ABSTRACT
Cell state transitions are often triggered by large changes in the concentrations of transcription factors and therefore large differences in their stoichiometric ratios. Whether cells can elicit transitions using modest changes in the ratios of co-expressed factors is unclear. Here, we investigate how cells in the Drosophila eye resolve state transitions by quantifying the expression dynamics of the ETS transcription factors Pnt and Yan. Eye progenitor cells maintain a relatively constant ratio of Pnt/Yan protein, despite expressing both proteins with pulsatile dynamics. A rapid and sustained twofold increase in the Pnt/Yan ratio accompanies transitions to photoreceptor fates. Genetic perturbations that modestly disrupt the Pnt/Yan ratio produce fate transition defects consistent with the hypothesis that transitions are normally driven by a twofold shift in the ratio. A biophysical model based on cooperative Yan-DNA binding coupled with non-cooperative Pnt-DNA binding illustrates how twofold ratio changes could generate ultrasensitive changes in target gene transcription to drive fate transitions. Thus, coupling cell state transitions to the Pnt/Yan ratio sensitizes the system to modest fold-changes, conferring robustness and ultrasensitivity to the developmental program.
Keywords: Drosophila, Differentiation, Eye, Photoreceptor
Summary: Using a biophysical model, we demonstrate how a modest change in the Pnt/Yan ratio affects the transition of eye disc cells to photoreceptor cell fates.
INTRODUCTION
Organismal development proceeds through a sequence of cell transitions that yield increasingly restricted cellular states or fates. A common feature of cells that undergo fate transitions is the apparent irreversibility of the transitions, even when triggered by transient stimuli. Dynamic modeling of these transitions often assumes multistability, in which cells can be resting in one of two or more stable states (Rand et al., 2021; Sáez et al., 2022). These stable states are typically a progenitor state and one or more derived states with lesser developmental potential.
Transcription factors are frequently used to define cell states, and transitions to a new state are often driven by varying their absolute or relative protein abundance (Laslo et al., 2006; Park et al., 2012; Yao et al., 2008). A considerable number of developmental transitions rely on mutually exclusive expression of two antagonistic transcription factors. Each factor programs a distinct cell state such that cells adopt one of two possible fates depending on whether they express high levels of one transcription factor or the other. Transitions are triggered when transient stimuli alter the balance between the two factors, resulting in expression of a new dominant factor. First documented in the early embryo of Drosophila (Jaeger et al., 2004), such regulatory switches built on mutual repression function in mouse embryonic germ-layer restriction, hematopoiesis, neural tube patterning, mesoderm development and pancreatic lineage restriction (Acloque et al., 2011; Briscoe and Ericson, 2001; De Kumar et al., 2017; Lagha et al., 2009; Schaffer et al., 2010; Schrode et al., 2014). A hallmark of such systems is the mutually exclusive expression of the two opposing transcription factor regulators, which is frequently controlled by a bi-stable mechanism and double-negative feedback loops connecting the two factors.
In Drosophila, two ETS-domain transcription factors, Pointed (Pnt) and Yan (also known as Aop, Anterior open), act downstream of signals mediated by receptor tyrosine kinases (RTKs) to regulate cell fate transitions in a wide variety of tissues across the body and across the life cycle (Flores et al., 2000; Gabay et al., 1996, 1997; Halfon et al., 2000; Boisclair Lachance et al., 2014; Melen et al., 2005; O'Neill et al., 1994; Rebay and Rubin, 1995; Rogge et al., 1995; Shwartz et al., 2013; Xu et al., 2000). Consistently across many different developmental transitions, loss of yan results in too many progenitors transitioning to a particular fate, whereas loss of pnt prevents fate transitions (Brunner et al., 1994; Flores et al., 2000; Gabay et al., 1996; Halfon et al., 2000; O'Neill et al., 1994; Xu et al., 2000). As both Pnt and Yan bind to a common GGA(A/T)-containing DNA sequence motif (Nitta et al., 2015; Wei et al., 2010; Zhu et al., 2011), competition for DNA occupancy and antagonistic regulation of common target genes is thought to orchestrate the transcriptional changes that drive particular transitions (Flores et al., 2000; Halfon et al., 2000; Hollenhorst et al., 2011; Boisclair Lachance et al., 2018; Nitta et al., 2015; Webber et al., 2013a,b, 2018; Wei et al., 2010; Xu et al., 2000; Zhu et al., 2011).
The opposing genetic phenotypes and transcriptional activities of Pnt and Yan, together with their mutually exclusive expression patterns in certain tissues, inspired development of a bi-stable model for cell fate specification based on mutual repression of the expression of one another (Gabay et al., 1996; Golembo et al., 1996a; Graham et al., 2010; Melen et al., 2005). For example, in the embryonic ventral ectoderm, ventral-most cells express Pnt whereas ventrolateral cells express Yan (Gabay et al., 1996). This pattern is established by secretion of a ligand for the EGF receptor (EGFR) from the ventral midline (Golembo et al., 1996b). EGFR activation in nearby cells induces the Ras pathway to express Pnt and degrade Yan (Gabay et al., 1996; Melen et al., 2005). Cells with insufficient EGFR activation express Yan, which represses Pnt. Dynamic modeling has described this as a bi-stable system in which cells assume different fates based on whether they are in a low Pnt/high Yan state versus a high Pnt/low Yan state (Graham et al., 2010; Melen et al., 2005).
Although the bi-stable model readily explains transitions in which the different states are marked by mutually exclusive Pnt and Yan expression, an unresolved paradox is that many tissues have extensive co-expression of Yan and Pnt (Boisclair Lachance et al., 2014). The third-instar larval eye disc is one such tissue. Qualitative imaging has shown that rather than adopting mutually exclusive expression states, eye progenitor cells co-express high levels of Pnt and Yan for prolonged periods. Furthermore, when progenitors transit to photoreceptor (R cell) fates, they continue to co-express both proteins, transiently increasing Pnt levels before initiating a parallel decay in both Pnt and Yan (Boisclair Lachance et al., 2014). These observations are at odds with the long-standing assumption that cross-repression within a bi-stable network creates mutually exclusive Yan and Pnt expression states during R cell fate specification (Graham et al., 2010; Lai and Rubin, 1992; Rebay and Rubin, 1995).
Here, we have explored how Yan and Pnt expression dynamics in the third instar eye disc impact progenitor to R-cell fate transitions. Quantitative fluorescence-based microscopy was coupled with automated high-throughput image analysis to measure Yan and Pnt protein levels at single cell resolution in thousands of eye disc cells. We find that despite the variation in Pnt and Yan levels, cells in the progenitor state maintain a fairly constant ratio of Pnt/Yan for up to 50 h of development. When specific progenitors transit to R-cell fates, the Pnt/Yan ratio rapidly increases, but only by at most twofold. Experimental perturbation of the ratio results in progenitor cells either prematurely transiting or failing to transit to R-cell fates, implicating the Pnt/Yan ratio as the driver of cell fate transitions. Using a biophysical model, we demonstrate how even a modest change in the Pnt/Yan ratio will cause a large change in the DNA occupancy of the two proteins on their common target genes. We conclude that the transition to photoreceptor fates is sensitive to the stoichiometric ratio of Pnt and Yan, and propose that ratiometric sensing mechanisms could confer ultrasensitivity to biological transitions regulated by multiple factors.
RESULTS
During the final 50 h of the larval phase of the Drosophila life cycle, a morphogenetic furrow (MF) moves across the eye disc epithelium at a constant velocity from posterior to anterior (Fig. 1A). As the MF moves, every 2 h a column of periodic R8 photoreceptor cells is formed almost simultaneously, immediately posterior to the MF. Each R8 cell in the column secretes ligands that activate the RTK-Ras-MAPK pathway in seven neighboring progenitor cells and causes them to transit from progenitor to R-cell fates (Freeman, 1996). These transitions occur stepwise every ∼2 h, first generating R2 and R5 cells, then R3 and R4 cells, then R1 and R6 cells, and finally the R7 cell (Fig. S1A; Wolff and Ready, 1993). As the MF repeatedly induces new columns of R-cell clusters or ommatidia, it ultimately forms a lattice pattern of ∼750 ommatidia in each eye. Although ∼6000 progenitor cells adopt R-cell fates, a pool of ∼14,000 progenitors remains uncommitted, and later adopts other retinal cell fates (Wolff and Ready, 1991).
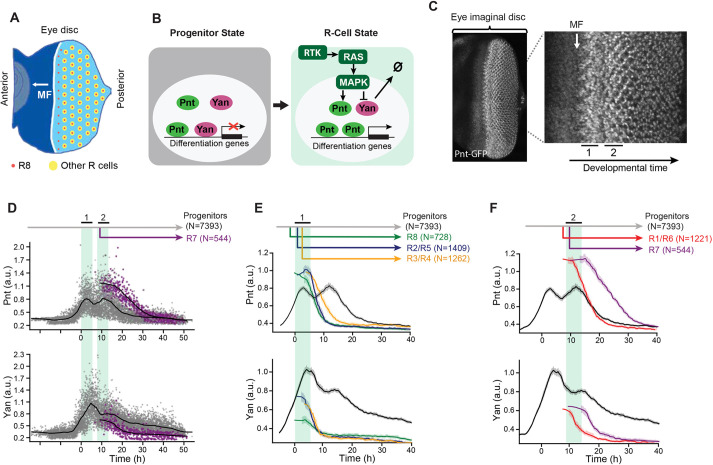
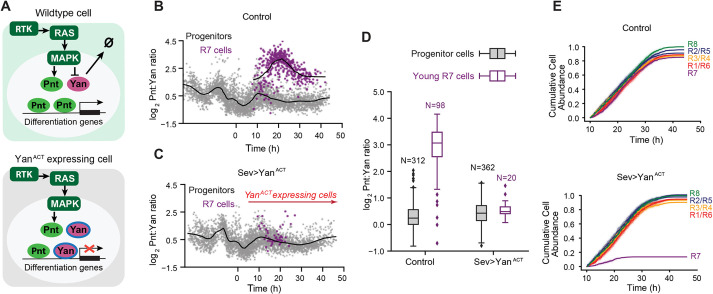
Fig. 1.
Pnt and Yan expression defines two waves of cell state transitions. (A) Schematic of a developing eye disc ∼100 h post-fertilization. The passage of the morphogenetic furrow (MF) (white arrow) initiates R cell fate specification. Specification of regularly spaced R8 cells (red dots) marks the start of ommatidial assembly and is followed by recruitment of additional R-cell types (yellow dots). Adapted from Peláez et al. (2015), where it was published under a CC-BY 4.0 license. (B) Schematic summary of RTK-mediated regulation of progenitor to R-cell fate transitions. Progenitor cells co-express Pnt and Yan (left). In response to RTK signals, cells increase Pnt and decrease Yan (right). (C) Maximum intensity projection of Pnt-GFP fluorescence in an eye disc oriented anterior left and dorsal up. Magnified view shows that the two zones where progenitor cells undergo fate transitions coincide with peaks of Pnt-GFP (black bars, labeled 1 and 2). (D) Pnt-GFP and Yan levels in individual progenitor (gray) and R7 (purple) cells over developmental time. N is the number of cells analyzed in each group. Progenitor cells are present across all times (gray arrow), while R7 cells arise later in time (purple arrow). Solid black lines are smoothed moving averages across 250 and 75 individual nuclei for progenitor and R7 cells, respectively. Black bars labeled 1 and 2 indicate the two peaks of Pnt-GFP in progenitor cells. Shaded vertical stripes highlight these two regions, which coincide with the transition of progenitors to various R fates. Although time appears continuous in these plots (see Materials and Methods), temporal resolution is about 2 h, i.e. the time between specification of each column of R8 cells. (E,F) Line averages, with 95% confidence intervals shaded, for Pnt-GFP and Yan levels in progenitor (gray), R8 (green), R2/R5 (blue), R3/R4 (orange), R1/R6 (red) and R7 (purple) cells over developmental time. N is the number of cells analyzed in each group.
