Using the dbVar Browser
The dbVar Browser is a genome browser tool that allows users to view data from multiple studies in the same genome context.
- Getting to the browser page
- Setting up the browser page
- Configuring the browser page
- Custom tracks
- Creating links to a specific region in the browser
To report a problem or make a feature request, please use the 'Write to Help Desk' link at the bottom of the page.
Getting to the dbVar Browser
There are two ways to get to the dbVar Genome Browser Page.
From the dbVar Home page:
This link will take you to the dbVar browser entry page, which will allow you to select an organism and assembly, enter a location or search term, and upload your own data to the browser.
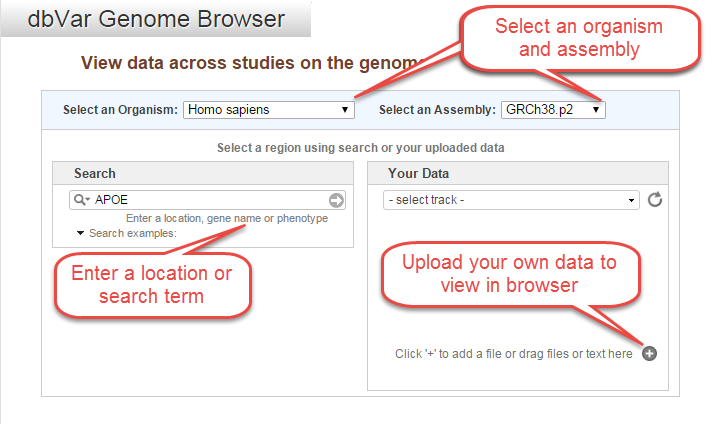
You can also get to the browser page from any of the variant pages within dbVar. In the genome view tab, you will see a new line that tells you how many variant regions from other studies overlap the variant described on the page. At the end of the line, there will be a link to the genome browser (labeled 'genome view').
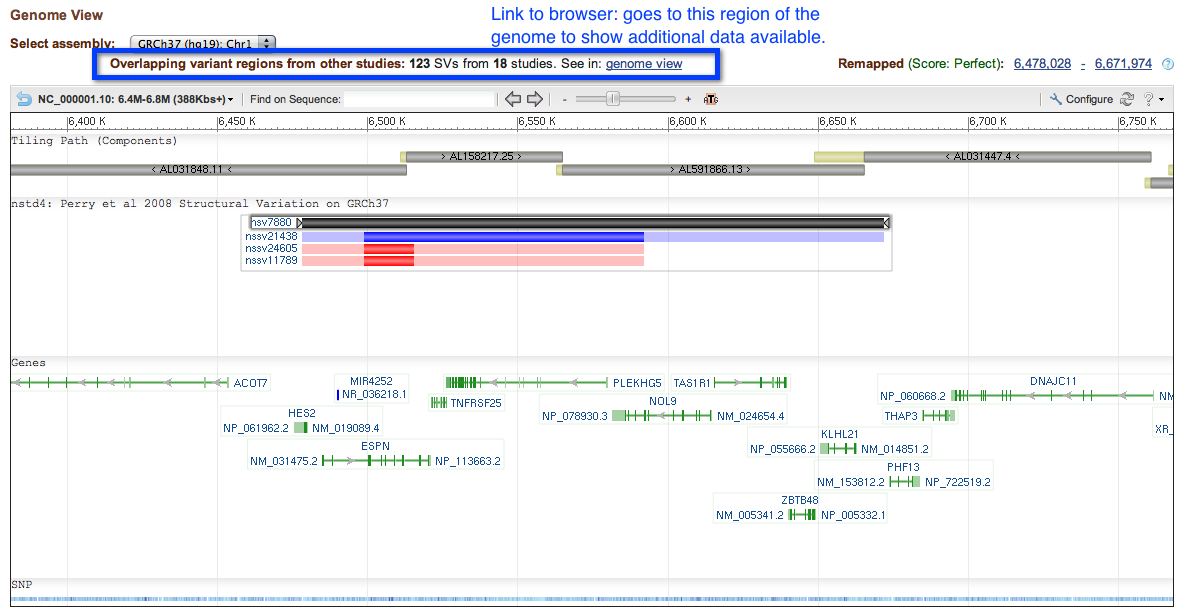 Clicking on this link will take you directly to the browser page.
