Abstract
Eukaryotic initiation factor 4E (eIF4E) is a pivotal protein involved in the regulatory mechanism for global protein synthesis in both physiological and pathological conditions. MicroRNAs (miRNAs) play a significant role in regulating gene expression by targeting mRNA. However, the ability of miRNAs to regulate eIF4E and its phosphorylation remains relatively unknown. In this study, we predicted and experimentally verified targets for miR-483-5p, including eukaryotic translation initiation factor eIF4E and its binding proteins, 4E-BPs, that regulate protein synthesis. Using the Web of Science database, we identified 28 experimentally verified miR-483-5p targets, and by the TargetScan database, we found 1818 predicted mRNA targets, including EIF4E, EIF4EBP1, and EIF4EBP2. We verified that miR-483-5p significantly reduced ERK1 and MKNK1 mRNA levels in HEK293 cells. Furthermore, we discovered that miR-483-5p suppressed EIF4EBP1 and EIF4EBP2, but not EIF4E. Finally, we found that miR-483-5p reduced the level of phosphorylated eIF4E (pSer209eIF4E) but not total eIF4E. In conclusion, our study suggests that miR-483-5p's multi-targeting effect on the ERK1/ MKNK1 axis modulates the phosphorylation state of eIF4E. Unlike siRNA, miRNA can have multiple targets in the pathway, and thereby exploring the role of miR-483-5p in various cancer models may uncover therapeutic options.
Subject terms: Cancer, Molecular biology
Introduction
Eukaryotic mRNA translation is an energy consuming process, and it occurs in three stages: initiation, elongation and termination. Among the well-understood mechanisms for mRNA translation initiation are: cap-dependent process (usually occurring under physiological conditions) and cap-independent process (usually occurring under stress/infection conditions), mediated by the internal ribosome entry site (IRES)1–3. In cap-dependent translation, eukaryotic translation initiation factor 4E (eIF4E) binds to the 7-methylguanosine 5′-triphosphate (m7GTP) at the 5′ end of mRNA4. eIF4E is a part of the multi-protein eIF4F complex that also comprises other proteins such as the scaffold protein eIF4G, and the RNA helicase eIF4A. eIF4F complex further recruits accessory 43S pre-initiation complex (containing 40S small ribosomal subunit, initiation factors such as eIF1, eIF1A, eIF3, eIF5, and the eIF2/Met-tRNAi/GTP ternary complex) that facilitates the detection of AUG start codon in the 5′UTR. Once 60S large ribosomal subunit binds to 43S pre-initiation complex, 80S ribosome is formed to initiate the peptide synthesis2,5.
In cap-dependent translation the binding of eIF4E to the m7GTP is a crucial step limiting the rate of mRNA translation. Therefore, to reduce the aberrant translation, eIF4E is regulated by several mechanisms. First, at the transcription level by the MYC transcription factor6,7. Second, at the level of eIF4F complex formation, through eIF4E binding proteins (4E-BPs) that compete with eIF4G for binding to eIF4E thus limiting the eIF4F complex formation and in consequence decelerating the cap-dependent translation8. Alternatively, active mTORC1 signalling phosphorylates 4E-BP and causes its dissociation from eIF4E, thus allowing the formation of eIF4F complex and subsequently accelerating translation (Fig. 1)8. Third, eIF4E activity is regulated by phosphorylation at the serine 2099. Phosphorylation of eIF4E at the serine 209 leads to the increased global protein translation and is observed in various cancer types. It is known to regulate several pathways, including proliferation, apoptosis and metastasis9–14. MKNK (MNK) is the upstream kinase that phosphorylates eIF4E, thus targeting the MKNK-eIF4E axis reduce the level of eIF4E phosphorylation15–20. Recently, a review compiled various MKNK inhibitors studied to limit the eIF4E phosphorylation15. Also, several independent studies showed that even inhibition of the upstream kinase ERK1/2 regulates the ERK-MKNK-eIF4E axis and results in the reduction of eIF4E phosphorylation21–23.
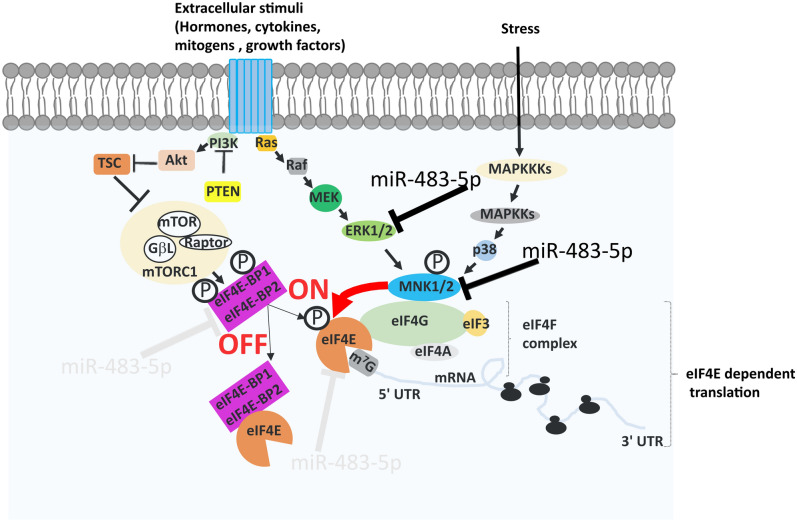
Figure 1.
Signalling pathways in cap-dependent mRNA translation. Activation of Ras/MAPK pathway kinases (in response to mitogenic and stress signals) such as ERK and p38 leads to MKNK1/2 activation, and further phosphorylation of eIF4E at Ser209. Activation of PI3K pathway kinases (in response to extracellular stimuli) such as mTORC1 complexes, leads to the phosphorylation of the translation repressors eIF4E-BP1/2, thereby allowing eIF4E to form the eIF4F complex. Net cumulative effect of these two pathways determines the on / off switching of the cap dependent translation. Inhibitory regulation by miR-483-5p in cap-dependent mRNA translation pathway is shown (black—experimentally verified targets, light gray—predictions yet to be verified).
microRNAs (miRNAs) are small noncoding RNAs (19–24 nucleotides) that regulate gene expression by post-transcriptional mechanisms and have been shown to reduce the overall protein synthesis by targeting the cap-dependent translation24,25. miRNAs are differentially expressed in several pathological conditions26,27. Previous studies by us and others have shown that miR-483-5p mediates ERK1 targeting28,29 and down-regulates the ERK1 phosphorylation in HEK293 cells. In a separate study, it was found that miR-483-5p targeted MKNK1 in Wilms’ tumour cells30. Because eIF4E phosphorylation was not examined in these studies, we wanted to explore the potential of miRNA to regulate eIF4E phosphorylation. We hypothesised that multi-targeting binding of miR-483-5p would synergistically affect the phosphorylation of both ERK1 and MKNK1, and lead to the repression of eIF4E phosphorylation (Fig. 1). In addition, in the current study, we explored the potential of miR-483-5p for regulation of 4E-BPs.
