NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Journal Article Tag Suite Conference (JATS-Con) Proceedings 2019 [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2019.
Europe PMC, one of the leading databases for life sciences literature, currently contains 3.0 million full-text journal articles tagged in JATS XML, as provided by PubMed Central. Integration of this literature with related research data is a key part of Europe PMC’s mission to support data discovery and re-use. The standard format provided by JATS is central to this endeavour, especially in section tagging and text-mining efforts. We present the methodology and analysis of two recent core developments in these areas: section tagging of data availability statements, and a text-mining project to extract funding organisation names and grant identifiers from acknowledgement and funding sections.
Introduction
Europe PMC (https://europepmc.org) is a comprehensive repository that provides access to worldwide life sciences journal abstracts and full-text articles, preprints, books, patents and clinical guidelines, and offers users advanced tools for search, retrieval, and interaction with the literature. As a partner in PubMed Central International (PMCi) (https://www.ncbi.nlm.nih.gov/pmc/about/pmci), Europe PMC receives full-text JATS XML content from PMC on a daily basis, as well as processing and sharing author manuscripts funded by the Europe PMC funders group (https://europepmc.org/Funders/) comprising twenty-nine international research funders. Europe PMC currently contains 3.0 million JATS XML articles, in addition to 2.3 million articles tagged using pre-NISO NLM DTDs. XMLs for full-text articles published as part of the PMC Open Access (OA-PMC) subset (~2.4 million as of April 2019) are available for bulk download on the Europe PMC FTP site (https://europepmc.org/ftp/oa/) and are served in full via RESTful and SOAP APIs.
A key part of Europe PMC’s mission is to create solutions that make it easier for users to effectively navigate the data-rich literature. Towards this ambition, we report the methodology and results from two recent core developments. Firstly we discuss the section tagging of data availability statements (DASs) within the full-text XML content to facilitate keyword searching via the Europe PMC Advanced Search tool. This allows us to investigate trends in data availability statements, for example by looking at how DAS content correlates with data accessibility. Secondly, we report on a significant expansion of the Europe PMC text-mining pipeline for funding attributions to encompass almost all of the Europe PMC funding group organisations.
Data availability section tagging
Background
To promote reproducibility and re-use of research datasets, many scientific journals have introduced data availability statements (DASs) – a distinct section of the article containing data access guidelines. A DAS should include information on both how the data directly underpinning the study can be accessed (e.g. by providing a URL, DOI, or other persistent identifier) and under what conditions they can be reused (e.g. a Creative Commons license, Data Sharing agreement, or other third party restriction). In an effort to improve access to scientific data reported in research articles and to facilitate analysis of data sharing practices, we have extended the Europe PMC section tagging pipeline to identify DASs contained within the full-text XML received from PubMed Central (PMC).
Granular searching within distinct full-text article sections has been a key feature of Europe PMC since 2014 [1]. Briefly, we employ a rule-based section tagger to categorise <sec> elements within the XML based on the contents of the associated <title> element (e.g. <title>Methods</title> is categorised as type “Materials&Methods”). We additionally categorise specific, non-<sec> elements such as <funding-statement>, and implement this tagger as part of the Europe PMC full-text XML ingest pipeline. A total of 16 pre-selected categories are utilised, with the category “Other” used for sections that do not meet any of the categorisation rules.
In this recent work we created an additional categorisation rule for DASs based on a manual analysis of previously uncategorised sections with <title> elements containing the word “data”. Table 1 lists the ten most common combinations of <title> element contents and XML path based on the OA-PMC subset as of June 2018. The full dataset (comprising combinations with at least 50 occurrences in the subset) is available in Supplementary File 1.