Progenitor cells posterior to the MF co-express Pnt and Yan, and are poised to adopt R-cell fates (Fig. 1B). When progenitors transition to R-cell fates, RTK-Ras-MAPK signals regulate the opposing transcriptional inputs from Pnt and Yan on their shared target genes. The signals concomitantly downregulate Yan activity and upregulate Pnt activity, which is thought to shift the state of the progenitor cell towards an R-cell state by reprogramming target gene expression.
To measure Pnt protein abundance in individual eye cells, we used a genomic transgene of the pnt locus in which fast-fold green fluorescent protein (GFP) was inserted in-frame at the C terminus of the coding sequence, tagging all Pnt isoforms. The Pnt-GFP transgene fully complemented null mutations in the endogenous pnt gene and completely restored normal eye development (Boisclair Lachance et al., 2014), demonstrating that the GFP tag does not compromise Pnt function and that all essential genomic regulatory sequences are included. To measure Yan protein abundance, we used a monoclonal antibody that elicits immunofluorescence linearly proportional to native Yan protein concentration (Peláez et al., 2015). This enabled us to measure nuclear Pnt-GFP and Yan protein abundance simultaneously in individual cells; however, it is important to note that our approach does not measure absolute protein concentrations. Using an established computational pipeline (Peláez et al., 2015), we segmented all eye cell nuclei and then calculated Pnt-GFP and Yan fluorescence levels, and exact 3D positions in each eye disc (Fig. S1B-E). A unique feature of eye development allowed us to map the spatial position of each cell in the plane of the eye disc onto developmental time. Because the MF moves at a fixed velocity, forming one column of R8 cells every 2 h (Basler and Hafen, 1989; Campos-Ortega and Hofbauer, 1977; Gallagher et al., 2022), the distance between the MF and any cell posterior to it is linearly proportional to the time elapsed since the MF passed. This mapping was applied to the thousands of segmented nuclei in each eye disc and repeated for multiple eye discs.
A second feature of eye development enabled us to annotate cell states without relying on marker genes but instead on reproducible changes in cell morphology and apical-basal nuclear position. These stereotyped morphological changes are observable before the earliest known marker genes begin to be expressed, allowing accurate annotation of progenitor and R-cell states (Fig. S1A,F) (Peláez et al., 2015; Tomlinson and Ready, 1987; Wolff and Ready, 1993). Cross-referencing with cell-type-specific markers confirmed that we could make cell state assignments with >97% accuracy using only morphology (Table S1). Combining these state assignments with the measurements of Pnt-GFP expression, Yan expression and developmental time for many thousands of cells generated a composite picture of Yan and Pnt expression dynamics (Fig. 1D). Although this approach cannot track the behavior of any individual cell over time, it nevertheless provides a dynamic view of eye cells as they develop. From this composite picture, individual cell behaviors can be inferred.
Focusing first on progenitor cells, we found that both Pnt-GFP and Yan expression dramatically increased coincident with passage of the MF (Fig. 1C,D). This rapid increase culminated in two successive peaks of Pnt-GFP expression and a single peak of Yan expression at a time period in between the two Pnt-GFP peaks (Fig. S2). The peaks of Pnt-GFP in progenitor cells coincided with the time periods when they transition to R-cell fates (Fig. 1C-F). Transitions to R8, R2/R5 and R3/R4 fates occurred during the first peak of expression (Fig. 1E), while transitions to R1/R6 and R7 fates occurred during the second peak (Fig. 1F). After their peaks, both Pnt-GFP and Yan decayed to a basal level in progenitors (Fig. 1D). Despite the alternating peaks of Pnt and Yan expression, an overall trend of induction and decay was apparent for both proteins.
Focusing next on Pnt-GFP and Yan dynamics in cells that had transitioned to R-cell fates, we found that these transitions coincided with a rapid increase in Pnt-GFP and rapid decrease in Yan (Fig. 1D-F). The Pnt-GFP increase was so rapid that the youngest cells we could classify as R-cells already had a 25-50% higher level of Pnt-GFP than progenitor cells at comparable time points (Fig. 1D). Thereafter, the R-cells maintained this elevated Pnt-GFP for 2-4 h before downregulating Pnt-GFP to a basal level of expression (Fig. 1E,F). Similarly, the decrease in Yan in transitioning cells was so rapid that the youngest cells classified as R-cells already had a 25-50% lower level of Yan than progenitors at comparable time points (Fig. 1D). Thereafter, R-cells also maintained this diminished level of Yan for 2-4 h before further downregulating its expression (Fig. 1E,F).
Two key conclusions emerged from this analysis. First, Pnt and Yan expression appear positively correlated in progenitor cells, confirming previous qualitative observations (Boisclair Lachance et al., 2014). Second, transitions to R-cell fates coincide with concomitant but modest up- and downregulation of Pnt and Yan expression, respectively. This suggests that cell fate specification in the eye does not rely on a classic bi-stable mechanism in which mutual repression between Pnt and Yan produces large changes in expression to distinguish the two states (Graham et al., 2010; Melen et al., 2005).
Pnt and Yan positively regulate the expression of one another
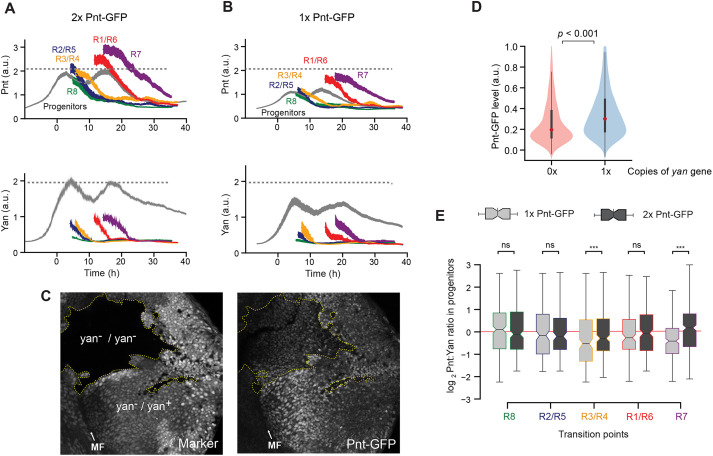
The positive correlation in expression of Yan and Pnt in progenitor cells led us to hypothesize that they might positively regulate the expression of one another. To test this possibility, we first manipulated Pnt protein levels by varying the copy number of the pnt gene. If Pnt positively regulates Yan expression, then we predicted Yan levels should change in parallel to altered Pnt levels. Cells with two copies of the pnt gene expressed ∼50% more Pnt protein than cells with one copy (Fig. 2A,B). This difference was observed both in progenitor cells and R cells. In spite of the different levels of Pnt protein in cells with one or two copies of the pnt gene, progenitors transited to R-cell fates at similar times (Fig. 2A,B), and produced adult eyes with wild-type morphologies (Fig. S3). When we measured Yan, we found that Yan levels also differed between the two genotypes, despite both having two copies of the yan gene (Fig. 2A,B). Yan output was 25-50% greater in progenitor cells with two copies of the pnt gene than in cells with one copy, consistent with a role for Pnt in positively regulating Yan expression.
Fig. 2.
Pnt and Yan activate the expression of one another. (A,B) Line averages, with 95% confidence intervals shaded, for Pnt-GFP and Yan levels in progenitor and R cells containing two copies (A) or one copy (B) of the pnt-GFP gene. The number of cells analyzed for each cell class ranged from 312 (R7 cells) to 3127 (progenitors) in A, and from 271 (R7 cells) to 2834 (progenitors) in B. (C) Confocal sections showing progenitor cell nuclei in an eye disc containing several patches (clones) of homozygous yan mutant cells, marked by lack of RFP and outlined by dotted yellow lines. Right panel: Pnt-GFP fluorescence signal is lower in the yan mutant clones. (D) Pnt-GFP abundance is significantly lower in homozygous yan mutant (0×) versus heterozygous (1×) progenitor cells. Violin plots contain red dots that indicate the median of each distribution and gray lines that indicate the interquartile range. P<0.001, Mann–Whitney U-test. (E) Comparison of Pnt/Yan ratios in progenitor cells with one copy (light gray) versus two copies (dark gray) of the pnt-GFP gene. Cells were differentially sampled across five time-windows of cell fate transitions. These time windows were defined in each disc by the time spanned by the first ten identifiable R cells of a given class. Progenitor cells were sampled from these time windows. Median ratios, boxed by the quartile ratios, are shown. Whiskers indicate the upper- and lower-most quartile ratios. The ratio in progenitor cells is statistically indistinguishable between cells bearing different gene doses during the R8, R2/R5 and R1/R6 cell fate transitions (Kolmogorov-Smirnov two-sample test; ns, P>0.1; ***P<0.001).
To ask whether this positive regulatory relationship might be mutual, we next assessed the effect of Yan levels on Pnt-GFP expression by varying the copy number of the yan gene. We conducted this experiment by generating clones of yan homozygous mutant cells in an eye where other cells have one or two copies of the yan gene. Qualitatively, cells with different yan gene copy numbers exhibited Pnt-GFP levels that positively correlated with yan gene dose (Fig. 2C). Using a published pipeline to segment and measure cells within eye disc clones (Bernasek et al., 2020), we quantified and compared Pnt-GFP protein levels in progenitor cells with zero versus one wild-type copy of the yan gene (Fig. 2D). Pnt-GFP expression was ∼30% greater in progenitor cells with one copy of yan than in cells with zero copies of yan (Fig. 2D). This result suggests that Pnt expression is positively dependent on Yan.
The parallel behaviors of Pnt and Yan in these experiments prompted us to quantitatively assess the extent to which Pnt and Yan levels are coordinated within individual cells. To do this, we calculated the Pnt/Yan ratio in progenitor cells containing one or two copies of the pnt gene. The Pnt/Yan ratio was invariant to the pnt gene copy number in progenitor cells that were sampled at discrete time points over their developmental trajectory (Fig. 2E). It was striking that the ratio measured in the progenitors was fairly constant between the sampled time points. To examine this more closely, we calculated the ratio for all progenitor cells over the entire 50 h of eye development (Fig. 3A). The average Pnt/Yan ratio in progenitor cells remained dynamically stable about a fixed value over time (Fig. 3A). In conclusion, these results demonstrate that Yan and Pnt expression are tightly and positively coordinated in progenitor cells.
