How to Request New Alleles for Beta-Lactamase, MCR, and Qnr Genes
The spread of beta-lactamases, which confer resistance to beta-lactam antibiotics, and of mobile quinolone resistance genes (Qnr), which confer resistance to quinolone antibiotics, is a serious clinical and public health problem. Genotypic and functional characterization of these genes is critical. The curation of many of these medically-important enzymes has in the past been performed solely by Drs. Karen Bush, George Jacoby, and Timothy Palzkill and has been hosted at the Lahey Clinic. At the request of the community, NCBI is also assigning novel MCR alleles, which are mobile colistin resistance genes (for further information about MCR nomenclature, see Partridge et al., 2018). After discussion with the above experts and additional experts that initially convened at the ICAAC 2015 conference, NCBI has agreed to host the data to ensure the long-term stability of these important resources which will include:
- The storage of current beta-lactamase, Qnr, and MCR alleles (the specific nucleotide and protein accessions associated with each allele) along with links to any publication when available.
- The curation (assignment of) new beta-lactamase, Qnr, and MCR alleles.
- Enabling the retrieval of beta-lactamase, Qnr, and MCR nucleotide and protein accessions along with the antibiotic susceptibility profile of a transconjugant/transformant bearing the beta-lactamase gene(s).
Following the guidelines established by the ASM Virtual β-lactamase Workshop held in November 2021 (for further information, see Bradford et al., 2022), allele numbers should be requested when the shorthand that results improves scientific communication. However, many beta-lactamase families may be chromosomally encoded as a rule, may occur primarily in non-pathogens, and may have little significance to clinical practice or microbial surveillance. Such beta-lactamases typically would not receive allele designations, unless the literature has already established a system for that family and its readers benefit from their familiarity with it.
NCBI therefore offers the following guidance.
NCBI assigns alleles for all beta-lactamase and Qnr families previously administered through the Lahey Clinic pages of Dr. George Jacoby and Karen Bush. We will continue to do so. NCBI assigns alleles for several additional families. These families are described in the table below.
NCBI encourages researchers who describe founding members of novel, clinically relevant beta-lactamase families to alert us regarding their efforts to name these families and alleles. We invite those researchers to request that NCBI assign future alleles for those families.