Table 1
The ten most frequently occuring combinations of <title> content containing the word “data” and the corresponding XML path within the OA-PMC subset as of June 2018.
| Title | XML path | Frequency |
|---|---|---|
| Data Availability | article:front:notes | 90,928 |
| Data accessibility | article:back:sec | 2,694 |
| Data Availability | article:back:sec:fn-group | 2,580 |
| Data | article:body:sec | 2,265 |
| Availability of supporting data | article:body:sec | 1,593 |
| Major datasets | article:back:sec:sec | 1,074 |
| Database survey | article:body:sec | 986 |
| Extended Data | article:body:sec | 851 |
| Data availability | article:body:sec | 795 |
| Extended Data Figure 1 | article:body:sec:SecTag:fig | 689 |
Rule creation and front-end implementation
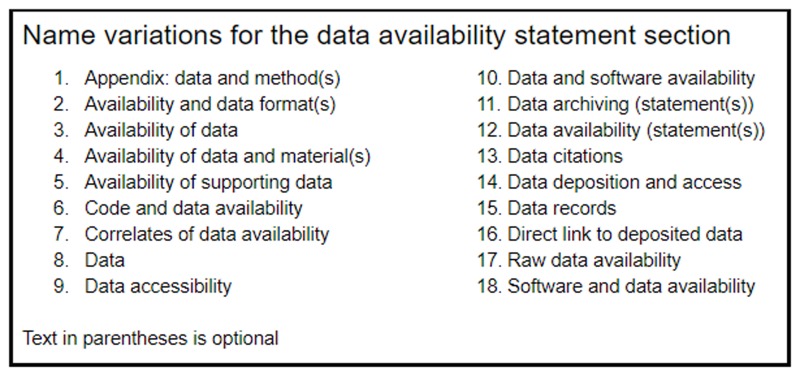
We see significant variation in both the titles used and paths within the XML for DASs. For example, the title “Data availability” appears most frequently within the <notes> element in the front matter, but can also be located within a <fn-group> in the back matter, as a stand-alone <sec> element in the body, or as a second-level <sec> element (e.g. contained within the “Methods” section, see Supplementary File 1). Further variations in the title of the DAS also hamper discoverability across multiple journals. Accordingly we developed a comprehensive regular expression (regex) to encompass the title variation, and implement this regex in a manner that is insensitive to the XML path. The corresponding title variations that are identified by the new rule are shown in Fig. 1. For further details, please refer to the section tagger source code written in the Perl scripting language, which is publicly available at: http://europepmc.org/ftp/oa/SectionTagger/.
Fig. 1
A list of all possible <title> name variations that match the DAS rule. Words and letters enclosed in parentheses are optionally included.
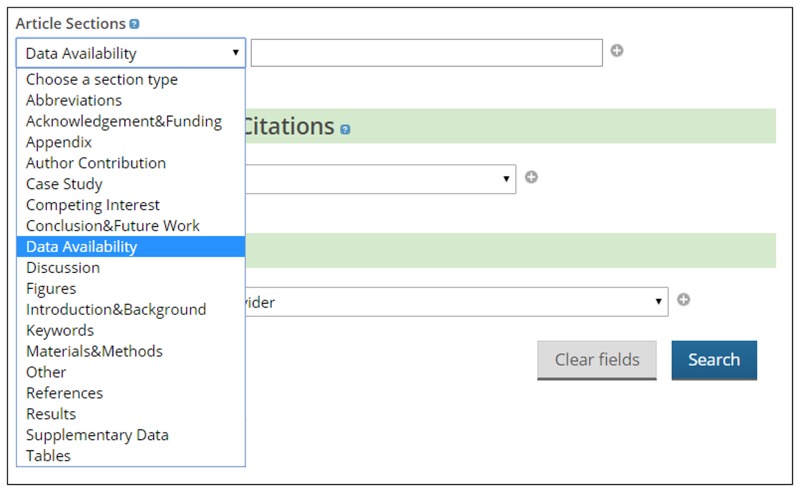
The DAS tagging is implemented on the front-end as a search filter within the Europe PMC Advanced Search (https://europepmc.org/advancesearch). This provides users with a drop-down menu to restrict text searching of full-text articles to within a particular section of interest as categorised by the section tagger (Fig. 2). This can be combined with other elements within the Advanced Search, such as publication dates and open access status, through the use of Boolean operators. This feature can also be accessed directly from the main search bar using the syntax, DATA_AVAILABILITY:keyword. At the time of writing, a total of 288,000 publications in Europe PMC contain a dedicated DAS.
