Abstract
So far, four RNA:pseudouridine (Ψ)-synthases have been identified in yeast Saccharomyces cerevisiae. Together, they act on cytoplasmic and mitochondrial tRNAs, U2 snRNA and rRNAs from cytoplasmic ribosomes. However, RNA:Ψ-synthases responsible for several U→Ψ conversions in tRNAs and UsnRNAs remained to be identified. Based on conserved amino-acid motifs in already characterised RNA:Ψ-synthases, four additional open reading frames (ORFs) encoding putative RNA:Ψ-synthases were identified in S.cerevisiae. Upon disruption of one of them, the YLR165c ORF, we found that the unique Ψ residue normally present in the fully matured mitochondrial rRNAs (Ψ2819 in 21S rRNA) was missing, while Ψ residues at all the tested pseudouridylation sites in cytoplasmic and mitochondrial tRNAs and in nuclear UsnRNAs were retained. The selective U→Ψ conversion at position 2819 in mitochondrial 21S rRNA was restored when the deleted yeast strain was transformed by a plasmid expressing the wild-type YLR165c ORF. Complementation was lost after point mutation (D71→A) in the postulated active site of the YLR165c-encoded protein, indicating the direct role of the YLR165c protein in Ψ2819 synthesis in mitochondrial 21S rRNA. Hence, for nomenclature homogeneity the YLR165c ORF was renamed PUS5 and the corresponding RNA:Ψ-synthase Pus5p. As already noticed for other mitochondrial RNA modification enzymes, no canonical mitochondrial targeting signal was identified in Pus5p. Our results also show that Ψ2819 in mitochondrial 21S rRNA is not essential for cell viability.
INTRODUCTION
Pseudouridine (Ψ) is the most abundant and universally found modified nucleoside in RNA. Indeed, numerous Ψ residues are present at specific positions in rRNAs and tRNAs from Archaea, Bacteria and Eukarya and in UsnRNAs from Eukarya (for reviews see 1–3). They are all formed post-transcriptionally by a family of enzymes designated as RNA:Ψ-synthases. The detailed mechanism of this reaction however is still unknown, yet it is thought that the carboxyl group of a conserved aspartate residue present in all RNA:Ψ-synthases identified so far plays an essential role (4–7, see below).
Present knowledge of RNA:Ψ-synthases is largely based on their identification in Escherichia coli. Two tRNA:Ψ-synthases were characterised in E.coli and other bacteria: PSU-I (recently renamed TruA) is specific for the formation of Ψ at positions 38, 39 and/or 40 in several tRNAs (8,9), while TruB catalyses the formation of the conserved Ψ residue at position 55 (10). Among the characterised E.coli rRNA:Ψ-synthases, RluC and RluD catalyse the formation of Ψ residues at six distinct sites in E.coli 23S rRNA (5,11,12), whereas RsuA and RluE catalyse each a unique U→Ψ conversion (6,13,14). In addition, RluA displays a dual-substrate specificity since it is implicated in the formation of both Ψ746 in 23S rRNAs and Ψ32 in tRNAs (15,16).
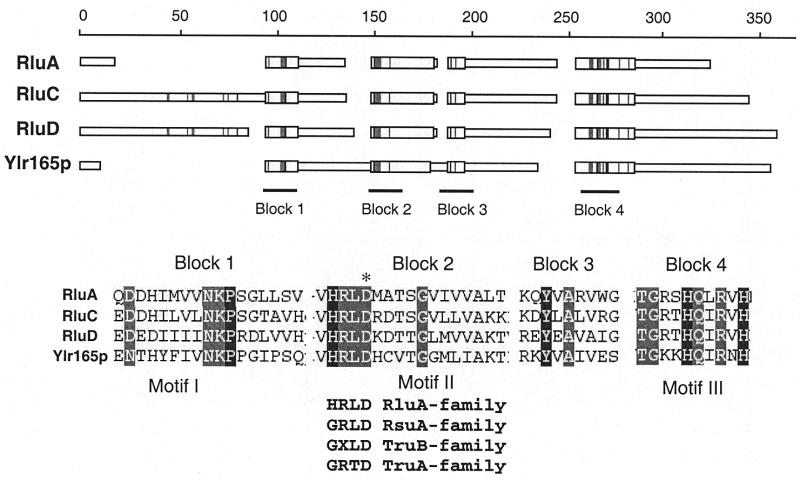
Analysis of the sequence similarity between already characterised RNA:Ψ-synthases showed that these enzymes belong to four distinct families: TruA, TruB, RsuA and RluA (17). The TruB, RsuA and RluA families share two short conserved motifs (Fig. 1, motifs I and II) (17), while only the GRTD tetra-amino acid sequence found in motif II is conserved in the TruA family (6,17). Site-directed mutagenesis of the aspartate residue (D) in motif II showed that this amino acid is essential for catalysis (4–7). Extensive searches in DNA databases identified more than 100 genes coding for putative Ψ-synthases in 47 different organisms covering altogether the three domains of living organisms (Archaea, Bacteria and Eukarya) (6).
Figure 1.