Results
Prediction and experimental investigation of miR-483-5p targets: EIF4E, EIF4EBP1 and EIF4EBP2
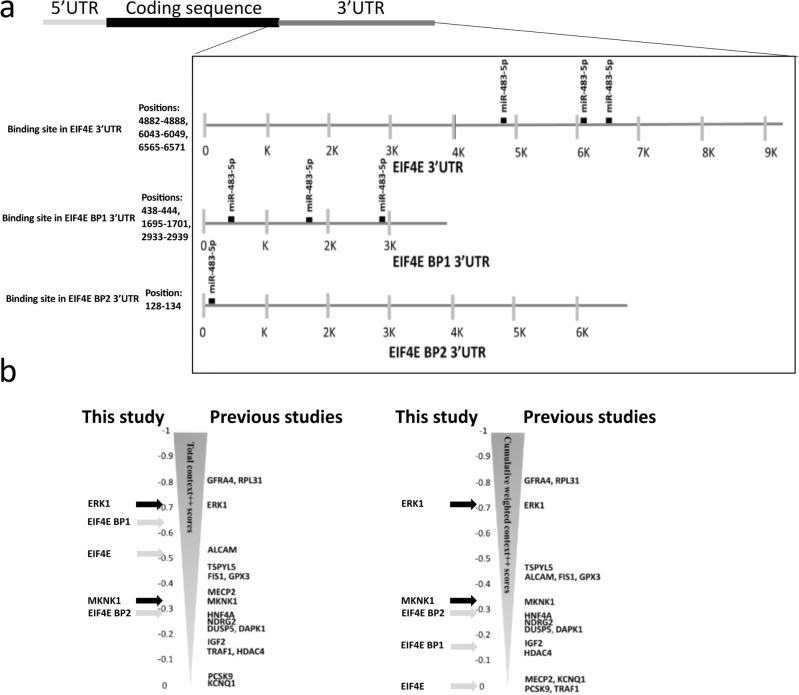
By searching the TargetScan database with an algorithm that finds binding sites for miRNA in the 3′ UTR of the mRNA, we found 1818 target mRNAs with varying strength of predicted binding to miR-483-5p. Among them 28 targets have been experimentally verified according to the Web of Science database by May 2023 [GFRA4, ERK1, RPL31, ALCAM, TSPYL5, FIS1, GPX3, MeCP2, MKNK1, HNF4A, NDRG2, DUSP5, DAPK1, IGF2, TRAF1, HDAC4, PCSK9, KCNQ1, SRF, SOCS3, RAI16, TBL1X, RhoGDI1, CKB, NOTCH3, TGFB1, RBM5, and PIAS1] that are known to be repressed by miR-483-5p (Fig. 2b). Interestingly, the experimental targets had varying total context++ scores, with the highest score (− 0.81) for GFRA4 and RPL31 and the lowest score (− 0.05) for KCNQ1, indicating the necessity for the experimental evaluation of predicted targets.
Figure 2.
Prediction of miR-483-5p targets in cap-dependent mRNA translation pathway. (a) miR-483-5p predicted to bind 3′UTR of eIF4E mRNAs (3 sites), of EIFE 4BP1 mRNAs (3 sites) and EIF4E BP2 mRNAs (1 site). (b) Relative positions of targets out of 1818 targets based on predicted computational scores (total context++ score and cumulative weighted context++ score) from TargetScan database. Black arrows and gray arrows represent verified and novel targets investigated in this study, respectively. Other experimentally verified targets were positioned accordingly.
Within the listed 28 targets, present are those involved in the regulation of the translation process, kinases ERK1 and MKNK1/2. Affecting these kinases could impair the downstream targets of miR-483-5p, including EIF4E. Therefore, we decided to further investigate genes encoding eIF4E and eIF4E-binding proteins (4E-BP1, 4E-BP2) as targets of miR-483-5p. In the 3′UTR of EIF4E we found three binding sites for miR-483-5p at positions 4882–4888, 6043–6049, 6565–6571. These are poorly conserved 7mer-m8 type sites (exact match to positions 2–8 of the mature miRNA) with the cumulative weighted context++ score 0 and total context++ score − 0.53 (Fig. 2a,b). In the 3′UTR of EIF4EBP1 we found three binding sites for miR-483-5p at positions 438–444, 1695–1701, 2933–2939. These are poorly conserved 7mer-m8 type sites (exact match to positions 2–8 of the mature miRNA) with the cumulative weighted context++ score − 0.16 and total context++ score − 0.65 (Fig. 2a,b). In the 3′UTR of EIF4EBP2 we found one binding site for miR-483-5p at the position 128–134. This is poorly conserved 7mer-m8 type site (exact match to positions 2–8 of the mature miRNA) with the cumulative weighted context++ score − 0.29 and total context++ score − 0.29 (Fig. 2a, b).
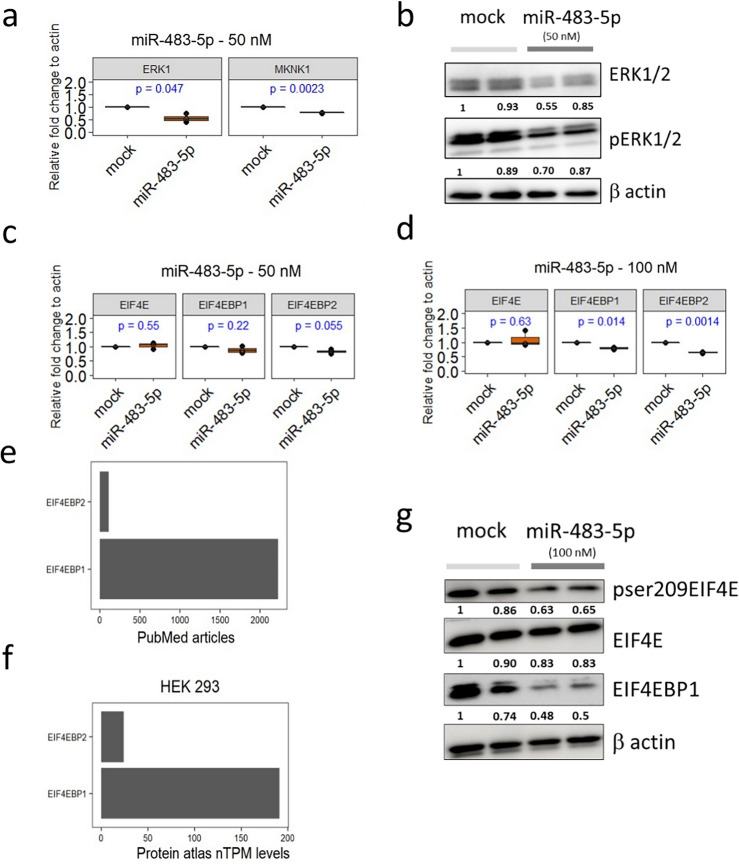
At first, we wanted to recapitulate the repression for the previously verified targets (ERK1 and MKNK1) in HEK293. As expected, we observed that 50 nM miR-483-5p mimic significantly reduced ERK1 and MKNK1 mRNA levels in comparison to mock-transfected cells (Fig. 3a). Further we noticed decreased ERK1/2 and pERK1/2 protein levels in these conditions (Fig. 3b). We then asked whether miR-483-5p at 50 nM could repress hitherto not experimentally verified targets EIF4E, EIF4EBP1 and EIF4EBP2. We noticed no change of the transcript level for EIF4E, slight but non-significant reduction for EIF4EBP1 and significant reduction of EIF4EBP2 (Fig. 3c). Increasing the miR-483-5p mimic to 100 nM, resulted in no change in transcript levels for EIF4E, but significantly reduced transcript levels for EIF4EBP1 and EIF4EBP2 (Fig. 3d). Overall, our data indicate that transcript of both EIF4EBP1 and EIF4EBP2 is suppressed by miR-483-5p, but not that of EIF4E.