Clicking on this link will take you directly to the browser page.
Setting up the browser page
To start using the dbVar Genome Browser, you need to define an organism, an assembly and a region of interest. If you link to the browser from a variant page, all of these are defined by the link and you go directly to the region of interest. If you start at the home page, you will configure these yourself. Data upload is optional.

Location searching
For all organisms, if you know the sequence coordinates you want to view, you can just type them in the text box labeled 'Go'. We take a variety of formats including:
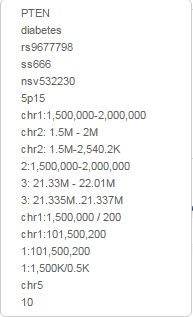
- chr1:10,000-20,000
- chr1:1M-2M
- chr1:100..500
- chr1:1500250
- 1:101,500,200
- NC_000001.11:101M
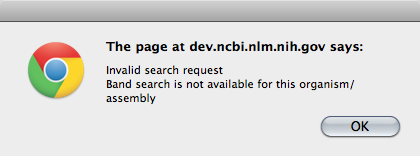
A few organisms also support a search by cytogenetic band. Cytogenetic band search must be tied to a specific assembly because we need to be able to make the conversion between cytogentic bands and sequence coordinates. That is why some organisms will support cytogenetic band searching for some assemblies but not others.
Included file 'browser_help-table1.inc' not found If you try to do a band search for an organism that does not support this, you will get an alert warning you that this is not supported.
Term Searching
Many (but not all) combinations of organism and assembly support term searching. That is, you can type in a term (a gene name, STS id, rsID, phenotype) and a list of results will be returned. Many older assemblies are not supported, and some organisms have no support but we will attempt to add this as we acquire more data or map the data onto newer assemblies. Below is a list of organism and assembly combinations that support term searching.
Included file 'browser_help-table2.inc' not found If your search returns results, a new table will appear on the page that provides a list of the matching features and their locations.
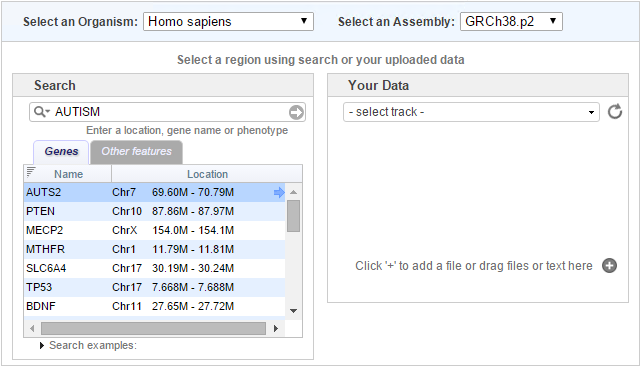
Placing your mouse over a row will cause the color to darken and an arrow will appear in the last column. If you click on the arrow, the browser page will open to this location. The Search term will be imported into the browser, but the results table will not be visible. Clicking on the arrow in the search bar to the right of the Search term enables you to repeat your search within the browser. Your history of previous searches can be accessed by clicking the magnifying glass icon in the Search bar.
NOTE: You may see some differences in the term search results you obtain using the dbVar browser as compared to the dbVar main site. The dbVar main site uses a search strategy that is based on the dbVar variant calls and variant regions and the submitted data that is submitted with them. The dbVar browser search uses the same search as the Map Viewer, which is based on indexing features that have been annotated on the genome. We are working on reconciling the two search back ends.
Configuring the browser page
Below is a figure showing an overview of the browser page. Individual page widgets are highlighted and will be described below.
