NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
SNP FAQ Archive [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2005-.
This publication is provided for historical reference only and the information may be out of date.
What kind of studies will GST allow me to perform with Genome-Wide Association (GWAS) Data?
Loading GWAS Data to GST
GWAS File Types defined
Loading Private GWAS Data to GST
What kind of studies will GST allow me to perform with Genome-Wide Association (GWAS) Data?
There are a number of planned features we intend to include in the GST, such as generation of LD-graphs based on uploaded GWAS data, but these features will not be available anytime soon. For now, the only thing that GST will allow you to do is to display pre-computed results in Gbench. (04/23/08)
Loading GWAS Data to GST
GWAS File Types defined
What is the difference between GWAS CHPB, Analysis files, and CHPA files, and which of these would I load into Gbench for study with GST?
GWAS CHPB files: CHPB files, or chip batch files, contain the SNPs of the GWAS chip for a particular build. These files are generated when dbSNP processes the data submissions of GWAS chips from DNA Chip manufacturers. CHPB files are available online at the dbSNP FTP site, and the rules for the CHPB file format are located in Box 1 of this document.
Box 1:
CHPB FILE Format Rules.
GWAS Analysis files: dbGAP maintains analysis files of submitted studies, whose file formats vary depending on analysis type. These files are available at the dbGap FTP site organized by submitting institution.
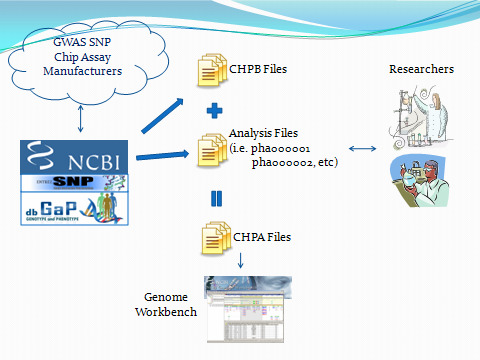
GWAS CHPA files: CHPA files, or chip analysis files, can be loaded into GBench. The file format follows the same format as CHPB, except the file extension is .chpa, and an extra column for pvalue is appended at the end. CHPA files are created by taking the CHPB files and the pvalues of an analysis file and merging them using another tool (perl, awk, etc). [see the figure below describing “GWAS Data Flow (File Types)”]
Please Note: As of this date, CHPA files are not published by NCBI and instead the user must separately generate a CHPA file. PERL script examples of how to do generate CHPA files are located in Box 2 of this document. In the future, dbGaP plans to release CHPA files in the same directory as the analysis files.
Box 2:
Example PERL Scripts for Merging Analysis File p-values and CHPB Files.
GWAS DATA Flow (File Types):
(05/19/08)
Loading Private GWAS Data to GST
How do I load private GWAS data (GWAS data my lab has generated) to GBench so I can study it using GST?
- 1.
Create a CHPA file by merging the p-values from your laboratory’s analysis files with the corresponding CHPB file(s):
- a.
Access your laboratory’s analysis files
- b.
Download the corresponding CHPB files from the GWAS array folder located in the human _9606 directory of the dbSNP FTP site.
- c.
Use an external program, such as PERL, to merge the p-values of your analysis files with the CHPB files to create CHPA files. Click here to see an example PERL scripts for this purpose.
Please Note: the CHPA files you generate will be organized by chromosome
2. Modify the “Name” attribute of your new CHPA file
You will need to create a name for your CHPA file. This name will appear above the data track in the graphical view that represents the data in the CHPA file. Make sure this name is meaningful to you so that you can discern the difference between the graphical track created by this CHPA file, and other tracks you may have open with it in the Graphical View.
To modify the “name” field in your CHPA file, all you have to do is open up the CHPA file in a text editor (like notepad or wordpad) and add the comment line “# Name:” followed by your “meaningful” name.
3: Create a View of your Loaded Data
Once loading is complete, your new project will appear in the GBench Project Tree (located in the upper left of the GBench pane) that contains the GWAS data. Each study will generate two nodes in the data tree: one node has a green arrow icon; the other node has a blue mountain icon: There is no discernable difference between the data contained within these nodes.
- a.
Double click on the green arrow icon for the data for your study.
- b.
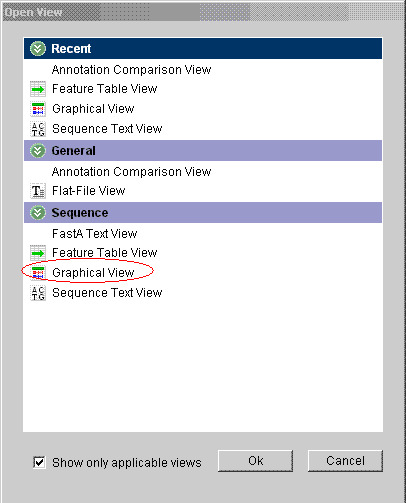
In the “Open View” dialogue box that appears, choose “Graphical View”:
c. In the “Graphical View” dialogue box that appears, select the data of your choice and click “OK”: The load operation may take a moment to finish.
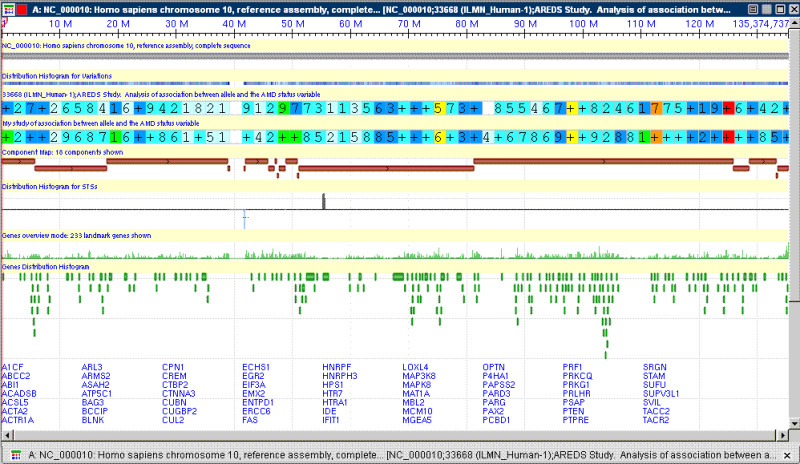
d. The view that is generated should look similar to the following if you have both a track of your private data and a track of public data displayed:
(05/22/08)
- Using GST to Study Genome Wide Association (GWAS) Data - SNP FAQ ArchiveUsing GST to Study Genome Wide Association (GWAS) Data - SNP FAQ Archive
Your browsing activity is empty.
Activity recording is turned off.
See more...