Summary
The NCBI Bookshelf at the National Library of Medicine is an online archive of books and documents in life science and healthcare. Its growing collection comprises over 5,000 titles, the majority of which are stored as full text XML. In the fall of 2014, Bookshelf began work to adopt the Book Interchange Tag Suite (BITS) DTD, replacing the NCBI Book Tag Set Version 2.3 as its XML format of choice. It became immediately apparent that Bookshelf could not simply perform a one-time “switch” to BITS. It needed to support the new schema alongside the old one. The complexity of the project would have required Bookshelf to focus so much energy on the transition to BITS, thereby bringing regular production workflows to a complete halt. This was particularly inconceivable, as Bookshelf judged the benefits of adopting BITS to be mostly long-term rather than immediate. Released only in December 2013, BITS was still very new. While there was no doubt that the format is superior to the NCBI Book DTD, the prospect of further revisions to the Tag Suite cautioned against acting too quickly. Therefore, adoption of BITS was conceived as a longer term project of small, incremental steps designed to neither disrupt the regular production cycle nor consume all resources. By the time version 2.0 of BITS was released in December 2015, Bookshelf had the ability to load, render, and index books tagged as per the BITS. A number of in-house XML converters were updated to output BITS, and the first titles in BITS were released. While the majority of new content was still tagged as per the NCBI Book Tag Set v2.3, Bookshelf now had a solid foundation to complete adoption using the new version of BITS. By the end of 2016, all workflows had switched to BITS, including Bookshelf’s Word authoring program, external vendors providing BITS XML, and over 20 in-house XML converters. This paper describes Bookshelf’s experience in adopting BITS: the challenges Bookshelf faced, the solutions it developed, and the lessons learned along the way. Special emphasis is placed on issues related to markup and XML conversion.
Background
XML at the NCBI Bookshelf
The NCBI Bookshelf, an online archive of books and documents, contains over 5,000 full text books and reports, to which about 70 titles are added each month [1,2]. Until 2015, all of those documents were tagged as per the NCBI Book Tag Set, Version 2.3 [3,4]. Bookshelf receives books data in three main formats:
- 1.
XML. Participants who wish to make their content publicly available via Bookshelf submit XML tagged in a mutually agreed upon DTD. Bookshelf converts the XML to its schema of choice using XSLT converters. Currently, active converters handle about 12 to 15 different DTDs and data from more than 20 submitters.
- 2.
PDFs. Bookshelf converts between 4,000 to 5,000 pages of content per month from PDF to XML using external vendors. These vendors produce the XML exactly to Bookshelf’s specifications.
- 3.
Microsoft Word documents. Bookshelf supports an XML authoring program. It enables participants to author content in MS Word, which is converted to XML using Inera’s eXtyles product. It encompasses a comparatively small subset of continuously updated content.
Once in XML format, all content follows roughly the same production cycle (see Figure 1):
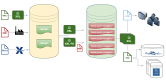
Figure 1.
Illustration of the books XML dataflow at NCBI (simplified).
- 1.
The XML is stored with images, PDFs, and other supplementary data files in a Content Management System (CMS). Each book or report is tagged as a <book> document.
- 2.
From the CMS, the content is loaded to a production database. Ingest involves splitting the single <book> document into several chapters or chapter-like documents: <book-part>s as per the NCBI Tag Set and <book-part-wrapper>s in BITS. The conversion also generates a Table of Contents that links the parts together. For a subset of titles, Bookshelf creates PDFs using XSL-FO.
- 3.
These smaller chapter documents are served from the production database as web pages to the end users. Rendering to HTML occurs at request time. Content delivery is online only, and the Bookshelf does not produce print versions of the books.
- 4.
The Bookshelf further utilizes and exchanges the XML. Examples include submission to other repositories such as PubMed, or to Entrez, NCBI’s search and retrieval system. If the participant permits it, then the XML itself, and not just respective renditions, is made publicly available on NCBI’s FTP servers.
