Working with JSON Lines data reports
NCBI data packages contain metadata in one or more data report files in JSON Lines format. Here are some frequently asked questions about how to work with them.
Working with JSON Lines data reports
NCBI Datasets tools provide data in zip files that we call “data packages.” These data packages contain metadata in one or more data report files in JSON Lines (pronounced “jason-lines”) format. For all JSON Lines data reports, each line represents a single record. But the number and type of JSON Lines data reports varies depending on the type of data package. For example, gene data packages only include a single gene data report , where each line of the data report represents a single gene record. In contrast, genome data packages include two types of data reports:
- a single genome assembly data report , where each line represents one genome assembly record, and
- one genome sequence data report per genome assembly record, where each line represents one nucleotide sequence record that comprises that assembly
Data report schemas describe each type of data report, including the available fields, with descriptions, examples, and mnemonic terms that can be used with the dataformat CLI tool .
Here are some frequently asked questions about how to work with JSON Lines data reports.
How do I make the JSON Lines data report more readable?
Make the JSON Lines data report more readable by using jq to pretty-print or alternatively, see the question below to generate a table.
First, download a gene data package for a set of NCBI GeneIDs and unzip it.
datasets download gene gene-id 1,2,9 --filename genes.zip
Downloading: genes.zip 68.8kB done
unzip genes.zip
Archive: genes.zip
inflating: README.md
inflating: ncbi_dataset/data/gene.fna
inflating: ncbi_dataset/data/rna.fna
inflating: ncbi_dataset/data/protein.faa
inflating: ncbi_dataset/data/data_report.jsonl
inflating: ncbi_dataset/data/data_table.tsv
inflating: ncbi_dataset/data/dataset_catalog.json
Now use jq to pretty-print the data report to make it more readable.
jq . ncbi_dataset/data/data_report.jsonl | head --lines=10
{
"annotations": [
{
"assembliesInScope": [
{
"accession": "GCF_000001405.40",
"name": "GRCh38.p14"
}
],
"releaseDate": "2021-11-19",
For a complete pretty-printed gene data report, see the Sample report in the gene data report schema .
How do I convert a JSON Lines data report to a table?
You can generate a table from the JSON Lines data report using the NCBI Datasets dataformat command line tool .
First, download a gene data package for a set of NCBI GeneIDs.
datasets download gene gene-id 1,2,9 --filename genes.zip
Downloading: genes.zip 68.8kB done
Then, generate a table using dataformat.
dataformat tsv gene --fields gene-id,symbol,tax-name,gene-type --package genes.zip
NCBI GeneID Symbol Taxonomic Name Gene Type
1 A1BG Homo sapiens PROTEIN_CODING
2 A2M Homo sapiens PROTEIN_CODING
9 NAT1 Homo sapiens PROTEIN_CODING
How do I find metadata for a single gene described in the JSON Lines data report?
Because each line of the data report represents a single gene, you can use grep to get the metadata describing that gene.
After downloading and unzipping a gene data package, use grep to pull out the line matching the desired gene symbol, then use jq to pretty-print and tail to show the last 10 lines.
grep A1BG ncbi_dataset/data/data_report.jsonl | jq . | tail --lines=10
"accessionVersion": "NP_570602.2",
"ensemblProtein": "ENSP00000263100.2",
"length": 495,
"name": "alpha-1B-glycoprotein precursor"
},
"type": "PROTEIN_CODING"
}
],
"type": "PROTEIN_CODING"
}
How do I view a JSON Lines data report without using the command line?
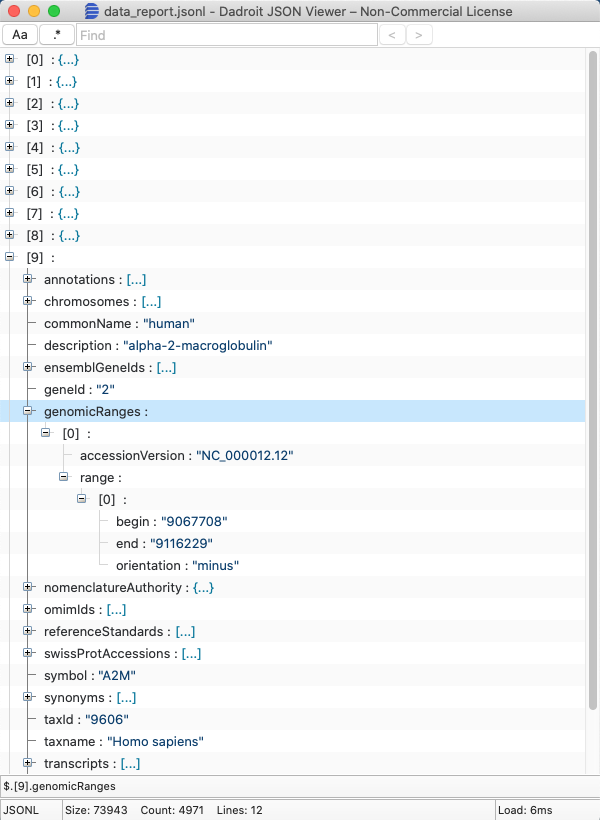
If you prefer not to use the command line to view a JSON Lines data report, you may want to use the Dadroit JSON Viewer .
Open the JSON Lines data report to show the report as a collapsed tree. Click on the + plus symbol to expand any of the nodes and view the contents.
In this example, you can see the genomic range (location on the genome) for the human alpha-2-macroglobulin gene:
Why use JSON Lines format for the data report?
The JSON Lines format offers offers advantages over conventional JSON that stem from the fact that each line of a JSON lines format file represents a single valid JSON object. Two notable advantages for users are:
- JSON Lines works well with UNIX tools including
grep,sed,headandtail - JSON Lines enables stream processing, which allows handling of large data reports that can’t fit into memory