JATS4R recommendation
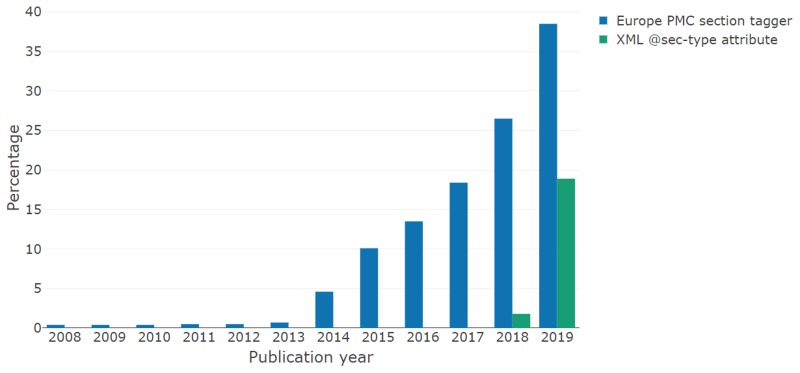
A much simpler alternative to the manual regex approach outlined above would be to identify <sec> elements containing the @sec-type=“data-availability” attribute, in line with the JATS4R recommendation for DASs [2]. Fig. 3 shows the percentage of all full-text articles in Europe PMC (open and closed access) published each year since 2008 that contain a <title> element matching the developed rule, as well as the percentage with a <sec> element with the JATS4R-recommended attribute. As the recommendation was only formally published on 28 December 2018, it is unsurprising that we see limited use of the attribute in articles published in 2018, and negligible usage prior to this. Moreover we see approximately double the coverage of DASs following this rule-based approach based on the section titles compared to presence of the attribute for articles published between January and March 2019. Accordingly we cannot rely on the presence of this attribute for broad coverage of DASs at present, however, the recent uptake is very encouraging.
Data DOIs within DASs
Moving the investigation into the content of DASs, one interesting avenue to explore is where the data underpinning articles are hosted. Within the life sciences domain, accession numbers are routinely used as persistent identifiers for particular datasets hosted in a specialised database. For example, UniProt [3] and ENA [4], hosting protein and nucleotide sequences respectively, have accession numbers with multiple complex patterns. Surfacing these accessions forms part of Europe PMC’s core text-mining and annotations pipeline [5]. For very standardised data DOIs however, we can use a single, relatively simple regular expression (?i)(10[.]\\d{4,9})(?=/)(?=[-._;()/:A-Z0-9]+) to identify DOI citations within DASs. For this initial analysis, we extracted DOI prefixes from DASs in articles in the OA-PMC subset published between January and March 2019. 6.2% of DASs in the studied dataset contained one or more DOIs.
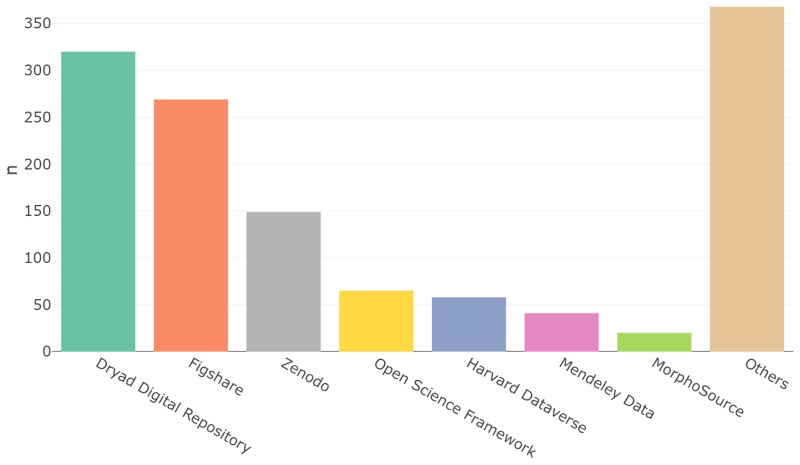
These DOI prefixes were then queried against the DataCite and Crossref REST APIs to retrieve the corresponding registrant names. As some registrant names (particularly publishers) were found to have multiple associated prefixes, these were grouped together prior to tallying the number of DOI citations associated with a particular registrant. Multiple citations to a DOI prefix within a single DAS were included in the citation count. The results plotted in Fig. 4 show the top seven most cited repositories, with all remaining repositories grouped under “Others”. The full list of repositories with the number of citations can be obtained via the R Markdown document in Supplementary File 1. The most cited repositories are perfectly aligned with the “generalist” data repositories as recommended by the journal Scientific Data published by the Nature Publishing Group [6].