XML is thus absolutely central to Bookshelf’s operation. The fact that all content is archived in a single, durable format forms the basis for leveraging it and adding to it. The NCBI Book Tag Set was designed specifically with the needs of the Bookshelf in mind. While those needs evolved over time, it had served Bookshelf rather well for almost a decade. Adopting a new DTD was therefore a consequential decision.
Rationale and Approach for Switch to BITS
Bookshelf began the discussion of adopting BITS [5,6] in 2014, soon after version 1.0 was released. The decision to adopt BITS was fairly easy:
- 1.
The content model was deemed superior to a degree that justified adoption; many of the features of BITS, especially metadata and structural elements, had been sorely missed when working with NCBI Tag Set.
- 2.
Based on the Journal Article Tag Suite (JATS) and evolving along with it made BITS a more resilient model for the future. Bookshelf had already skipped version 3.0 of the NCBI Book Tag Set, having completed a migration from the in-house book DTD only a few years earlier. Adherence to NCBI Book DTD v2.3 would result Bookshelf missing out on important current and future developments.
- 3.
Many objects, especially lower level elements such as tables, paragraphs, but also sections or book-parts, did not change radically compared to the NCBI Book DTD. Thus a relatively high degree of familiarity with BITS could be assumed, both in terms of supporting technologies as well as for staff or vendors handling the XML.
- 4.
Given the success of JATS, Bookshelf was optimistic that other organizations would also see the benefits of BITS, which would eventually find fairly widespread use. Adopting BITS would then make data exchange with other parties easier and thus cheaper.
The disadvantages of moving to BITS, however, were considerable also:
- 1.
There was no tangible, short-term benefit, or any sort of improvement that Bookshelf users would immediately feel. On the contrary, it meant a burden on some consumers of Bookshelf’s raw book XML data, because they now had to cope with a new DTD.
- 2.
It was clear from the outset that the cost of adoption was very high; despite all the similarities between BITS and the NCBI DTDs, a major problem remained. The root element of the XML documents as stored in the production database had to change from <book-part> to <book-part-wrapper>. This alone meant that virtually every process involving XML had to be updated. Well over one hundred XSL transformations, programs, scripts, and supporting tools needed to be altered. Touching every component of the XML ecosystem made this a costly endeavor that came with great risks, as plenty of opportunities for new bugs arose.
- 3.
Despite all optimism regarding the future of BITS, the tag set was still very new. Adopting it early meant betting on its success. At a minimum, revisions were to be expected, and there was a risk that work to support BITS version 1.0 elements would be futile.
Given the complexity of the project, Bookshelf could not conceive of a way to perform a one-time “switch” to BITS. To develop a parallel system and then migrate all the legacy data into it seemed unrealistic. It would have either consumed all available resources, bringing regular production to complete halt for a considerable amount of time, or taken too long to be a manageable project. Additionally, there was no active external pressure or immediate requirement to adopt BITS. There was, however, the expectation that Bookshelf would continue to meet its regular production capacity. A switch to BITS should not impact users or the growth of Bookshelf negatively. Adoption was therefore conceived as an incremental process. Rather than building a new “BITS system” and migrating data into it at the end, existing components were modified step by step to support BITS alongside the NCBI Book DTD. The benefits of this approach were apparent:
- 1.
It allowed for enough resources to support the main production processes and to address more pressing and time-sensitive issues when needed.
- 2.
It allowed for the tag set to further mature before playing a central role in Bookshelf’s operation.
- 3.
Problems and challenges would emerge incrementally and would thus have less of an impact on the project and on the operation as a whole.
The main problems with the approach were:
- 1.
For the foreseeable future, Bookshelf would need to live with a more complex XML infrastructure supporting two DTDs, as BITS would not fully replace the NCBI Tag Set.
- 2.
The project of adopting BITS would be at constant risk of neglect at the expense of other priorities.
Lacking a real alternative, Bookshelf decided it had to tolerate the first problem. The risk of constant delays could be mitigated by committing to milestones and setting priorities with some flexibility. Delays due to other projects would be deemed acceptable as long as they did not lead to cost-increasing inefficiencies. The work to support BITS at NCBI thus started in earnest in the fall of 2014.
