Abstract
Myocardial infarction (MI) leading to heart failure (HF) is a major cause of death worldwide. Previous studies revealed that the circadian system markedly impacts cardiac repair post-MI, and that light is an important environmental factor modulating the circadian influence over healing. Recent studies suggest that gut physiology also affects the circadian system, but how it contributes to cardiac repair post-MI and in HF is not well understood. To address this question, we first used a murine coronary artery ligation MI model to reveal that an intact gut microbiome is important for cardiac repair. Specifically, gut microbiome disruption impairs normal inflammatory responses in infarcted myocardium, elevates adverse cardiac gene biomarkers, and leads to worse HF outcomes. Conversely, reconstituting the microbiome post-MI in mice with prior gut microbiome disruption improves healing, consistent with the notion that normal gut physiology contributes to cardiac repair. To investigate a role for the circadian system, we initially utilized circadian mutant ClockΔ19/Δ19 mice, revealing that a functional circadian mechanism is necessary for gut microbiome benefits on post-MI cardiac repair and HF. Finally, we demonstrate that circadian-mediated gut responses that benefit cardiac repair can be conferred by time-restricted feeding, as wake time feeding of MI mice improves HF outcomes, but these benefits are not observed in MI mice fed during their sleep time. In summary, gut physiology is important for cardiac repair, and the circadian system influences the beneficial gut responses to improve post-MI and HF outcomes.
Keywords: Cardiovascular, Circadian, Myocardial Infarction, Inflammation, Microbiome, Remodeling
1. Introduction
Cardiovascular disease (CVD) is a leading cause of death worldwide [1–4]. Coronary artery disease frequently leads to myocardial infarction (MI; heart attack), and subsequently the development of heart failure (HF), for which there is no cure. Previously, clinical studies revealed that the circadian system underlies healthy cardiovascular physiology, for example, driving 24-hour day and night rhythms in heart rate [5], blood pressure [6], and timing of onset of adverse cardiovascular events [7–11]. Most recently, experimental studies by our group and others reveal that the circadian system also plays a critical role in remodeling in cardiovascular disease. For example, the circadian mechanism drives rhythms in gene expression and protein production important for heart and blood vessels [12–23], diurnal efficacy of angiotensin converting enzyme inhibitor chronotherapy [24], oxidative stress pathways [25, 26], cell death pathways [27], cardiac metabolism [28, 29], cardiac electrophysiology [30–33], heart-brain pathophysiology [34, 35], sex and gender differences in the healing heart [36–38], and cardiac inflammation involved in repair post-MI and in HF [39–41], and has been extensively reviewed [11, 42–46]. Collectively, these studies reveal that the circadian mechanism is fundamentally important not only for healthy cardiovascular physiology, but also for how the heart heals during disease.
Light is a primary zeitgeber, or daily time giving cue, that entrains / resets the circadian system. Briefly, light is detected by dedicated melanopsin containing retinal ganglionic cells, which transmit signals to a small region of the hypothalamus termed the suprachiasmatic nucleus (SCN). The SCN is considered the master pacemaker in mammals, which is critical for orchestrating temporal control over physiologic processes, in part through synchronizing local circadian clocks throughout the organism (via specific neurohormonal outputs). The cellular circadian clock mechanism consists of 24-hour transcription-translation feedback loops. The positive limb includes two factors termed CLOCK and BMAL1, which heterodimerize and bind to E-box enhancer elements to promote the transcription of proteins PER, CRY, and other core circadian elements [47–49]. PER and CRY represent the negative feedback loop, and inhibit CLOCK:BMAL1 activity and therefore, their own transcription [50–52]. This circadian mechanism that is present in virtually all cells in the body, including in the cardiomyocytes [12, 17, 53]. In addition to light synchronization via the SCN, peripheral tissue circadian clocks can be entrained by common behaviors, such as physical activity and food intake [54, 55]. The importance of enforcing a normal circadian environment / lifestyle is exemplified in shiftwork; inappropriate timing of light exposure, physical activity, and food intake common in night shift workers increases risk of heart disease, and exacerbates underlying cardiovascular conditions (e.g. reviewed in [11, 45, 46]).
Intriguingly, gut physiology has recently also been shown to be circadian regulated, both at the level of the host, as well as the microbiome. Interestingly, disruption of normal day and night rhythms in healthy gut physiology [56–62] alters the gut microbial composition with metabolic consequences [58, 60, 63], and differential injury response outcomes (e.g. spinal cord injury [64, 65]). However, the importance of normal circadian influences on gut physiology for cardiac repair post-MI and for HF outcomes are not known. Addressing such a question may ultimately improve patient outcomes, especially given the association between circadian and/or microbiome disruption on cardiovascular disease outcomes.