Figure 3.
Reduction of pSer209eIF4E, 4E-BP1 by miR-483-5p. (a) RT-qPCR analysis of previously validated targets ERK1 and MKNK1 mRNA levels upon transfection with 50 nM of miR-483-5p mimic. (b) Representative immunoblots for pERK1/2 and ERK1/2 protein levels upon transfection with mock and miR-483-5p mimic at 50 nM. (c,d) RT-qPCR analysis of novel targets EIF4E, EIF4EBP1 and EIF4EBP2 mRNA levels upon transfection with 50 nM (c) and 100 nM (d) of miR-483-5p mimic. (e) Number of PubMed articles retrieved by May 2023 on 4E-BP1 and 4E-BP2. (f) Relative expression of 4E-BP1 and 4E-BP2 in HEK293 based on protein atlas database (proteinatlas.org). (g) Representative immunoblots for pSer209eIF4E, eIF4E and 4E-BP1 protein levels upon transfection with mock and miR-483-5p mimic at 100 nM. β-Actin (ACTB) was used as a reference for normalization in a-d and g. Uncropped blots are provided in supplementary material.
miR-483-5p mediated reduction of pSer209eIF4E but not eIF4E levels
ERK1/2 and MKNK1 are kinases that phosphorylate their downstream targets including eIF4E. Therefore, we decided to investigate the level of phosphorylated protein eIF4E after transfection of HEK293 cells with miR-483-5p. For this purpose, we used antibody against total eIF4E and another antibody against phosphorylated eIF4E, specific to serine 209 (pSer209eIF4E). Indeed, immunoblotting experiments showed that at concentration of 100 nM miR-483-5p mimic reduced pSer209eIF4E protein level in comparison to mock transfected cells (Fig. 3g). On the other hand, transfection with miR-483-5p mimic had no effect on the levels of eIF4E (Fig. 3g). This observation is consistent with RT-qPCR analysis that showed unchanged levels of EIF4E mRNA (Fig. 3c,d) after miR-483-5p mimic transfection, while ERK1 and MKNK1 mRNA levels significantly decreased (Fig. 3a). Collectively, our results indicate that miR-483-5p indirectly reduces the level of phosphorylated eIF4E by downregulating the expression of ERK1/2 and MKNK1 kinases.
miR-483-5p mediates reduction of 4E-BP1 protein
The other miR-483-5p targets we have selected were mRNAs encoding 4E-BP1 and 4E-BP2, two proteins involved in the regulation of cap-dependent translation process. 4E-BP1 is a well-studied protein that binds to eIF4E and thus inhibits the formation of eIF4F, which in consequence affects cap-dependent translation. PubMed search revealed 20 times more articles on 4E-BP1 compared to those on 4E-BP2 as of May 15th 2023 (Fig. 3e). Moreover, Protein atlas database indicated that the relative expression of 4E-BP1 is eightfold higher than 4E-BP2 in HEK293 cells (Fig. 3f). Due to low expression levels in HEK293 (Fig. 3f), limited literature showing insufficient characterization in translational regulation, and a lack of well-annotated antibodies against 4E-BP2 (https://www.citeab.com/antibodies/search?q=4ebp1,https://www.citeab.com/antibodies/search?q=4e+bp2 ), we did not explore protein levels of 4E-BP2 in this study, only mRNA levels (Fig. 3c,d). However, we evaluated 4E-BP1 protein levels in HEK293 cell lysates, using well-validated antibody against 4E-BP1. We found that miR-483-5p mimic at 100 nM reduced mRNA levels of both 4E-BP1 and 4E-BP2 (Fig. 3c,d) and lowered 4E-BP1 protein level in comparison to mock transfected cells (Fig. 3g). Overall, upon miR-483-5p transfection we found reduction of protein levels of pSer209eIF4E and of 4E-BP1.
Multi targeting miR-483-5p vs single targeting siERK1: Different consequences on C-MYC and CCND1 expression
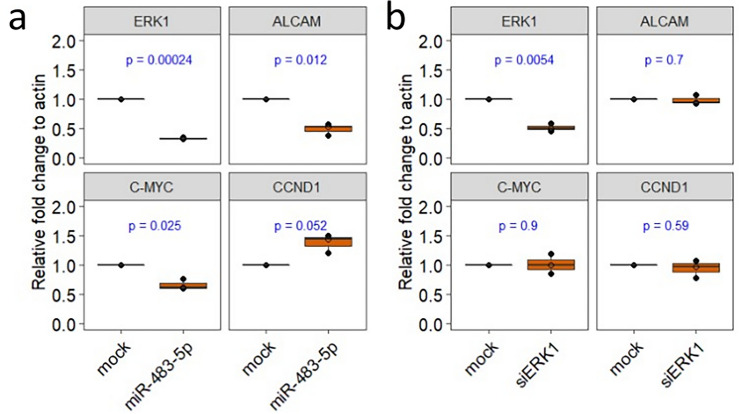
Next, we evaluated the cellular consequences of miR-483-5p mediated down regulation of ERK1 and resulting reduced level of pSer209eIF4E. In these experiments we used ERK1 and another experimentally validated target of miR-483-5p, Activated Leukocyte Cell Adhesion Molecule, ALCAM as positive reference controls to show the multi targeting potential of miR-483-5p (Fig. 2b). A recent study demonstrated that S209A substitution in eIF4E decreased the level of phosphorylated eIF4E and in consequence reduced the levels of both C-MYC mRNA and C-MYC protein14. Also in our case, after transfection of HEK293 cells with miR-483-5p, the level of C-MYC mRNA was reduced, which may be due to the decreased levels of pSer209eIF4E (Fig. 4a). We previously reported miR-483-5p mediated reduction of pERK1 in HEK29328, however its nuclear and cytoplasmic distribution is unknown. Interestingly, a study showed miR-483-5p mediated regulation of nucleus resident DUSP531. Moreover, it is documented that DUSP5 loss increases nuclear ERK1/2 phosphorylation and expression of its nuclear targets32. Therefore, further we evaluated one such nuclear ERK target, cyclin D1 (CCND1), whose expression is enhanced by nuclear ERK activity33,34. We asked if miR-483-5p transfection would increase CCND1 expression. We noticed that it increases the CCND1 mRNA levels upon miR-483-5p transfection (Fig. 4a). To further confirm that such observation is specific to the multi-targeting effect of miR-483-5p, we used siRNA specific to ERK1 (siERK1) as a control to evaluate the levels of C-MYC and CCND1. As expected, in siERK1 experiments, only ERK1 mRNA levels were reduced but not ALCAM, C-MYC, and CCND1 (Fig. 4b). Taken together, in HEK293 cells upon miR-483-5p transfection, multi targeting regulation by miR483-5p results in the reduced mRNA levels of C-MYC and enhanced mRNA levels of CCND1.