Phases of adopting BITS
Basic NCBI Book to BITS converters
The first step of the project was to write a converter that transforms XML in the NCBI Book DTD to BITS. Bookshelf could not commit to a process that would obligate it to convert all books legacy data to BITS. It did anticipate, however, that some content would need to be converted, and wanted to be able to efficiently migrate any book at any time. It quickly became clear that more than a simple converter was needed, since a number of different transformation scenarios had to be covered:
- 1.
Bookshelf would need to handle legacy <book> documents, marked up per its specific tagging flavor.
- 2.
Bookshelf stores whole book documents in its content management system and smaller book-parts in its production database. Therefore, it also needed a converter to chunk a BITS <book> into several smaller documents, the <book-part-wrapper>s.
- 3.
The Bookshelf anticipated eventually combining these two conversions, chunking <books>s in the NCBI DTD directly into BITS documents during ingest. This would reduce maintenance cost, as such a chunking transformation is a lot more complex than it may appear. Crosslinks need to be resolved, footnotes and floating objects may need to be re-located, or hanging content needs to be handled. Generating only BITS documents meant that Bookshelf could retire the old code that produced chunks in the NCBI Book DTD.
- 4.
Many content providers who deposited <book> or <book-part> XML used the NCBI Tag Set. Those external submissions could come in a different DTD version, 3.0, or in a completely different tagging style. Thus, a whole different set of conversion challenges were to be expected.
Very different conversion processes would thus share basic elements of NCBI Book to BITS conversion. For example, <custom-meta-wrap> needed to be turned into <custom-meta-group> in all scenarios. Bookshelf handled this by producing modular XSLTs; dedicated entry-point files would import a mix of respective common modules (see Figure 2). The main challenge was to share and re-use code, but to avoid coupling unrelated processes too strongly and overcomplicating individual conversions. Writing the transforms of course helped in becoming more intimately familiar with the BITS. Completing the first two converters enabled Bookshelf to very quickly generate any test data that was needed for the next phases.
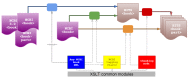
Figure 2.
Different conversion scenarios and use of common modules.
Loading, Rendering, Secondary Processes, and Tools
Another phase and the first major milestone of adopting BITS was to enable loading and rendering of the BITS XML data. The ability to store, view, and exchange the new XML was the main requirement for public release of the first BITS books.
The loader not only saves the XML documents as a blob to a database, but also extracts information from the XML for storage in SQL tables. Therefore, it needed to learn the additional location of key metadata, such as DOIs or dates. Even though most element names remain the same between the NCBI Book DTD and BITS, almost all XPaths to the data of interest changed and all exported fields had to be remapped. Adjusting the loader also meant finalizing some longer term tagging decisions; it is obvious that DOIs in BITS should be stored as book-id[@book-id-type = 'doi'] and as book-part-id[@book-part-id-type = 'doi']. Other decisions, however, were less clear. Where, for example, should the main internal id of the document be placed for the purpose of loading? Should the loader use the book-part-id element or the ID attribute of the book-part-wrapper? How should the pub-type attribute values of our dates map to date-type and publication-format? Finalizing these decisions also meant cycling back to the converters and making necessary adjustments.
Next came work on the HTML renderer, a rather complex XSLT library of over 200 stylesheets. At this stage, the intention was to make existing BITS content viewable and to cover a large percentage of rendering features. It was not the goal to address every last detail found in select books. There was also no intention to support rendering for every new BITS structure, for example for indices, for question and answers, or for publication history events. Fine-tuning and especially enhancing the HTML render was seen as an ongoing process, and the current goal was limited to enable release of first books in BITS. As expected, it was the new root element that caused the most work. It made the task a rather tedious exercise of combing through a lot of code and updating simple XPaths.