In this study we investigate the relationship between gut physiology, circadian biology, and cardiac repair post-MI and leading to HF. We used a well-documented murine myocardial infarction (MI) model [34, 39, 41], and established protocols for gut microbial disruption [64, 66, 67], to simulate the clinical scenarios in humans. Here we report that disruption of the gut microbiome impairs the coordinated responses involved in cardiac repair, thereby exacerbating HF progression. Conversely, microbiome reconstitution rescues beneficial healing responses, leading to better outcomes. Moreover, we found that a functional circadian clock mechanism in the host is critical for the beneficial effects of the microbiome on cardiac repair. Finally, restricting mice to consume food only during the light (sleep) period (which disrupts peripheral circadian clocks) worsened HF outcomes. Moreover, the circadian influence on gut responses benefiting cardiac repair can be conferred by time-restricted feeding, as wake time - but not sleep time - feeding improves HF outcomes. In summary, we demonstrate that normal gut physiology in the first few days after MI is important for cardiac repair, and that the circadian system influences these beneficial healing responses in cardiovascular disease.
2. Materials and Methods
2.1. Experimental animals and protocol
Male C57Bl/6 mice were obtained from Charles River (Quebec, Canada). Male ClockΔ19/Δ19 mice on a C57Bl/6 background and wild type (WT) control littermates were bred at the University of Guelph Central Animal Facility. ClockΔ19/Δ19 mice are homozygous for an A-T transversion mutation which translates to a 51 amino acid deletion in exon 19 [47, 68]. ClockΔ19/Δ19 mice were genotyped by allele-specific PCR and characterized by actigraphy, as described previously [34, 36]. All animals were housed under a normal 12-hour light (L):12-hour dark (D) cycle, with standard chow and water provided ad libitum, unless otherwise specified. For determination of the circadian period by wheel-running actigraphy, mice were individually housed and entrained to a diurnal 12:12 LD cycle for at least 2 weeks, followed by transfer to constant darkness (DD) for 2 weeks, and data analyzed by ClockLab (ActiMetrics) as described [13, 14, 37]. All experiments were performed in accordance with the Canadian Council on Animal Care, and the Guide for the Care and Use of Laboratory Animals, National Institutes of Health. The Animal Care and Use Committee at the University of Guelph approved all studies.
2.2. DNA extraction and 16S rRNA gene sequencing
Mice were euthanized with CO2 and cervical dislocation, and then cecal contents were collected at specific zeitgeber (ZT) times across the diurnal cycle (ZT 03, 07, 11, 15, 19, 23, or as described in the applicable figures), snap frozen in liquid nitrogen, and stored at −80˚C until processing. DNA was extracted using the FastDNA Spin Kit for Soil (MP Biomedicals) in accordance with the manufacturer’s specifications, then quantified using Qubit (Thermo Scientific), and stored in 10 mM Tris buffer (pH 8.5) at −20˚C. For 16S rRNA gene sequencing, DNA libraries were prepared using the Nextera XT Index Kit (Illumina), according to the manufacturer’s instructions. Briefly, DNA (5ng/μL) was amplified with barcoded primers annealing to the V4 region of the 16S rRNA gene with overhang adapter sequences, Illumina indices and sequencing adaptors were added by PCR, then samples were sequenced by MiSeq. Data were analyzed using Mothur (v.1.39.3) [69], in accordance with the standard operating procedure for MiSeq-derived sequence data [70]. Sequences were classified using the Ribosomal Database Project Classifier implemented in Mothur and the Ribosomal Database Project taxonomy training set [71]. Relative abundance data were imported to Microsoft Excel for figure plots.
2.3. Antibiotic induced gut microbiome disruption
An antibiotic cocktail prepared with streptomycin (2 g/L; Sigma), gentamicin (0.17 g/L), ciprofloxacin (0.125 mg/L), and bacitracin (1g/L) was given via drinking water as described, with maple syrup added to make the mixture more palatable [64]. Mice received antibiotics starting 7 days before experimentation, or untreated controls, and were randomized to protocols as needed. Effects of antibiotic treatment on gut bacteria were evaluated using two complementary methods. First, feces were collected at ZT03, in 1.5 mL of phosphate-buffered saline (PBS; 8 g NaCl, 0.2 g KCl, 1.44 g Na2HPO4, 0.24 g KH2PO4, pH7.4), and gut microbial ecosystem derangement was demonstrated by decreased colony counts on LB agar plates that had been incubated at 37˚C for 36 hours. To evaluate broader effects on the microbiome, including anaerobic bacteria, we also collected cecal samples at ZT03, on different days of antibiotics (AN), and quantified bacteria using a Bacteria Counting Kit (Molecular Probes) on an Accuri C6 flow cytometer (BD Biosciences).
2.4. Left anterior descending (LAD) coronary artery ligation
Myocardial infarction (MI) leading to heart failure (HF) was induced in 8-week-old mice by LAD ligation, as described [39, 41]. All surgeries were performed between ZT01 - ZT04, as this is consistent with prior studies and findings that MI tolerance is time-of-day dependent [18, 39, 41, 72]. Briefly, mice were anesthetized with isoflurane, intubated, and ventilated (Harvard Apparatus model 687) throughout the surgery. Prior to the incision, a 50:50 bupivacaine/lidocaine mix was administered to the site. To expose the heart and LAD, an incision was made in the left 3rd intercostal space and a prolene 7–0 suture (Ethicon) was passed at 1 mm below the left auricle under the LAD and ligated. Sham animals underwent the same procedures, but the LAD was not ligated. Buprenorphine (0.1 mg/kg) was administered upon recovery, and at 8- and 24-hours post-surgery.