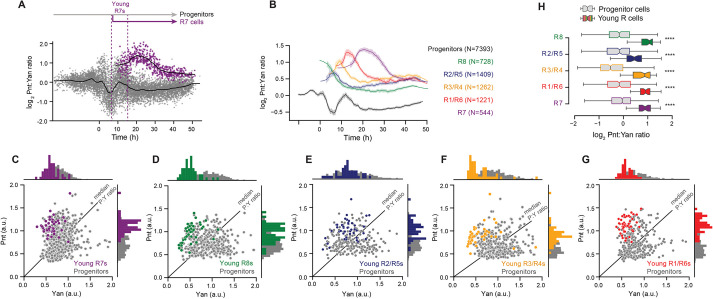
Fig. 3.
A twofold shift in the Pnt/Yan ratio accompanies R cell fate transitions. (A) The log2-transformed Pnt/Yan ratio in individual progenitor (gray, 7393 cells) and R7 (purple, 544 cells) cells, and their moving line averages (solid black lines). Cells marked as ‘Young R7s’ are the initial cohort of R7 cells that are present in the timespan of the first ten identifiable R7 cells (‘young’ R7s become ‘mature’ Elav-positive R7s in about 2 h). (B) Line averages, with 95% confidence intervals shaded, of the log2-transformed Pnt/Yan ratio for all annotated cell types over time. N is number of cells sampled. (C-G) Joint distributions of Pnt-GFP and Yan levels in young R7 (C), young R8 (D), young R2/R5 (E), young R3/R4 (F) and young R1/R6 (G) cells. Progenitor cells (gray) present in the timespan of the young R cells were analyzed for comparison. Black lines indicate median Pnt/Yan ratios among progenitor cells. Histograms of the data at the top and side of each scatterplot highlight the overlap between R cells and some of the concurrent progenitors. (H) Comparison of the Pnt/Yan ratios in young R cells (colored boxes and whiskers) versus progenitor cells (gray boxes and whiskers) sampled over comparable time-periods. Forty cells were analyzed for each group of young R cells, and 153-487 cells were analyzed for each group of progenitors. Median ratios boxed by the quartile ratios are shown. Whiskers indicate the upper- and lower-most quartile ratios. ****P<0.001, one-tailed Mann–Whitney U-test.
An increase in the Pnt/Yan ratio accompanies cell state transitions
We next determined the Pnt/Yan ratio for R cells. Because we are not measuring absolute Pnt and Yan protein concentrations, and because even after normalizing by nuclear intensity the constant of proportionality may be different for Pnt (measured with GFP) and Yan (measured with antibody), our measurements are limited to assessing how the ratio values are changing relatively between sampled populations and between time points. We first measured the ratio in R7 cells along their developmental trajectory. Transitions to the R7 fate coincided with a rapid increase in the ratio (Fig. 3A). The increase was so rapid that the youngest cells we could classify as R7 cells already had a twofold higher ratio than progenitor cells at comparable time points. Thereafter, the ratio rose and then gradually fell over time, but always remained at a higher value than that of progenitor cells. Examination of the Pnt/Yan ratio in all other R-cell types revealed a similar and consistent rapid increase as they transitioned from a progenitor state, followed by a gradual decline to a new steady value that was higher than that of progenitor cells (Fig. 3B and Fig. S4).
The rapid shift in the Pnt/Yan ratio during cell state transitions suggested that the progenitor ratio becomes destabilized at the onset of these transitions. If so, then cell-to-cell heterogeneity of the ratio should rise during the time-windows of state transitions, and then subside as cell states are resolved. Indeed, greater cell-to-cell variation in the ratio coincided with the times when Pnt and Yan levels peaked (Fig. 3A), corresponding to the two waves of R-cell differentiation. To further characterize this instability, we used an established method (Peláez et al., 2015) to quantify the cell-to-cell heterogeneity of Pnt, Yan and the Pnt/Yan ratio over time (Fig. S5). Progenitor cells exhibited maximal ratio heterogeneity during the two time periods that coincide with cell state transitions. Thereafter, heterogeneity diminished in the remaining progenitor cells. R cells exhibited an abrupt drop in heterogeneity immediately after their fate specification, and heterogeneity descended further thereafter. These results support the idea that the Pnt/Yan ratio becomes specifically destabilized when progenitor cells are poised to undergo state transitions.
If cells undergoing state transitions experience a rise in the Pnt/Yan ratio, we should observe cells at transition points that cannot be morphologically classified as R cells but that possess an elevated ratio. This prediction was verified as there was an overlap in ratio values between the youngest cells we could classify as R cells and a subset of other cells sampled at comparable time points (Fig. 3C-G). Nevertheless, the median ratio values were significantly greater in R cells and in all other cells (Fig. 3H, P<0.001, one-tailed Mann–Whitney U-test). Together, these measurements suggest that, in response to inductive signaling, a subset of progenitor cells elevates their Pnt/Yan ratio by approximately twofold relative to the average progenitor pool ratio, and that this increase is sufficient to confer molecular competency for transitioning to a photoreceptor state. From this pool of competent progenitors, only the cell that sustains the elevated ratio will complete the transition to an R state.
Perturbation of the Pnt/Yan ratio biases cells towards or against R-cell fates
The previous model for eye development suggested that Yan and Pnt antagonize one another to maintain cells in a progenitor state defined by high Yan and to move cells towards a R-cell state defined by high Pnt (Graham et al., 2010). Our results demonstrate that the cell states are not distinguished by large differences in the Pnt/Yan ratio. Instead, we hypothesize that a twofold change in the ratio is sufficient to trigger R-cell differentiation. We tested this by experimentally altering the ratio in progenitor cells and then asking whether this perturbation altered the normal course of R-cell fate transitions.
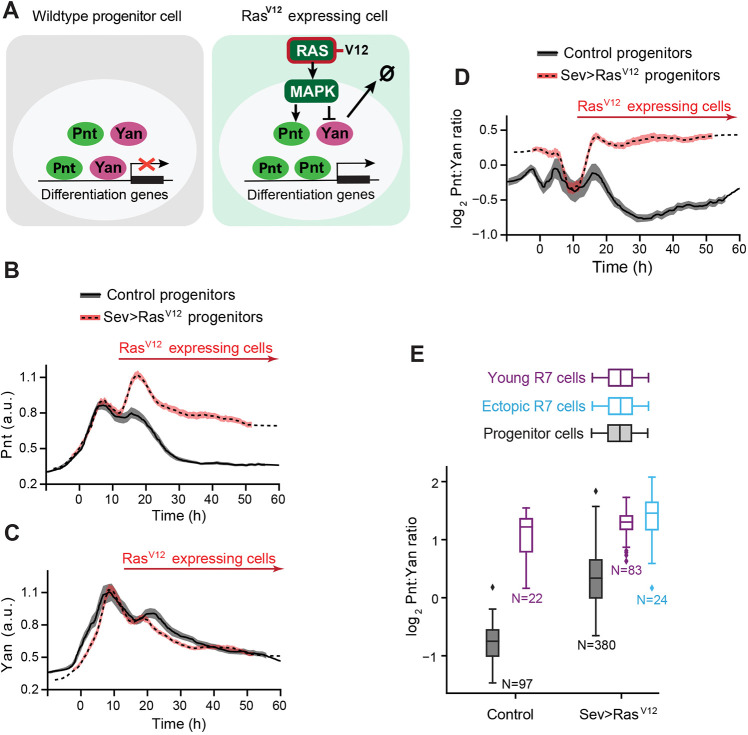
Ras-dependent MAPK phosphorylation of Yan and Pnt has previously been found to stimulate turnover of Yan and synthesis of Pnt (Brunner et al., 1994; Gabay et al., 1996; O'Neill et al., 1994; Rebay and Rubin, 1995). Therefore, we expressed a mutant allele of the Ras1 gene encoding a constitutively active version of the Ras GTPase, reasoning that the increased signaling would artificially increase the Pnt/Yan ratio (Fig. 4A). RasV12 was transiently expressed in subsets of progenitor cells during the R3/R4 and R7 transition points by driving it with the sevenless (sev) promoter (Fig. S6; Fortini et al., 1992). In addition to driving expression in subsets of progenitors during the second wave of R fate transitions, the sev promoter also drove stable expression in R3/R4 and R7 cells (Fig. S6). As predicted, sev>RasV12 progenitor cells had an elevated second wave of Pnt expression, which coincides with the second time-window of cell state transitions (Fig. 4B). Thereafter, progenitors maintained abnormally high levels of Pnt. Yan levels were weakly reduced in progenitor cells during the second transition period (Fig. 4C). Although Ras/MAPK signaling promotes Yan degradation (Fig. 4A) (Rebay and Rubin, 1995), the close to normal Yan levels suggests that the system ‘tried’ to compensate for the increase in Pnt levels by increasing Yan production (Fig. 2), but ‘failed’ because the sustained MAPK activation induced by RasV12 prevented Yan protein accumulation. The end result was a twofold elevation in the Pnt/Yan ratio in progenitor cells at the second transition period and thereafter (Fig. 4D). When we looked at R-cell fate specification, we observed supernumerary R7 cells in the sev>RasV12 eye discs (Fig. 4E), consistent with the known ability of sev>RasV12 expression to induce ectopic R7 photoreceptors (Fortini et al., 1992). These ectopic R7 cells were typically located beside R7 cells, expressed the R7 marker Prospero (Fig. S7A,B,D) and had comparable Pnt/Yan ratios to R7 cells (Fig. 4E). Thus, just as a twofold ratio increase accompanies progenitor to R7 fate transitions in a wild-type eye disc, artificially inducing a twofold elevation in the Pnt/Yan ratio in the progenitor pool causes additional progenitor cells to inappropriately transit to R-cell fates.
Fig. 4.
An artificial increase in the Pnt/Yan ratio biases progenitor cells towards an R fate. (A) Schematic showing the effect of RasV12 on signal transduction and Yan and Pnt expression in eye cells. (B-D) Line averages, with 95% confidence intervals shaded, of Pnt-GFP (B), Yan (C) and log2-transformed ratio of Pnt/Yan ratio (C) in wild-type (black) and sev>RasV12 (red) progenitor cells over developmental time. Red arrows indicate the time during which cells express RasV12. We have previously reported a modest increase in the duration of Yan-YFP expression in sev>RasV12 progenitor cells (Peláez et al., 2015), but this difference was not detected using the Yan antibody (B). 1381 (wild type) and 4038 (sev>RasV12) progenitors were analyzed. (E) The log2-transformed ratio of Pnt/Yan in wild-type and sev>RasV12 young R7 cells (purple). Progenitor cells (gray) present in the same timespan were also analyzed. In sev>RasV12 eye discs, there were ectopic R7 cells (blue). Box plots show the median (horizontal line) and quartiles (upper and lower edges of the box). Whiskers indicate the upper- and lower-most quartile ratios. N is the number of cells analyzed in each group.
We next asked whether reducing the ratio would prevent R-cell state transitions. It has previously been shown that MAPK phosphorylation of Yan strongly stimulates Yan turnover (Rebay and Rubin, 1995). If the serines and threonines targeted by MAPK are mutated in Yan, then signaling-induced Yan turnover is blocked (Fig. 5A) and photoreceptor fates are not induced (Rebay and Rubin, 1995). We transiently expressed this mutant YanACT protein in progenitor cells and R3, R4 and R7 cells using the sev promoter. The Pnt/Yan ratio in progenitor cells was not detectably altered in sev>YanACT eye discs (Fig. 5B-D and Fig. S8A-C). This was not surprising as the RTK-Ras-MAPK pathway is low in progenitors, and therefore YanACT protein is not abnormally stable in these cells. Nor was the ratio altered in R8, R2, R5, R1 and R6 cells of sev>YanACT eye discs, as the sev promoter does not induce YanACT expression in these cells (Fig. S9A-D,G,H). However, in R3, R4 and R7 cells, which all receive a strong RTK signal and express YanACT, there was a reduction in the Pnt/Yan ratio. The reduction was weak in R3/R4 cells (Fig. S9E,F) but was larger in R7 cells, which had a ratio value similar to progenitor cells sampled at comparable times (Fig. 5B-D). Consistent with the experimental design, the reduction in the Pnt/Yan ratio in R cells was primarily driven by higher levels of Yan protein (Fig. S8D).
