NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
McEntyre J, Ostell J, editors. The NCBI Handbook [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2002-.
This publication is provided for historical reference only and the information may be out of date.
Summary
PubMed Central (PMC) is the National Library of Medicine's digital archive of full-text journal literature. Journals deposit material in PMC on a voluntary basis. Articles in PMC may be retrieved either by browsing a table of contents for a specific journal or by searching the database. Certain journals allow the full text of their articles to be viewed directly in PMC. These are always free, although there may be a time lag of a few weeks to a year or more between publication of a journal issue and when it is available in PMC. Other journals require that PMC direct users to the journal's own Web site to see the full text of an article. In this case, the material will always be available free to any user no more than 1 year after publication but will usually be available only to the journal's subscribers for the first 6 months to 1 year.
To increase the functionality of the database, a variety of links are added to the articles in PMC: between an article correction and the original article; from an article to other articles in PMC that cite it; from a citation in the references section to the corresponding abstract in PubMed and to its full text in PMC; and from an article to related records in other Entrez databases such as Reference Sequences, OMIM, and Books.
A PubMed Central (PMC) Site Guide
The PMC homepage has a list of all journals available in PMC and the earliest available issue for each journal (Figure 1). From here, a table of contents for the latest available issue of a journal or a list of all issues of the journal available through PMC can be viewed.
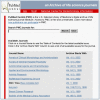
Figure 1
The PMC journal list.
Every article citation in a table of contents includes one or more links (Figure 2). Articles for which the full text is available directly in PMC generally have links to an Abstract view, a Full Text view, and a PDF (printable view). Where applicable, they also have links to Corrections and to supplementary data that may be available for the article. In cases where the full text is available only at the journal publisher's site, there is only one link, to a PubLink page (described below).

Figure 2
PMC table of contents.
In addition to the header information for the article itself, the upper part of a Full Text or PubLink page contains a variety of links, including links to other forms of the article, to related information in PubMed and other Entrez databases, and to corrections or “cited-in” lists where these apply (Figure 3). The sidebar in the body of a Full Text page (Figure 4) has links to tables and thumbnail images of any figures in the article, which when selected will display the full figure. Figures and tables may also be opened directly from the point in the text where they are referenced. Citations in the References section of an article frequently include a link to the corresponding PubMed abstract and sometimes also have a link to the full text of the referenced article in PMC (Figure 5).
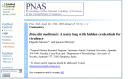
Figure 3
Header of Abstract and Full Text pages.

Figure 4
Body of Full Text page.

Figure 5
References.
An Abstract page is identical to a Full Text page that has been cut off at the end of the abstract.
A PMC PubLink page (Figure 6) is similar to an Abstract page, except that it does not have links to alternate forms (Full Text or PDF) of the article in PMC. Instead, it contains a link to the full text at the publisher's site and information about when it will be freely available.
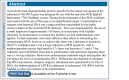
Figure 6
PubLink page.
When an article has been cited by other articles in PMC, a “cited-in” link displays just under the article header information on both the Abstract and Full Text pages. Selecting this cited-in link gives you a list of the articles that have referenced the subject article (Figure 7).
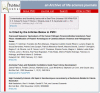
Figure 7
Cited-in list.
PMC article citations may also be retrieved by doing a search in PMC or through a PubMed search. (In PubMed, use the subsets limit if you want to find only articles that are available in PMC.)
Participation in PMC
Participation by publishers in PMC is voluntary, although participating journals must meet certain editorial standards. A participating journal is expected to include all of its peer-reviewed primary research articles in PMC. Journals are encouraged to also deposit other content such as review articles, essays, and editorials. Review journals, and similar publications that have no primary research articles, are also invited to include their contents in PMC. However, primary research papers without peer review are not accepted.
Journals that deposit material in PMC may make the full text viewable directly in PMC or may require that PMC link to the journal site for viewing the complete article. In the latter case, the full text must be freely available at the journal site no more than 1 year after publication. In the case of full text that is viewable directly in PMC, which by definition is free, the journal may delay the release of its material for more than 1 year after publication, although all current journals have delays of 1 year or less.
In either case, the journal must provide SGML or XML for the full text, along with any related high-resolution image files. All data must meet PMC standards for syntactically correct and complete data.
The rationale behind the insistence on free access is that continued use of the material, which is encouraged by open access, serves as the best test of the durability and utility of the archive as technology changes over time. PMC does not claim copyright on any material deposited in the archive. Copyright remains with the journal publisher or with individual authors, whichever is applicable.
Refer to Information for Publishers to learn more about participating in PMC.
Links to Other NCBI Resources
From Abstract and Full Text pages in PMC are links to related articles in PubMed and to related records in other Entrez databases, such as Nucleotides or Books. These are identical to the links between databases that you can find in any Entrez record.