Lastly, we encourage researchers who discover less clinically relevant beta-lactamases to name the beta-lactamase family, and refer to individual alleles through their protein accession numbers.
The following table lists which families of beta-lactamases are curated by either NCBI or other groups:
| Beta-lactamase, Qnr, or MCR family | Curator |
|---|---|
| AAK | NCBI |
| ACC | NCBI |
| ACT | NCBI |
| ADC | NCBI |
| AFM | NCBI |
| AIM | NCBI |
| AXC | NCBI |
| BAT | NCBI |
| BEL | NCBI |
| BIC | NCBI |
| BIM | NCBI |
| BKC | NCBI |
| BPU | NCBI |
| BSU | NCBI |
| CAE | NCBI |
| CARB | NCBI |
| CDD | NCBI |
| CFE | NCBI |
| CIB | NCBI |
| CMH | NCBI |
| CMY (includes CFE and LAT) | NCBI |
| CRH | NCBI |
| CTX-M | NCBI |
| CVI | NCBI |
| DHA | NCBI |
| DYB | NCBI |
| EBR | NCBI |
| FOX | NCBI |
| FRI (includes FLC) | NCBI |
| GES | NCBI |
| GIM | NCBI |
| GMA | NCBI |
| GMB | NCBI |
| GOB | NCBI |
| HMB | NCBI |
| IDC | NCBI |
| IMI (includes NMC-A) | NCBI |
| IMP | NCBI |
| IND (includes CGB) | NCBI |
| KBL | NCBI |
| KHM | NCBI |
| KLUC | NCBI |
| KPC | NCBI |
| LAQ | NCBI |
| LAT | NCBI |
| LCR | NCBI |
| LEN | Institut Pasteur |
| MIR | NCBI |
| MOX | NCBI |
| MUN | NCBI |
| NDM | NCBI |
| NPS | NCBI |
| NWM | NCBI |
| OKP-A | Institut Pasteur |
| OKP-B | Institut Pasteur |
| OXA | NCBI |
| OXY | Institut Pasteur |
| PAM | NCBI |
| PAU | NCBI |
| PDC | NCBI |
| PER | NCBI |
| PFM | NCBI |
| PJM | NCBI |
| POM | NCBI |
| PRC | NCBI |
| PSE | NCBI |
| PST | NCBI |
| RAA | NCBI |
| RAD | NCBI |
| RASA | NCBI |
| RATA | NCBI |
| ROB | NCBI |
| RSD1 | NCBI |
| RSD2 | NCBI |
| RTG | NCBI |
| SFC | NCBI |
| SFDC | NCBI |
| SHV | NCBI |
| SIM | NCBI |
| SME | NCBI |
| TEM | NCBI |
| TLA | NCBI |
| VEB | NCBI |
| VHH | NCBI |
| VHW | NCBI |
| VIM | NCBI |
| WUS | NCBI |
| ampS | NCBI |
| blaB | NCBI |
| cfiA | NCBI |
| mcr-1 | NCBI |
| mcr-10 | NCBI |
| mcr-2 | NCBI |
| mcr-3 | NCBI |
| mcr-4 | NCBI |
| mcr-5 | NCBI |
| mcr-6 | NCBI |
| mcr-7 | NCBI |
| mcr-8 | NCBI |
| msiOXA | NCBI |
| qnrA | NCBI |
| qnrB | NCBI |
| qnrD | NCBI |
| qnrE | NCBI |
| qnrS | NCBI |
| qnrVC | NCBI |
Note: Because our NARMS collaborators found mcr-9 did not confer resistance to colistin in over 100 natural mcr-9+ isolates (Tyson et al., 2020, despite laboratory evidence that mcr-9 is capable of conferring resistance to colistin (Carroll et al., 2020, Kieffer et al., 2019), we are not curating mcr-9 alleles at this time, though we still will offer nomenclature advice to prevent nomenclature collisions, if requested.