Sequence alignment of E.coli proteins from the RluA family (RluA, RluC and RluD) and S.cerevisiae YLR165c-encoded protein. The scale in amino acid residues is presented at the top. The three motifs (I, II and III) conserved in the RluA and RsuA families of RNA:Ψ-synthases are shown at the bottom. Identical and conserved residues are shaded. Aspartic acid residue mutated to alanine in the conserved motif II is indicated by an asterisk. Amino acid sequences of Motif II in different RNA:Ψ-synthase families are indicated at the bottom.
Such analysis revealed nine genes encoding putative RNA:Ψ-synthases in the Saccharomyces cerevisiae genome (6). Five of them correspond to the already characterised RNA:Ψ-synthases (Pus1p, Pus2p, Pus3p, Pus4p and Cbf5p) (18–22). Pus1p, Pus2p and Pus3p are members of the TruA family, whereas Pus4p and Cbf5p belong to the TruB family. The targets of the Pus1p, Pus3p, Pus4p and Cbf5p enzymes were identified. Pus3p catalyses Ψ formation at positions 38 and 39 in the anticodon loop of cytoplasmic and mitochondrial tRNAs (22), while Pus1p is implicated in Ψ formation at eight different positions in cytoplasmic tRNAs (18,20) and also at position 44 in U2 snRNA (23). Pus4p, like TruB in E.coli, is responsible for the site-specific formation of the highly conserved Ψ residue at position 55 in both cytoplasmic and mitochondrial tRNAs (19). Finally, Cbf5p, a protein present in the H/ACA snoRNPs (21,24), catalyses the formation of most, if not all, Ψ residues in the yeast 18S and 25S rRNAs at the precursor level (35S pre-rRNA) in the nucleolus (21,25). Selection of the pseudouridylation sites in rRNAs is mediated through direct base pairing between the snoRNAs and the nucleolar pre-rRNA (26,27).
Previous examination of 34 cytoplasmic and 17 mitochondrial tRNAs so far sequenced in yeast (28) revealed that Ψ can be found at 15 distinct positions in cytoplasmic tRNAs and at eight positions in mitochondrial tRNAs (for review see 1). Altogether, the Pus1p, Pus3p and Pus4p RNA:Ψ-synthases mentioned above are responsible for Ψ formation at 11 out of 15 positions in cytoplasmic tRNAs and 5 out of 8 positions in mitochondrial tRNAs (18–20,22). Only the enzymes catalysing the formation of Ψ residues at positions 1, 13, 31 and 32 in cytoplasmic tRNAs and at positions 31, 32 and 72 in mitochondrial tRNAs remain to be identified. Moreover, while yeast Pus1p is capable of forming Ψ35 in tRNATyr in vitro, the disruption of the corresponding PUS1 gene does not affect the formation of Ψ35 in vivo, attesting for the existence of another enzyme acting on U35 with overlapping specificity with Pus1p (20).
Six Ψ residues have been identified in UsnRNAs from S.cerevisiae (23). So far, only the enzyme responsible for the formation of residue Ψ44 in U2 snRNA was identified (Pus1p) (23). The enzymes responsible for Ψ formation at the five other sites in UsnRNAs have still to be identified.
Finally, the S.cerevisiae mitochondrial 21S LSU rRNA was found to contain one Ψ residue at position 2819 (29) and the corresponding Ψ-synthase had not been identified.
In this work, we focused our attention on one of the four S.cerevisiae genes encoding putative RNA:Ψ-synthases with unknown substrate, the YLR165c open reading frame (ORF). The encoded protein displays the two characteristic motifs I and II common to proteins of the three distinct RNA:Ψ-synthases families TruA, RsuA and RluA, and also an additional motif III common to the RsuA and RluA families (Fig. 1). To identify the substrate of the YLR165c ORF protein product, the effect of the deletion of this ORF on the formation of Ψ residues at positions 13, 31 and 32 in cytoplasmic tRNAs, 31 and 32 in mitochondrial tRNAs, 5 and 6 in U1 snRNA, 35 and 42 in U2 snRNA, 99 in U5 snRNA and 2819 in mitochondrial 21S rRNA was tested. The results obtained reveal that the S.cerevisiae YLR165c ORF encodes the mitochondrial rRNA:Ψ-synthase.
MATERIALS AND METHODS
Disruption of YLR165c ORF
The YLR165c ORF (accession number in TREMBL, Q06244) was disrupted in the S.cerevisiae strains BMA64 (MAT a/α ura 3-1 ade 2-1 leu 2-3, 112 his3-11, 15 trp1D can 1-100) and BMA64-1A (MAT a ura 3-1 ade 2-1 leu 2-3, 112 his3-11, 15 trp1D can 1-100), by replacement of the entire coding region with the TRP1 auxotrophic marker. Replacement was done by the PCR-targeting method using the TRP1 gene flanked by short regions homologous to the target locus of the S.cerevisiae DNA (30,31). The TRP1 gene was amplified from plasmid pFL35. Both haploid and diploid strains were transformed with 3 or 6 µg of PCR-generated DNA fragments using the lithium acetate method (32). Positive transformants (ΔYLR165c) were selected on YNB plates lacking tryptophan. The disruption was confirmed by PCR amplification on yeast colonies.
Saccharomyces cerevisiae RNA preparation
Total RNA from the wild-type and the ΔYLR165c yeast strains was prepared as described previously (23).
