Microbiology and Infection. 2010;16(6):527–532. [PubMed: 20569263]
- Vancanneyt M., Lombardi A., Andrighetto C., Knijff E., Torriani S., Björkroth K. J., et al. Intraspecies genomic groups in Enterococcus faecium and their correlation with origin and pathogenicity. Applied and Environmental Microbiology. 2002;68(3):1381–1391. [PMC free article: PMC123736] [PubMed: 11872491]
- Vancanneyt M., Snauwaert C., Cleenwerck I., Baele M., Descheemaeker P., Goossens H., et al. Enterococcus villorum sp. nov. an enteroadherent bacterium associated with diarrhoea in piglets. International Journal of Systematic and Evolutionary Microbiology. 2001;51(Pt 2):393–400. [PubMed: 11321084]
- Vancanneyt M., Zamfir M., Devriese L. A., Lefebvre K., Engelbeen K., Vandemeulebroecke K., et al. Enterococcus saccharominimus sp. nov. from dairy products. International Journal of Systematic and Evolutionary Microbiology. 2004;54(Pt 6):2175–2179. [PubMed: 15545454]
- Vandamme P., Vercauteren E., Lammens C., Pensart N., Leven M., Pot B., et al. Survey of enterococcal susceptibility patterns in Belgium. Journal of Clinical Microbiology. 1996;34(10):2572–2576. [PMC free article: PMC229320] [PubMed: 8880522]
- Vollaard E. J., Clasener H. A. Colonization resistance. Antimicrobial Agents and Chemotherapy. 1994;38(3):409–414. [PMC free article: PMC284472] [PubMed: 8203832]
- Voznyakovskaya Y. L., Khudyakov Y. P. Species composition of the epiphytic microflora. Mikrobiologiya. 1960;29:73–76.
- Ward D. M., Weller R., Bateson M. M. 16S rRNA sequences reveal numerous uncultured microorganisms in a natural community. Nature. 1990;345(6270):63–65. [PubMed: 1691827]
- Werner G., Coque T. M., Hammerum A. M., Hope R., Hryniewicz W., Johnson A., et al. Emergence and spread of vancomycin resistance among enterococci in Europe. Eurosurveillance. 2008;13(47) [PubMed: 19021959]
- Whitman R. L., Shively D. A., Pawlik H., Nevers M. B., Byappanahalli M. N. Occurrence of Escherichia coli and enterococci in Cladophora (Chlorophyta) in nearshore water and beach sand of Lake Michigan. Applied and Environmental Microbiology. 2003;69(8):4717–4719. [PMC free article: PMC169104] [PubMed: 12902262]
- Willems R. J., Top J., van den Braak N., van Belkum A., Endtz H., Mevius D., et al. Host specificity of vancomycin-resistant Enterococcus faecium. The Journal of Infectious Diseases. 2000;182:816–823. [PubMed: 10950776]
- Willems R. J., Top J., van Santen M., Robinson D. A., Coque T. M., Baquero F., et al. Global spread of vancomycin-resistant Enterococcus faecium from distinct nosocomial genetic complex. Emerging Infectious Diseases. 2005;11(6):821–828. [PMC free article: PMC3367597] [PubMed: 15963275]
- Willems R. J., Top J., van Schaik W., Leavis H., Bonten M., Sirén J., et al. Restricted gene flow among hospital subpopulations of Enterococcus faecium. mBio. 2012;3(4):e00151–e00112. [PMC free article: PMC3413404] [PubMed: 22807567]
- Woodford N., Adebiyi A.-M. A., Palepou M.-F. I., Cookson B. D. Diversity of vanA glycopeptide resistance elements in enterococci from human and nonhuman sources. Antimicrobial Agents and Chemotherapy. 1998;42(3):502–508. [PMC free article: PMC105489] [PubMed: 9517923]
- Wunderlich P. F., Braun L., Fumagalli I., D'Apuzzo V., Heim F., Karly M., et al. Double-blind report on the efficacy of lactic acid-producing Enterococcus SF68 in the prevention of antibiotic-associated diarrhoea and in the treatment of acute diarrhoea. Journal of International Medical Research. 1989;17:333–338. [PubMed: 2676650]
- Yamahara K. M., Layton B. A., Santoro A. E., Boehm A. B. Beach sands along the California coast are diffuse sources of fecal bacteria to coastal waters. Environmental Science & Technology. 2007;41(13):4515–4521. [PubMed: 17695890]
- Yamahara K. M., Walters S. P., Boehm A. B. Growth of enterococci in unaltered, unseeded sands subjected to tidal wetting. Applied and Environmental Microbiology. 2009;75(6):1517–1524. [PMC free article: PMC2655449] [PubMed: 19151188]
- Yoshimura H., Ishimaru M., Endoh Y. S., Kojima A. Antimicrobial susceptibilities of enterococci isolated from faeces of broilers and layer chickens. Letters in Applied Microbiology. 2000;31(6):427–432. [PubMed: 11123550]
- Zhang X., Vrijenhoek J. E., Bonten M. J., Willems R. J., van Schaik W. A genetic element present on megaplasmids allows Enterococcus faecium to use raffinose as carbon source. Environmental Microbiology. 2011;13(2):518–528. [PubMed: 20946531]
- Review Genomic transition of enterococci from gut commensals to leading causes of multidrug-resistant hospital infection in the antibiotic era.[Curr Opin Microbiol. 2013]Review Genomic transition of enterococci from gut commensals to leading causes of multidrug-resistant hospital infection in the antibiotic era.Gilmore MS, Lebreton F, van Schaik W. Curr Opin Microbiol. 2013 Feb; 16(1):10-6. Epub 2013 Feb 5.
- Review The Physiology and Metabolism of Enterococci.[Enterococci: From Commensals t...]Review The Physiology and Metabolism of Enterococci.Ramsey M, Hartke A, Huycke M. Enterococci: From Commensals to Leading Causes of Drug Resistant Infection. 2014
- Review Enterococci as Indicators of Environmental Fecal Contamination.[Enterococci: From Commensals t...]Review Enterococci as Indicators of Environmental Fecal Contamination.Boehm AB, Sassoubre LM. Enterococci: From Commensals to Leading Causes of Drug Resistant Infection. 2014
- Enterococci from Wild Magellanic Penguins (Spheniscus magellanicus) as an Indicator of Marine Ecosystem Health and Human Impact.[Appl Environ Microbiol. 2020]Enterococci from Wild Magellanic Penguins (Spheniscus magellanicus) as an Indicator of Marine Ecosystem Health and Human Impact.Prichula J, Van Tyne D, Schwartzman J, Sant'Anna FH, Pereira RI, da Cunha GR, Tavares M, Lebreton F, Frazzon J, d'Azevedo PA, et al. Appl Environ Microbiol. 2020 Sep 17; 86(19). Epub 2020 Sep 17.
- Ready-to-eat dairy products as a source of multidrug-resistant Enterococcus strains: Phenotypic and genotypic characteristics.[J Dairy Sci. 2020]Ready-to-eat dairy products as a source of multidrug-resistant Enterococcus strains: Phenotypic and genotypic characteristics.Chajęcka-Wierzchowska W, Zadernowska A, García-Solache M. J Dairy Sci. 2020 May; 103(5):4068-4077. Epub 2020 Mar 18.
- Enterococcus Diversity, Origins in Nature, and Gut Colonization - EnterococciEnterococcus Diversity, Origins in Nature, and Gut Colonization - Enterococci
Your browsing activity is empty.
Activity recording is turned off.
See more...