2.5. Cardiac gene expression
For RNA isolation and Real Time Polymerase Chain Reaction (RTPCR) analyses of cardiac remodeling genes, mice were euthanized with CO2 and cervical dislocation. Hearts were collected at ZT03, and total RNA was purified by the TRIZOL method as described [15, 21, 73]. RNA quality was assessed by Nanodrop (Thermo Scientific). Amplification using an RNA-to-Ct one step PCR kit (Applied Biosystems) was performed on a VIIA7 system (Life Technologies) as follows: reverse transcription at 48˚C for 30 min then 95˚C for 10 min (1 cycle), amplification at 95˚C for 15 sec then 60˚C for 1 min (40 cycles), then room temperature. Primers used: atrial natriuretic factor (Anf) 5F’-GGGTAGGATTGACAGGATTGG-3’, 5R’-AGAATCGACTGCCTTTTCCTC-3’; brain derived natriuretic factor (Bnp) 5F’-GCGAGACAAGGGAGAACAC-3’, 5R’-GCGGTGACAGATAAAGGAAAAG-3’; regulator of calcineurin1 (Rcan1) 5F’-CCCGTGAAAAAGCAGAATGC-3’, 5R’-TCCTTGTCATATGTTCTGAAGAGGG-3’; NACHT, LRR, PYD domain-containing protein3 (Nlrp3) 5F’-CATGTTGCCTGTTCTTCCAGAC-3’, 5R’-CGGTTGGTGCTTAGACTTGAGA-3’; interleukin-1 beta (Il-1β) 5F’-GCCACCTTTTGACAGTGATGAG-3’, 5R’-GTTTGGAAGCAGCCCTTCATC-3’; Il-18 5F’-TCCAGCATCAGGACAAAG-3’, 5R’-ACGCAAGAGTCTTCTGAC-3’; chemokine C-C motif ligand 2 (Ccl2) 5’F-GTCCCTGTCATGCTTCTGG-3’, 5R’-TCTTGCTGGTGAATGAGTAGC-3’; chemokine C-C motif ligand 7 (Ccl7) 5F’-TCTCTCACTCTCTTTCTCCAC-3’, 5R’-GGATCTTTTGTTTCTTGACATAGC-3’; histone 5F’-GCAAGAGTGCGCCCTCTACTG-3’, 5’R-GGCCTCACTTGCCTCCTGCAA-3’. Data were normalized to histone using the delta delta CT method, as described previously [12, 21, 39, 41].
2.6. Echocardiography and in vivo hemodynamics
To assess cardiac function, mice were followed by echocardiography for up to 10 weeks post-infarction as they progressed to HF. All echocardiography was done at ZT07 – ZT09. Briefly, mice were placed under light anesthesia (1.5% isoflurane), and cardiac imaging was performed using a GE Vivid 7 Dimension ultrasound machine (GE Medical Systems) with the i13L 14 MHz linear-array transducer. Images (at least 5 per animal) were acquired at mid-papillary level to determine left ventricular (LV) internal dimensions at diastole (LVIDd) and at systole (LVIDs), % ejection fraction (%EF), % fractional shortening (%FS), and heart rate (HR). For pressure-volume in-vivo hemodynamics, mice were deeply anesthetized (4% isoflurane) and intubated. A 1.2F pressure-volume catheter (Transonic Scisense) was inserted into the right carotid artery and advanced to the LV for measurements (PowerLab, ADInstruments). All in-vivo hemodynamics were done at ZT07 – ZT09. Following hemodynamics measurements, the mice were sacrificed under 5% isoflurane by cervical dislocation. Measurements taken include end systolic and diastolic pressure (LVESP, LVEDP), end systolic and diastolic volume (LVESV, LVEDV), and maximum and minimum first derivatives of LV pressure (dP/dtmax; dP/dtmin). Systolic and diastolic blood pressures (SBP, DBP) were also recorded, and mean arterial pressure (MAP) was calculated as (1/3 x SBP) + (2/3 x DBP). Data were analyzed using Lab Chart 7 (ADInstruments).
2.7. Evans Blue and 2,3,5-Triphenyltetrazolium chloride (TTC) Staining
At 6-hours post-infarction, mice were anesthetized, intubated and ventilated (Harvard Apparatus model 687). To visualize ischemic tissue, a 1% Evans Blue solution was infused via the inferior vena cava. The heart was excised and rinsed in 0.9% saline before being sectioned into 1 mm sections using a heart matrix (Zivic Instruments). Sections were incubated in 1% TTC solution for 10 minutes, transferred to 10% neutral buffered formalin for 90 minutes and imaged. Infarct area and area at risk (AAR) as a percentage of LV were determined using Adobe Photoshop CS4 as previously described [39, 40]. Briefly, samples were normalized to each slice weight using the following: weight (total AAR) = [weight (slice 1) x % AAR (slice 1)] + [weight (slice 2) x % AAR (slice 2)] + [weight (slice n) x % AAR (slice n)]. Infarct area was calculated in a similar manner. Absolute infarct size was calculated as a ratio of weight (total infarct area)/weight (total AAR).