Ψ residues analysis in UsnRNAs and mitochondrial 21S LSU rRNA
Ψ residues in the UsnRNAs and mitochondrial 21S LSU rRNA of the ΔYLR165c and wild-type S.cerevisiae strains were identified by the CMCT-RT approach (33), with the modifications described previously (23). In brief, total RNA fractions were treated by CMCT, followed by alkaline treatment to remove the CMC group from U. The remaining bulky CMC-Ψ were identified by reverse transcription using various oligonucleotides complementary to snRNAs or 21S rRNA. The oligonucleotides used for Ψ identification in U1, U2 and U5 snRNAs were previously described (23). The oligonucleotide complementary to nt 2846–2866 of the mitochondrial 21S LSU rRNA was used to detect the formation of a Ψ residue at position 2819 in this RNA.
Test for RNA:Ψ-synthase activities acting at positions 13, 31, 32 or 35 in tRNAs
The preparation of cell-free extracts from wild-type and ΔYLR165c yeast strains was performed as described earlier (34). The enzymatic activity of RNA:Ψ-synthases acting at positions 13 or 32 and at position 35 in T7 transcripts of internally 32P-labelled tRNA was tested using as substrates yeast tRNAAsp (anticodon GUC) and Arabidopsis thaliana pre-tRNATyr (anticodon GUA, including the intron). ATP and UTP labelling of tRNAAsp transcript followed by nearest neighbour approach allows the selective detection of Ψ13 and Ψ32 respectively (20). The tRNA:Ψ35-synthase activity was tested using heterologous [α-32P]ATP radiolabelled pre-tRNATyr from the plant A.thaliana. Quantification of Ψ formation in these tRNA was performed after complete T2 RNase digestion and separation of the digest by 2D-TLC, as described by Motorin et al. (20).
The presence of pseudouridine residues at positions 31 and 32 in the yeast mitochondrial tRNAMet and tRNASer was tested using the CMCT-RT approach as described for analysis of mitochondrial 21S rRNA. The oligonucleotides complementary to mitochondrial tRNAMet (anticodon CAU) (nt 46–68) and to tRNASer (anticodon GCU) (nt 44–57) were used as primers. The presence of Ψ at position 31 in cytoplasmic tRNAMet (anticodon CAU) was tested by the same approach, using the oligonucleotide complementary to bases 40–57.
Complementation of the ΔYLR165c S.cerevisiae strain by plasmid p413TEF-YLR165c
The YLR165c ORF was amplified by PCR from the genomic DNA of strain BMA64-1A using an oligonucleotide corresponding to the 5′ end of the ORF (CAAGCGGAATTCGGatgtcaaaaaagcag) and an oligonucleotide complementary to the 3′ end of the ORF (GTTCGCCTTAAGTTAttactggtcccagttttc). Sense and antisense sequences of the YLR165c ORF are shown in lower case. The 5′-oligonucleotide generated an EcoRI restriction site (underlined). The amplified PCR product was subcloned in the pT-Adv cloning vector (Clontech, CA, USA). The resulting construct was cleaved by EcoRI and the fragment containing ORF YLR165c was inserted at the unique EcoRI site of the shuttle vector p413TEF (35), downstream from the TEF promoter. The sequence of the insert was verified by DNA sequencing. Point mutation D→A at position 71 was introduced by PCR-mediated site-directed mutagenesis using the Quick Change Kit (Stratagene, WI, USA). The haploid BMA64-1A-ΔYLR165c strain was transformed with plasmids containing the wild-type or the mutated YLR165c ORF using the standard lithium acetate procedure (36).
Protein alignment and prediction of cell sorting signals
The multiple sequence alignment of RNA:Ψ-synthases was constructed with the ClustalW (37) and MACAW (38) software and further refined manually. Presence of putative cellular targeting signals in the Pus5p sequence was tested with the PSORT Web site (39), http://psort.nibb.ac.jp , and NNPSL software Web site (40), http://predict.sanger.ac.uk/nnpsl/
RESULTS AND DISCUSSION
Disruption of the YLR165c ORF does not affect tRNA or UsnRNA pseudouridylation
To identify the RNA substrate(s) of the protein encoded by the YLR165c ORF, we performed a one-step gene replacement of this ORF by the gene encoding n-(5′-phosphoribosyl)-anthranilate isomerase (TRP1 auxotrophic marker). The resulting ΔYLR165c yeast cells did not show any growth phenotype when plated on rich or minimal media and grown at 20, 30 or 37°C. The cell-free extract and total RNA fraction obtained from the strain with the ΔYLR165c ORF were used to determine the consequences of the gene deletion on the pseudouridylation pattern of tRNAs, UsnRNAs and mitochondrial rRNA. The activity towards positions 13 and 32 in cytoplasmic tRNAs was tested on an in vitro produced yeast tRNAAsp as described in Materials and Methods. To test for a putative tRNA:Ψ35-synthase activity the action of extracts from wild-type and ΔYLR165c yeast cells on an [α-32P]ATP radiolabelled pre-tRNATyr from the plant A.thaliana was compared. Indeed, previous data showed that the purified yeast Pus1p enzyme does not modify A.thaliana tRNATyr at position 35 in vitro, whereas the putative tRNA:Ψ35-synthase present in the yeast cell extract does catalyse Ψ35 formation. As evidenced by 2D-TLC of the tRNA RNase T2 digest, the presence of Ψ13, Ψ32 or Ψ35 in the various tRNA substrates was not affected by deletion of the YLR165c ORF (Table 1).
