GenBank (GCA) and RefSeq (GCF) genome assemblies
Understanding the differences between GenBank (GCA) and RefSeq (GCF) genome assemblies
GenBank (GCA) and RefSeq (GCF) genome assemblies
What is the difference between a GenBank (GCA) and RefSeq (GCF) genome assembly?
A GenBank (GCA) genome assembly contains assembled genome sequences submitted by investigators or sequencing centers to GenBank or another member of the International Nucleotide Sequence Database Collaboration (INSDC). The GenBank (GCA) assembly is an archival record that is owned by the submitter. In some cases NCBI identifies potential contaminants in a GenBank assembly after it has been publicly released and contacts the submitter to request permission to remove the contaminating sequences. GenBank (GCA) assemblies may include user-submitted or NCBI-generated annotation. User-submitted annotation can include annotation generated using NCBI’s Prokaryotic Genome Annotation Pipeline (PGAP). PGAP annotation can be requested by the submitter during submission of the genome to GenBank or can be generated using the PGAP standalone software package.
A RefSeq (GCF) genome assembly represents an NCBI-derived copy of a submitted GenBank (GCA) assembly. RefSeq (GCF) assembly records are maintained by NCBI. In some cases the RefSeq (GCF) assembly may not be completely identical to the GenBank (GCA) assembly because NCBI staff may (1) remove short sequences or reported contaminants from the assembly or (2) add non-nuclear genome sequences (for example, mitochondrial or chloroplast genomes) to the assembly. All RefSeq (GCF) genome assemblies include annotation. In the majority of cases, this annotation is generated by the NCBI prokaryotic or eukaryotic genome annotation pipelines. In some cases, annotation is provided by the assembly submitter.
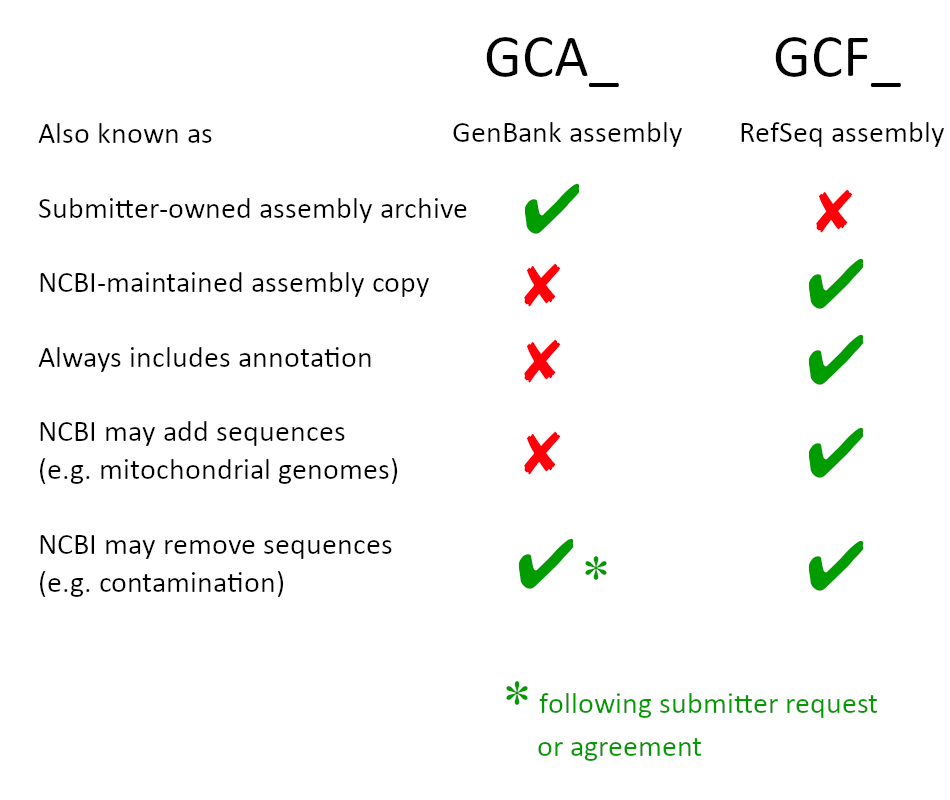
Table 1. Key differences between GenBank (GCA) and RefSeq (GCF) Genome Assemblies