2.8. Cardiac histopathology
Mice were euthanized with isoflurane and cervical dislocation at ZT07 – ZT09. Body weight (BW), heart weight (HW) and tibia length (TL) were collected for each animal. Hearts were perfused with 1M KCl, fixed in 10% neutral buffered formalin (Sigma), and stored in 70% ethanol. For histology, hearts were paraffin embedded and 5 μm sections were taken every 600 μm starting from the apex. Sections were stained with Masson’s Trichrome. Absolute scar size was determined using Image J 1.51 (NIH), from the sum of the epicardial and endocardial circumferences of the infarct, divided by the sum of the total LV epicardial and endocardial circumferences. Infarct thickness was determined from 5 measurements over equidistant points.
2.9. Enzyme-linked immunosorbent assays (ELISA)
Mice were euthanized with isoflurane and cervical dislocation at ZT03, and hearts collected. To prepare heart protein lysates, CelLytic MT Cell Lysis Reagent (Sigma) was used, and heart protein lysates were assayed according to the manufacturer’s instructions for IL-1β (BMS6002, Invitrogen), or IL-18 (BMS618/3, Invitrogen).
2.10. Flow cytometry analyses
A separate set of hearts was used to quantify inflammatory cell responses in infarcted myocardium of sham, normal-HF, and mice with antibiotic induced gut microbiome disruption (disrupted-HF). Briefly, mice were sacrificed by isoflurane and cervical dislocation on day 0, 1, 2, 3, 5, or 7 post-MI (at ZT03), timepoints were selected consistent with other studies and findings demonstrating circadian rhythms in the immune system [74]. Hearts were perfused with ice-cold heparin (Fisher BioReagents) saline, the LV was collected and finely minced, then digested in 10 mL of RPMI-1640 media (Gibco) containing collagenase type II (1 mg/mL, Worthington Biochemical Corporation) and DNase (60 U/mL, Roche) for 1 hour at 37˚C with gentle agitation. Samples were triturated, filtered through a 70 μm cell strainer, and pelleted by centrifugation (400 x g, 4˚C, 5 min). Pellets were washed with RPMI-1640 and cells were resuspended in PBS for further analysis. Cell count and viability (>90%) were determined using the trypan blue exclusion method in a separate aliquot. Aliquots (1×106 cells) were incubated with anti-CD16/32 antibody (BioLegend) at 4˚C for 5 minutes to block Fcγ receptors. Cells were then incubated with fluorochrome-conjugated antibodies to specific leukocyte markers. Total leukocytes were characterized using APC anti-mouse CD45 (BioLegend). Macrophages were characterized using PE anti-mouse CD11b (BioLegend) and FITC anti-mouse F4/80 (BioLegend). T lymphocytes were characterized using APC anti-mouse CD45 (BioLegend), FITC anti-mouse CD3 (BioLegend), and PE anti-mouse CD4 (BioLegend). Samples were analyzed using the Accuri C6 (BD Biosciences) and FlowJo software (Tree Star).
2.11. Myeloperoxidase (MPO) assay
Neutrophil infiltration to infarcted hearts was assessed using a separate set of hearts and the MPO assay, as previously described [39]. Briefly, following sacrifice at ZT06, ventricular tissue was isolated and homogenized in 1 mL of 50 mM potassium phosphate buffer (pH 6.0) with 0.5% hexadecyltrimethylammonium bromide (Sigma), sonicated, freeze thawed, then centrifuged at 40,000 x g for 15 min (Beckman Ultracentrifuge LE-80K). Supernatant containing MPO was quantified by EnzChek Myeloperoxidase Activity Assay Kit (Life Technologies, Molecular Probes) as per manufacturer’s directions. MPO concentration were normalized to heart tissue weight.
2.12. 1H nuclear magnetic resonance spectroscopy
Mice were sacrificed as described above (at ZT03), and cecal contents were prepared for NMR analysis for targeted metabolomics [75, 76]. Briefly, 50 mg cecal samples were suspended in 500 μL of phosphate buffer (0.1 M K2HPO4/NaH2PO4, pH 7.4) and centrifuged (16,000 x g, 4˚C, 10 min). Supernatant containing the metabolites was diluted by 10% with an internal Chenomx standard (99.9% deuterium oxide (D2O) with 5 mM 4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS) and 0.2% (w/v) sodium azide), and then transferred into 3 mm Wilmad Lab Glass tubes for NMR analyses. 1H NMR spectra were acquired at 298 K on a Bruker Avance III 600 MHz spectrometer with a 5 mm triple resonance cryoprobe at the NMR Centre (University of Guelph). NMR spectra were acquired using the first increment of the NOESY pulse sequence with mixing time of 100 millisecs, 3 secs acquisition time, 2 secs relaxation delay, and a spectral width of 14 ppm. A pre-saturation pulse (80 Hz field strength) was applied during the relaxation delay to suppress water resonance. Following acquisition, spectra were imported into Chenomx NMR Suite 8.3 for processing, targeted metabolite profiling, and quantification using the known concentration of DSS [77].
2.13. Treatment to reconstitute gut microbiota
Mice that had been pretreated with antibiotics were immediately rehoused for 1 week in cages containing feces from healthy mice, starting on the day of MI surgery. Coprophagic behavior of mice provides a relatively simple experimental approach to fecal transplantation. The benefits of a reconstituted microbiome that accrue to cardiac remodeling were demonstrated by echocardiography, in vivo hemodynamics, and histology. In addition, we used another set of mice to evaluate the reconstituted gut microbiota profile in reconstituted-HF mice and compared to that of normal-HF and disrupted-HF. Cecal samples were collected on day 3 post-infarction from all groups, sequenced using 16S rRNA gene sequencing and analyzed using Mothur. Graph plots were generated using Microsoft Excel.