Figure 4.
Multi-targeting potential of miR-483-5p compared to mono-targeting by siERK1. RT-qPCR analysis of ERK1, ALCAM, C-MYC, and CCND1 mRNA levels upon transfection with 100 nM of miR-483-5p mimic (a) and 50 nM of siERK1 (b). β-Actin (ACTB) was used as a reference for normalization in RT-qPCR experiments.
Discussion
miRNAs are known to target multiple mRNAs through complementary base pairing26,27. This miRNA-mRNA interaction is complex and not all miRNA mediated mRNA targeting impacts the cellular phenotype. Some of the interactions counterbalance each other (mRNA targets considered weak) whereas other interactions (mRNA targets considered strong) consistently determine the cellular phenotype35,36. Furthermore, this complexity is enhanced by intrinsic feedback loops37 and dose specific response38. Therefore, there is still a need for extensive characterization of miRNA and its effect on predicted mRNA targets in different cell types.
Here we showed reduction in pSer209eIF4E and 4E-BP1 levels upon transfection with miR-483-5p mimic in HEK293 cells. The rationale for selecting miR-483-5p was based on our previous research, which highlighted its capability to target ERK128. This finding aligns with other studies, establishing a consensus on miR-483-5p's regulation of ERK129,39. Through the exploration of possible targets, we identified the robust potential of miR-483-5p to bind MKNK1 (Fig. 2b), a discovery supported by both our investigations (Fig. 3a) and those of others30. However, the impact of miR-483-5p on the downstream relays of the ERK1/MKNK1 axis, due to a cumulative effect on these two kinases, remains unclear. The ERK1/MKNK1 axis plays a crucial role in regulating the eIF4E signalling pathway, determining global protein levels. Under normal physiological conditions, the ERK pathway is activated by factors such as hormones, growth factors, and differentiation factors40. In pathological conditions, such as those associated with tumor-promoting factors, the ERK pathway is also activated. While the individual effects of the cascading activation signals are extensively studied for each kinase41,42, the well-established function of the ERK1/MKNK1 axis is to phosphorylate the mRNA cap-binding protein eIF4E15,22,43,44.
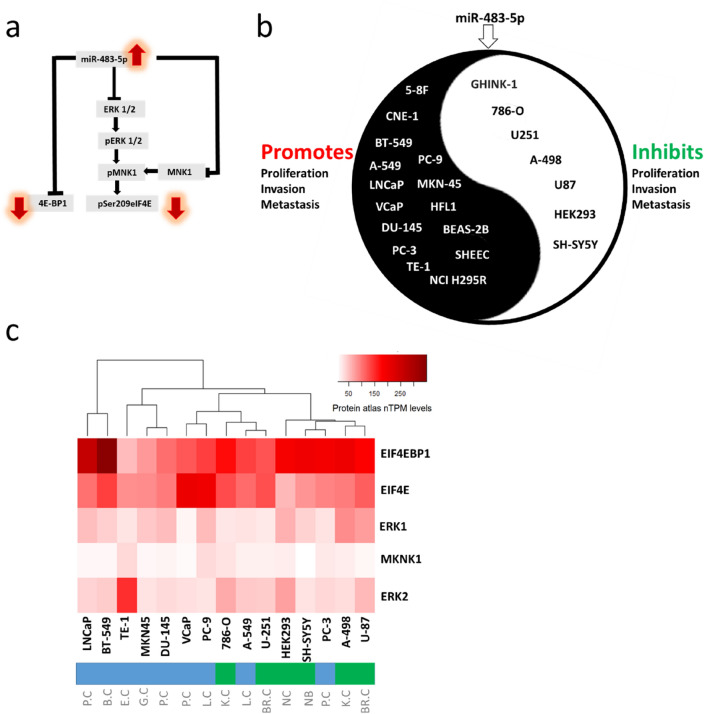
In physiological contexts, miR-483-5p plays a role in preserving the function and identity of pancreatic β-cells, promoting adipogenesis in adipose tissue, and alleviating hyperlipidemia-associated fatty liver disease45. Dysregulation of miR-483-5p is associated with various diseases, including cancer46,47, type 2 diabetes, fatty liver disease, diabetic nephropathy, and neurological injury45. Recognizing that miR-483-5p regulates the ERK1/MKNK1 axis could be a crucial information for modulating both physiological and pathological conditions. This is the first study to report how miR-483-5p affects cellular levels of key proteins involved in global protein translation. The proposed mechanism of these effects are schematically shown in Fig. 5a. Notably, miR-483-5p is a microRNA dysregulated in various cancers, and the levels of both pSer209eIF4E and 4E-BP1 are also increased several-fold in different cancers15,48. Therefore this is of particular interest to further investigate the role of miR-483-5p in the regulation of pSer209eIF4E and 4E-BP1 in various cells. Such studies are important because they can aid to explain the basic mechanism of miRNA action and define the differences in cancer cells, allowing for the identification of novel therapeutic targets.
Figure 5.
Cell to cell variability upon miR-483-5p transfection. (a) The scheme shows the pathway for the reduction of pSer209eIF4E and 4E-BP1 in HEK293. (b) Evidence from the literature for phenotypic variability among cell lines (promotion/inhibition of proliferation, invasion and metastasis) upon miR-483-5p transfection. (c) Protein atlas data showing variance in the relative protein levels targeted by miR-483-5p among different cell lines. P.C Prostate cancer, B.C Breast cancer, E.C Esophageal cancer, G.C Gastric cancer, L.C Lung cancer, K.C Kidney cancer, NC Non-cancerous, NB Neuroblastoma, BR.C Brain cancer.
It is currently unknown if the observed miR-483-5p-driven repression of the levels of these two proteins is part of a buffering noise or a significant repression that determines the cellular phenotype. In order to determine the exact role of this mechanism in proliferation, invasion and metastasis, different cell lines need to be investigated especially that divergent phenotypes are observed upon miR-483-5p transfection of various cancer cell lines, as shown in Fig. 5b. In certain cell lines miR-483-5p promoted46,49 whereas in other cell lines it inhibited the proliferation, invasion and metastasis29,47,50. One possible mechanism for such divergent phenotypes could be due to the net outcome of the collective miRNA-mRNA interactions, probably stemming from the differences in available mRNA targets and their competition for miR-483-5p binding (Fig. 5b). Intriguingly, the protein atlas data is in line with literature evidence showing two different clusters (shown in green and blue) among the cell lines based on the protein signatures of miR-483-5p targets, at least those involved in the cap-dependent translation process (Fig. 5c). Nevertheless, such data should be validated both in vitro and in vivo across various cancer models to further examine phenotypic variability upon miR-483-5p overexpression.