Having the ability to store BITS in the production database also meant that secondary processes could be adjusted in a next step. This included a number of QA tools running specific checks or reports. Secondly and more importantly, all processes to further utilize and exchange the production XML needed work. Examples include transformations to produce XML for submission to PubMed, for indexing in NCBI’s search and retrieval system, or for cataloging in the NLM Catalog. Again, work consisted mainly of updating XSLTs to support BITS. Overall, the updates were more limited in scope than rendering, because many of the processes and conversions were primarily concerned with metadata. The system was in principal ready to release BITS data once this step was completed.
XML Submissions and Consolidation
The first content released in BITS was data submitted in XML and converted in-house. Upgrading the XSLT-based converters to output BITS was more labor-intensive than simply instructing vendors to supply BITS. However, the process was completely under Bookshelf’s control. It was possible to move the 20 different projects gradually and incrementally to BITS, to learn lessons and make corrections quicker. Temporarily rolling back problematic developments was less consequential. This is why XML submissions were moved to BITS first. Upgrading a converter usually proceeded as follows:
- 1.
For a number of test cases, the production output in the NCBI Book DTD v2.3 was converted to BITS using the above-mentioned generic converter, which by now was sufficiently employed and tested. This provided a baseline for testing.
- 2.
The project-specific XSL transforms were updated and the BITS output compared against the baseline. Adjustments were made until the BITS output and the baseline matched or until the differences were expected and acceptable.
- 3.
The XSLT code and the source schema were reviewed for tagging in which direct BITS output could contain higher quality markup.
- 4.
For more complex or difficult projects, staff performed additional human QA.
As content was moved to BITS, development continuously cycled back to previous stages in the process in order to perform fixes and adjustments. Bookshelf permanently improved and re-fined the processes, especially rendering. In the fall of 2015, about a year after work had started, Bookshelf released the first BITS book. As anticipated, other priorities had intervened, and actual time spent on BITS amounted to approximately two months. The end of 2015 also saw the release of BITS version 2.0. Bookshelf again iterated over previous steps in order to accommodate this later version. Depending on the nature of the changes, this could have invalidated some of the previous work. However, this was not the case, and the adjustments were fairly quick and easy.
Word Conversion, PDF build, and Conversion Vendors
Switching many XML converters and releasing a growing number of titles in BITS provided confidence that the system was sufficiently stable and tested. It was now appropriate to ask external partners and vendors to switch to BITS. Moving the Word authoring program and Inera’s eXtyles build to BITS constituted a real, permanent switch. It was not practical in the long run to support both the creation of NCBI Book XML and BITS. Legacy documents existed for which future updates were to be expected. However, the overall number was limited and the one-time investment of migrating all content to BITS was cheaper than the recurring cost of two word converters, one for BITS and one for the NCBI Book DTD. More importantly, the Word authoring program was used for many projects to add new chapters to existing books and resources. If new chapters were to be in BITS, then old ones had to convert to BITS also. This left very few titles that could conceivably remained tagged as per the NCBI Book DTD. Bookshelf thus decided to convert all Word authored content to BITS.
Bookshelf maintains an in-house PDF build based on XSLT-FO. It needed to decide whether to update the existing code to support BITS or whether to create a brand new set of XSLT-FO. The existing code was by now very old. Most of the PDFs were built for Word-authored content, which had to be converted to BITS in any case. The Bookshelf therefore decided to also convert the remaining titles, to create a new PDF build exclusively for BITS, and to retire the old code handling the NCBI Book DTD. While the creation of the XSLTs came at a considerable cost in development and QA, future maintenance work was expected to be much less expensive. More importantly, improved PDF versions of the content directly benefited Bookshelf’s users and thus yielded immediate return on the investment.
In last stage of the process, Bookshelf asked third-party taggers to switch to BITS. This was one of the smoothest steps in the process. Bookshelf converted sample documents previously supplied in the NCBI DTD to BITS. The output was reviewed to ensure that the markup was appropriate given a direct tagging scenario. Simultaneously, Bookshelf updated tagging documentation and file submission specifications. BITS was now officially established as the XML schema of choice. This meant that not just vendors, but also other partnering organizations and participants, started to supply XML in the new DTD. By then end of 2016, NCBI had completed the adoption of BITS.