Fig. 5.
An artificial decrease in the Pnt/Yan ratio biases progenitor cells against an R fate. (A) Schematic showing the effect of YanACT on signal transduction, and Yan and Pnt expression in eye cells. (B) The log2-transformed Pnt/Yan ratio in wild-type control progenitor (gray, 3796 cells) and R7 (purple, 413 cells) cells, and their moving line averages (solid black lines). (C) The log2-transformed Pnt/Yan ratio in sev>YanACT progenitor (gray) and R7 (purple) cells, and their moving line averages (solid black lines). Red arrow indicates the time during which cells express YanACT. (D) The log2-transformed ratio of Pnt/Yan in wild-type control and sev>YanACT young R7 cells. Progenitor cells (gray) that were present in the timespan of these young R7 cells (purple) were also analyzed. Box plots show the median (horizontal line) and quartiles (upper and lower edges of the box). Whiskers indicate the upper- and lower-most quartile ratios. N is number of cells analyzed in each group. (E) Cumulative abundance of each R cell type as a function of developmental time in wild-type (top) and sev>YanACT (bottom) eye discs. Counts are normalized by the total number of R8 cells detected across the time-course, and R2, R5, R3, R4, R1 and R6 cells are each tallied separately. Shading indicates 95% confidence intervals.
Changing the Pnt/Yan ratio to make it Yan biased in young R7 cells induced a remarkable reduction in the total number of R7 cells that developed (Fig. 5B,C and Fig. S7A,C). Only a few R7 cells could be identified shortly after fate specification, and more mature R7 cells could not be found. Overall, the progressive fate specification of the R7 cells was reduced seven-fold in the sev>YanACT eye discs (Fig. 5E). Thus, preventing the ratio increase that normally occurs in transitioning R7 cells was sufficient to disrupt the transition, even though the ratio was unchanged in the progenitor pool. In contrast, R3/R4 cell fate specification was not impaired, even though the Pnt/Yan ratio was slightly lower than normal (Fig. 5E and Fig. S9I,J). Together, these results suggest that cell state transitions can withstand weak or transient variation in the ratio, but not sustained larger changes.
In conclusion, two independent approaches to artificially alter the Pnt/Yan ratio resulted in cells making errors in state transitions. Increasing the ratio caused cells to undergo ectopic state transitions, while decreasing the ratio caused cells to fail to undergo transitions. The results suggest that progenitor cells can sense a twofold change in the Pnt/Yan stoichiometric ratio and respond by transitioning to R-cell fates.
A biophysical model explains how small changes in the stoichiometric ratio can have a large effect on Yan and Pnt DNA occupancy
The effect of transcription factors on their target genes is highly correlated with their occupancy on DNA elements within these genes. When transcription factors do not compete for binding sites, occupancy is driven by total nuclear concentration of the transcription factor and its binding affinity. In systems with transcription factors that mutually repress the expression of one another expression, the stoichiometry of the factors can be very different in cells. Thus, DNA occupancy by antagonistic transcription factors reflects the large difference in their relative abundance. As Pnt and Yan have overlapping sequence specificity for DNA binding both in vitro and in vivo, they are thought to compete for common binding sites in target genes (Flores et al., 2000; Halfon et al., 2000; Boisclair Lachance et al., 2018; Nitta et al., 2015; Webber et al., 2018; Wei et al., 2010; Xu et al., 2000; Zhu et al., 2011). However, the differences in Pnt/Yan stoichiometry that we measured in eye cells are only about twofold. How could such modest differences in stoichiometry profoundly affect relative DNA occupancy by Yan and Pnt to alter transcription of target genes?
Measuring DNA occupancy at single cell resolution is not yet experimentally feasible, so we instead investigated this question in silico. We first created a simple biophysical model for equilibrium binding of two transcription factors that compete for common DNA-binding sites. If the proteins have a similar binding affinity for DNA and if the binding sites are saturated, then at equilibrium, occupancy will be sensitive to the relative concentration of each factor across a broad range of absolute concentrations (Fig. S10).
However, the situation with Pnt and Yan is more complex. Yan binds to common binding sites with higher affinity than Pnt (Xu et al., 2000). Moreover, recent experiments suggest Yan and Pnt differentially interpret the structural syntax of enhancers and promoters (Boisclair Lachance et al., 2018). This complexity is a consequence of cooperative interactions between DNA-bound Yan molecules. Yan monomers are able to self-associate via their sterile alpha motifs (SAM), enabling DNA-bound Yan monomers to stabilize the recruitment of additional Yan monomers to adjacent sites (Hope et al., 2017; Boisclair Lachance et al., 2018; Qiao et al., 2004). As Pnt does not self-associate (Mackereth et al., 2004; Meruelo and Bowie, 2009), this Yan-Yan cooperativity will bias its competitiveness for common binding sites.
Therefore, we adapted a modeling framework developed to probe the effects of cis-regulatory syntax on Yan binding site occupancy (Hope et al., 2017). The model considers an ensemble of microstates, each defined by a unique configuration of vacant or Yan-bound sites. Each microstate is assigned a thermodynamic potential based on the cumulative influence of strong sequence-specific binding to consensus ETS sites, weak non-specific DNA binding and SAM-SAM polymerization. We generalized the earlier model by incorporating Pnt as a second transcription factor that competes for occupancy of the same binding sites but does not self-associate (Fig. S11A,B). Using this model, we explored how Yan-Yan cooperativity affects Pnt binding site occupancy.
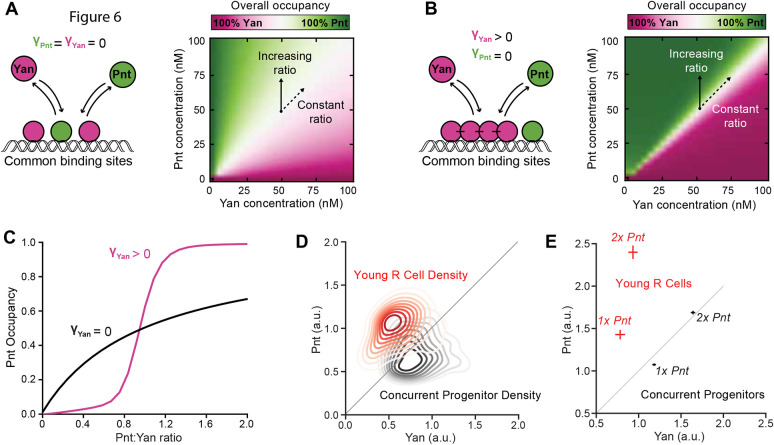
We first considered a scenario in which neither Yan nor Pnt exhibits cooperativity (Fig. 6A). In the absence of stabilizing SAM-SAM interactions, the landscape of overall binding site occupancy was identical to that obtained with the simple equilibrium model described earlier (Fig. 6A, Figs S10 and S11). Increasing the Pnt/Yan ratio resulted in a gradual increase in Pnt occupancy for all individual binding sites, producing a titration contour closely resembling a Michaelian curve (Fig. 6C and Fig. S11C).
Fig. 6.
Cooperative binding by Yan greatly sensitizes DNA occupancy to the Pnt/Yan ratio. (A) Left: schematic showing that Pnt and Yan compete for occupancy of mutual DNA-binding sites when Yan is unable to self-polymerize. Right: overall binding site occupancy calculated as a function of transcription factor abundance. We use a diverging scale because all sites are fully saturated at total transcription factor concentrations above 1 nM. Under the range of conditions shown, this implies that Yan occupies all sites left vacant by Pnt. Simultaneous proportional increases in absolute abundance of both species have minimal impact on Pnt occupancy (dashed arrow), while a varying ratio confers gradual change (solid arrow). (B) Left: schematic of Pnt and Yan competition for occupancy of mutual DNA binding sites when Yan can self-polymerize. Right: overall binding site occupancy calculated as a function of transcription factor abundance. The sharpness of the transition between all-Pnt occupancy and all-Yan occupancy is increased compared with A. (C) Average Pnt occupancy across all binding sites as a function of the Pnt/Yan ratio when Yan is unable to self-polymerize (black line) versus when Yan can self-polymerize (pink line). Contours correspond to vertical paths traversed across the phase-plots in A and B at a fixed Yan concentration of 50 nM. (D) Probability density function of experimentally measured Pnt-GFP and Yan levels for the combined populations of all young R cells (red) and concurrent progenitor cells (black). Contours decrease in opacity with decreasing density and are not shown for values below 0.1. Density function is estimated using Gaussian kernels with a bandwidth selected using Scott's rule. A thin diagonal line shows a constant Pnt/Yan ratio to aid interpretation. (E) Experimentally measured average Pnt-GFP and Yan levels for the combined populations of all young R cells (red) and concurrent progenitor cells (black), sampled from eye discs containing either one (1x Pnt) or two (2x Pnt) copies of the pnt-GFP gene. Horizontal and vertical lines through each point indicate s.e.m. A thin diagonal line shows a constant Pnt/Yan ratio to aid interpretation.
We then introduced stabilizing SAM-SAM interactions for Yan, which dramatically sharpened the differences between Yan and Pnt occupancy (Fig. 6B). The model output resembles a phase diagram in which there is a sharp boundary that separates the state in which all sites are bound by Yan from the state in which all sites are bound by Pnt. This phase boundary lies along a region in which Pnt and Yan are in 1:1 stoichiometry, regardless of total protein concentration. Weighting the energetic contributions of binding strength and polymerization by the statistical frequency of each microstate revealed that the transition was driven by an abrupt change in the dominant binding mechanism. Polymerization-associated cooperativity effects dominate binding site occupancy when the Pnt/Yan ratio is low, while binding strength dominates when the ratio is high (Fig. S11D).
Increasing the Pnt/Yan ratio revealed nonlinear transitions from low to high Pnt occupancy for each individual binding site (Fig. 6C and Fig. S11E). These transitions resembled Hill function forms, with sharp thresholds delimiting distinct outputs, suggesting that transitions from low to high Pnt occupancy are ultra-sensitive to changes in the ratio. At low Pnt/Yan ratios, Yan occupancy dominated, while at some critical ratio, Pnt-bound molecules prevented Yan-Yan interactions, ultimately allowing Pnt to outcompete Yan as the ratio increased further. If transcription factor levels are sufficient to saturate binding sites, then relative occupancy by Pnt and Yan was agnostic to changes in the absolute abundance of either factor, as long as the ratio remained constant.