Table 1. Ψ formation in the extracts of wild-type and ΔYLR165c yeast strains.
| Position tested in tRNA | tRNA transcript/labelling | Wild-type strain | ΔYLR165c strain |
|---|---|---|---|
| 13 | yeast tRNAAsp/ATP | 0.75 | 0.65 |
| 32 | yeast tRNAAsp/UTP | 0.85 | 0.85 |
| 35 | A.thaliana tRNATyr/ATP | 0.55 | 0.65 |
Radiolabelled transcripts were incubated with yeast extract at 30°C for 60 min and the Ψ content was analysed by 2D-TLC after RNase T2 digestion.
Influence of the disruption of the YLR165c ORF on the pseudouridylation of yeast cytoplasmic tRNA at position 31 and mitochondrial tRNAs at positions 31 and 32 was tested with the CMCT-RT approach as described in Materials and Methods. As demonstrated by identification of the alkaline resistant CMC-Ψ adducts, Ψ31 and Ψ32 in the mitochondrial tRNAMet and tRNASer, as well as Ψ31 in the cytoplasmic tRNAMet, were retained upon gene disruption (data not shown).
Hence, altogether these results strongly suggested that the protein encoded by the YLR165c ORF is not a tRNA:Ψ-synthase. To investigate its possible involvement in UsnRNAs pseudouridylation, the presence of residues Ψ5 and Ψ6 in U1 snRNA, Ψ35 and Ψ42 in U2 snRNA and Ψ99 in U5 snRNA was tested in the ΔYLR165c haploid strain using the CMCT modification approach and appropriate oligonucleotides for primer extension analysis. As shown in Figure 2, all the Ψ residues present in UsnRNAs were conserved upon deletion of the YLR165c ORF, indicating that the corresponding protein is not required for modification of UsnRNAs in vivo.
Figure 2.
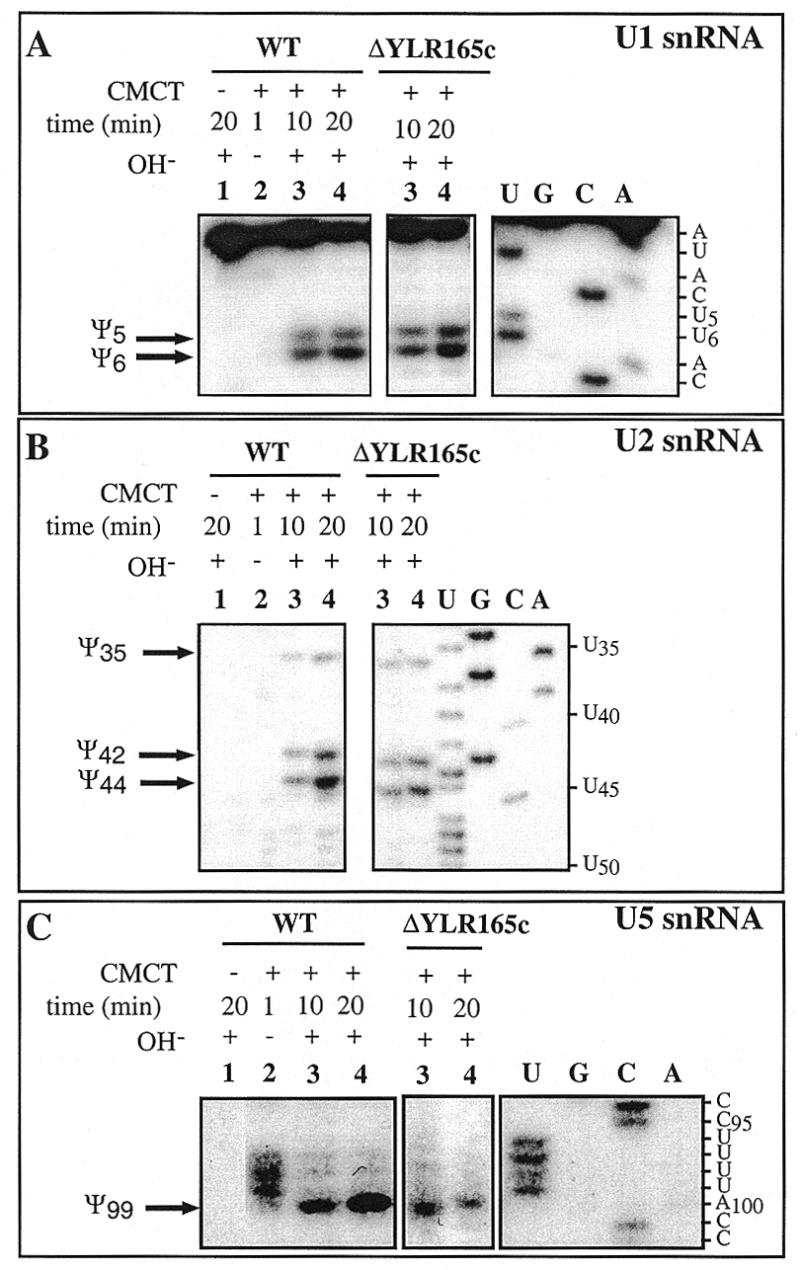
UsnRNA pseudouridylation pattern in the ΔYLR165c S.cerevisiae strain. Primer extension analysis was performed for U1 (A), U2 (B) and U5 (C) snRNAs from a total RNA fraction of the wild-type (WT) and ΔYLR165c S.cerevisiae strains. CMCT modifications were done for 1, 10 and 20 min (lanes 2, 3 and 4 respectively), experimental conditions are described in Materials and Methods. In lanes 3 and 4, the CMCT-modified RNA was subjected to an alkaline treatment at pH 10.4. A control extension experiment was made without addition of CMCT (lane 1). Lanes U, G, C and A correspond to the RNA sequencing ladder. Nucleotide positions, starting from the 5′-terminal nucleotide, are indicated on the right. The reverse transcription stops, corresponding to Ψ residues in U1, U2 and U5 snRNAs, are indicated by arrows on the left.