2.14. Circadian System Influence on Gut Physiology and Benefits for Cardiac Repair
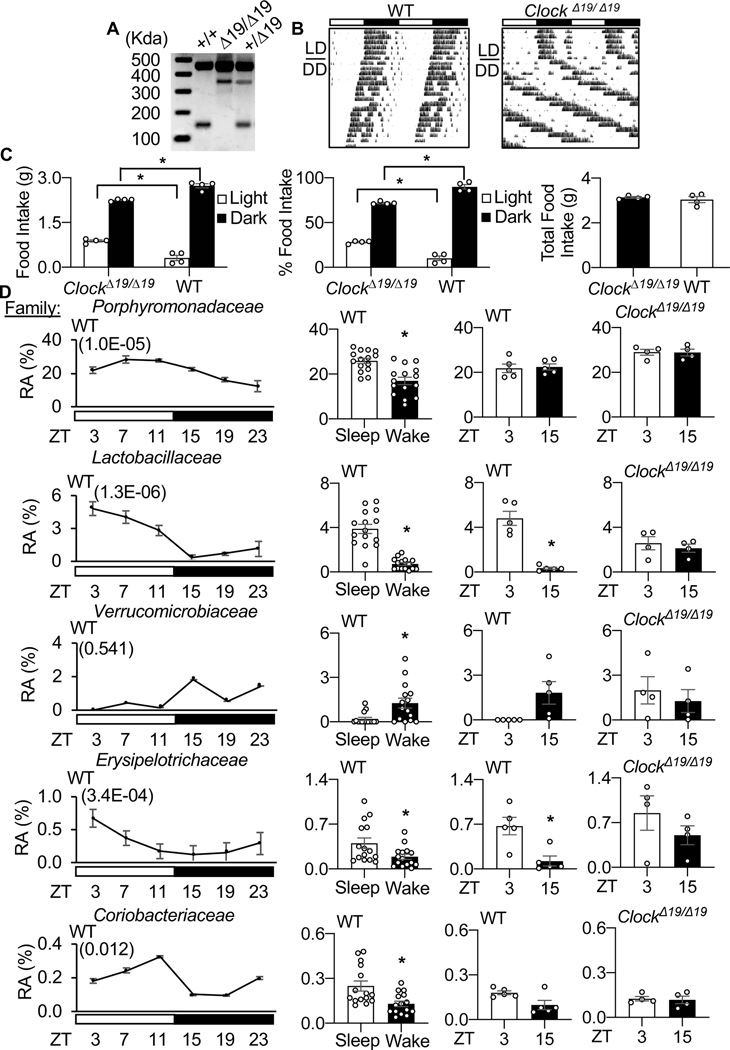
To investigate a role for the circadian mechanism in post-MI cardiac repair, we first used ClockΔ19/Δ19 mice. Diurnal food intake in ClockΔ19/Δ19 and WT mice was measured immediately before the onset of the dark period (ZT12) and immediately after the onset of the light period (ZT0), over 3 consecutive days. ClockΔ19/Δ19 mice were randomized to receive either 1) antibiotics starting 7 days before MI surgery and continuing for 14 days after (ClockΔ19/Δ19+Disrupted-HF), or 2) controls given no antibiotics (ClockΔ19/Δ19+Normal-HF). Pathophysiology was determined by echocardiography and histology; methods are listed above. For immune cell infiltration to infarcted myocardium, mice were sacrificed by isoflurane and cervical dislocation at ZT03, and hearts collected for flow cytometry, at day 3 post-infarction. Quantification of cecal metabolites was determined by 1H NMR spectroscopy (using samples collected at ZT03). To further investigate a role for an intact host circadian system in cardiac repair, WT mice were given antibiotics to disrupt the gut microbiota and were re-housed for 1 week in cages containing feces from either healthy WT mice (WT to WT), or feces from ClockΔ19/Δ19 mice (ClockΔ19/Δ19 to WT) to promote the re-establishment of the gut microbiota immediately after MI surgery. Pathophysiology was determined by echocardiography, using the approaches described above. Next, a behavior-induced model of peripheral tissue circadian clock disruption was employed, to determine whether a non-genetic model of circadian disruption affected MI outcomes. To do this, we used time restricted feeding protocols, which are known to uncouple the central pacemaker from circadian rhythms in the gut [54, 58, 78]. Mice were maintained on a normal 24-hour LD cycle, and randomized to either light-phase restricted feeding (9AM to 9PM), or dark-phase restricted feeding (9PM to 9AM) for 9 days prior to MI surgery, then returned to ad libitum feeding afterwards; MI outcomes were followed using the same pathophysiological assessments described above.