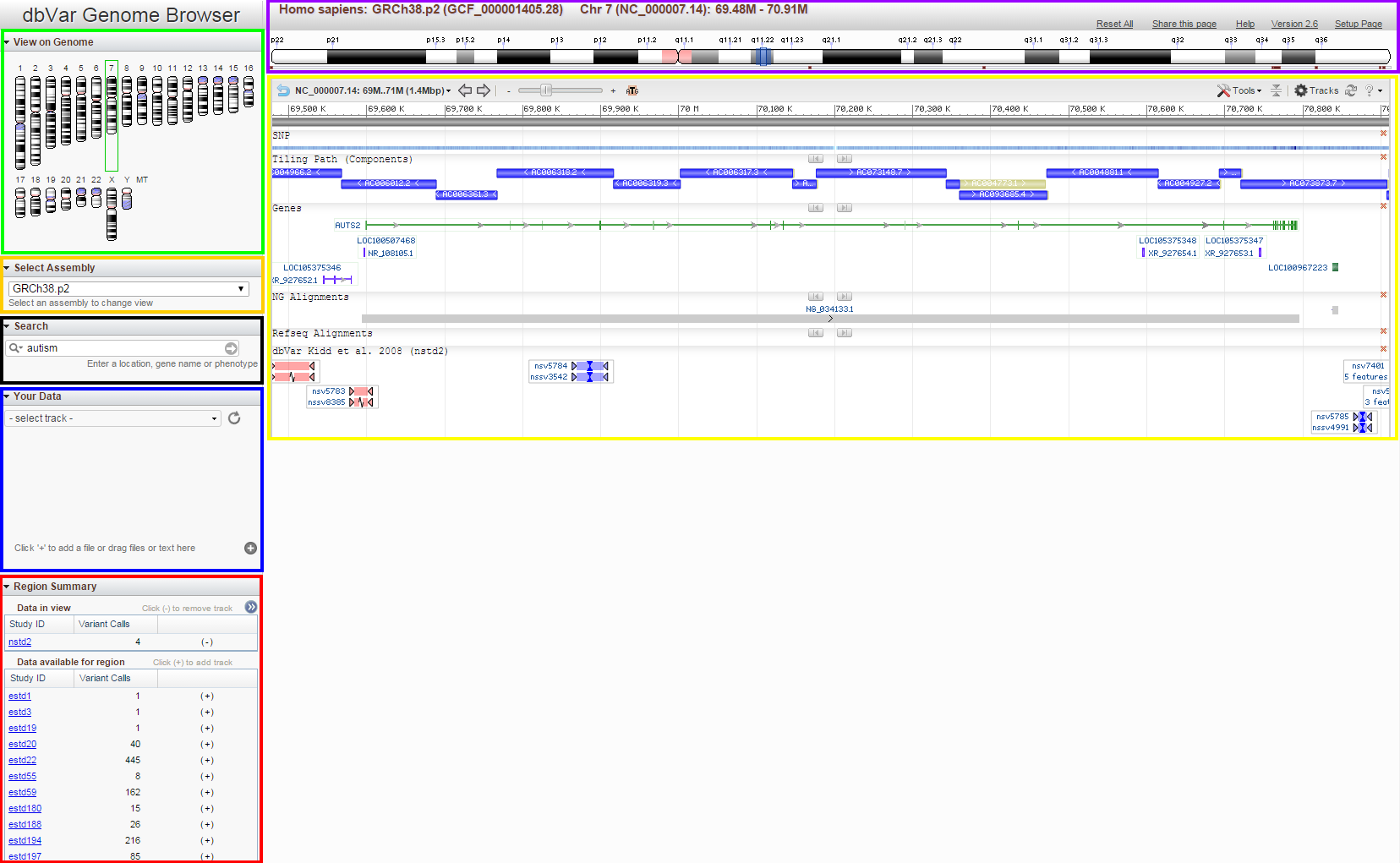
The page is set up so that the different widgets on the page interact to give you a consistent overview of the selected region. Additionally, the page is set up to facilitate moving from one region of the genome to another quickly and easily.
Genome Overview
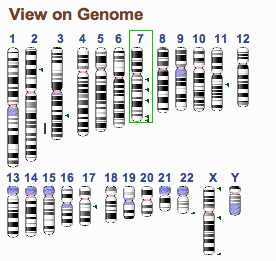
The Genome Overview widget provides you with both context and navigation. If you do a term search, the locations of the search results will be shown on the Genome (as in the figuure above). The chromosome containing the region being shown by the page will have a green highlight (chr 7 in the figure above). Clicking on any of the chromosomes will update the page so that the selected chromosome is shown.