Fig. 4
The number of DOI citations in DASs from articles in the OA-PMC subset published between January and March 2019.
We find that the most frequently cited repository is the Dryad Digital Repository with DOI prefix 10.5061, closely followed by Figshare. The most popular non-generalist repository in the dataset is MorphoSource, a data repository for 3D computerised tomography (CT) scans hosted at Duke University. It is worth noting however that this represents only a single article which generated and cited all 20 of the entries. As is clear from the number of citations to repositories outside the top seven (i.e. the “Others” bar in Fig. 4), there are a large number of infrequently cited repositories, suggesting a rather distributed system. In particular we see a significant number of university data repositories: 40 of the 128 distinct repositories appear to be university data repositories.
Categorising DASs using regular expressions
Several recent papers have categorised DASs for analysis of the availability and even reusability of the data. Federer et al. coded and analysed nearly 50,000 DASs from articles published in PLOS ONE between March 2014 and May 2016 using a joint automated and manual approach [7]. While compliance with the policy was shown to increase, only ~20% of DASs indicated that data were deposited in a public repository (the preferred method according to PLOS’s policy). A substantial increase in DASs was similarly noted in the Elsevier journal Cognition following the introduction of a mandatory open data policy [8]. However, the authors raised concerns regarding reusability of the data. McDonald et al. looked at clinical studies published in the British Medical Journal and found that 63% of studies published between 2015 and 2017 included a DAS that implied that data used in the study could not be shared [9]. Likewise, many DAS templates provided in publisher guidelines involve a lack of data, data already being available in the article/supplementary files/from authors on request, or restrictions to data access (see, for example, DAS templates from Taylor & Francis [10]).
While it would be impractical to manually categorise individual DASs at the scale of the OA-PMC subset (~2.4 million articles), we can use carefully constructed regular expressions to give a reasonable initial indication of the nature of a DAS. As an initial survey, we used the R “tidytext” package [11] to tokenize the DASs contained with OA-PMC articles published in 2019 to date. The most frequently occuring words are shown as a word cloud in Fig. 5. Markedly high frequencies are found for the words “corresponding”, “author”, and “request” (dark green font in Fig. 5), indicating the likely prevalence of DASs which indicate that requests for the data should be made to the corresponding author.
Fig. 5
A word cloud showing the top 50 most common words contained within 2019 data availability statements in the OA-PMC subset. Font size and colour reflect the word frequency.
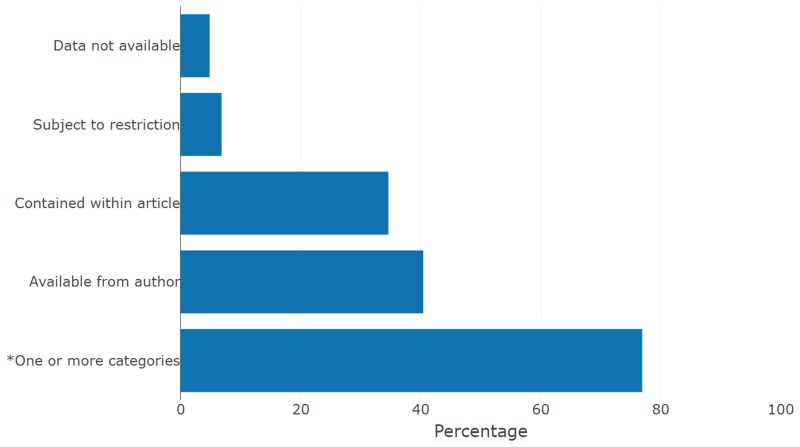
Four regular expressions were then developed to categorise statements into: i) data not available (or no data generated); ii) data subject to restriction; iii) data available within the article; and iv) data available on request to the author(s). The percentages of DASs matching these regular expressions are shown in Fig. 6.