Markup Issues and Lessons Learned
Bookshelf learned many lessons over the course of adopting BITS. The focus of this section is on markup and data issues. By no means does it constitute an exhaustive list of all differences between the NCBI Book DTD and BITS. The section only highlights select conversion problems and XML-related lessons that NCBI encountered in adopting BITS.
Top Level Structure
One of the most consequential changes for Bookshelf was that BITS changed the root element and top-level structure of the documents as NCBI stores them in its production database.
In terms of conversion, this does not present an interesting challenge. A few lines of code will achieve this change even in a language as verbose as XSLT. Yet it cannot be overstated that this difference in top-level structure is highly consequential. As mentioned, virtually every script, program, and process at NCBI had to be adjusted to accommodate the new structure. This meant a lot of “busy work” wading through code and making simple adjustments. Having to perform a very large number of very simple changes tends to lead to a lot of bugs as some details are inevitably overlooked. Only extensive testing and repeat iterations can guarantee that all necessary adjustments were completed. Bookshelf foresaw this problem prior to starting the work, and all expectations were fully met. The new root element was the main challenge in adopting BITS. It makes moving from the NCBI Book DTD to BITS a project very different from switching the NLM Journal Article Suite to JATS where top-level structure remains the same.
Enhancing mark-up
Converting NCBI Book XML to BITS presents opportunities to improve the markup of some structural and textual elements even without human intervention. Front-matter sections, for example, can be tagged with more specific element names, such as <preface> or <dedication>. Automatic conversion can enhance mark-up by creating these elements based on the title:
The new <index> and <toc> elements provide another opportunity for improved markup. It is not uncommon for respective content to be tagged as lists:
This example already bears potential problems: <toc> is not allowed in all the places that a list can be found in. The former is a section-level element, while the latter is a paragraph-level object. Attempting to improve markup of legacy data can thus quickly lead to a whole set of unexpected problems. Even a seemingly simple enhancement to create elements based on titles has the potential to snowball, if, for example, content comes in different languages. NCBI focused quite strictly on converting data, not on enhancing it. In order to avoid mission creep, it used new elements or structures only when they were very obvious, easy, and safe.
Citations
Like JATS, BITS, as well as the NCBI Book Tag Set 3.0, have two ways to tag citations: As <element-citation>, which contains only descriptive elements, and as <mixed-citation>, which contains also untagged text. [7] The former leaves formatting completely up to the rendering application, while the latter intends to preserve the citation exactly as found in the source. Precursor elements in version 2.3 and earlier are <citation> allowing untagged text and <nlm-citation> as the element-only model.
Bookshelf employed only <citation> for tagging bibliographic references. Both functions of element-citation and mixed-citation were combined in one element; it could contain only the individual components without any punctuation and spacing, but it could also contain text if necessary. The rendering application had to detect which flavor is used and to what degree the citations needed formatting. Using <citation> this way and ignoring <nlm-citation> is not atypical.
Conversion of <citation> to BITS is not very difficult. When it contains text or formatting elements, it becomes <mixed-citation>. When it consists only of citation elements, then it becomes <element-citation>. Focusing merely on the mechanics of conversion, however, is misguided for an element as prominent as a reference citation. Faced with new elements, citation tagging should be revisited for new data. Shall current tagging simply transfer to <mixed-citation>? Should tagging standardize around <element-citation>? Or shall there be a mix of the two elements? Legacy data likely has to be supported either way, but moving to BITS presents an opportunity to reevaluate citation tagging and possibly change course for new content. Focused primarily on conversion, Bookshelf realized this quite late in the process; it was only after asking vendors to switch to BITS that Bookshelf consciously rethought its citation-tagging strategy and decided on exclusive use of <mixed-citation> for new content. There are some tagging questions that can probably be deferred to a later stage when switching to BITS, for example whether and how to employ the new question and answer model. Citation mark-up is not one of those things. Focusing on conversion is right, but it must not lead to overlooking major strategic tagging questions.