YLR165c ORF is responsible for Ψ2819 formation in mitochondrial 21S rRNA
Finally, we tested the possibility that the protein encoded by the YLR165c ORF acts on the S.cerevisiae mitochondrial LSU rRNA that is known to contain a single Ψ residue at position 2819 (33). Modification by CMCT was done on total RNA fractions from the ΔYLR165c haploid strain and the isogenic yeast strain, and the presence of Ψ2819 in the mitochondrial LSU 21S rRNA was tested as above by primer extension analysis. As shown in Figure 3, the results obtained with the ΔYLR165c strain differed from those obtained with the wild-type strain by the absence of the Ψ residue at position 2819 in the mitochondrial 21S rRNA.
Figure 3.
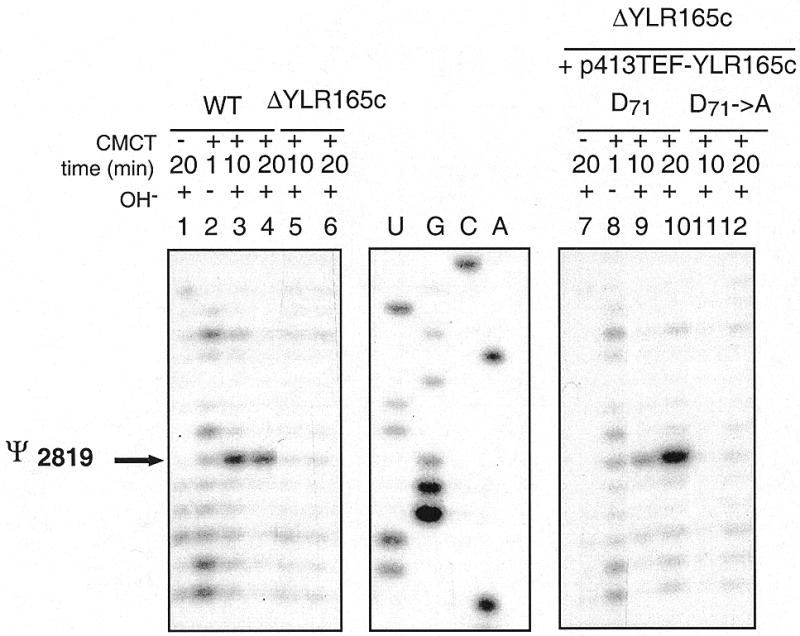
Ψ at position 2819 in yeast mitochondrial 21S rRNA disappears upon deletion of the YLR165c ORF. Primer extension analysis of the CMCT-modified S.cerevisiae mitochondrial 21S rRNA in a total RNA fraction from wild-type (WT) or ΔYLR165c S.cerevisiae strains, for 1 (lanes 2 and 8), 10 (lanes 3, 5, 9, 11) and 20 min (lanes 4, 6, 10, 12). A control extension experiment was made without addition of CMCT (lanes 1 and 7). The experimental conditions are the same as in Figure 2. The reverse transcription stop, corresponding to residue Ψ2819, is indicated by an arrow on the left. Absence of this stop in the reverse transcription pattern of the 21S rRNA from the ΔYLR165c strain (lanes 5 and 6) indicates the absence of residue Ψ2819. Primer extension analysis of the CMCT-modified S.cerevisiae mitochondrial 21S rRNA in a total RNA fraction from the ΔYLR165c strain transformed with plasmid p413TEF-YLR165c (lanes 9 and 10) and with a plasmid bearing mutated YLR165c ORF (lanes 11 and 12).
In order to verify that the YLR165c protein is indeed involved in Ψ2819 formation, the ΔYLR165c strain was complemented by the p413TEF-YLR165c plasmid, containing the wild-type ORF under the control of the TEF promoter. As shown in Figure 3 (lanes 7 and 8), Ψ formation at position 2819 in the mitochondrial LSU 21S rRNA was restored in the transformed yeast strain. To confirm the direct implication of the YLR165c-encoded protein in U→Ψ conversion at position 2819, we substituted the aspartate residue (D71) by an alanine in the GRTD sequence of the YLR165c ORF motif II (Fig. 1) and tested in vivo the complementation capacity of the mutated protein. The results presented in Figure 3 (lanes 9 and 10) show that the point mutation abrogated the complementation capacity of the cloned gene. Altogether, these results confirmed a mitochondrial rRNA:Ψ-synthase activity of the YLR165c-encoded protein. For homogeneity of RNA:Ψ-synthase nomenclature in yeast we propose to rename the YLR165c ORF to PUS5 and the corresponding protein to Pus5p.