Chromosome Overview
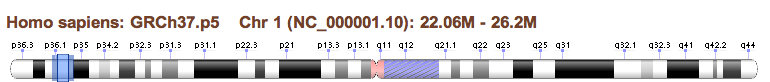
The Chromosome Overview provides context and navigation for the page. The blue overlay shown on the ideogram covers the amount of the chromosome shown in the Sequence Viewer (described below). The sides of this box (outside of the dark blue line) can be selected with the mouse and moved to adjust the size of the box (thus adjusting the size of the region being shown). Making the box smaller is the equivalent of 'zooming' in on a region. You can also select the center of the blue box and drag it to another location on the chromosome. If you adjust the location using another widget on the page, the blue box will automatically adjust to reflect the new location.
Select Assembly
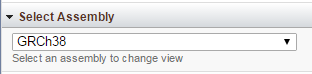
The Select Assembly widget on the browser page provides you with the option to change assembly for the specified organism, if more than one assembly is available. If you want to view data for another organism, you will need to return to the browser home page ('Setup Page' link in the upper right side of the page).
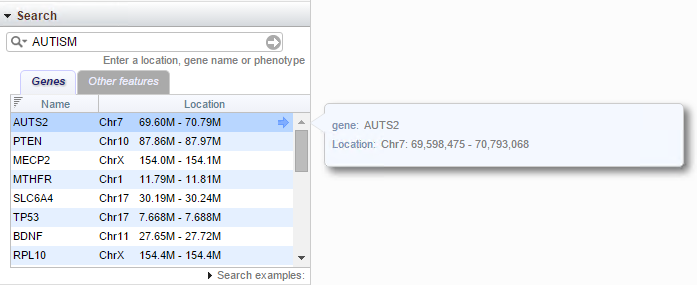
Search
The Search widget on the browser page operates exactly like the Search widget on the browser home page. Clicking on any row in the results table will update the browser to the corresponding location.
Region Summary
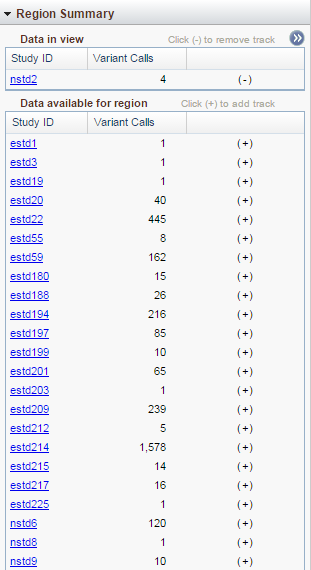
The Region Summary widget provides information concerning the dbVar studies that have data for this region. The 'Data in view' table shows the studies that are displayed in the Sequence Viewer (described below). The 'Data available for region' table shows a list of studies with data in this region. To see more information about the available studies, click on the white arrows in the blue circle (at top right in the figure above) and the table will expand.
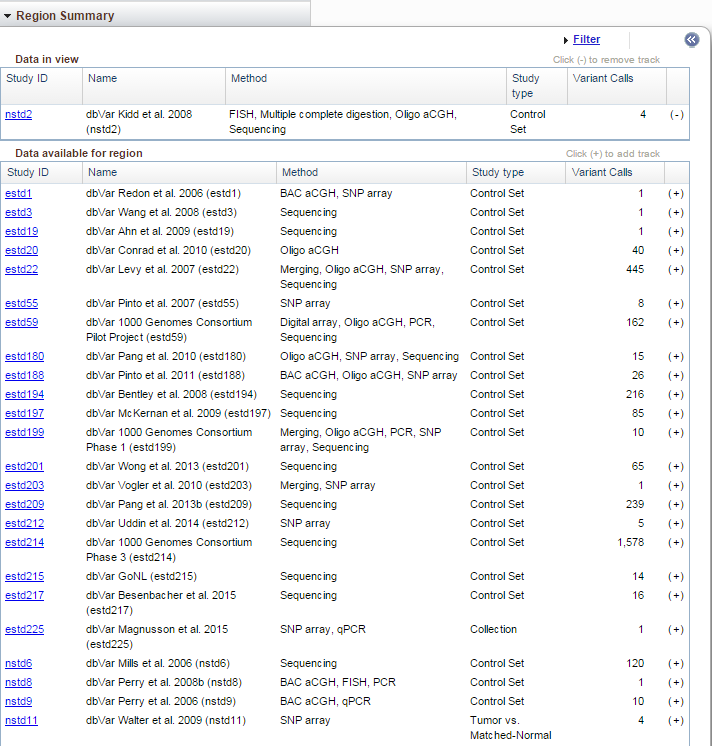
To close the table, just click on the blue icon again. To add a study to the view, click on the (+) in the last column of the 'Data available for region' table. To remove a study from the view, click on the (-) in the last column of the 'Data in view' table.