List IDs
Version 3.0 of the NCBI Book Tag Set corrected a bug present all in earlier versions. The data type of the id attribute on list and list-item was defined as CDATA and not as ID. This means that duplicate IDs on those elements may exist in legacy data. The attribute may also contain syntax disallowed for the ID type, for example if it starts with a number. When converting to BITS such cases flush up as validation issues. Many instances of problematic list IDs had accumulated over the years in Bookshelf data. Addressing the IDs themselves can be done easily during conversion, for example by adding an alpha prefix to an integer value or by adding suffixes to make duplicate IDs unique. However, it is more problematic to identify possible pointers referencing the ID that need to be changed accordingly in order to avoid broken links. Pointers could come, for example, in the form of a related-object:
Or, they may come in the form of an xlink:href:
The more controlled general tagging practices are, the easier it is to deal with this issue. For an archive with multiple data suppliers like Bookshelf, the task can quickly get problematic and involve broader data surveys. Since pointers to lists are overall rather rare, this can become the unfortunate task of finding a needle in a haystack. Despite being aware of the bug, Bookshelf did not anticipate this problem at all.
MathML
Both the NCBI Book DTDs and BITS allow equations or other mathematical expressions to be captured as graphics, text, TeX, or LaTeX, or in the Mathematical Markup Language (MathML). All versions of the NCBI Book Tag Set include MathML Version 2.0 in their standard distribution, and BITS includes Version 3.0 [8,9]. Dealing with MathML is involved and therefore expensive, as Usdin rightly emphasized [7]. It is important to realize that switching from the NCBI DTD to BITS also means switching to Version 3.0 of MathML. That version is not backward compatible and is much stricter than its predecessor. Up-conversion is easy in theory. In practice, however, tagging errors and data issues must be factored in. Bookshelf had to deal with the fact that problems in MathML2 tagging, some of which were even forgiven by rendering applications, became apparent as DTD validation errors when converting to BITS. The <msup> element, for example, should consist of exactly two children: the base and the superscript. MathML3 enforces this, while MathML2 does not. Bookshelf encountered a number of incorrect <msup>s, <msub>s, and <msubsup>s after upgrading its converters to BITS. While flushing up data issues is generally a welcome development, this added an unexpected cost. For the most part, the errors had to be fixed manually, which was difficult and slow. The books content at Bookshelf does not include a large number of equations. MathML is therefore not a focus of concern and expertise. When planning for BITS, MathML was not a consideration. However, Bookshelf discovered that there was still enough MathML to cause some headaches in upgrading to version 3.
Alternatives
Both the NCBI Book Tag Set as well as BITS allow for grouping or linking of alternative representations of an object, such as a table or a graphic. The mechanisms to do so, however, are very different. The NCBI Tag Set, up to version 2.3, employs the alternate-form-of attribute and uses the ID/IDREF mechanism to link objects. The emphasis is on identifying a primary format. Version 3.0 and BITS, in contrast, wrap the different formats within <alternatives>. Marking a primary object is not a concern, as objects are grouped and not linked. Those two fundamentally different mechanisms make the handling of alternatives difficult, because alternate-form-of can reference any element with an ID, while <alternatives> restricts potential partners to a much smaller set of allowed child elements. A PowerPoint version of an illustration, for example, could link in to a figure in the NCBI Tag Set. In BITS, however, it would need to be paired with the figure’s graphic because <fig> is not allowed under <alternatives>:
This conversion is problematic if the slide included the whole figure and was thus really more than just an alternative to the graphic. It would be even more difficult, if the figure contained multiple graphics. Grouping several graphics together with one media element would suggest that the graphics were alternative forms of each other. Not only are there more options for linking alternatives in the NCBI Tag Set than in BITS, it is also possible to link “remote” objects in terms of document order, for example linking a graphic in the narrative to the corresponding plate in an appendix at the end of the book. If alternate-form-of is used heavily in the data, then this will likely present the most difficult conversion challenge when moving from the NCBI Tag Set 2.3 to BITS. It is possible that proper conversion cannot be achieved automatically and without human intervention.