2.15. Statistics
All values are mean ± SEM. Statistical comparisons were done using an unpaired Student’s t-test, or two-way ANOVA with Tukey post hoc for multiple comparisons, as applicable. Analyses were done using SigmaPlot 12 (Systat Software Inc.), or Excel (Microsoft). Unless otherwise noted, throughout all figures, data are presented as mean ± SEM with values of *p<0.05 considered statistically significant. Statistical parameters, including n-values, are noted in figure legends. Statistical analysis of 24-hour rhythmic profiles in 16S rRNA gene sequencing data was performed using JTK_CYCLE version-3 as described [36, 41, 79]. Briefly, the JTK_CYCLE algorithm is used to estimate the period, phase, amplitude, JTK p-value (estimated probability of rejecting the null hypothesis that the dataset is not rhythmic), and JTK q-value (estimates the false discovery rate for considering the profile to be cyclic).
3. Results
3.1. Circadian Influence on the Gut Microbiome
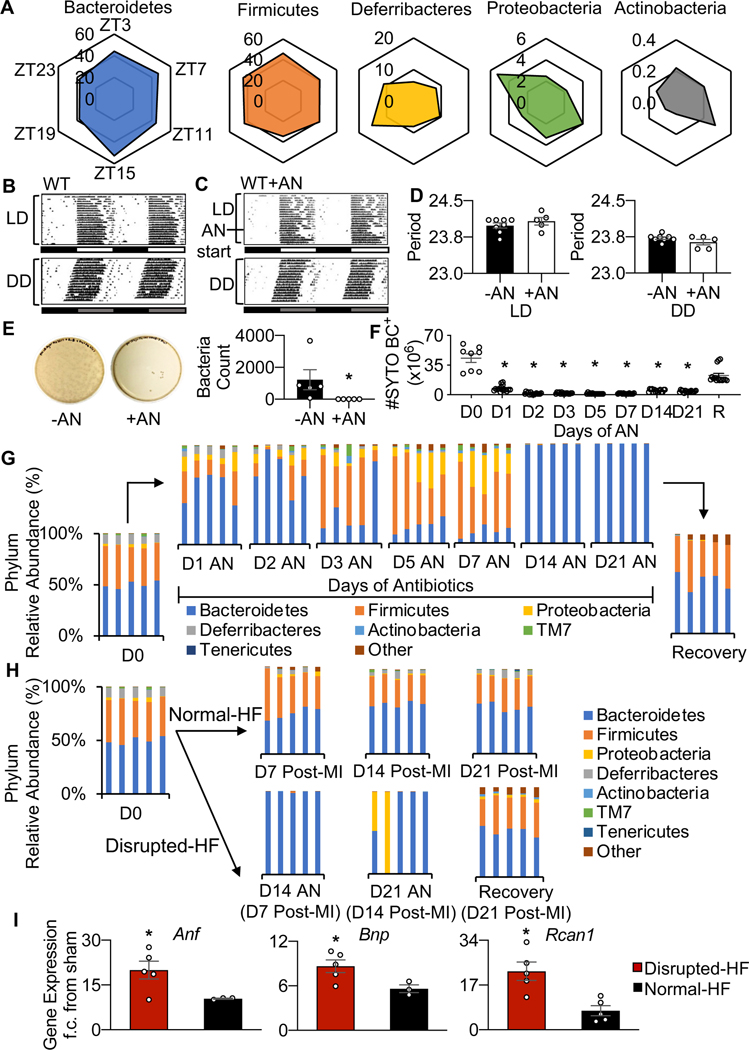
We first investigated a circadian influence on gut physiology by determining the time-of-day rhythmicity of the gut microbiome composition. To do this, we profiled the murine cecal microbiome over a 24-hour cycle, using 16S rRNA gene sequencing. We found time dependent profiles in the relative abundance of all the major phyla (Fig. 1A; Supplementary Table 1). Next, we investigated the effects of disrupting the gut microbiome. We administered oral antibiotics, as a known model for disrupting the composition and quantity of bacteria, and found that antibiotic treatment itself did not obviously affect daily behavior under normal light-dark (LD) or in constant darkness (DD) circadian conditions, as shown by running wheel actigraphy (Fig. 1B, 1C, 1D). However, the antibiotics did lead to gut microbial derangements, as fecal contents cultured on LB agar showed a significant decrease (p<0.0001) in colony forming units by the first day of treatment (Fig. 1E). Moreover, microbiome quantity remained very low throughout all 21 days of antibiotic treatment, as measured by flow cytometry of SYTO BC+ cells (Fig. 1F). Furthermore, changes in the relative abundance of gut microbial taxa were evident following antibiotic treatment, including a loss of diversity and almost complete dominance by Bacteroidetes, as shown by 16S rRNA gene sequencing data. The loss of gut microbiota abundance and diversity rapidly resolved after the antibiotic treatment period ended (recovery) (Fig. 1G, Supplementary Table 2). Thus, these findings reveal significant daily rhythms in gut microbial physiology, and that antibiotic-induced disruption of the gut microbiome does not influence circadian behavior.
Fig. 1.