PMC Architecture
PubMed Central is an XML-based publishing system for full-text journal articles. All journal content in the archive was either supplied in, or has been converted to, a Document Type Definition (DTD) written at NCBI for the publication and storage of full-text articles.
The content is displayed dynamically on the PMC site by journal, volume, and issue (if applicable). XML, Web graphics, PDFs, and supplemental data are stored in a Sybase database. When a reader requests an article, the XML is retrieved from the database, and it is converted to HTML using XSLT stylesheets. The look of the HTML pages is controlled further by using Cascading Style Sheets (CSS), which allow manipulation of colors, fonts, and typefaces.
Data Flow: 1. SGML/XML Processing
We receive journal content either directly from publishers or from publishers' vendors. This content includes:
- SGML or XML of the articles to be deposited
- High-resolution images
- Supplemental data associated with the articles
- PDF versions of the articles
All of the text is converted to a central DTD, the PMC DTD, and the images are converted to Web format (GIF and JPEG). These files, along with any supplemental data or PDFs, are loaded into the database for linking, indexing, and retrieval (Figure 8). The source text in SGML or XML format is parsed against the source DTD. If the source files do not conform to the DTD, they are returned to the publisher or vendor for correction.
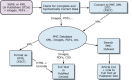
Figure 8
Data flow.
Once all of the files in an issue or batch have been validated, they are converted to PMC XML (referred to as a PMC XML file or PXML) using XSLT (Figure 9). Because XSLT is an XML conversion tool, SGML source files must first be converted to XML. This is done using SX, which is available from James Clark. The transformation will close any empty elements and insert ending tags for any element that is not closed.
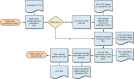
Figure 9
Text conversion flow.
For each publisher that submits SGML, an XML version of the DTD is created. This is used to parse the output of SX before the XML conversion is started. The XML version of the publisher's DTD is not used to validate the source data (because this has already been done using the original DTD).
XSLT requires that the input file be valid XML. If a DTD is not available for validation, the parser will check the syntax; it will also replace all of the character entities with the appropriate UTF-8 representation. This can cause a problem because the relationship between the characters in the input file and their UTF-8 representations may not be one to one. This means that characters translated to UTF-8 might not translate back to the original character entity accurately.
After the XSLT conversion, the original character entities are converted to character entities that are valid under the PMC DTD. Character translation tables for each source DTD regulate this conversion.
The resulting XML is then validated against the PMC DTD.
Several other items are created along with the PXML file. These are:
1. An entity file (articlename.ent). This file lists all of the character entities (from the PMC DTD-defined entity sets) that are in the article. One entity file is created for each article. This information is loaded into the database and is used to prepare the final HTML file for display. A sample is below:
agr
deg
Dgr
ldquo
lsqb
lt
pmc811
mdash
2. A PubMedID file (articlename.pmid). This file includes a set of reference citations in the format
Journal_Title|year|volume|first_page|AuthorName|Refno|pubmedid
When the article is converted, this information is collected from each journal citation in the bibliography and sent in a query to PubMed (using the Citation Matcher utility). If a value is returned, it is written in the last field. If no value is returned, an error message is written in this field. This information is saved so that if the article ever needs to be reconverted, the PubMed IDs will not need to be looked up again. A sample is below:
Nucleic Acids Res|1992|20|2673|Murray JM|13:2:493:16|1319571 J Biol Chem|1999|274|35297|Naumann M|13:2:493:17|10585392 EMBO J|2000|19|3475|Osaka F|13:2:493:18|10880460 Nature|2000|405|662|Osterlund MT|13:2:493:19|0 FASEB J|1998|12|469|Seeger M|13:2:493:20|9535219
3. A source node file (articlename.src). When an article is passed through the XSLT conversion, a list is made of each node, or named piece of information, that is included in the file. As the conversion is running, each node that is being processed is recorded. When the conversion is complete, the processed node list is compared with the list of nodes in the source file, and any piece of information that was not processed is reported in a conversion log. A sample is below:
/ART /ART/@AID /ART/@DATE /ART/@ISS /ART/BM/FN/P/EXREF /ART/BM/FN/P/EXREF/@ACCESS /ART/BM/FN/P/EXREF/@TYPE /ART/FM /ART/FM/ABS /ART/FM/ABS/P /ART/FM/ABS/P/EMPH /ART/FM/ACC /ART/FM/ATL /ART/FM/ATL/EMPH /ART/FM/AUG /ART/FM/PUBFRONT/CPYRT/CPYRTNME/COLLAB /ART/FM/PUBFRONT/CPYRT/DATE /ART/FM/PUBFRONT/CPYRT/DATE/YEAR /ART/FM/PUBFRONT/DOI /ART/FM/PUBFRONT/EXTENT /ART/FM/PUBFRONT/FPAGE /ART/FM/PUBFRONT/ISSN /ART/FM/PUBFRONT/LPAGE /ART/FM/RE /ART/FM/RV
Image Processing
To accommodate the archiving requirements of the PubMed Central project, it is important that figures be submitted in the greatest resolution possible, in TIFF or EPS format. Figures in these formats will be available for data migration when formats change in the future, and PubMed Central will be able to keep all of the figures current.