Sequence Viewer
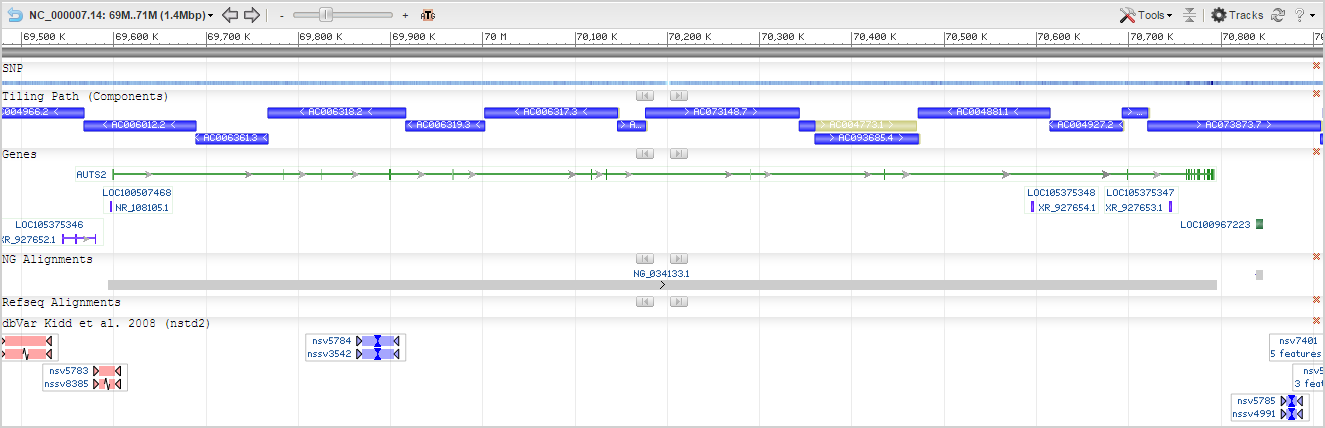
The NCBI Sequence Viewer is an embeddable widget that provides graphical representation of features annotated on individual sequences. Access to other NCBI tools (such as BLAST) are also available within this widget. The 'Tracks' button (in the upper right hand corner of the widget) allows you to add or remove tracks from the display. Note: dbVar tracks in the 'Data available for region' table will not be in the Sequence Viewer 'Tracks' dialog, but dbVar tracks in the 'Data in view' table will be. You can adjust how these tracks are displayed using the Sequence Viewer 'Tracks' dialog.
Additional information on using the Sequence Viewer (which is embedded on many NCBI pages) can be found here: Sequence Viewer.
There are also some videos available on the NCBI YouTube channel that can give you a quick introduction to using the Sequence Viewer.
For information on rendering options for particular tracks and feature types, please see this legend.
Custom Tracks
Your Data
The 'Your Data' widget allows you to add custom tracks for display in the Sequence Viewer alongside NCBI-provided tracks, by either uploading files or streaming data from remotely-hosted files. To start, click the "+" button. Uploaded tracks will expire 60 days after they are last touched in the browser; streamed tracks will persist until you remove them. If you are logged in with your My NCBI account and uploading/streaming human data, your custom tracks will also be available to you in other NCBI browsers displaying the same assembly version as your data (e.g. Variation Viewer or 1000 Genomes Browser).