Another web site for TEM, SHV, and class B enzymes can be found at http://www.laced.uni-stuttgart.de. If you have questions whether a particular beta-lactamase family is supported or recommendations for families to support, please contact NCBI at [email protected].
For more information on general antibiotic susceptibility profiles and submitting phenotypic results linked to sample metadata see the Pathogen submission page, or the BioSample antibiogram documents.
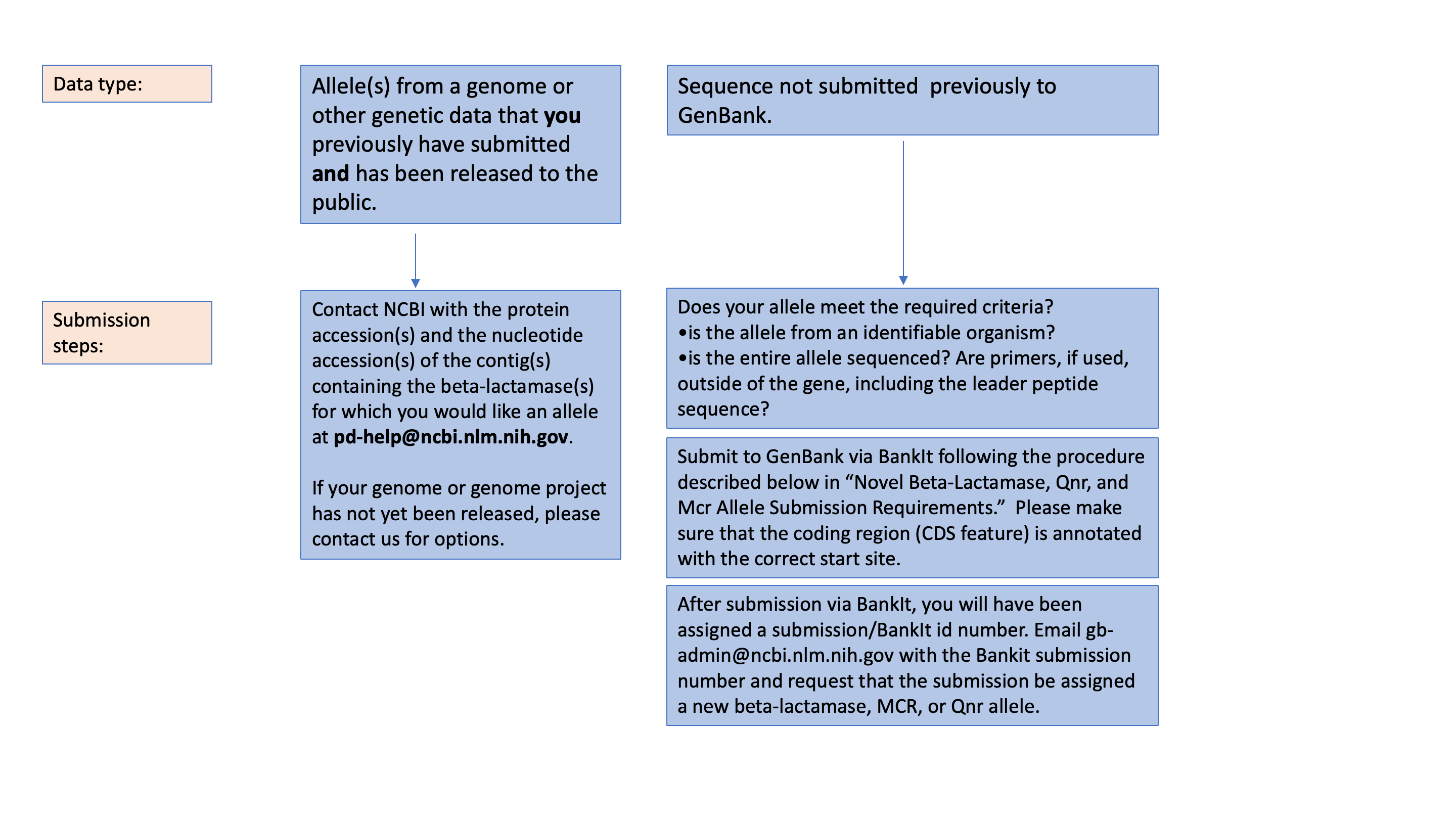
NCBI is accepting three different data types related to beta-lactamase, Qnr, and MCR genes:
- Whole genome sequence data submissions. NCBI is building a database linking genome sequence and antimicrobial susceptibility tests, the National Database of Antibiotic Resistant Organisms, which is described here. To submit a genome sequence or sequencing reads that also has antibiotic susceptibility testing data associated with it, please see the Pathogen submission page, or the BioSample antibiogram documents.
- A novel beta-lactamase, Qnr, or MCR protein sequence. Requirements are described in the following section.
- A novel beta-lactamase protein sequence that also has been characterized phenotypically. Providing resistance phenotypes of novel beta-lactamase alleles expressed in a sensitive background furthers our understanding of how genotype is related to resistance phenotypes. This is not required to receive an allele designation , but these data are extremely useful. Requirements are described in Phenotypic Characterization of Beta-Lactamase Alleles section below.
Note: we currently are not accepting allele submissions for novel alleles from metagenomic samples, unless that sequence has been verified by cloning and sequencing to ensure sequence quality.
Novel Beta-Lactamase, Qnr, and Mcr Allele Submission Requirements
If wish you to receive an allele assignment for a sequence, such as one found in a genome assembly that you have submitted previously to GenBank and that is currently publicly available, contact NCBI at [email protected] with the protein accession and nucleotide or contig accession (e.g., AAWUNZ010000001.1) of the sequence. If you have not submitted the sequence previously or the sequence is not publicly available at the time of the request, to receive assignment of a new allele designation, NCBI requires the complete nucleotide and protein coding sequences submitted in GenBank compliant format using BankIt (for more information about BankIt see the BankIt page). If sequencing primers were used to obtain the sequence, they must be external to the entire protein sequence, including the leader peptide. The organism from which the gene was originally isolated should be included in the BankIt file as well (i.e., species, strain name).