Fig. 6
The percentage of DASs in the dataset matching each of the four regular expressions, or where one or more of the regular expressions were matched.
In line with previous studies, the results suggest that the majority of DASs do not indicate that data are publicly available, with 76.9% (14,117/18,369) of DASs matching one or more the four categories. The full data and regular expressions used to categorise the DASs are available as part of an R Markdown file in Supplementary File 1.
Finally we noted some examples where the authors appear to have misunderstood the purpose of the DAS. Three such statements were: i) “We agree with the statement”; ii) “Available”; and iii) “Yes”. This suggests that publisher guidance may not have been clear to these authors, and there is potential room for improvement in editorial processes to identify and correct such statements. These DASs appear to be the exception however.
Text-mining for funding attributions
Background
Funding agencies have a need to identify and track research outcomes in order to assess the impact of their funding, potentially among different disciplines and geographical areas. In the life sciences, articles are the core currency of research assessment, and therefore identifying articles that have been supported by a given funder, and through a particular grant or funding stream, is vitally important. There is also the need to monitor compliance with, for example, open access policies.
Grant–article associations are made available in Europe PMC through several routes: metadata supplied by PubMed, data upload from ResearchFish (https://www.researchfish.net/), a tool within the Europe PMC plus manuscript submission system and spreadsheets supplied directly to Europe PMC by the funders. In the past, MEDLINE indexers at the USA National Library of Medicine also identified articles funded by the founding group of, at that time, UK PubMed Central (UKPMC) funders, however this was never extended to the wider funder group that now exists, and the practice was discontinued for the founding group in 2016.
Ideally, journal production workflows would tag funding information within the XML, allowing the funding information to be processed automatically in the same way as other elements of a document, for example, references, figure legends or author information. Although supplying full funding information in machine-readable format is on the rise, this best practice is not currently widespread, and frequently only contains the funder name and not the specific grant ID. Often the only source of funding information associated with an article is the content of the acknowledgement and/or funding sections within the XML. Text-mining therefore has a role in identifying more articles that can be attributed to specific grants.
We previously developed a text-mining pipeline exclusively for European Research Council (ERC) grants in 2015. This pipeline runs daily on all incoming full-text XML articles from PMC. The purpose of this project was to significantly extend this service to all Europe PMC funders (https://europepmc.org/Funders/). The work occurred in two phases: (1) develop the text-mining dictionaries to identify specific grant awards in full text articles; (2) extend the daily text-mining workflows to run the dictionaries on all incoming full-text content.
Approach taken
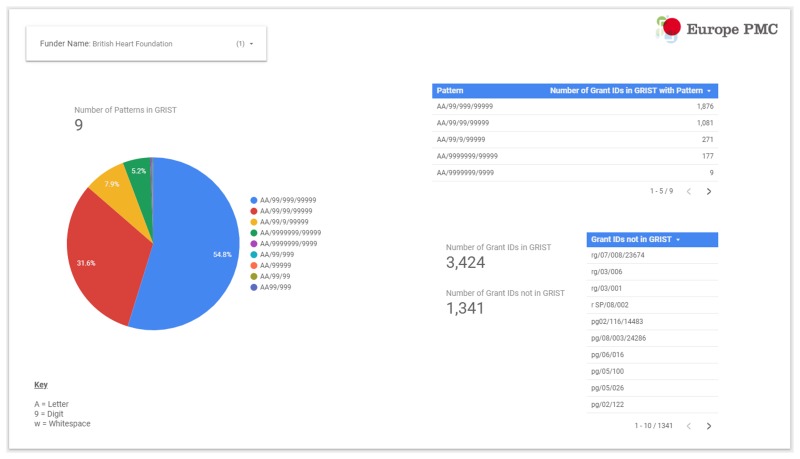
First, we undertook a pattern analysis of all the grant IDs available in the Europe PMC grants database (known as GRIST). As GRIST contains grant data supplied to the Europe PMC Helpdesk staff directly from the funders, this served as a good starting point. A pattern in this context describes the format of the grant IDs, which ranged from simple numerical strings (e.g. “999999”, where “9” represents any digit), to more complex sequences containing letters, hyphens, slashes, parentheses, whitespaces etc. A total of 544 (505 distinct) patterns were identified corresponding to 78,216 grant IDs as of December 2017. The results were presented to funders in a Google Data Studio report (Fig. 7) and discussed in detail in a series of webinars. We considered that the proportion of grants IDs associated with a particular pattern would be a fair measure of the importance of the pattern (i.e. the likelihood it would appear in acknowledgement sections) and so asked the funders to concentrate their review efforts on these patterns (for one funder 137 patterns were identified).