Mitochondrial targeting of YLR165c-encoded protein
Since mitochondrial 21S rRNA is synthesised and maturated exclusively in mitochondria, the corresponding rRNA:Ψ-synthase has to enter into mitochondrial compartments by a mechanism which involves some kind of addressing signal. So far, only a few mitochondrial rRNA and tRNA modifying enzymes were characterised; all of them are encoded by nuclear DNA, synthesised by cytoplasmic ribosomes and then targeted to mitochondria (41,42). The analysis of the YLR165c protein using the PSORT prediction software (39), which takes into account only the first 20 amino acids, did not reveal any plausible mitochondrial leader peptide. Another approach for determination of possible subcellular localisation is based on the global amino acid composition of proteins (NNPLS software, 40). Using this prediction system the protein, encoded by the YLR165c ORF, can be classified as mitochondrial enzyme. Thus, despite the absence of pronounced N-terminal leader peptide, the YLR165c encoded protein is targeted into mitochondria.
The YLR165c encoded protein is likely dedicated only to mitochondrial rRNA modification since the residue at position 2945 in yeast cytoplasmic 26S rRNA (counterpart of position 2819 in the mitochondrial 21S rRNA) is not converted into a Ψ residue. In addition, even if a modification had to occur at this position, it would likely depend upon the Cbf5p/snoRNA-guided system.
U→Ψ conversion at position 2819 of the S.cerevisiae mitochondrial 21S rRNA is not essential for growth
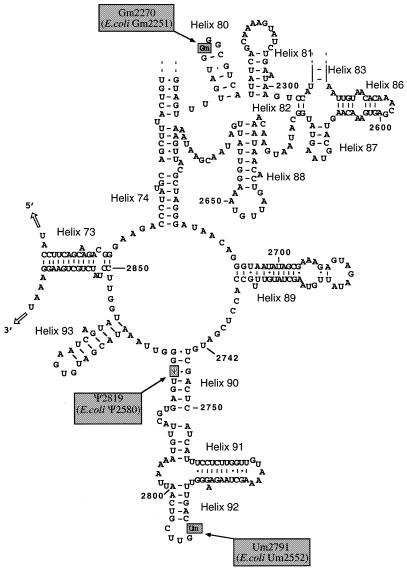
Several previous studies revealed a low content of post-transcriptional modifications in mitochondrial rRNAs. The mitochondrial S.cerevisiae LSU rRNA contains only three modified nucleosides: two 2′-O-methylated residues (Gm at position 2270 and Um at position 2791) and one Ψ residue at position 2819 (43–45, reviewed in 46). The two 2′-O-methylation sites found in the S.cerevisiae mitochondrial 21S rRNA are strongly conserved in cytoplasmic rRNAs from Eukarya and Bacteria (47–49). In contrast, the unique Ψ residue present in yeast mitochondrial 21S rRNA is conserved only in mitochondrial LSU rRNAs from some other species (for example human and Mus musculus) and in Gram-negative bacteria such as E.coli (29,33). It is absent in the LSU rRNAs from the Gram-positive bacteria Bacillus subtilis (29), in 26S rRNA of eukaryotic cytoplasmic ribosomes (45,49) and also in the archaea Halobacterium halobium (29). This is in agreement with our observation that the presence of Ψ2819 is not essential for growth on fermentable carbon source. Similarly, in E.coli, inactivation of RluC that forms residue Ψ2580 (the counterpart of the yeast mitochondrial Ψ2819 residue) and two other Ψ residues in 23S rRNA does not affect growth rate (11,12).
Nevertheless, because of its location in a 23S rRNA region considered to be located at the ribosome peptidyl transfer centre, residue Ψ2819 in the S.cerevisiae mitochondrial 21S rRNA may modulate the efficiency or the fidelity of the translation machinery. Indeed, the single-stranded sequences flanking helix 90 that contains Ψ2819 (helices are numbered according to nomenclature of Leffers et al. 50) (Fig. 4), are the most highly conserved regions in the LSU rRNA (47,48,50). Several mutations in these single-stranded sequences confer resistance of bacterial or mitochondrial ribosomes to various antibiotics affecting the peptidyl transferase activity (for review see 51). The second modified nucleotide found in 21S mitocondrial rRNA of S.cerevisiae (Um2791) is also located in the vicinity of the peptidyl transferase centre. Note that the terminal loop of helix 90 (Fig. 4) bearing Um2791 was shown to crosslink with puromycin derivatives (47,52) and to base-pair to the CCA terminal sequence of the tRNA located at the ribosomal A site (52).
Figure 4.