New allele requests can be based on predicted amino acid changes in the leader sequence; however, DNA sequences that differ by a single nucleotide that does not encode a different amino acid (synonymous change) will not be given a new allele number. Only sequences from natural sources will receive allele assignments, not laboratory constructs. Without some form of characterization to verify function, metagenomic sequences likely will not receive allele assignments, unless there is overwhelming scientific need for assignment.
To submit, use BankIt to submit novel sequences, using the following additional instructions:
- In the "Nucleotide" tab (tab #4), "Molecule Type" should be entered as "genomic DNA".
- In the "Features" tab (tab #7), you can choose "Add features by completing input forms", and then "Coding region".
- Once you begin to add features, you can assign each sequence a gene name; this will not be the final name for the sequence (e.g., OXA-1234). NCBI will determine that name.
- In the final tab, "Review and Correct", add "AMR" to "Submission Title (Optional)", then complete the BankIt process, and submit the file (tab #8)
- You will have been assigned a submission/BankIt id number. Email [email protected] with the Bankit submission number and request that the submission be assigned a new beta-lactamase, MCR, or Qnr allele.
For existing alleles (i.e., those that already have publicly available Genbank accessions), please provide NCBI at [email protected] with a list of nucleotide accessions, and we will determine the allele number, if appropriate. For any questions about these requirements, please contact NCBI at [email protected] .
Phenotypic Characterization of Beta-Lactamase Alleles
NCBI is also accepting data related to phenotypic characterization of beta-lactamase alleles expressed in sensitive backgrounds as a primary purpose of allele assignment is to provide a framework for functional characterization. While these requirements are not required for allele assignment, to enable comparisons among alleles, we have the following requirements for phenotypic characterization.
Mandatory requirements:
- MICs of transformants or transconjugants (strain with no other active beta-lactamase present) against:
- One or both of the following: cephalothin or cefaclor
- One or more of the following: benzylpenicillin, amoxicillin, ampicillin, piperacillin
- Cefotaxime with and without clavulanic acid
- Ceftazidime with and without clavulanic acid
- note both drugs do not have to be tested with clavulanic acid
- Cefoxitin
- One or more of the following: imipenem, doripenem, ertapenem, meropenem
- Aztreonam
- One or both of the following: cloxacillin or oxacillin
- CLSI standard testing methodology is required
- MICs or Etest can be used
- Carbapenemases should be tested for activity in the presence of EDTA
- The transconjugant/transformant strain should be described (i.e., strain name)
- Complete nucleotide and amino acid sequence
- The organism from which the gene was originally isolated should be described (i.e., species, strain name)
Optional information:
- MICs of transformants or transconjugants against carbenicillin or ticaricillin
- If the enzyme is a carbapenemase, we recommend including additional carbapenems (e.g., imipenem, doripenem, ertapenem, meropenem)
These requirements are also described at Biosample beta-lactamase antibiogram documents. To submit these phenotypic data, register a Biosample for this sequence and then submit the Bankit file as described above (along with an antibiogram if available). When registering a Biosample, please select the "Beta-lactamase" package on the Sample Type tab. Do not use the Biosample accession of the isolate from which the sequence was derived. (For isolate data, use either the Pathogen affecting public health or One Health Enteric Package Biosample packages). Your Biosample will be linked automatically to the Beta-Lactamase Alleles Bioproject , so you should not register a Bioproject for this Biosample. Once your Bankit submission has been approved by GenBank, email [email protected] a completed antibiogram and also request that your Bankit submission be linked to this Biosample.
Note: if you are not submitting AST data with your allele request, we encourage submitters to use the procedure described in the previous section "Novel Beta-Lactamase, Qnr, and Mcr Allele Submission Requirements".
For any questions or concerns about any of these requirements, please contact NCBI at [email protected].