Fig. 7
An example initial report page for the funder “British Heart Foundation” created in Google Data Studio. The pattern “AA/99/999/99999” matches over half the grant IDs assigned by this funder.
Funders were requested to confirm we had identified the most significant patterns and provide any variations or abbreviations of their organisation name, as well as any conflicting funder names that should be excluded (e.g. “Burroughs Wellcome” as an exclusion for “Wellcome Trust”).
Dictionary generation and algorithm refinement
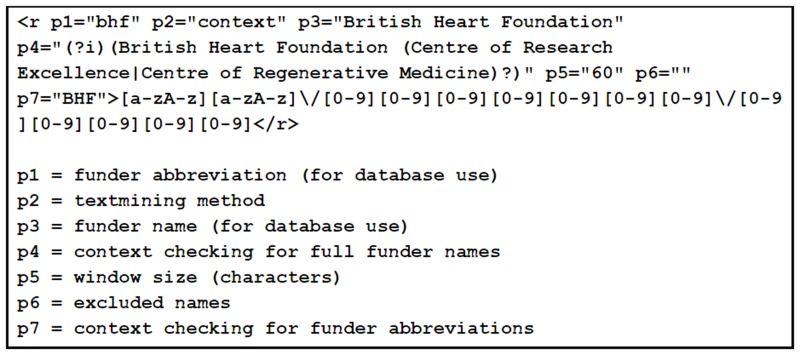
Once grant ID patterns and funder name inclusions/exclusions were collected, the dictionary files were created (see Fig. 8 for an example). For this task, the dictionary was designed to encode two main types of information: 1) funder names, abbreviations and exclusions, and 2) grant ID patterns. We also apply a custom window size (total number of characters within which a funder name and grant ID are matched) for each funder, given the variation in length of funder names (the longest Europe PMC funder name is “National Centre for the Replacement, Refinement and Reduction of Animals in Research”).
The basic elements of the algorithm are: (1) identify the Acknowledgements and Funding sections of articles using the section tagger discussed earlier; (2) using patterns of grant IDs, find matches of these patterns within sentences in these sections; (3) search the sentence text (within the window size) preceding any matched pattern for the name or abbreviation of the funder (avoiding excluded names); (4) apply boundary checks and resolve any conflicts for patterns matching multiple funders. To perform the text-mining we use monq.jfa, an in-house developed class library for fast and flexible text filtering with regular expressions [12].
To test the algorithm we manually identified appropriate examples for each funder and pattern from full-text XML articles. The initial testing gave us the opportunity to select appropriate window sizes for each funder, and also raised an unforeseen sub-pattern issue where, in particular, DOIs were matching short digit-based grant ID patterns (e.g. 10.13039.501100007903 matched a 5-digit pattern). Accordingly boundary checks were implemented in a validation procedure to define the limits of a grant ID string. Frequently grant IDs are contained within parentheses or followed by a comma or semicolon, and so such cases were permitted, whereas in the example above, periods are not permitted to immediately precede the grant ID string. Finally we took steps to resolve conflicts where multiple Europe PMC funders with the same grant patterns were co-located within the same sentence.
We then iteratively improved the algorithm and dictionaries to minimise false positive results by manually checking the sentences outputted from the text-mining process. This included the following procedures:
- Removing too generic patterns with either no or acceptably few true positives
- Making certain grant ID patterns more restrictive (e.g. limiting to specific letters rather than any letter if the grant format allowed for this)
- Removing short funder name abbreviations with either no or acceptably few true positives
- Adding more funder name exclusions. This was particularly relevant for the funder “Multiple Sclerosis Society”, where upon checking many country variations were found (Italian MSS, Swiss, Danish, Canadian, etc.)