The three modified nucleotides of the S.cerevisiae mitochondrial 21S rRNA and their counterpart in E.coli 23S rRNA. Only the part of Domain V containing the three modified residues is represented according to the 2D structure drawn by Sirum-Connolly et al. (44). The positions of the three modified nucleotides are shown, the numbers of their counterparts in E.coli are indicated in brackets (43–45). Nucleotides are numbered starting from 5′-terminal. Helices are numbered according to Leffers et al. (50).
In conclusion, this study demonstrated that one of the four putative yeast RNA:Ψ-synthases (Pus5p) of the RluA-family is implicated in the modification of yeast mitochondrial rRNA. The functional characterisation and the identification of RNA targets for the three other putative RNA:Ψ-synthases are in progress in our laboratory.
Acknowledgments
ACKNOWLEDGEMENTS
We thank M. Minet and F. Lacroute (CNRS, Gif-sur-Yvette, France) for providing us with plasmid pFL35 and the BMA64 and BMA64-1A S.cerevisiae strains, Z. Szweykowska-Kulinska (Mickiewicz University, Poznan, Poland) for the plasmid containing the A.thaliana pre-tRNATyr gene and R. Giegé (CNRS, Strasbourg, France) for the plasmid containing the yeast tRNAAsp gene. This work was supported by laboratory funds from the ‘Ministère de la Recherche et de l’Enseignement Supérieur’, the French ‘Centre National de la Recherche Scientifique’ (Programme Physique et Chimie du Vivant 1997–99) and the ‘Association pour la Recherche sur le Cancer’. I.A. and S.M. are pre-doctoral fellows supported by a fellowship from the ‘Ministère de la Recherche et de l’Enseignement Supérieur’. Y.M. is the recipient of an Associated researcher position at the CNRS (‘Poste Rouge’).
REFERENCES
- 1.Grosjean H., Sprinzl,M. and Steinberg,S. (1995) Biochimie, 77, 139–141. [DOI] [PubMed] [Google Scholar]
- 2.Massenet S., Mougin,A. and Branlant,C. (1998) In Grosjean,H. and Benne,R. (eds), The Modification and Editing of RNA. ASM Press, Washington DC, pp. 201–228.
- 3.Ofengand J. and Fournier,M.J. (1998) In Grosjean,H. and Benne,R. (eds), The Modification and Editing of RNA. ASM Press, Washington DC, pp. 229–254.
- 4.Huang L., Pookanjanatavip,M., Gu,X. and Santi,D.V. (1998) Biochemistry, 37, 344–351. [DOI] [PubMed] [Google Scholar]
- 5.Raychaudhuri S., Conrad,J., Hall,B.G. and Ofengand,J. (1998) RNA, 4, 1407–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conrad J., Niu,L., Rudd,K., Lane,B.G. and Ofengand,J. (1999) RNA, 5, 751–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramamurthy V., Swann,S.L., Paulson,J.L., Spedaliere,C.J. and Mueller,E.G. (1999) J. Biol. Chem., 274, 22225–22230. [DOI] [PubMed] [Google Scholar]
- 8.Arena F., Ciliberto,G., Ciampi,S. and Cortese,R. (1978) Nucleic Acids Res., 5, 4523–4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kammen H.O., Marvel,C.C., Hardy,L. and Penhoet,E.E. (1988) J. Biol. Chem., 263, 2255–2263. [PubMed] [Google Scholar]
- 10.Nurse K., Wrzesinski,J., Bakin,A., Lane,B.G. and Ofengand,J. (1995) RNA, 1, 102–112. [PMC free article] [PubMed] [Google Scholar]
- 11.Huang L., Ku,J., Pookanjanatavip,M., Gu,X., Wang,D., Greene,P.J. and Santi,D.V. (1998) Biochemistry, 37, 15951–15957. [DOI] [PubMed] [Google Scholar]
- 12.Conrad J., Sun,D., Englund,N. and Ofengand,J. (1998) J. Biol. Chem., 273, 18562–18566. [DOI] [PubMed] [Google Scholar]
- 13.Wrzesinski J., Bakin,A., Nurse,K., Lane,B.G. and Ofengand,J. (1995) Biochemistry, 34, 8904–8913. [DOI] [PubMed] [Google Scholar]
- 14.Niu L. and Ofengand,J. (1999) Biochemistry, 38, 629–635. [DOI] [PubMed] [Google Scholar]
- 15.Wrzesinski J., Nurse,K., Bakin,A., Lane,B.G. and Ofengand,J. (1995) RNA, 1, 437–448. [PMC free article] [PubMed] [Google Scholar]
- 16.Raychaudhuri S., Niu,L., Conrad,J., Lane,B.G. and Ofengand,J. (1999) J. Biol. Chem., 274, 18880–18886. [DOI] [PubMed] [Google Scholar]
- 17.Koonin E.V. (1996) Nucleic Acids Res., 24, 2411–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simos G., Tekotte,H., Grosjean,H., Segref,A., Sharma,K., Tollervey,D. and Hurt,E.C. (1996) EMBO J., 15, 2270–2284. [PMC free article] [PubMed] [Google Scholar]