The intriguing aspect of the results obtained in this study lies in the fact that miR-483-5p exerts control over two distinct routes: (1) the ERK1/MKNK1 axis and (2) EIF4EBP1. These routes, in essence, may counteract each other, leading to a delicate balance of pEIF4E levels. This balance, in turn, may play a pivotal role in determining the fate of translation processes. The intricate regulation of these pathways by miR-483-5p underscores the complexity and nuanced control exerted by this microRNA on protein translation. Additionally, miR-483-5p has multiple potential target mRNAs, and its multi-targeting effect can extend beyond the ERK1/MKNK1 axis. However, a limited number of miR-483-5p targets, as illustrated in Fig. 2, have undergone experimental verification. Notably, these validated targets are recognized players in cancer-related processes. The intricate network of pathways involving these targets suggests a high likelihood of interplay among them, potentially contributing to a discernible phenotype across diverse cancer types.
Previous studies showed that miR-455-3p, miR-15a and miR-141 mediated direct repression of eIF4E and suggested the tumour suppressing role of miRNAs in prostate cancer, renal cell carcinoma, and non-small cell lung cancer, respectively24,51,52. However, the previous studies have not addressed the issue of pSer209eIF4E levels. Collectively, in our study we show for the first time that miR-483-5p mediates reduction of pSer209eIF4E without affecting total eIF4E levels via direct regulation of the ERK1 and MKNK1.
At mRNA levels we noticed a reduction of C-MYC and an increase of CCND1 after miR-483-5p transfections, but not after siERK1 transfections. These results emphasize the multi targeting potential of a miRNA, not achieved with siRNA. More importantly, it is yet to be investigated in different cells whether reduced eIF4E phosphorylation could have compensating effect in affecting the cap dependent translation of CCND1. Furthermore, to obtain deeper mechanistic insights, the cross talk between 4E-BP1-C-MYC53 and 4E-BP1-CCND154 needs to be deciphered in individual cell types.
In our study, we used the standard HEK293 cell line, which is a model of an unaltered cell. This was to understand the physiological basis of miR-483-5p's influence on the phosphorylation of the cap-binding protein eIF4E by targeting the ERK1/MKNK1 axis. Our results provide a basis for the characterization of differences in the molecular processes responsible for pathological alterations. It is essential to expand on this finding, particularly in to the context of cancer, where the phosphorylation of the cap-binding protein eIF4E is notably high11 and dysregulation of miR-483-5p has been previously reported55. Taken together our results indicate that miR-483-5p mediated reduction of pSer209eIF4E and 4E-BP1 in HEK293. However, the underlying mechanisms differ significantly. The decrease in pSer209eIF4E levels can be attributed to the diminished kinases in the eIF4E phosphorylation pathway, a consequence of miR-483-5p binding to the mRNA of these kinases (ERK1 and MKNK1). In contrast, the reduction in 4E-BP1 levels is a direct result of miR-483-5p binding to 4E-BP1 mRNA. Moreover, we suggest the importance to evaluate pSer209eIF4E and 4E-BP1 levels upon miR-483-5p transfections among different cancer cell lines to understand the mechanisms behind the cellular phenotype, such as proliferation, invasion and metastasis. Furthermore, deciphering co-regulated targets of miR-483-5p, in conjunction with phosphorylated Ser209 eIF4E and 4E-BP1, is needed for unbiased understanding. Additionally, the assessment of off-target effects of miR-483-5p and the exploration of delivery modes for miR-483-5p in vivo are crucial steps towards comprehending its therapeutic applications. Before making conclusive statements about the potential therapeutic implications of miR-483-5p, it is essential to systematically classify the cell types or cancer models where miR-483-5p exerts inhibitory or promotional effects on proliferation and metastasis. Given that this observation is based on existing literature, it is equally important to reproduce the findings using different dosage titrations to gain comprehensive insights into the effects on the relevant targets and pathways. It's noteworthy that the dosage of miRNA can lead to varying effects, as evidenced by existing research38.Another important investigation is analysis of the patient prognosis associated with miR-483-5p levels. However, it is noteworthy that several studies have already been conducted to assess the prognostic significance of miR-483-5p across various cancer types56–61. The continuation of this research can provide an insight into prognostic biomarker potential of miR-483-5p.
Methods
Cell culture
Human embryonic kidney 293 (HEK293) obtained from the American Type Culture Collection (ATCC) were grown in Eagle’s Minimum Essential Medium (EMEM) (Sigma-Aldrich) supplemented with 10% foetal bovine serum (FBS) (Sigma-Aldrich), 2 mM L-glutamine and 100 mg/mL penicillin/streptomycin. Cells were cultured at 37 °C in a humidified atmosphere of 5% CO2.
miRNA and siRNA transfection
miR-483-5p mirVana miRNA mimic (#4464066) was used (Ambion). Mock transfections were performed with only Opti-MEM medium and Lipofectamine 2000 without mimics. In miRNA transfection experiments, mock was used as baseline for evaluating the effect of miR-483-5p mimic. In siRNA transfection experiments, siERK1 (#AM16708) (Ambion) was used to silence ERK1. In siRNA transfection experiments, mock was used as baseline for evaluating the effect of siERK1. In all these experiments, cells were seeded at 1 × 106 cells/well in 12-well plates 24 h prior to transfection. Cells were grown to 60–80% confluence and transfected with miRNA mimics (up to 100 nM) or siERK1 (50 nM). Cells were harvested at 48 h post-transfection for the quantitative real-time polymerase chain reaction (RT-qPCR) and immunoblotting.
RNA isolation and RT-qPCR
Total RNA was isolated from cells using the Total RNA Mini isolation kit (A&A Biotechnology) according to the manufacturer’s instructions. RNA was quantified spectrophotometrically and its quality was analysed using A260/A280 ratio (DeNovix). Up to 150 ng of RNA was used to obtain cDNA using the High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific). Quantitative PCR was performed on LightCycler 480 II System (Roche). Briefly, 2 μl of cDNA (obtained from 150 ng of RNA), 4 μl of each mRNA specific primers (5 pmol of forward and reverse) (Supplementary material), 10 μl of Maxima SYBR Green qPCR Master Mix (2X) (Thermo Fisher Scientific) were mixed in a 20 μl reaction and run with a thermal profile of an initial 10 min melting step at 95 °C, followed by 45 cycles at 95 °C for 10 s, 60 °C for 10 s and 72 °C for 10 s. The relative fold change of mRNAs was normalized to β-actin mRNA by 2−ΔΔCt method62.