Once the number of false positives reached an acceptable level, the dictionaries and code were finalised with a total of 126 regular expressions run against all full-text XML content in Europe PMC as of January 2019. For more details on the algorithm and dictionaries, please refer to the source code available in a public GitHub repository: https://github.com/EuropePMC/EuropePMC-Identifier-Extractor
Results
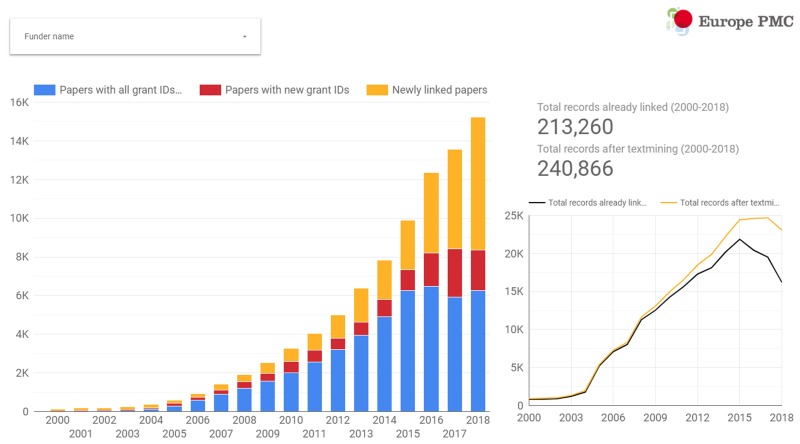
We identified Europe PMC funder grant IDs in ~85,000 full-text articles, of which ~26,000 are newly identified as being linked to a Europe PMC funder, and ~12,000 were enriched by either adding a grant ID to an existing attribution of a funder name only, or discovering additional grant IDs (Fig. 9). The largest gains are made for more recently published articles, reflecting perhaps both the time lag in reporting outputs of grants via tools such as ResearchFish, as well as the cessation of adding grant IDs for the founding Europe PMC funders by the PubMed indexers.
Fig. 9
Final report page for the text-mining results created in Google Data Studio. The graph on the left displays, aggregated for all funders, the number of articles newly identified as belonging to a Europe PMC funder (yellow bars), the number of articles with additional grant IDs associated (red bars) and the number of articles where grant IDs had been previously identified by other means (blue bars). The graph on the right shows the number of articles attributed to Europe PMC funders before (black line) and after (yellow line) the text-mining process.
While the overall growth of attributed papers for the Europe PMC funders group was close to 13%, the benefits of grants text-mining for individual funders varied widely. For example, a significant number of newly identified articles were identified for the Swiss National Science Foundation (SNSF; ~10,000 new articles with a funding attribution, representing a ~20-fold increase), whereas for the World Health Organisation (WHO) only a single new grant link was discovered. The SNSF are a large funding organisation who recently joined Europe PMC in April 2018, and appear to have a good policy of inclusion of text in funding statements. This is not the case for the WHO who have a limited number of grant IDs in GRIST which don’t appear to be cited frequently in the funding section, even if the organisation name is mentioned. One limitation of the approach here therefore is the requirement for both a funder name and grant ID pattern.
Implementation and distribution of grant attributions
The grants mining algorithm has been deployed in the Europe PMC daily pipelines. The algorithms have also been applied to all back-dated content. Text-mined grant IDs that resolve to a valid grant record in the GRIST database are also shared with PubMed. Grant attributions for a respective article are displayed on the front-end (see Fig. 10) and searchable via an indexed field: GRANT_AGENCY_ID:"CH/1996001/9454_agency_British Heart Foundation".
Fig. 10
a) An example funding section from an article matching the grant ID pattern (red) and full funder name context (blue) from the dictionary entry in Fig. 8. b) The front-end display of this grant on an article abstract page in Europe PMC with a hyperlink to the corresponding data hosted in GRIST.