The gut microbiome displays time-of-day rhythmicity. (A) Diurnal rhythms in microbiome abundance as determined by 16S rRNA sequencing, from cecal samples taken at 6 serial time points (ZT03, 07, 11, 15, 19, 23) over 24 hours. The top 5 murine phyla are plotted to show relative differences in abundance across the diurnal cycle. Each corner of the plot represents one time point, and each plot represents 24 hours (n=5 mice per time point). See data values in Supplementary Table 1. (B) Antibiotics do not appear to affect circadian activity as running wheel actigraphy under a normal light dark cycle (LD, top) and in constant darkness (DD, bottom) is comparable for wildtype (WT) mice, and (C) mice given antibiotics (AN) for 3 weeks (WT+AN), and D) they show similar period profiles (n≥5 mice per group, mean ± SEM). (E) Gut microbial disruption due to antibiotics was validated by culture on LB agar plates (fecal samples from healthy WT mice (AN-, left), 1 day of antibiotics (AN+, middle), and quantified (right), and (F) long term quantified by flow cytometry (antibiotics administered for up to 21 days, followed by recovery (R), all samples were collected at ZT03, n=5 animals per group, *p<0.0001 by Student’s t-test, mean ± SEM. (G) Relative abundance of cecal microbial phyla at day 0, 1, 2, 3, 5, 7, 14, 21 of antibiotics, and at recovery (day 7 of no antibiotics), and (H) and at day 7, 14, 21 post-MI (+/−AN), all samples were collected at ZT03, n=5 animals per time point, see data values in Supplementary Table 2. (I) Increased gene expression of biomarkers of heart failure Anf, Bnp, and Rcan1 in disrupted-HF versus normal-HF hearts (n≥3 hearts per group, *p<0.05 by Student’s t-test, mean ± SEM).
3.2. Gut Microbiome and Heart Failure (HF) Outcomes
Next, we investigated a role for gut microbial physiology in cardiovascular disease, and specifically heart failure outcomes. To do this, mice were subjected to left anterior descending (LAD) coronary artery ligation (HF model). Notably, the disease condition (normal-HF mice) did not significantly alter the gut microbiome post-MI, in the manner noted in Figure 1H, and compared to non-infarcted controls. However, antibiotics before MI (disrupted-HF mice) induced gut microbial derangements until antibiotic treatment ceased (Fig. 1H). Antibiotic treatment in MI mice also led to significant changes in the expression of biomarker genes involved in cardiac remodeling. Cardiac mRNA levels were increased for these pathophysiological biomarkers in the disrupted-HF mice, including atrial natriuretic factor (Anf), brain natriuretic factor (Bnp), and regulator of calcineurin 1 (Rcan1) (Fig. 1I), suggesting that gut microbial derangements may alter healing. Collectively these findings are consistent with the notion that the intact gut microbiome composition contributes to normal healthy cardiac repair, and that altering the gut microbiome adversely affects the remodeling heart.