This model suggests a molecular mechanism for how even a twofold change in the Pnt/Yan ratio could shift DNA occupancy and therefore regulation of common target genes from an all-Yan to an all-Pnt regime. Therefore, the model provides guidance on how to interpret the experimental data. Although our fluorescence measurements do not provide us with absolute concentration estimates for Yan and Pnt, nevertheless the Pnt/Yan ratio measured in progenitor and R cells can be viewed as a parallel representation to the phase spaces calculated in the model (Fig. 6B,D). In this way, the small changes in the Pnt/Yan ratio we measured could elicit large changes in DNA binding site occupancy, while significant changes in absolute Pnt and Yan concentration would have little effect as long as the relative ratio remains constant (Fig. 6E). Together this ratio-based mechanism provides both ultrasensitivity and robustness to retinal cell state transitions.
DISCUSSION
The Drosophila retina has an extraordinary track record as a tractable genetic system for elucidating the conserved molecular mechanisms that direct cell state transitions. By introducing single-cell resolution quantitative analysis of Yan and Pnt expression dynamics, and then computationally modeling how they might impact DNA binding complexes and hence transcriptional output, our study has uncovered important and unanticipated mechanistic features of this well-studied system. We find that eye cells in the progenitor pool maintain a dynamically stable Pnt/Yan ratio predicted to favor the Yan-dominated enhancer occupancy necessary to sustain the progenitor state. In transitioning R cells, RTK signaling disrupts the progenitor ratio set-point and produces a sustained increase in the ratio to now favor Pnt DNA occupancy. In this way, transcriptional programs associated with the progenitor state are shifted to those appropriate to the R-cell state. We suggest that sensitivity to small but sustained relative changes makes this ratiometric system robust to fluctuations in absolute Pnt and Yan protein levels while enabling rapid ultrasensitive responses to inductive signaling.
For a transition to occur, the Pnt/Yan ratio set point must be shifted. However, before undergoing a state transition, the ratio set-point of the progenitor is dynamically stable over time even while cells vary in Yan and Pnt expression over a fourfold range. This suggests a stabilizing mechanism that monitors the ratio of Pnt and Yan, and takes corrective action when the ratio deviates from a specified reference value. The results of experiments in which we manipulated pnt or yan gene copy number suggest the stabilizing mechanism includes a weak positive-feedback loop between Pnt and Yan. As a result of this positive feedback, any fluctuation in the level of one factor will bring about a compensatory change in the other factor, maintaining dynamic but approximately constant stoichiometry of the two transcription factors. Overall, this allows cell fate transitions to occur reliably over a wide range of absolute protein concentrations and prevents transient fluctuations in either Yan or Pnt from inappropriately triggering or blocking differentiation.
Our results show that experimental manipulation of Ras and MAPK activity can change the Pnt/Yan ratio, most likely by regulating the synthesis and degradation of Pnt and Yan (Graham et al., 2010). Presumably, the ratio shift that naturally occurs at state transitions is due to inductive signals coming from R8 cells, as the signals are received by RTKs and transduced by the Ras-MAPK pathway. The ratio adopts a new set-point in R cells, twice the value of the old set-point, and is sustained thereafter. If the positive-feedback loop between Yan and Pnt regulates the set-point, then the strength of their mutual interactions must be differentially altered in order for the expression ratio to maintain a new value. Alternatively, feedback might remain the same but new regulatory inputs could tune Yan and Pnt expression to the new ratio in R cells. Investigating the contributions of the multiple regulatory interactions that have been documented within the Pnt/Yan network (Graham et al., 2010; Rohrbaugh et al., 2002; Webber et al., 2013b, 2018; Wu et al., 2020) to maintenance and modulation of the ratio will be an interesting future direction.
Previous studies have shown how cells can sense relative concentration changes of individual molecular species using fold-change detection (Adler et al., 2017; Alon et al., 1999; Barkai and Leibler, 1997; Frick et al., 2017; Goentoro and Kirschner, 2009; Lee et al., 2014; Mesibov et al., 1973; Shoval et al., 2010). To compute fold-change of a single protein, regulatory circuits first ‘measure’ and establish a molecular memory of the state of one variable, and later compare it with its background level or with a new measurement of the same variable (Frick et al., 2017; Lyashenko et al., 2020). In contrast, rather than measuring the individual fold-change in Pnt or Yan over time, eye cells likely rely on concurrent comparison of Pnt and Yan levels or, as suggested more speculatively by the biophysical model, on concurrent comparison of the transcriptional outcome of Yan and Pnt competition for DNA-binding sites.
Progenitor cells exhibit pulsatile expression of Yan and Pnt in which their expression levels decay to a basal state by 50 h of developmental time. If cells transit to a R-cell fate, this decay in expression is accelerated. Nevertheless, the Pnt/Yan ratio remains stable around a fixed set-point that is specific for cells in either progenitor or R-cell states. This is in contrast to many developmental systems in which the ratio of antagonistic transcription factors remains stable because expression of the factors remains static when cells exist in a particular developmental state (Laslo et al., 2008). A ratiometric system with modest variation may provide several advantages not only to the eye, but potentially to other systems.
First, such a system can permit fast response times. In the eye, a cascade of four inductive events occurs over a brief 8 h window of time to recruit R1-R7 cells. If a classical mechanism of mutual transcriptional repression existed between Yan and Pnt expression, then the time to resolve the ratio change would likely be too slow. By using a system in which a twofold shift in the ratio triggers cells to switch states, transient RTK inputs are sufficient to change the relative levels of the two proteins, thereby robustly and rapidly achieving the objective.
Second, a ratio-sensing system can define precise temporal windows of competence for differentiation. In the eye, the competence of cells to respond to RTK signals is not only dependent on the transduction machinery but also on the presence of Yan and Pnt. Eye progenitor cells anterior to the MF do not express Yan and Pnt, likely to prevent these cells from inappropriately responding to RTK activity. Over a 50 h time window as the MF moves across the eye disc, progenitor cells undergo the first phase of cell fate specification during which R cells and cone cells are recruited into ommatidia. As fate specification occurs as a wave, posterior ommatidia complete the first specification program ∼50 h before anterior ommatidia do. Yet posterior progenitor cells do not initiate further fate transitions until the anterior ommatidia complete the initial induction of R cells and cone cells (Cagan and Ready, 1989). It is only then that progenitor cells resume undergoing RTK-induced fate transitions – forming the pigment cells (Freeman, 1996; Miller and Cagan, 1998). We suggest that Yan and Pnt are shut off after the R-cell specification phase in order to create a multi-hour gap in progenitor competence that separates the two phases of fate specification.
Finally, a ratio-based mechanism can confer ultrasensitivity to modest changes in molecular stoichiometry when it is coupled to additional molecular mechanisms, e.g. competition and cooperativity in enhancer occupancy. Although the ratio of Pnt and Yan shifts only by twofold when cell state transitions occur, this change is sufficient to produce robust regulation of the transit to differentiation. Our biophysical model shows how the composition of transcription factor complexes on Yan/Pnt target genes could dramatically change when a twofold ratio change occurs, going from a repressive to an activating composition. Stabilizing interactions between Yan molecules bound to adjacent DNA sites sensitize enhancer occupancy to the relative abundance of the competing transcription factors. Indeed, previous work has shown that DNA-bound Yan monomers enhance recruitment of Yan to adjacent binding sites through cooperative protein-protein interactions (Boisclair Lachance et al., 2018; Qiao et al., 2004). Given that cooperativity and competition are features of many biological systems, ratiometric control mechanisms analogous to the one we have described in the Drosophila eye could prove to be a broadly used regulatory strategy.
MATERIALS AND METHODS
Genetics
Unless otherwise noted, all experiments were performed with animals reared at 25°C on standard cornmeal-molasses Drosophila culture medium. Fly stocks from the Bloomington Stock Center were as follows: pnt-GFP BAC transgene, BL42680; pnt2, BL2222; H2Av-mRFP BL23650, pntΔ88 (O'Neill et al., 1994); sev>RasV12 (Fortini et al., 1992); sev>YanACT (Rebay and Rubin, 1995); and sev>Gal4 (Jemc and Rebay, 2006). Measurements of wild-type dynamics of Pnt-GFP were made in eye discs from w1118; pnt-GFP; pntΔ88/pnt2, H2Av-mRFP. The effect of pnt gene dose on Pnt/Yan ratio was measured in w1118; pnt-gfp/+ ; pntΔ88/pnt2 (1× pnt) and w1118; pnt-gfp; pntΔ88/pnt2 (2× pnt) discs. In experiments with RasV12 or YanACT, Pnt-GFP and Yan levels were measured in eye discs from animals of the following genotypes: w1118; pnt-gfp/sev>RasV12; pntΔ88/+ relative to w1118; pnt-gfp/+; pntΔ88/+; and w1118; pnt-gfp/sev>YanACT; pntΔ88/+ relative to w1118; pnt-gfp/+; pntΔ88/+. Yan mutant eye clones were generated using the yan833 null allele, ey>FLP and the FRT40 crossover point. Yan+ cells were labeled using the clonal marker Ubi>mRFPNLS (Bloomington Stock 34500). Developing eyes were dissected from white prepupae carrying w, ey>FLP ; pnt-gfp, yan833, FRT40A/ pnt-gfp, Ubi>mRFPNLS, FRT40A.
Immunohistochemistry
All eye-antennal discs were dissected from white prepupae. For experiments in which 4′,6-diamidino-2-phenylindole (DAPI) was used as the nuclear marker, samples were fixed in 4% paraformaldehyde (w/v)/PBS-Triton X-100 0.1% (v/v) for 25 min at room temperature. After fixation, eye discs were blocked in PBS-Triton X-100 0.1% (v/v) and 1% (v/v) normal goat serum for 30 min at room temperature. Primary and secondary antibodies were incubated each for 2 h at room temperature with DAPI. Including DAPI during primary and secondary antibody staining was important for its even penetration. Antibodies used were as follows: mouse anti-Yan 8B12 (DHSB, 1:200) and goat anti-mouse Cy3 (1:2000, Jackson Immunoresearch). Discs were washed for 20 min in PBS-Triton X-100 0.1% (v/v), and mounted in VectaShield Antifade mounting medium (Vector Laboratories). To avoid rapid changes in cell volume, which would affect relative fluorescence measurements, the substitution of PBS-Triton X-100 with VectaShield was carried out gradually using increments of ∼33% in VectaShield concentration. For experiments in which H2Av-mRFP was used as the nuclear marker, discs were fixed in 4% (w/v) paraformaldehyde/PBS for ∼45 min and goat anti-mouse Pacific Blue antibody (Life Technologies, 1:200 dilution) was used as the secondary antibody.
Samples were imaged within 18-24 h after fixation. 1024×1024 16-bit images were captured using either a Zeiss LSM880 or a Leica SP5 confocal microscope equipped with 40× oil objectives. Discs were oriented with the equator (dorsal-ventral boundary) parallel to the x-axis of the image. Optical slices were set at 0.8 μm (45-60 optical slices per disc) with an additional digital zoom of 1.2-1.4. Images were recorded a region of at least six rows of ommatidia on each side of the equator. All discs for a given perturbation were fixed, mounted and imaged in parallel with control discs to reduce measurement error. Sample preparation, imaging and analysis were not performed under blind conditions, nor was sex of the animals noted at the time of dissection.