Conclusions
Europe PMC is a comprehensive resource for accessing the life sciences literature, an appreciable proportion of which is available as full-text JATS XML. As part of the aim to establish Europe PMC as a platform for innovation, we focus on linking literature and data and provide powerful APIs in order to support complex researcher workflows. The interoperability and structural consistency afforded by JATS assist greatly in this endeavour, facilitating both the development and sharing of tools such as the ones described in this paper.
Supplementary materials
Supplementary File 1. A .zip file containing an R Markdown document and associated datasets for reproduction of the data availability analysis.
Download file (1.4M)
Funding
Funding for Europe PMC is provided by twenty-nine funders of life science research (https://europepmc.org/Funders/) under Wellcome Trust grants 098321 and 108758, awarded to EMBL-EBI, and an ELIXIR-EXCELERATE grant, funded by the European Commission within the Research Infrastructures programme of Horizon 2020 (676559 to A.V.).
References
- 1.
- Kafkas S, Pi X, Marinos N, Talo’ F, Morrison A, McEntyre JR.: Section level search functionality in Europe PMC. J Biomed Semantics. 2015; 6: 7. 10
.1186/s13326-015-0003-7 . [PMC free article: PMC4359544] [PubMed: 25774284] - 2.
- JATS4R Data availability statements – JATS4R [Internet]. [cited 27 Mar 2019]. Available: https://jats4r
.org/data-availability-statements . - 3.
- Uniprot accession numbers [Internet]. [cited 8 Apr 2019]. Available: https://www
.uniprot.org /help/accession_numbers . - 4.
- EMBL-EBI ENA accession numbers [Internet]. [cited 8 Apr 2019]. Available: https://www
.ebi.ac.uk /ena/submit/accession-number-formats . - 5.
- Levchenko M, Gou Y, Graef F, Hamelers A, Huang Z, Ide-Smith M, et al. Europe PMC in 2017. Nucleic Acids Res. 2018; 46: D1254–D1260. 10
.1093/nar/gkx1005. [PMC free article: PMC5753258] [PubMed: 29161421] - 6.
- Recommended Data Repositories | Scientific Data [Internet]. Springer Nature; [cited 3 Apr 2019]. Available: https://www
.nature.com /sdata/policies/repositories#general . - 7.
- Federer LM, Belter CW, Joubert DJ, Livinski A, Lu Y-L, Snyders LN, et al. Data sharing in PLOS ONE: An analysis of Data Availability Statements. PLoS One. 2018; 13: e0194768. 10
.1371/journal.pone.0194768 . [PMC free article: PMC5931451] [PubMed: 29719004] - 8.
- Hardwicke TE, Mathur MB, MacDonald K, Nilsonne G, Banks GC, Kidwell MC, et al. Data availability, reusability, and analytic reproducibility: evaluating the impact of a mandatory open data policy at the journal. R Soc Open Sci. 2018; 5: 180448. 10
.1098/rsos.180448. [PMC free article: PMC6124055] [PubMed: 30225032] - 9.
- McDonald L, Schultze A, Simpson A, Graham S, Wasiak R, Ramagopalan SV. A review of data sharing statements in observational studies published in the BMJ: A cross-sectional study. F1000Res. 2017; 6. 10
.12688/f1000research.12673.2. [PMC free article: PMC5676190] [PubMed: 29167735] - 10.
- Data availability statement templates - Author Services [Internet]. 29 Nov 2017 [cited 8 Apr 2019]. Available: https:
//authorservices .taylorandfrancis.com /data-availability-statement-templates/ - 11.
- Silge J, Robinson D. tidytext: Text Mining and Analysis Using Tidy Data Principles in R. JOSS. 2016; 1: 37. 10
.21105/joss.00037. - 12.
- Monq.jfa —a DFA implementation for Java [Internet]. [cited 5 Apr 2019]. Available: http://www
.pifpafpuf.de/Monq.jfa/
- How JATS supports data integration: extracting data availability statements and ...How JATS supports data integration: extracting data availability statements and funding information from research articles in Europe PMC - Journal Article Tag Suite Conference (JATS-Con) Proceedings 2019
Your browsing activity is empty.
Activity recording is turned off.
See more...

 1
1