Datasets downloads for Pathogen Detection isolates 
The Datasets command-line tool allows you to download large amounts of data using the "dehydrated download" feature. To download large numbers of assemblies using this tool first you need to Install datasets.
Here's an example of how to use this feature to download all 68,000+ Listeria monocytogenes genomes available in Pathogen Detection and included in GenBank.
1. Select the isolates you want to download using the Isolates Browser
In this example we will download all Listeria monocytogenes genomes using the query taxgroup_name:"Listeria monocytogenes" in the Isolates Browser and click on the Download link.
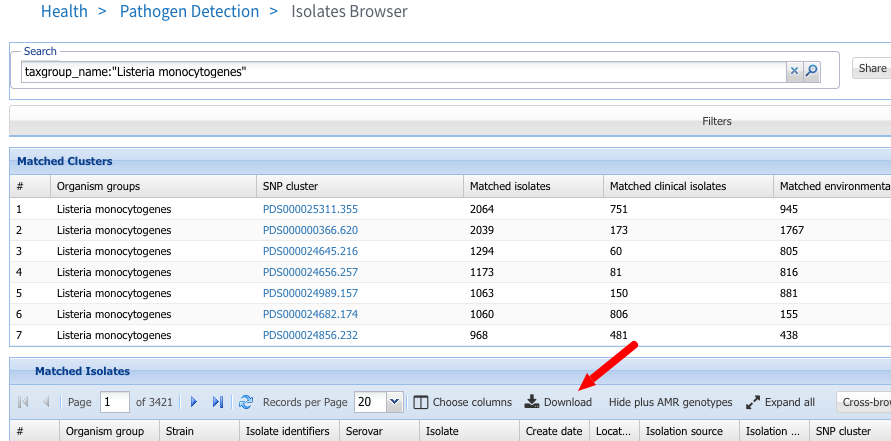
2. Select Assembly accessions and click Download to download.
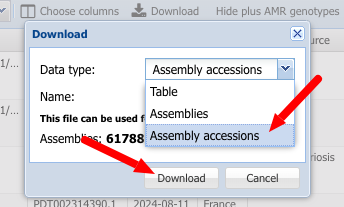
3. Download a "dehydrated data package" using the datasets CLI.
Here we will download the assembly, annotation GFF3 file, and annotated proteins
datasets download genome accession --dehydrated --inputfile accessions.txt --include genome,gff3,protein
4. Unzip the downloaded zip archive to a directory
unzip ncbi_dataset.zip -d listeria_genomes
5. "Rehydrate" the extracted zip archive to retrieve the data
datasets rehydrate --directory listeria_genomes
For more information on the datasets CLI see the datasets CLI documentation.