Quantification of expression levels
A minimum of three experimental replicate eye discs were imaged for each experimental condition, with the exception of the sev>RasV12 condition for which only one control disc was used. For each wild-type eye disc, ∼160 R8, ∼320 R2/5, ∼300 R3/4, ∼260 R1/6, ∼220 R7 and >1000 progenitor cells were manually selected from a set of computationally 2D-segmented nuclei using Silhouette. The fluorescence intensity of each nucleus was measured by our established procedure (Peláez et al., 2015). Our new pipeline includes Silhouette, an open-source package for macOS that integrates our image segmentation algorithm with a GUI (graphical user interface) for cell type annotation (available at: https://www.silhouette.amaral.northwestern.edu/). Subsequent analysis and visualization procedures were carried out using the accompanying FlyEye python package, which is freely available on GitHub. The resulting expression dynamics were inferred from confocal image stacks using an updated version of the segmentation and annotation pipeline described previously (Peláez et al., 2015). Empirically, the replicate number of three eye discs provided reproducible composite dynamics. No outlier eye discs were found for any experimental condition, so all samples were pooled, temporally aligned and included in the analysis. The sample size was not pre-computed before data collection.
Nuclear segmentation was performed using either H2Av-mRFP (Figs 1, 3, 4, Figs S1-S2, S4 and S5) or DAPI (Figs 2, 5 and Figs S7-S9) as a reference channel to identify individual nuclear boundaries. Nuclei expressing H2Av-mRFP were segmented using the Silhouette app's optimized implementation of the GraphCut algorithm (Qi, 2013), while DAPI-stained discs were segmented using an open-source implementation of the watershed algorithm designed to mitigate the effect of bright spots caused by DAPI accumulation in nucleoli (Bernasek et al., 2020). Each layer of the confocal stack was segmented in 2D independently in both cases. For each nucleus measured, a single contour was manually selected and assigned a cell type (cell state) using Silhouette, with care taken to avoid annotating nucleolar contours rather than complete nuclear contours in DAPI-stained discs. As each nucleus spans multiple optical sections, contours were always selected in the middle of the nucleus. The population of progenitor cells was measured in multiple layers to include cells whose nuclei were at different positions in the apical-ventral axis of the eye disc. In all cases, each nucleus was measured only in one layer to avoid measurement pseudoreplication.
Cells were identified and annotated by nuclear position in xyz space and shape using the His2Av-mRFP channel with the other channels turned off and the marker protein channel turned off. To validate the accuracy of this method of cell identification/annotation, cells were also immunostained for various cell-type-specific marker proteins, as noted in Table S1. Annotations made with tie His2Av-mRFP channel were then compared with the cell-type specific markers visualized in other channels.
For each annotated nucleus contour, expression measurements were obtained by normalizing the mean fluorescence of the Pnt-GFP and Yan antibody channels by the mean fluorescence of the reference channel used for segmentation (DAPI or His2Av-mRFP). This normalization mitigates variability due to potentially uneven sample illumination, segment area, nuclear volume and, in the case of His-RFP, differences in protein expression capacity between cells.
Conversion of distance to time
Cell positions along the anterior-posterior axis were mapped to developmental time as described previously (Peláez et al., 2015). This is predicted based on two assumptions: the furrow proceeds at a constant rate of one column of R8 neurons per 2 h; and minimal cell migration occurs. For each disc, Delaunay triangulations were used to estimate the median distance between adjacent columns of R8 neurons (Fortune, 1992). We used the median rather than the mean distance because it minimized the influence of non-adjacent R8s that were falsely identified by the triangulation (Peláez et al., 2015). Dividing the furrow velocity of 2 h per column by this median distance yields a single conversion factor from position along the anterior-posterior axis to developmental time. This factor was applied to all cell measurements within the corresponding disc, yielding expression time series. Notably, these are not single cell dynamics, but rather aggregate dynamics across the developmental time course of a spatially organized cell population.
Computation of moving averages and confidence intervals
Moving averages were computed by first-order Savitzky-Golay filtration (Savitzky and Golay, 1964). This method augments the simple windowing approach used by Peláez et al. (2015) by enabling visualization of expression trends at early time-points that are otherwise obscured by large window sizes. A secondary first-order filtration with one-fifth the original window size was applied to smooth lines for visualization purposes. None of our conclusions is sensitive to our choice of filtration or smoothing method. Primary window sizes of 250 and 75 cells were used for reporting the expression of multipotent and differentiated cells, unless noted otherwise. Confidence intervals for the moving average were inferred from the 2.5th and 97.5th percentile of ten-thousand-point estimates of the mean within each window. Point estimates were generated by bootstrap resampling with replacement of the expression levels within each window.
Alignment of expression data
To align multiple eye disc samples, cells of each sample were aligned with a reference population by shifting them in time. The magnitude of this shift was determined by maximizing the cross-correlation of progenitor Pnt-GFP expression Y(t) with the corresponding reference time series X(t). Rather than raw measurements, moving averages within a window of ten cells were used to improve robustness against noise. This operation amounts to:
where μ and σ are the mean and standard deviation of each time series, and dt is the time shift by which the population should be shifted.
For each experimental treatment, a disc was randomly chosen and shifted in time, such that time zero corresponds to the first annotated R8 neuron. This disc then served as the reference population for the alignment of all subsequent biological replicates within the treatment. Similarly, different experimental treatments (e.g. control and perturbation) were aligned by first aligning the discs within each treatment, then aggregating all cells within each treatment and repeating the procedure, with the first treatment serving as the reference.
This approach differs from the previous implementation of our pipeline in which discs were manually aligned by the inflection point of their Yan-YFP expression profiles (Peláez et al., 2015). Manual alignment entails arbitrary prioritization of certain dynamic features over others. Our revised protocol yields consistent, reproducible alignment of expression time series that equally weighs the entire time course. The automated approach is more principled but less robust than the manual approach. Specifically, it fails when dynamic forms qualitatively differ between experimental treatments. In these instances, we revert to manual alignment using the inflection point of Pnt-GFP induction as a reference.
Analysis of yan clones
We used ey>FLP and FRT40A to generate yan833 null clones within 23 eye discs carrying the Pnt-GFP transgene (see Genetics section). The chromosome carrying the wild-type yan allele was marked with a Ubi>mRFPNLS transgene. Discs were dissected, fixed and co-stained with DAPI before confocal imaging of RFP and GFP. Images of 36 unique vertical cross-sections spanning non-overlapping cells were collected in total. Images were analyzed using our previously published method called Fly-QMA that quantitatively analyzes mosaic imaginal discs (Bernasek et al., 2020). Briefly, cell nuclei were identified by watershed segmentation of the DAPI channel. For each segment, Ubi-mRFPNLS and Pnt-GFP fluorescence was quantified by normalizing the average intensity of all pixels within the respective fluorescence channel by the average DAPI fluorescence. Fluorescence bleed-through between the GFP and RFP channels was corrected as described previously (Bernasek et al., 2020). Bleed-through correction successfully eliminated any detectable difference in Pnt-GFP expression in mosaic eye discs in which all cells were wild type for yan. The same correction procedure was therefore applied to all measurements of mosaic eye discs containing yan mutant clones.
Mitotic recombination between the mutant and wild-type chromosomes yields cell populations exhibiting low, medium and high levels of RFP fluorescence, which correspond to cells with 0, 1 and 2 copies of the wild-type allele, respectively (Bernasek et al., 2020). Segmented nuclei were assigned to one of three groups using a k-means classifier. The procedure was validated through comparison with manual annotation of ∼2500 cells. The overall classification rate was ∼95%, Cells residing on the border of clones were excluded from all analyses to mitigate edge effects. The remaining measurements were aggregated across all eye discs for comparison of cells with zero versus one copy of the wild-type yan gene.
Simple competitive binding model
Fig. S10 presents results for an equilibrium model of two species, Yan (Y) and Pnt (P), competing for a finite pool of shared binding sites, S:
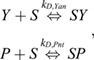 |
where KD,Yan and KD,Pnt are equilibrium association constants and SY and SP denote the bound species. Applying a mass balance to the total protein and binding site (S0) abundances:
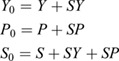 |
yields an analytically tractable system of nonlinear equations (Wang, 1995). For each pair of absolute protein abundances (Y0, P0), the Pnt binding site occupancy is simply SP/S0.
Competitive binding model with cooperativity
The model presented in Fig. 6 and Fig. S11 expands upon the work of Hope et al. (2017). The model is based on a single cis-regulatory element consisting of n adjacent binding sites, each of which may be designated as ETS or non-ETS. Each binding site may exist in one of three binding states: bound by a single copy of Yan, bound by a single copy of Pnt or unbound. Thermodynamic potentials were assigned to each binding state using two parameters for each transcription factor. The parameter αX defines the free energy of transcription factor X binding to an ETS site, while βX defines the free energy of binding to a non-ETS site. A unique configuration of binding states for all n binding sites constitutes a single microstate, k. The thermodynamic potential of each microstate was taken to be the sum of thermodynamic potentials for each of its constituent binding sites. For each microstate, the stabilizing effect of polymerization was incorporated via a third parameter, γX, that defines the free energy of SAM-SAM binding between a pair of similar transcription factors bound to adjacent sites. The net result is a total thermodynamic potential, ΔGk, for each microstate. An example enumeration of all possible microstates for an element consisting of one ETS site preceding two non-ETS sites is provided in Fig. S11B. The statistical frequencies of each microstate were evaluated by constructing a canonical ensemble:
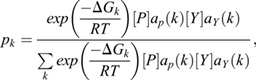 |
in which pk is the statistical frequency of microstate k, [P] is the Pnt concentration, [Y] is the Yan concentration, aP(k) and aY(k) are functions representing the number of bound molecules of P and Y within microstate k, T is a fixed temperature set to 300 K, and R is the gas constant. Fractional occupancies for each binding site correspond to the cumulative statistical frequency of all microstates in which the site is occupied by a given transcription factor. Overall fractional occupancies are similarly evaluated across all sites within the element.
We considered regulatory elements comprising 12 binding sites in which only the first site carries the ETS designation. We retained the same parameterization of Yan binding used by Hope et al. (2017): αY=−9.955 kcal mol−1, βY=−5.837 kcal mol−1 and γY=−7.043 kcal mol−1. We parameterized Pnt-binding thermodynamics to provide balanced competition between Pnt and Yan in the absence of any SAM-mediated polymerization of Pnt. That is, we set Pnt-binding affinities such that the transition from Pnt to Yan occupancy occurs when Pnt and Yan concentrations are approximately equal. The model used to generate Fig. 6B assumes that Pnt binds individual sites with elevated affinities αP=0.96(αY+γY) and βP=0.96(βY+γY). The model used to generate Fig. 6A uses these same elevated binding affinities for Yan, while setting γY=0 kcal mol−1. Qualitatively, our results are not sensitive to this parameterization.
Statistical frequencies of all microstates were enumerated using a recursive graph-traversal algorithm implemented in python, where each node in the graph represents an individual binding site and each edge reflects occupancy by a specific transcription factor. An open-source python implementation of this modeling framework has been published on GitHub (https://doi.org/10.5281/zenodo.5520493).
Supplementary Material
Acknowledgements
We thank members of the Amaral, Carthew and Rebay labs for helpful discussions during the course of this project, and C. LaBonne and M. Glotzer for helpful comments on the manuscript. We thank Kevin White's lab and the MODEncode Consortium for recombineering the Pnt-GFP transgene. the Bloomington Drosophila Stock Center for flies, Laura Nilson for the use of computational resources, the Developmental Studies Hybridoma Bank for antibody reagents and the Northwestern Biological Imaging Facility (BIF) for technical imaging support.