Immunoblotting
HEK293 lysates were obtained using RIPA lysis buffer (Sigma Aldrich) with phosphatase and protease inhibitor cocktails (Roche). The protein concentrations in the samples were determined using the Pierce BCA protein assay kit (Thermo Fisher Scientific). A total of 20 µg of protein samples diluted in sample buffer containing 2-mercaptoethanol (Sigma-Aldrich), denatured for 10 min at 95 °C, were loaded in each well and separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) in a 10 or15% polyacrylamide gel cast using the TGX FastCast acrylamide kit (Bio-Rad). After electrophoretic separations, proteins were transferred to polyvinylidene fluoride (PVDF) membrane using a transfer unit (Bio-Rad) at 80 V for 1 h. After transfer, the PVDF membranes were blocked with 5% bovine serum albumin (BSA) for 2 h and incubated overnight at 4 °C with primary antibodies. These include: anti-pSer209eIF4E (Cell Signaling Technology #9741), anti- eIF4E (Cell Signaling Technology, #9742, anti-4E-BP1(Cell Signaling Technology #9452) and anti-β-actin (Cell Signaling Technology #8H10D10). Detection of the immunoreaction was performed with a Clarity western enhanced chemiluminescence substrate kit (Bio-Rad) using ChemiDoc MP imaging system (Bio-Rad).
Software and statistical analysis
To retrieve predicted targets for miR-483-5p, we used TargetScan v7.263. To retrieve relative protein expression levels, we used protein atlas (proteinatlas.org). Immunoblot densitometry was performed using Fiji software64. All statistical analyses were performed with R software v3.6.2 (cran.r-project.org). For RT-qPCR experiments unpaired student t test was used. Raw p value was mentioned, and the data are shown as mean ± standard error.
Supplementary Information
Acknowledgements
This study was supported with funds from the University of Warsaw the “Excellence Initiative – Research University Program,” action No. II.2.1—Tandems for Excellence and the Polish National Science Centre Grants 2018/29/B/NZ7/02757 and UMO-2019/33/B/NZ1/01322.
Author contributions
S.N. and A.S.-D. performed the experiments. S.N., E.D., U.W. and R.G. designed the study, analyzed the data, and wrote the manuscript.
Data availability
All data generated or analyzed during this study are included in this published article and its Supplementary information files.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Urszula Wojda, Email: u.wojda@nencki.edu.pl.
Renata Grzela, Email: rgrzela@fuw.edu.pl.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-54154-1.
References
- 1.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell. 2009;136(4):731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang Y, Wang Z. IRES-mediated cap-independent translation, a path leading to hidden proteome. J. Mol. Cell Biol. 2019;11(10):911–919. doi: 10.1093/jmcb/mjz091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braunstein S, Karpisheva K, Pola C, Goldberg J, Hochman T, Yee H, Cangiarella J, Arju R, Formenti SC, Schneider RJ. A hypoxia-controlled cap-dependent to cap-independent translation switch in breast cancer. Mol. Cell. 2007;28(3):501–512. doi: 10.1016/j.molcel.2007.10.019. [DOI] [PubMed] [Google Scholar]
- 4.Niedzwiecka A, Darzynkiewicz E, Stolarski R. Deaggregation of eIF4E induced by mRNA 5' cap binding. Nucleosides Nucleotides Nucleic Acids. 2005;24(5–7):507–511. doi: 10.1081/ncn-200061784. [DOI] [PubMed] [Google Scholar]
- 5.Fischer PD, Papadopoulos E, Dempersmier JM, Wang ZF, Nowak RP, Donovan KA, Kalabathula J, Gorgulla C, Junghanns PPM, Kabha E, et al. A biphenyl inhibitor of eIF4E targeting an internal binding site enables the design of cell-permeable PROTAC-degraders. Eur. J. Med. Chem. 2021;219:113435. doi: 10.1016/j.ejmech.2021.113435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmidt EV. The role of c-myc in regulation of translation initiation. Oncogene. 2004;23(18):3217–3221. doi: 10.1038/sj.onc.1207548. [DOI] [PubMed] [Google Scholar]
- 7.Cowling VH, Cole MD. Mechanism of transcriptional activation by the Myc oncoproteins. Semin. Cancer Biol. 2006;16(4):242–252. doi: 10.1016/j.semcancer.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 8.Siddiqui N, Sonenberg N. Signalling to eIF4E in cancer. Biochem. Soc. Trans. 2015;43(5):763–772. doi: 10.1042/BST20150126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furic L, Rong L, Larsson O, Koumakpayi IH, Yoshida K, Brueschke A, Petroulakis E, Robichaud N, Pollak M, Gaboury LA, et al. eIF4E phosphorylation promotes tumorigenesis and is associated with prostate cancer progression. Proc. Natl. Acad. Sci. USA. 2010;107(32):14134–14139. doi: 10.1073/pnas.1005320107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liang S, Guo R, Zhang Z, Liu D, Xu H, Xu Z, Wang X, Yang L. Upregulation of the eIF4E signaling pathway contributes to the progression of gastric cancer, and targeting eIF4E by perifosine inhibits cell growth. Oncol. Rep. 2013;29(6):2422–2430. doi: 10.3892/or.2013.2397. [DOI] [PubMed] [Google Scholar]
- 11.Robichaud N, del Rincon SV, Huor B, Alain T, Petruccelli LA, Hearnden J, Goncalves C, Grotegut S, Spruck CH, Furic L, et al. Phosphorylation of eIF4E promotes EMT and metastasis via translational control of SNAIL and MMP-3. Oncogene. 2015;34(16):2032–2042. doi: 10.1038/onc.2014.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Truitt ML, Ruggero D. New frontiers in translational control of the cancer genome. Nat. Rev. Cancer. 2017;17(5):332. doi: 10.1038/nrc.2017.30. [DOI] [PubMed] [Google Scholar]
- 13.Carroll M, Borden KL. The oncogene eIF4E: Using biochemical insights to target cancer. J. Interferon Cytokine Res. 2013;33(5):227–238. doi: 10.1089/jir.2012.0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruan H, Li X, Xu X, Leibowitz BJ, Tong J, Chen L, Ao L, Xing W, Luo J, Yu Y, et al. eIF4E S209 phosphorylation licenses myc- and stress-driven oncogenesis. Elife. 2020 doi: 10.7554/eLife.60151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang X, Zhong W, Cao R. Phosphorylation of the mRNA cap-binding protein eIF4E and cancer. Cell Signal. 2020;73:109689. doi: 10.1016/j.cellsig.2020.109689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Proud CG. Mnks, eIF4E phosphorylation and cancer. Biochim. Biophys. Acta. 2015;1849(7):766–773. doi: 10.1016/j.bbagrm.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 17.Dreas A, Kucwaj-Brysz K, Pyziak K, Kulesza U, Wincza E, Fabritius CH, Michalik K, Gabor-Worwa E, Gołas A, Milik M, et al. Discovery of indazole-pyridinone derivatives as a novel class of potent and selective MNK1/2 kinase inhibitors that protecting against endotoxin-induced septic shock. Eur. J. Med. Chem. 2021;213:113057. doi: 10.1016/j.ejmech.2020.113057. [DOI] [PubMed] [Google Scholar]