For display on the PMC site, two copies of each figure are made: a GIF thumbnail (100 pixels wide) and a JPEG file that will be displayed with the figure caption when the figure is requested.
Supplemental Data Processing
Supplemental data include any supporting information that accompanies the article but is not part of the article. They may be text files, Word document files, spreadsheet files, executables, video, and others.
Sometimes a journal has a Web site where all of this supplemental information is stored. In this case, PubMed Central establishes links from the article to the supplemental information on the publisher's site.
In other cases, the supplemental data files are submitted with the article to be loaded into the PMC database. Either way, the information concerning this supplemental data is collected in a Supplemental Data file, which includes the location of the supplemental file(s), the type of information that is available, and how the link should be built from the article. PMC does not validate any of the supplemental data files that are supplied.
Mathematics
Mathematical symbols and notations can be difficult to display in HTML because of built-up expressions and unusual characters. For the most part, expressions that are simple enough to display using HTML are not handled as math unless they are tagged as math specifically. Publishers that supply content to PMC handle math expressions in one of two ways: supplied images or encoding in SGML.
1. Math Images
Any expression that cannot be tagged by the source DTD is supplied as an image. In this case, PMC will pass the image callout through to the PXML file and display the supplied image in the HTML file.
2. Math in SGML
Several of the Source DTDs used by publishers to submit data to PMC are robust enough to allow coding of almost any mathematical expression in SGML. Most of these were derived from the Elsevier DTD; therefore, many of the elements are similar.
During article conversion, any items that are recognized as math are translated into Teχ. This would include any expression tagged specifically as a "formula" or "display formula," as well as any free-standing expression that cannot be represented in HTML. These expressions include radicals, fractions, and anything with an overbar (other than accented characters). For example:
"x + y = 2z" would not be recognized as a math expression, but "<formula>x + y = 2z</formula>" would be.
"1/2" would not be recognized as a math expression, but <fraction><numerator>1</numerator><denominator>2 </denominator></fraction> would be.
"<radical>2x</radical> would be recognized as a math expression, as would "<overbar>47X</overbar>."
The SGML:
<FD ID="E2">I<SUP><UP>o</UP> </SUP><INF><UP>f</UP></INF>&cjs1134;I<INF><UP>f</UP></INF>=&phgr; <SUP><UP>o</UP></SUP><INF><UP>f</UP></INF>&cjs1134;&phgr;<INF> <UP>f</UP></INF>=1+K<INF><UP>sv</UP></INF>[<UP>Q </UP>]</FD>
Converts to:
<fd id="E2"><math mathtype="tex"
id="M2">\documentclass[12pt]{minimal}
\usepackage{wasysym}
\usepackage[substack]{amsmath}
\usepackage{amsfonts}
\usepackage{amssymb}
\usepackage{amsbsy}
\usepackage[mathscr]{eucal}
\usepackage{mathrsfs}
\DeclareFontFamily{T1}{linotext}{}
\DeclareFontShape{T1}{linotext}{m}{n} { <-> linotext }{}
\DeclareSymbolFont{linotext}{T1}{linotext}{m}{n}
\DeclareSymbolFontAlphabet{\mathLINOTEXT}{linotext}
\begin{document}
\[
I^{o}_{f}/I_{f}={\phi}^{o}_{f}/{\phi}_{f}=1+K_{sv}[Q]
\]
\end{document}
</math></fd>
When the articles are loaded into the database, the equation markup is written into an Equation table. This table will also include the equation image, which will be created from the Teχ markup.
The image for the equation shown above in SGML and PXML (with Teχ) is:
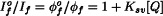
Data Flow: 2. Loading the Database
Because all of the content in PubMed Central is in the same format—PMC DTD—loading articles into the database is relatively straightforward. Once an article is loaded into the production (public) database, it will retain its ArticleId (article ID number) in perpetuity. On loading, each article is validated against the PMC DTD. Also, any external files that are referenced by the XML are checked. If any file, such as a figure, is missing, the loading will be aborted.
The database loading software and daily maintenance programs perform several other tests to ensure the accuracy and vitality of the archive:
- 1.