Conclusion
Many of the conversion problems described above are not really a result of moving to BITS. NCBI encountered issues with list IDs, alternatives, or citation tagging because it skipped NCBI Tag Set version 3.0. The underlying lesson transcends any specific DTD version. Starting the project and gauging the scope of work, NCBI was confident that features like MathML, list IDs, or alternatives would not be a problem. It deemed a more methodological data survey unnecessary. That assumption was erroneous, in that there were sufficient problems to slow down the process of adopting BITS. This extends to general data issues and tagging errors completely unrelated to any particular DTD migration. Typically the process of converting at least some legacy data will flush up unexpected problems. The quality of data is probably not as good as one thinks it is—what you think you know about your data is probably wrong! Under a more rigid schedule, this has the potential to become a serious problem. Planning a project such as the adoption of a new DTD should allow for some surprises. In this regard, NCBI’s approach of incremental adoption was the right one. Having converters that could generate test data, approaching external partners only after having gained considerable experience, and applying fixes to previous stages after each new step prevented any larger setbacks or costly consequences.
Acknowledgements
We wish to credit our colleagues at the National Center for Biotechnology Information for their contributions to the Bookshelf project, especially members of the Bookshelf team, Sergey Krasnov, Jeff Beck, Karanjit Siyan, and the PMC team of developers. We thank Jim Ostell, Chief, Information Engineering Branch, NCBI, and David Lipman, Director, NCBI, for their vision and support of the Bookshelf project.
References
- 1.
- Hoeppner M, Latterner M, Siyan K. Bookshelf. 2013 Mar 18 [Updated 2013 Nov 4]. In: The NCBI Handbook [Internet]. 2nd edition. Bethesda (MD): National Center for Biotechnology Information (US); 2013-. Available from: https://www
.ncbi.nlm .nih.gov/books/NBK169440/ - 2.
- Hoeppner MA. NCBI Bookshelf: books and documents in life sciences and health care. Nucleic Acids Res. 2013 Jan;41(Database issue):D1251–60. 10
.1093/nar/gks1279 doiPubMed Central PMCID: PMC3531209. [PMC free article: PMC3531209] [PubMed: 23203889] - 3.
- Latterner M, Hoeppner M. Bookshelf: Leafing through XML. 2010 Oct 12. In: Journal Article Tag Suite Conference (JATS-Con) Proceedings 2010 [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2010-. Available from: https://www
.ncbi.nlm .nih.gov/books/NBK47113/. - 4.
- NCBI Book Tag Set Version 2.3. Available from: https://dtd
.nlm.nih.gov/book/2 .3/index.html. - 5.
- Book Interchange Tag Set. Available from: https://jats
.nlm.nih .gov/extensions/bits/ - 6.
- Beck J. What JATS Users Should Know about the Book Interchange Tag Suite (BITS). In: Journal Article Tag Suite Conference (JATS-Con) Proceedings 2013 [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2013. Available from: https://www
.ncbi.nlm .nih.gov/books/NBK159737/ - 7.
- Usdin BT. So You Want to Adopt JATS. What Decisions Do You Need To Make? In: Journal Article Tag Suite Conference (JATS-Con) Proceedings 2016 [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2016. Available from: https://www
.ncbi.nlm .nih.gov/books/NBK350380/ - 8.
- World Wide Web Consortium (W3C) Recommendation, Mathematical Markup Language (MathML) Version 2.0 (Second Edition). 21 October 2003. Available from: http://www
.w3 .org/TR/2003 /REC-MathML2-20031021/ - 9.
- World Wide Web Consortium (W3C) Recommendation, Mathematical Markup Language (MathML) Version 3.0 (Second Edition). 10 April 2014. Available from: http://www
.w3 .org/TR/2014 /REC-MathML3-20140410/
Publication Details
Author Information and Affiliations
Authors
Martin Latterner and Marilu Hoeppner, Ph.D.Affiliations
Copyright
Publisher
National Center for Biotechnology Information (US), Bethesda (MD)
NLM Citation
Latterner M, Hoeppner M. Adoption without Disruption: NCBI’s Experience in Switching to BITS. In: Journal Article Tag Suite Conference (JATS-Con) Proceedings [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2017.