- 18.Hay N. Mnk earmarks eIF4E for cancer therapy. Proc. Natl. Acad. Sci. USA. 2010;107(32):13975–13976. doi: 10.1073/pnas.1008908107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin X, Yu R, Wang X, Proud CG, Jiang T. Progress in developing MNK inhibitors. Eur. J. Med. Chem. 2021;219:113420. doi: 10.1016/j.ejmech.2021.113420. [DOI] [PubMed] [Google Scholar]
- 20.Bell JB, Eckerdt FD, Alley K, Magnusson LP, Hussain H, Bi Y, Arslan AD, Clymer J, Alvarez AA, Goldman S, et al. MNK inhibition disrupts mesenchymal glioma stem cells and prolongs survival in a mouse model of glioblastoma. Mol. Cancer Res. 2016;14(10):984–993. doi: 10.1158/1541-7786.MCR-16-0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sawicka K, Pyronneau A, Chao M, Bennett MV, Zukin RS. Elevated ERK/p90 ribosomal S6 kinase activity underlies audiogenic seizure susceptibility in fragile X mice. Proc. Natl. Acad. Sci. USA. 2016;113(41):E6290–E6297. doi: 10.1073/pnas.1610812113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu S, Zha J, Lei M. Inhibiting ERK/Mnk/eIF4E broadly sensitizes ovarian cancer response to chemotherapy. Clin. Transl. Oncol. 2018;20(3):374–381. doi: 10.1007/s12094-017-1724-0. [DOI] [PubMed] [Google Scholar]
- 23.Wang X, Flynn A, Waskiewicz AJ, Webb BL, Vries RG, Baines IA, Cooper JA, Proud CG. The phosphorylation of eukaryotic initiation factor eIF4E in response to phorbol esters, cell stresses, and cytokines is mediated by distinct MAP kinase pathways. J. Biol. Chem. 1998;273(16):9373–9377. doi: 10.1074/jbc.273.16.9373. [DOI] [PubMed] [Google Scholar]
- 24.Wang D, Ma J, Ji X, Xu F, Wei Y. miR-141 regulation of EIF4E expression affects docetaxel chemoresistance of non-small cell lung cancer. Oncol. Rep. 2017;37(1):608–616. doi: 10.3892/or.2016.5214. [DOI] [PubMed] [Google Scholar]
- 25.Mathonnet G, Fabian MR, Svitkin YV, Parsyan A, Huck L, Murata T, Biffo S, Merrick WC, Darzynkiewicz E, Pillai RS, et al. MicroRNA inhibition of translation initiation in vitro by targeting the cap-binding complex eIF4F. Science. 2007;317(5845):1764–1767. doi: 10.1126/science.1146067. [DOI] [PubMed] [Google Scholar]
- 26.Bartel DP. Metazoan microRNAs. Cell. 2018;173(1):20–51. doi: 10.1016/j.cell.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagaraj S, Zoltowska KM, Laskowska-Kaszub K, Wojda U. microRNA diagnostic panel for Alzheimer's disease and epigenetic trade-off between neurodegeneration and cancer. Ageing Res. Rev. 2019;49:125–143. doi: 10.1016/j.arr.2018.10.008. [DOI] [PubMed] [Google Scholar]
- 28.Nagaraj S, Want A, Laskowska-Kaszub K, Fesiuk A, Vaz S, Logarinho E, Wojda U. Candidate Alzheimer's disease biomarker miR-483-5p lowers TAU phosphorylation by direct ERK1/2 repression. Int. J. Mol. Sci. 2021 doi: 10.3390/ijms22073653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang L, Shi M, Hou S, Ding B, Liu L, Ji X, Zhang J, Deng Y. MiR-483-5p suppresses the proliferation of glioma cells via directly targeting ERK1. FEBS Lett. 2012;586(9):1312–1317. doi: 10.1016/j.febslet.2012.03.035. [DOI] [PubMed] [Google Scholar]
- 30.Liu K, He B, Xu J, Li Y, Guo C, Cai Q, Wang S. miR-483-5p targets MKNK1 to suppress Wilms' tumor cell proliferation and apoptosis in vitro and in vivo. Med. Sci. Monit. 2019;25:1459–1468. doi: 10.12659/MSM.913005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang CL, Zuo B, Li, Zhu JF, Xiao F, Zhang XL, Chen XD. The long noncoding RNA H19 attenuates force-driven cartilage degeneration via miR-483-5p/Dusp5. Biochem. Biophys. Res. Commun. 2020;529(2):210–217. doi: 10.1016/j.bbrc.2020.05.180. [DOI] [PubMed] [Google Scholar]
- 32.Kidger AM, Keyse SM. The regulation of oncogenic Ras/ERK signalling by dual-specificity mitogen activated protein kinase phosphatases (MKPs) Semin. Cell. Dev. Biol. 2016;50:125–132. doi: 10.1016/j.semcdb.2016.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sharrocks AD. Cell cycle: Sustained ERK signalling represses the inhibitors. Curr. Biol. 2006;16(14):R540–542. doi: 10.1016/j.cub.2006.06.038. [DOI] [PubMed] [Google Scholar]
- 34.Roovers K, Davey G, Zhu X, Bottazzi ME, Assoian RK. Alpha5beta1 integrin controls cyclin D1 expression by sustaining mitogen-activated protein kinase activity in growth factor-treated cells. Mol. Biol. Cell. 1999;10(10):3197–3204. doi: 10.1091/mbc.10.10.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ecsedi M, Rausch M, Großhans H. The let-7 microRNA directs vulval development through a single target. Dev. Cell. 2015;32(3):335–344. doi: 10.1016/j.devcel.2014.12.018. [DOI] [PubMed] [Google Scholar]
- 36.Pinzón N, Li B, Martinez L, Sergeeva A, Presumey J, Apparailly F, Seitz H. microRNA target prediction programs predict many false positives. Genome Res. 2017;27(2):234–245. doi: 10.1101/gr.205146.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lai X, Wolkenhauer O, Vera J. Understanding microRNA-mediated gene regulatory networks through mathematical modelling. Nucleic Acids Res. 2016;44(13):6019–6035. doi: 10.1093/nar/gkw550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shu J, Xia Z, Li L, Liang ET, Slipek N, Shen D, Foo J, Subramanian S, Steer CJ. Dose-dependent differential mRNA target selection and regulation by let-7a-7f and miR-17-92 cluster microRNAs. RNA Biol. 2012;9(10):1275–1287. doi: 10.4161/rna.21998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen K, He H, Xie Y, Zhao L, Zhao S, Wan X, Yang W, Mo Z. miR-125a-3p and miR-483-5p promote adipogenesis via suppressing the RhoA/ROCK1/ERK1/2 pathway in multiple symmetric lipomatosis. Sci. Rep. 2015;5:11909. doi: 10.1038/srep11909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang S, Liu G. Targeting the Ras/Raf/MEK/ERK pathway in hepatocellular carcinoma. Oncol. Lett. 2017;13(3):1041–1047. doi: 10.3892/ol.2017.5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martin-Vega A, Cobb MH. Navigating the ERK1/2 MAPK cascade. Biomolecules. 2023 doi: 10.3390/biom13101555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu W, Kannan S, Verma CS, Nacro K. Update on the development of MNK inhibitors as therapeutic agents. J. Med. Chem. 2022;65(2):983–1007. doi: 10.1021/acs.jmedchem.1c00368. [DOI] [PubMed] [Google Scholar]