Footnotes
Author contributions
Conceptualization: S.M.B., N.P.-R., L.A.N.A., N.B., I.R., R.W.C.; Methodology: S.M.B., S.S.J.H., N.P.-R., H.T.N.; Software: S.M.B., H.T.N., N.S.-L.; Formal analysis: S.M.B., S.S.J.H., N.P.-R., J.-F.B.L., I.R., R.W.C.; Investigation: S.S.J.H., N.P.-R., J.-F.B.L., R.B.; Data curation: S.M.B.; Writing - original draft: R.W.C.; Writing - review & editing: S.M.B., S.S.J.H., N.P.-R., J.-F.B.L., R.B., H.T.N., N.S.-L., L.A.N.A., N.B., I.R.; Visualization: S.S.J.H., N.P.-R.; Supervision: L.A.N.A., N.B., I.R., R.W.C.; Project administration: L.A.N.A., N.B., I.R., R.W.C.; Funding acquisition: I.R., R.W.C.
Funding
This work was funded by the Howard Hughes Medical Institute’s Hanna H. Gray Fellowship program (N.P.-R.), the Chicago Biomedical Consortium (N.P.-R.), the Robert H. Lurie Comprehensive Cancer Center of Northwestern University (N.P.-R.), the National Institutes of Health (EY025957 to I.R. and R.W.C., EY032233 to R.W.C., GM118144 to R.W.C., and GM080372 to I.R.), the National Science Foundation (1764421 to L.A.N.A., N.B. and R.W.C.) and the Simons Foundation (597491 to L.A.N.A., N.B. and R.W.C.). Deposited in PMC for release after 12 months.
Data availability
All segmented and annotated eye disc data have been deposited in the Northwestern University open-access ARCH system (https://doi.org/10.21985/n2-fkxb-x241). A series of Jupyter notebooks that use these data to reproduce all analysis, simulations and figures presented in this manuscript has also been made available on GitHub (https://github.com/sbernasek/binding).
The Silhouette app used to segment nuclei in eye discs expressing His-2Av-mRFP and annotate nuclei in all eye discs is freely available via the macOS App Store, while the FlyEye python package used to generate, align and visualize expression dynamics has been published on GitHub (https://doi.org/10.5281/zenodo.5520468).
Contributor Information
Ilaria Rebay, Email: irebay@uchicago.edu.
Richard W. Carthew, Email: r-carthew@northwestern.edu.
Peer review history
The peer review history is available online at https://journals.biologists.com/dev/lookup/doi/10.1242/dev.201467.reviewer-comments.pdf.
References
- Acloque, H., Ocaña, O. H., Matheu, A., Rizzoti, K., Wise, C., Lovell-Badge, R. and Nieto, M. A. (2011). Reciprocal repression between Sox3 and Snail transcription factors defines embryonic territories at gastrulation. Dev. Cell 21, 546-558. 10.1016/j.devcel.2011.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler, M., Szekely, P., Mayo, A. and Alon, U. (2017). Optimal regulatory circuit topologies for fold-change detection. Cell Syst. 4, 171-181.e8. 10.1016/j.cels.2016.12.009 [DOI] [PubMed] [Google Scholar]
- Alon, U., Surette, M. G., Barkai, N. and Leibler, S. (1999). Robustness in bacterial chemotaxis. Nature 397, 168-171. 10.1038/16483 [DOI] [PubMed] [Google Scholar]
- Barkai, N. and Leibler, S. (1997). Robustness in simple biochemical networks. Nature 387, 913-917. 10.1038/43199 [DOI] [PubMed] [Google Scholar]
- Basler, K. and Hafen, E. (1989). Dynamics of Drosophila eye development and temporal requirements of sevenless expression. Development 107, 723-731. 10.1242/dev.107.4.723 [DOI] [PubMed] [Google Scholar]
- Bernasek, S. M., Peláez, N., Carthew, R. W., Bagheri, N. and Amaral, L. A. N. (2020). Fly-QMA: automated analysis of mosaic imaginal discs in Drosophila. PLoS Comput. Biol. 16, e1007406. 10.1371/journal.pcbi.1007406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisclair Lachance, J.-F., Peláez, N., Cassidy, J. J., Webber, J. L., Rebay, I. and Carthew, R. W. (2014). A comparative study of Pointed and Yan expression reveals new complexity to the transcriptional networks downstream of receptor tyrosine kinase signaling. Dev. Biol. 385, 263-278. 10.1016/j.ydbio.2013.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisclair Lachance, J.-F. B., Webber, J. L., Hong, L., Dinner, A. R. and Rebay, I. (2018). Cooperative recruitment of Yan via a high-affinity ETS supersite organizes repression to confer specificity and robustness to cardiac cell fate specification. Gene Dev. 32, 389-401. 10.1101/gad.307132.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briscoe, J. and Ericson, J. (2001). Specification of neuronal fates in the ventral neural tube. Curr. Opin. Neurobiol. 11, 43-49. 10.1016/S0959-4388(00)00172-0 [DOI] [PubMed] [Google Scholar]
- Brunner, D., Dücker, K., Oellers, N., Hafen, E., Scholz, H. and Klambt, C. (1994). The ETS domain protein pointed-P2 is a target of MAP kinase in the sevenless signal transduction pathway. Nature 370, 386-389. 10.1038/370386a0 [DOI] [PubMed] [Google Scholar]
- Cagan, R. L. and Ready, D. F. (1989). The emergence of order in the Drosophila pupal retina. Dev. Biol. 136, 346-362. 10.1016/0012-1606(89)90261-3 [DOI] [PubMed] [Google Scholar]
- Campos-Ortega, J. A. and Hofbauer, A. (1977). Cell clones and pattern formation: on the lineage of photoreceptor cells in the compound eye of Drosophila. Wilehm Roux Arch. Dev. Biol. 181, 227-245. 10.1007/BF00848423 [DOI] [PubMed] [Google Scholar]
- De Kumar, B., Parker, H. J., Parrish, M. E., Lange, J. J., Slaughter, B. D., Unruh, J. R., Paulson, A. and Krumlauf, R. (2017). Dynamic regulation of Nanog and stem cell-signaling pathways by Hoxa1 during early neuro-ectodermal differentiation of ES cells. Proc. Natl. Acad. Sci. USA 114, 5838-5845. 10.1073/pnas.1610612114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores, G. V., Duan, H., Yan, H., Nagaraj, R., Fu, W., Zou, Y., Noll, M. and Banerjee, U. (2000). Combinatorial signaling in the specification of unique cell fates. Cell 103, 75-85. 10.1016/S0092-8674(00)00106-9 [DOI] [PubMed] [Google Scholar]
- Fortini, M. E., Simon, M. A. and Rubin, G. M. (1992). Signalling by the sevenless protein tyrosine kinase is mimicked by Ras1 activation. Nature 355, 559-561. 10.1038/355559a0 [DOI] [PubMed] [Google Scholar]
- Fortune, S. (1992). Voronoi diagrams and delaunay triangulations. Computing in Euclidean Geometry 199-233. 10.1142/9789814355858_0006 [DOI] [Google Scholar]
- Freeman, M. (1996). Reiterative use of the EGF receptor triggers differentiation of all cell types in the Drosophila eye. Cell 87, 651-660. 10.1016/S0092-8674(00)81385-9 [DOI] [PubMed] [Google Scholar]
- Frick, C. L., Yarka, C., Nunns, H. and Goentoro, L. (2017). Sensing relative signal in the Tgf-β/Smad pathway. Proc. Natl. Acad. Sci. USA 114, E2975-E2982. 10.1073/pnas.1611428114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabay, L., Scholz, H., Golembo, M., Klaes, A., Shilo, B.-Z. and Klämbt, C. (1996). EGF receptor signaling induces pointed P1 transcription and inactivates Yan protein in the Drosophila embryonic ventral ectoderm. Development 122, 3355-3362. 10.1242/dev.122.11.3355 [DOI] [PubMed] [Google Scholar]
- Gabay, L., Seger, R. and Shilo, B.-Z. (1997). MAP kinase in situ activation atlas during Drosophila embryogenesis. Development 124, 3535-3541. 10.1242/dev.124.18.3535 [DOI] [PubMed] [Google Scholar]
- Gallagher, K. D., Mani, M. and Carthew, R. W. (2022). Emergence of a geometric pattern of cell fates from tissue-scale mechanics in the Drosophila eye. eLife 11, e72806. 10.7554/eLife.72806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goentoro, L. and Kirschner, M. W. (2009). Evidence that fold-change, and not absolute level, of β-catenin dictates Wnt signaling. Mol. Cell 36, 872-884. 10.1016/j.molcel.2009.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golembo, M., Schweitzer, R., Freeman, M. and Shilo, B. Z. (1996a). Argos transcription is induced by the Drosophila EGF receptor pathway to form an inhibitory feedback loop. Development 122, 223-230. 10.1242/dev.122.1.223 [DOI] [PubMed] [Google Scholar]
- Golembo, M., Raz, E. and Shilo, B.-Z. (1996b). The Drosophila embryonic midline is the site of Spitz processing, and induces activation of the EGF receptor in the ventral ectoderm. Development 122, 3363-3370. 10.1242/dev.122.11.3363 [DOI] [PubMed] [Google Scholar]
- Graham, T. G. W., Tabei, S. M. A., Dinner, A. R. and Rebay, I. (2010). Modeling bistable cell-fate choices in the Drosophila eye: qualitative and quantitative perspectives. Development 137, 2265-2278. 10.1242/dev.044826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfon, M. S., Carmena, A., Gisselbrecht, S., Sackerson, C. M., Jiménez, F., Baylies, M. K. and Michelson, A. M. (2000). Ras pathway specificity is determined by the integration of multiple signal-activated and tissue-restricted transcription factors. Cell 103, 63-74. 10.1016/S0092-8674(00)00105-7 [DOI] [PubMed] [Google Scholar]
- Hollenhorst, P. C., McIntosh, L. P. and Graves, B. J. (2011). Genomic and biochemical insights into the specificity of ETS transcription factors. Annu. Rev. Biochem. 80, 437-471. 10.1146/annurev.biochem.79.081507.103945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope, C. M., Rebay, I. and Reinitz, J. (2017). DNA occupancy of polymerizing transcription factors: a chemical model of the ETS family factor Yan. Biophys. J. 112, 180-192. 10.1016/j.bpj.2016.11.901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger, J., Blagov, M., Kosman, D., Kozlov, K. N., Manu, Myasnikova, E., Surkova, S., Vanario-Alonso, C. E., Samsonova, M., Sharp, D. H.et al. (2004). Dynamical analysis of regulatory interactions in the Gap gene system of Drosophila melanogaster. Genetics 167, 1721-1737. 10.1534/genetics.104.027334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemc, J. and Rebay, I. (2006). Characterization of the split ends-like gene spenito reveals functional antagonism between SPOC family members during Drosophila eye development. Genetics 173, 279-286. 10.1534/genetics.106.055558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagha, M., Brunelli, S., Messina, G., Cumano, A., Kume, T., Relaix, F. and Buckingham, M. E. (2009). Pax3:Foxc2 reciprocal repression in the somite modulates muscular versus vascular cell fate choice in multipotent progenitors. Dev. Cell 17, 892-899. 10.1016/j.devcel.2009.10.021 [DOI] [PubMed] [Google Scholar]