- 19.Becker H.F., Motorin,Y., Planta,R.J. and Grosjean,H. (1997) Nucleic Acids Res., 25, 4493–4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Motorin Y., Keith,G., Simon,C., Foiret,D., Simos,G., Hurt,E. and Grosjean,H. (1998) RNA, 4, 856–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lafontaine D.L.J., Bousquet-Antonelli,C., Henry,Y., Caizergues-Ferrer,M. and Tollervey,D. (1998) Genes Dev., 12, 527–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lecointe F., Simos,G., Sauer,A., Hurt,E.C., Motorin,Y. and Grosjean,H. (1998) J. Biol. Chem., 273, 1316–1323. [DOI] [PubMed] [Google Scholar]
- 23.Massenet S., Motorin,Y., Lafontaine,D.L., Hurt,E.C., Grosjean,H. and Branlant,C. (1999) Mol. Cell. Biol., 19, 2142–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Watkins N.J., Gottschalk,A., Neubauer,G., Kastner,B., Fabrizio,P., Mann,M. and Lührmann,R. (1998) RNA, 4, 1549–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zebarjadian Y., King,T., Fournier,M.J., Clarke,L. and Carbon,J. (1999) Mol. Cell. Biol., 19, 7461–7472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ni J., Tien,A.L. and Fournier,M.J. (1997) Cell, 89, 565–573. [DOI] [PubMed] [Google Scholar]
- 27.Ganot P., Bortolin,M.L. and Kiss,T. (1997) Cell, 89, 799–809. [DOI] [PubMed] [Google Scholar]
- 28.Sprinzl S., Horn,C., Brown,M., Ioudovitch,A. and Steinberg,S. (1998) Nucleic Acids Res., 26, 148–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ofengand J. and Bakin,A. (1997) J. Mol. Biol., 266, 246–268. [DOI] [PubMed] [Google Scholar]
- 30.Baudin A., Ozier-Kalogeropoulos,O., Denouel,A., Lacroute,F. and Cullin,C. (1993) Nucleic Acids Res., 21, 3329–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wach A., Brachat,A., Pohlmann,R. and Philippsen,P. (1994) Yeast, 10, 1793–1808. [DOI] [PubMed] [Google Scholar]
- 32.Gietz D., St Jean,A., Woods,R.A. and Schiestl,R.H. (1992) Nucleic Acids Res., 20, 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bakin A. and Ofengand,J. (1993) Biochemistry, 32, 9754–9762. [DOI] [PubMed] [Google Scholar]
- 34.Jiang H.Q., Motorin,Y., Jin,Y.X. and Grosjean,H. (1997) Nucleic Acids Res., 25, 2694–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mumberg D., Muller,R. and Funk,M. (1995) Gene, 156, 119–122. [DOI] [PubMed] [Google Scholar]
- 36.Adams A., Gottschling,D.E., Kaiser,C.A. and Stearns,T. (1997) Methods in Yeast Genetics, A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 37.Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lawrence C.E., Altschul,S.F., Boguski,M.S., Liu,J.S., Neuwald,A.F. and Wootton,J.C. (1993) Science, 262, 208–214. [DOI] [PubMed] [Google Scholar]
- 39.Nakai K. and Kanehisa,M. (1992) Genomics, 14, 897–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reinhardt A. and Hubbard,T. (1998) Nucleic Acids Res., 26, 2230–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martin N.C. and Hopper,A.K. (1994) Biochimie, 76, 1161–1167. [DOI] [PubMed] [Google Scholar]
- 42.Maden B.E.H. (1998) In Grosjean,H. and Benne,R. (eds), The Modification and Editing of RNA. ASM Press, Washington DC, pp. 421–440.
- 43.Sirum-Connolly K. and Mason,T.L. (1993) Science, 262, 1886–1889. [DOI] [PubMed] [Google Scholar]
- 44.Sirum-Connolly K., Peltier,J.M., Crain,P.F., McCloskey,J.A. and Mason,T.L. (1995) Biochimie, 77, 30–39. [DOI] [PubMed] [Google Scholar]
- 45.Bakin A., Lane,B.G. and Ofengand,J. (1994) Biochemistry, 33, 13475–13483. [DOI] [PubMed] [Google Scholar]
- 46.Mason T.L. (1998) In Grosjean,H. and Benne,R. (eds), The Modification and Editing of RNA. ASM Press, Washington DC, pp. 273–280.
- 47.Branlant C., Krol,A., Machatt,M.A., Pouyet,J., Ebel,J.P., Edwards,K. and Kossel,H. (1981) Nucleic Acids Res., 9, 4303–4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Veldman G.M., Klootwijk,J., de Regt,V.C., Planta,R.J., Branlant,C., Krol,A. and Ebel,J.P. (1981) Nucleic Acids Res., 9, 6935–6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maden B.E. (1990) Prog. Nucleic Acid Res. Mol. Biol., 39, 241–303. [DOI] [PubMed] [Google Scholar]
- 50.Leffers H., Kjems,J., Ostergaard,L., Larsen,N. and Garrett,R.A. (1987) J. Mol. Biol., 195, 43–61. [DOI] [PubMed] [Google Scholar]
- 51.Kirillov S., Porse,B.T., Vester,B., Woolley,P. and Garrett,R.A. (1997) FEBS Lett., 406, 223–233. [DOI] [PubMed] [Google Scholar]
- 52.Kim D.F. and Green,R. (1999) Mol. Cell, 4, 859–864. [DOI] [PubMed] [Google Scholar]