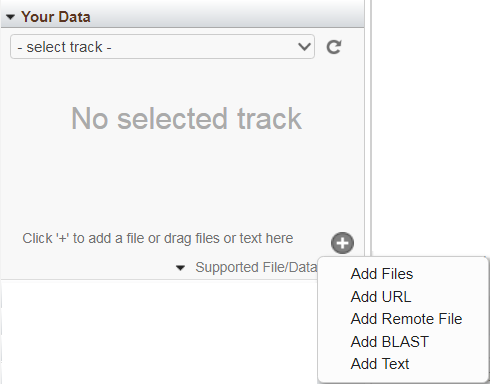
Uploading files: You can upload files by selecting one of the following menu options: "Add Files", "Add URL" or "Add Text", or by dragging the file into the widget. The following file types are supported: BED, GFF3, GTF, GVF, VCF, HGVS, ASN.1 (text and binary). Concurrent upload of multiple files is supported. The per-file upload limit is currently 4 GB. The file name will be used as the track's display name, unless an (optional) alternate name is provided. Once uploaded, the files will appear in the 'select tracks' drop-down menu in this widget.
Remote streaming files: BAM files hosted on HTTP can be streamed for display in the dbVar browser. To add these data as tracks, select “Add Remote Track” from supported files menu, and enter the corresponding URL in the display. Note that an index file with the .bai extension must be located at the same location as the BAM file. The file name will be used as the track’s display name, unless an (optional) alternate name is provided. A progress bar will indicate the status of the connection and validation processes. Once connected, remotely hosted files will appear in the graphical display and be listed in the 'select tracks' drop-down menu of the “Your Data” widget, designated by “(R)”. Other file types and BAM files hosted on FTP are not yet supported. To request streaming support for additional file types, click the "Support Center" link located at the lower right of the browser page.
BLAST: You can also enter the RID from a BLAST search into the Your Data widget. The results will appear as an alignment track in the display.
If your track has discrete features (e.g. SNPs or gene annotations, rather than graphs or alignments) on the currently displayed sequence, you can display those features in a paginated table if you select that track from the "Your Data" drop-down menu. Hovering the mouse over a table row opens a tool-tip with feature details. Click on a row to go to the location of that that feature in Sequence Viewer. Use the menus below the table for additional table naviation. To remove uploaded or remote tracks, select the track from this drop-down menu and then click on the 'minus' icon next to the track name. Note: A selected track will be marked by a check-mark in this menu, regardless of whether it has data suitable for tabular display.
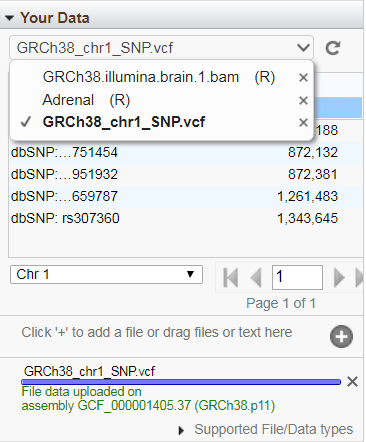
The progress bar at the bottom of the window indicates the status of the upload or initial connection to the data file. If validation detects the presence of target sequences in your uploaded or streamed remote file that are not part of the assembly displayed in the dbVar browser, those sequences will be reported as errors, but you will still be connected and able to view tracks for all target sequences in the file that are part of the assembly. To see any warning or error messages associated with uploading or streaming your custom track data, click on the "Details" option that will be a part of the status message.
File uploads, streaming of remotely hosted files and BLAST results can also be managed via the track configuration menu found under the 'Tracks' button at the top right of the Sequence Viewer display. Files uploaded or connected by this mechanism will also appear in the 'Your Data' widget.
Creating links to a specific region in the browser
You can create links directly into the browser using the following url and parameters:
http://www.ncbi.nlm.nih.gov/dbvar/browse/org/?$parameter_list
Parameter list
Included file 'browser_help-table3.inc' not found