- Lai, Z. C. and Rubin, G. M. (1992). Negative control of photoreceptor development in Drosophila by the product of theyangene, an ETS domain protein. Cell 70, 609-620. 10.1016/0092-8674(92)90430-K [DOI] [PubMed] [Google Scholar]
- Laslo, P., Spooner, C. J., Warmflash, A., Lancki, D. W., Lee, H.-J., Sciammas, R., Gantner, B. N., Dinner, A. R. and Singh, H. (2006). Multilineage transcriptional priming and determination of alternate hematopoietic cell fates. Cell 126, 755-766. 10.1016/j.cell.2006.06.052 [DOI] [PubMed] [Google Scholar]
- Laslo, P., Pongubala, J. M. R., Lancki, D. W. and Singh, H. (2008). Gene regulatory networks directing myeloid and lymphoid cell fates within the immune system. Semin. Immunol. 20, 228-235. 10.1016/j.smim.2008.08.003 [DOI] [PubMed] [Google Scholar]
- Lee, R. E. C., Walker, S. R., Savery, K., Frank, D. A. and Gaudet, S. (2014). Fold change of nuclear NF-kappaB determines TNF-induced transcription in single cells. Mol. Cell 53, 867-879. 10.1016/j.molcel.2014.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyashenko, E., Niepel, M., Dixit, P. D., Lim, S. K., Sorger, P. K. and Vitkup, D. (2020). Receptor-based mechanism of relative sensing and cell memory in mammalian signaling networks. eLife 9, e50342. 10.7554/eLife.50342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackereth, C. D., Schärpf, M., Gentile, L. N., MacIntosh, S. E., Slupsky, C. M. and McIntosh, L. P. (2004). Diversity in structure and function of the Ets family PNT domains. J. Mol. Biol. 342, 1249-1264. 10.1016/j.jmb.2004.07.094 [DOI] [PubMed] [Google Scholar]
- Melen, G. J., Levy, S., Barkai, N. and Shilo, B.-Z. (2005). Threshold responses to morphogen gradients by zero-order ultrasensitivity. Mol. Syst. Biol. 1, 2005.0028. 10.1038/msb4100036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meruelo, A. D. and Bowie, J. U. (2009). Identifying polymer-forming SAM domains. Proteins 74, 1-5. 10.1002/prot.22232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesibov, R., Ordal, G. W. and Adler, J. (1973). The range of attractant concentrations for bacterial chemotaxis and the threshold and size of response over this range. J. Gen. Physiol. 62, 203-223. 10.1085/jgp.62.2.203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, D. T. and Cagan, R. L. (1998). Local induction of patterning and programmed cell death in the developing Drosophila retina. Development 125, 2327-2335. 10.1242/dev.125.12.2327 [DOI] [PubMed] [Google Scholar]
- Nitta, K. R., Jolma, A., Yin, Y., Morgunova, E., Kivioja, T., Akhtar, J., Hens, K., Toivonen, J., Deplancke, B., Furlong, E. E. M.et al. (2015). Conservation of transcription factor binding specificities across 600 million years of bilateria evolution. eLife 4, e04837. 10.7554/eLife.04837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill, E. M., Rebay, I., Tjian, R. and Rubin, G. M. (1994). The activities of two Ets-related transcription factors required for Drosophila eye development are modulated by the Ras/MAPK pathway. Cell 78, 137-147. 10.1016/0092-8674(94)90580-0 [DOI] [PubMed] [Google Scholar]
- Park, B. O., Ahrends, R. and Teruel, M. N. (2012). Consecutive positive feedback loops create a bistable switch that controls preadipocyte-to-adipocyte conversion. Cell Rep. 2, 976-990. 10.1016/j.celrep.2012.08.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peláez, N., Gavalda-Miralles, A., Wang, B., Navarro, H. T., Gudjonson, H., Rebay, I., Dinner, A. R., Katsaggelos, A. K., Amaral, L. A. N. and Carthew, R. W. (2015). Dynamics and heterogeneity of a fate determinant during transition towards cell differentiation. eLife 4, e08924. 10.7554/eLife.08924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, J. (2013). Dense nuclei segmentation based on graph cut and convexity-concavity analysis: dense nuclei segmentation. J. Microsc. 253, 42-53. 10.1111/jmi.12096 [DOI] [PubMed] [Google Scholar]
- Qiao, F., Song, H. Y., Kim, C. A., Sawaya, M. R., Hunter, J. B., Gingery, M., Rebay, I., Courey, A. J. and Bowie, J. U. (2004). Derepression by depolymerization: structural insights into the regulation of Yan by Mae. Cell 118, 163-173. 10.1016/j.cell.2004.07.010 [DOI] [PubMed] [Google Scholar]
- Rand, D. A., Raju, A., Sáez, M., Corson, F. and Siggia, E. D. (2021). Geometry of gene regulatory dynamics. Proc. Natl. Acad. Sci. USA 118, e2109729118. 10.1073/pnas.2109729118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebay, I. and Rubin, G. M. (1995). Yan functions as a general inhibitor of differentiation and is negatively regulated by activation of the Ras1/MAPK pathway. Cell 81, 857-866. 10.1016/0092-8674(95)90006-3 [DOI] [PubMed] [Google Scholar]
- Rogge, R., Green, P. J., Urano, J., Horn-Saban, S., Mlodzik, M., Shilo, B. Z., Hartenstein, V. and Banerjee, U. (1995). The role of yan in mediating the choice between cell division and differentiation. Development 121, 3947-3958. 10.1242/dev.121.12.3947 [DOI] [PubMed] [Google Scholar]
- Rohrbaugh, M., Ramos, E., Nguyen, D., Price, M., Wen, Y. and Lai, Z.-C. (2002). Notch activation of yan expression is antagonized by RTK/pointed signaling in the Drosophila eye. Curr. Biol. 12, 576-581. 10.1016/S0960-9822(02)00743-1 [DOI] [PubMed] [Google Scholar]
- Sáez, M., Blassberg, R., Camacho-Aguilar, E., Siggia, E. D., Rand, D. A. and Briscoe, J. (2022). Statistically derived geometrical landscapes capture principles of decision-making dynamics during cell fate transitions. Cell Syst. 13, 12-28.e3. 10.1016/j.cels.2021.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitzky, A. and Golay, M. J. E. (1964). Smoothing and differentiation of data by simplified least squares procedures. Anal. Chem. 36, 1627-1639. 10.1021/ac60214a047 [DOI] [Google Scholar]
- Schaffer, A. E., Freude, K. K., Nelson, S. B. and Sander, M. (2010). Nkx6 transcription factors and Ptf1a function as antagonistic lineage determinants in multipotent pancreatic progenitors. Dev. Cell 18, 1022-1029. 10.1016/j.devcel.2010.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrode, N., Saiz, N., Di Talia, S. and Hadjantonakis, A.-K. (2014). GATA6 levels modulate primitive endoderm cell fate choice and timing in the mouse blastocyst. Dev. Cell 29, 454-467. 10.1016/j.devcel.2014.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoval, O., Goentoro, L., Hart, Y., Mayo, A., Sontag, E. and Alon, U. (2010). Fold-change detection and scalar symmetry of sensory input fields. Proc. Natl. Acad. Sci. USA 107, 15995-16000. 10.1073/pnas.1002352107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shwartz, A., Yogev, S., Schejter, E. D. and Shilo, B.-Z. (2013). Sequential activation of ETS proteins provides a sustained transcriptional response to EGFR signaling. Development 140, 2746-2754. 10.1242/dev.093138 [DOI] [PubMed] [Google Scholar]
- Tomlinson, A. and Ready, D. F. (1987). Cell fate in the Drosophila ommatidium. Dev. Biol. 123, 264-275. 10.1016/0012-1606(87)90448-9 [DOI] [PubMed] [Google Scholar]
- Wang, Z.-X. (1995). An exact mathematical expression for describing competitive binding of two different ligands to a protein molecule. FEBS Lett. 360, 111-114. 10.1016/0014-5793(95)00062-E [DOI] [PubMed] [Google Scholar]
- Webber, J. L., Zhang, J., Mitchell-Dick, A. and Rebay, I. (2013a). 3D chromatin interactions organize Yan chromatin occupancy and repression at the even-skipped locus. Genes Dev. 27, 2293-2298. 10.1101/gad.225789.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber, J. L., Zhang, J., Cote, L., Vivekanand, P., Ni, X., Zhou, J., Nègre, N., Carthew, R. W., White, K. P. and Rebay, I. (2013b). The relationship between long-range chromatin occupancy and polymerization of the Drosophila ETS family transcriptional repressor Yan. Genetics 193, 633-649. 10.1534/genetics.112.146647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber, J. L., Zhang, J., Massey, A., Sanchez-Luege, N. and Rebay, I. (2018). Collaborative repressive action of the antagonistic ETS transcription factors Pointed and Yan fine-tunes gene expression to confer robustness in Drosophila. Development 145, dev165985. 10.1242/dev.165985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, G.-H., Badis, G., Berger, M. F., Kivioja, T., Palin, K., Enge, M., Bonke, M., Jolma, A., Varjosalo, M., Gehrke, A. R.et al. (2010). Genome–wide analysis of ETS–family DNA–binding in vitro and in vivo. EMBO J. 29, 2147-2160. 10.1038/emboj.2010.106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff, T. and Ready, D. F. (1991). Cell death in normal and rough eye mutants of Drosophila. Development 113, 825-839. 10.1242/dev.113.3.825 [DOI] [PubMed] [Google Scholar]
- Wolff, T. and Ready, D. F. (1993). Pattern Formation in the Drosophila Retina: Cold Spring Harbor Laboratory Press. pp. 1277-1325. [Google Scholar]
- Wu, C., Boisclair Lachance, J.-F., Ludwig, M. Z. and Rebay, I. (2020). A context-dependent bifurcation in the Pointed transcriptional effector network contributes specificity and robustness to retinal cell fate acquisition. PLoS Genet. 16, e1009216. 10.1371/journal.pgen.1009216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, C., Kauffmann, R. C., Zhang, J., Kladny, S. and Carthew, R. W. (2000). Overlapping activators and repressors delimit transcriptional response to receptor tyrosine kinase signals in the Drosophila eye. Cell 103, 87-97. 10.1016/S0092-8674(00)00107-0 [DOI] [PubMed] [Google Scholar]
- Yao, J.-G., Weasner, B. M., Wang, L.-H., Jang, C.-C., Weasner, B., Tang, C.-Y., Salzer, C. L., Chen, C.-H., Hay, B., Sun, Y. H.et al. (2008). Differential requirements for the Pax6(5a) genes eyegone and twin of eyegone during eye development in Drosophila. Dev. Biol. 315, 535-551. 10.1016/j.ydbio.2007.12.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, L. J., Christensen, R. G., Kazemian, M., Hull, C. J., Enuameh, M. S., Basciotta, M. D., Brasefield, J. A., Zhu, C., Asriyan, Y., Lapointe, D. S.et al. (2011). FlyFactorSurvey: a database of Drosophila transcription factor binding specificities determined using the bacterial one-hybrid system. Nucleic Acids Res. 39, D111-D117. 10.1093/nar/gkq858 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.