Journal identity. The Journal title being loaded is verified against the ISSN number in the PXML to verify that the journal identity is correct.
- 2.
Duplicate articles. An article may not be loaded more than once. Any changes to the article must be submitted as a replacement article, which will use the same ArticleId.
- 3.
Publication date/delay. Rules for delay of publication embargo can be set up in the database to ensure that an issue will not be released to the public before a certain amount of time has passed since the publisher made the issue available.
- 4.
PubMed IDs. PubMedIDs for the article being loaded or any bibliographic citation in the article that are not defined in the PXML are looked up upon loading.
- 5.
Link updates. Links between related articles and from articles to external sources are updated daily.
The database has been designed to allow multiple versions of articles. In addition to article information, the database also stores information on content suppliers and publishers and journal-specific information.
Special Characters
PubMed Central uses a number of standard ISO character sets (8879 and 9573), along with a set of characters that has been defined to accommodate characters not in the standard set. The ISO Standard Character sets referenced are listed in Box 1.
Box 1
ISO Standard Character sets used by PMC.
Each publisher DTD defines a set of characters that may be used in their articles. Generally, these publisher DTDs use the same standard ISO character sets listed in Box 1. Any character that cannot be represented by the standard ISO sets is defined in a publisher-specific character set. These publisher-specified characters are converted into characters in the PMC entity list during conversion (see SGML/XML Processing). The PMC entity list is publicly available.
The supplied data also include groups of entities that are to be combined in the final document. Sometimes these are grouped in a tag such as:
<A><AC>α</AC><AC>´</AC></A>
and sometimes they are just positioned next to each other in the text. These combined entities must be mapped either to an ISO character or to a character in the PMC character set.
For the most flexibility in displaying characters across platforms, PMC uses UTF-8 encoding whenever possible. Because not all browsers support the same subset of UTF-8 characters and some characters cannot be represented in UTF-8, PMC displays characters as a combination of GIFs and UTF-8 characters, depending on the Browser/OS combination and the character to be displayed.
PMC DTD
History
In the first version of the PMC project, the SGML and XML were loaded into a database in its native format. The HTML rendering software was then required to convert content from different sources into normalized HTML on the fly when a reader requested an article.
This was slow and cumbersome on the rendering side and was not scaleable. At that time, PMC was receiving content for about five journals in two DTDs, the keton.dtd from HighWire Press and the article.dtd from BioMed Central. The set-up for a new journal was difficult, and it soon became obvious that this solution would not scale easily.
To satisfy the archiving requirement for the PMC project and to simplify the delivery of articles online, PubMed Central decided to convert all content into a centralized format. The normalized content is easier to render, allows enhanced value such as links to other NCBI databases to be added, and simplifies content archiving.
PMC created a new DTD, which was strongly influenced by the BioMedCentral article.dtd and the keton.dtd. The original emphasis was on simplicity. As more and more articles from more and more journals were converted to the PMC DTD, changes had to be made to accommodate the data. The PMC DTD is publicly available.
Review and Revision of the PMC DTD
Because the PMC DTD grew rapidly, it was feared that the original "simplicity" of its design would lead to confusing data structures. With more and more publishers inquiring about submitting content directly in the PMC DTD, PubMed Central decided that an independent review was necessary. Mulberry Technologies, Inc., an electronic publishing consultancy specializing in SGML- and XML-based systems, reviewed the DTD and created a modified version.
At approximately the same time, under the auspices of a Mellon Grant to explore ejournal archiving, Harvard University Library contracted with Inera, Inc. to review a variety of DTDs from selected publishers, PMC included. The study focused on two key questions:
- 1.
Can a common DTD be designed and developed into which publishers' proprietary SGML files can be transformed to meet the requirements of an archiving institution?
- 2.
If such a structure can be developed, what are the issues that will be encountered when transforming publishers' SGML files into the archive structure for deposit into the archive?
The requirement of the archival article DTD was defined as the ability to represent the intellectual content of journal articles. This study is available and suggestions from the study were used in the NLM Archiving DTD Suite.
The NLM Archiving DTD will not be backwards-compatible with the pmc-1.dtd. It should be publicly available by the end of 2002, along with complete documentation for publishers and authors. A draft version is available (http://www.pubmedcentral.nih.gov/pmcdoc/dtd/nlm_lib/0.1/documentation/HTML/index.html), along with a draft version of the documentation.
Frequently Asked Questions
Please refer to the PMC site for answers to frequently asked questions.
- PubMed Central (PMC): An Archive for Literature from Life Sciences Journals - Th...PubMed Central (PMC): An Archive for Literature from Life Sciences Journals - The NCBI Handbook
Your browsing activity is empty.
Activity recording is turned off.
See more...