- 43.Shi F, Len Y, Gong Y, Shi R, Yang X, Naren D, Yan T. Ribavirin inhibits the activity of mTOR/eIF4E, ERK/Mnk1/eIF4E signaling pathway and synergizes with tyrosine kinase inhibitor imatinib to impair Bcr-Abl mediated proliferation and apoptosis in Ph+ leukemia. PLoS ONE. 2015;10(8):e0136746. doi: 10.1371/journal.pone.0136746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pyronnet S. Phosphorylation of the cap-binding protein eIF4E by the MAPK-activated protein kinase Mnk1. Biochem. Pharmacol. 2000;60(8):1237–1243. doi: 10.1016/s0006-2952(00)00429-9. [DOI] [PubMed] [Google Scholar]
- 45.Matson K, Macleod A, Mehta N, Sempek E, Tang X. Impacts of microRNA-483 on human diseases. Noncoding RNA. 2023 doi: 10.3390/ncrna9040037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Song Q, Xu Y, Yang C, Chen Z, Jia C, Chen J, Zhang Y, Lai P, Fan X, Zhou X, et al. miR-483-5p promotes invasion and metastasis of lung adenocarcinoma by targeting RhoGDI1 and ALCAM. Cancer Res. 2014;74(11):3031–3042. doi: 10.1158/0008-5472.CAN-13-2193. [DOI] [PubMed] [Google Scholar]
- 47.Niture S, Gadi S, Qi Q, Gyamfi MA, Varghese RS, Rios-Colon L, Chimeh U, Vandana, Ressom HW, Kumar D. MicroRNA-483–5p inhibits hepatocellular carcinoma cell proliferation, cell steatosis, and fibrosis by targeting PPARα and TIMP2. Cancers. 2023 doi: 10.3390/cancers15061715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rutkovsky AC, Yeh ES, Guest ST, Findlay VJ, Muise-Helmericks RC, Armeson K, Ethier SP. Eukaryotic initiation factor 4E-binding protein as an oncogene in breast cancer. BMC Cancer. 2019;19(1):491. doi: 10.1186/s12885-019-5667-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Agosta C, Laugier J, Guyon L, Denis J, Bertherat J, Libé R, Boisson B, Sturm N, Feige JJ, Chabre O, et al. MiR-483-5p and miR-139-5p promote aggressiveness by targeting N-myc downstream-regulated gene family members in adrenocortical cancer. Int. J. Cancer. 2018;143(4):944–957. doi: 10.1002/ijc.31363. [DOI] [PubMed] [Google Scholar]
- 50.Niu ZY, Li WL, Jiang DL, Li YS, Xie XJ. Mir-483 inhibits colon cancer cell proliferation and migration by targeting TRAF1. Kaohsiung J. Med. Sci. 2018;34(9):479–486. doi: 10.1016/j.kjms.2018.04.005. [DOI] [PubMed] [Google Scholar]
- 51.Zhao Y, Yan M, Yun Y, Zhang J, Zhang R, Li Y, Wu X, Liu Q, Miao W, Jiang H. MicroRNA-455-3p functions as a tumor suppressor by targeting eIF4E in prostate cancer. Oncol. Rep. 2017;37(4):2449–2458. doi: 10.3892/or.2017.5502. [DOI] [PubMed] [Google Scholar]
- 52.Li G, Chong T, Xiang X, Yang J, Li H. Downregulation of microRNA-15a suppresses the proliferation and invasion of renal cell carcinoma via direct targeting of eIF4E. Oncol. Rep. 2017;38(4):1995–2002. doi: 10.3892/or.2017.5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Balakumaran BS, Porrello A, Hsu DS, Glover W, Foye A, Leung JY, Sullivan BA, Hahn WC, Loda M, Febbo PG. MYC activity mitigates response to rapamycin in prostate cancer through eukaryotic initiation factor 4E-binding protein 1-mediated inhibition of autophagy. Cancer Res. 2009;69(19):7803–7810. doi: 10.1158/0008-5472.CAN-09-0910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Averous J, Fonseca BD, Proud CG. Regulation of cyclin D1 expression by mTORC1 signaling requires eukaryotic initiation factor 4E-binding protein 1. Oncogene. 2008;27(8):1106–1113. doi: 10.1038/sj.onc.1210715. [DOI] [PubMed] [Google Scholar]
- 55.Ren J, Xu G, Sun H, Lin T, Xu S, Zhao Y. Inhibition of miR-483-5p improves the proliferation, invasion and inflammatory response of triple-negative breast cancer cells by targeting SOCS3. Exp. Ther. Med. 2021;22(4):1047. doi: 10.3892/etm.2021.10480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Soon PS, Tacon LJ, Gill AJ, Bambach CP, Sywak MS, Campbell PR, Yeh MW, Wong SG, Clifton-Bligh RJ, Robinson BG, et al. miR-195 and miR-483-5p identified as predictors of poor prognosis in adrenocortical cancer. Clin. Cancer Res. 2009;15(24):7684–7692. doi: 10.1158/1078-0432.CCR-09-1587. [DOI] [PubMed] [Google Scholar]
- 57.Xu H, Yang Y, Zhao H, Yang X, Luo Y, Ren Y, Liu W, Li N. Serum miR-483-5p: A novel diagnostic and prognostic biomarker for patients with oral squamous cell carcinoma. Tumour Biol. 2016;37(1):447–453. doi: 10.1007/s13277-015-3514-z. [DOI] [PubMed] [Google Scholar]
- 58.Xue L, Nan J, Dong L, Zhang C, Li H, Na R, He H, Wang Y. Upregulated miR-483-5p expression as a prognostic biomarker for esophageal squamous cell carcinoma. Cancer Biomark. 2017;19(2):193–197. doi: 10.3233/CBM-160506. [DOI] [PubMed] [Google Scholar]
- 59.Li XZ, Tu YJ, Zhou T, Zhang JB, Xiao RW, Yang DW, Zhang PF, You PT, Zheng XH. MicroRNA-483-5p predicts poor prognosis and promotes cancer metastasis by targeting. Front. Oncol. 2021;11:720835. doi: 10.3389/fonc.2021.720835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tang S, Chen Y, Feng S, Yi T, Liu X, Li Q, Liu Z, Zhu C, Hu J, Yu X, et al. MiR-483-5p promotes IGF-II transcription and is associated with poor prognosis of hepatocellular carcinoma. Oncotarget. 2017;8(59):99871–99888. doi: 10.18632/oncotarget.21737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oreglia M, Sbiera S, Fassnacht M, Guyon L, Denis J, Cristante J, Chabre O, Cherradi N. Early postoperative circulating miR-483–5p is a prognosis marker for adrenocortical cancer. Cancers. 2020 doi: 10.3390/cancers12030724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 63.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015 doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its Supplementary information files.