3.3. Intact Gut Microbiome Reduces (Benefits) Cardiac Remodeling
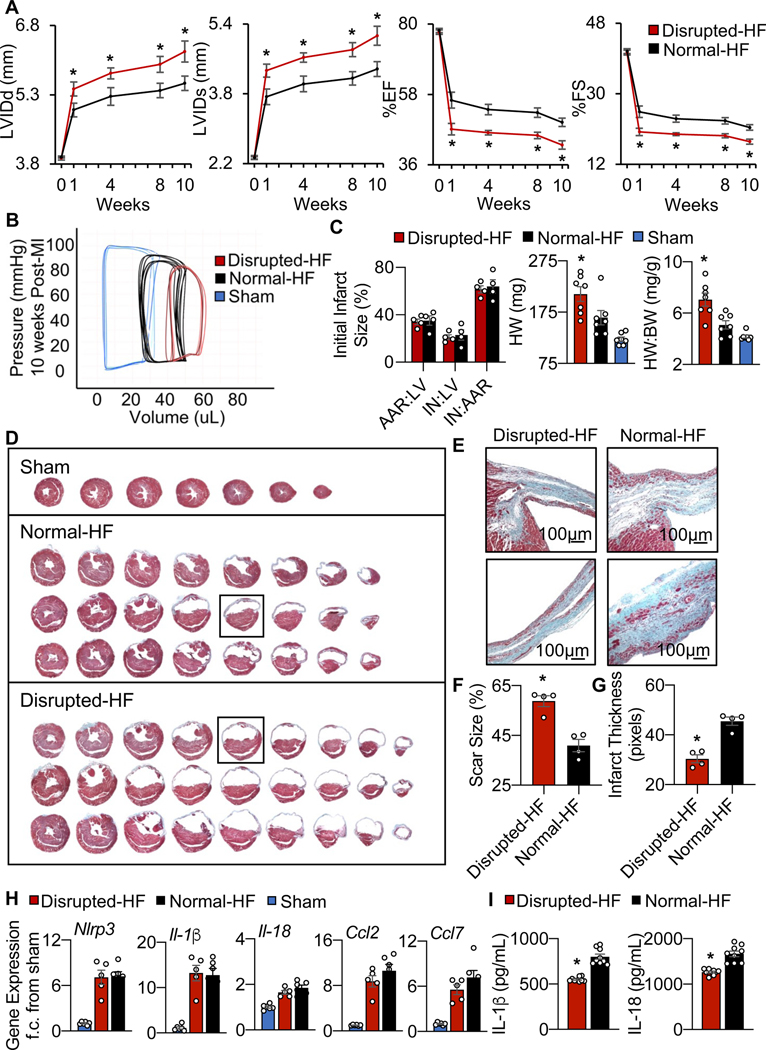
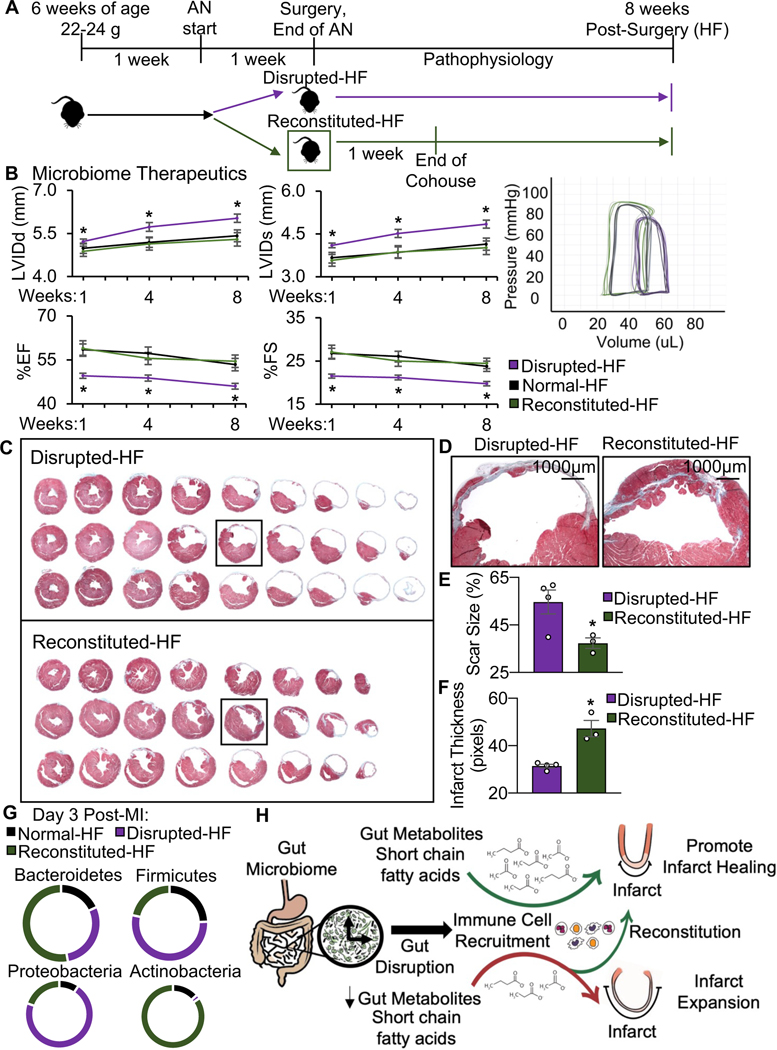
To further investigate whether the elevated HF biomarkers in the mice with gut disruption corresponded with adverse cardiac remodeling outcomes in vivo, we next performed echocardiography. Fig. 2A and Table 1 show that normal-HF mice (with an intact microbiome) have significantly (p<0.001) less adverse remodeling as compared to the disrupted-HF mice, with smaller left ventricular (LV) internal diastolic dimensions (LVIDd, 5.55±0.18 vs. 6.24±0.23 mm) and systolic dimensions (LVIDs, 4.37±0.17 vs. 5.13±0.22 mm), along with better % ejection fraction (%EF) and % fractional shortening (%FS) to 10 weeks HF. Importantly, there were no significant differences between our disrupted-sham and sham groups, implying that there were no adverse effects of this antibiotic regimen on cardiac structure or function independent of the effects on the gut (Table 1). Thus, mice with a healthy gut microbiome appear to be better protected against adverse cardiac remodeling and exhibit reduced progression to HF, as compared to mice with a depleted gut microbiome at the time of infarction.
Fig. 2.
Gut microbial ecosystem derangement affects HF outcomes. (A) Disrupted-HF mice had worse outcomes including increased left ventricular internal dimensions at diastole (LVIDd) and systole (LVIDs), and decreased % ejection fraction (%EF) and % fractional shortening (%FS), as measured by echocardiography for up to 10 weeks HF (*p<0.05 Disrupted-HF (3 weeks) vs. all other groups by two-way ANOVA, followed by a Tukey post-hoc test), and (B) shifted in vivo hemodynamic pressure-volume loops (representative image), and (C) although initial infarct size was the same at 6 hours post-infarction (left, initial infarct size (%) as quantified by area at risk:left ventricle (AAR:LV), infarct area:LV (IN:LV), and IN:AAR), the disrupted-HF mice developed bigger hearts (greater heart weight, HW) and increased HW: to body weight (BW) ratio by 10 weeks HF, (n=7 mice per group in each study, except 4 mice per group in the infarct size study, *p<0.05 Disrupted-HF (3 weeks) vs. all other groups by two-way ANOVA, followed by a Tukey post-hoc test, mean ± SEM, see data values in Table 1). (D) The disrupted-HF hearts also showed greater LV dilation by histology (more equidistant sections) and (E) close-up of infarct regions by Masson’s trichrome, and (F) quantification revealing greater absolute scar size and (G) thinner infarcts, (n=4 hearts per group, *p<0.002 by Student’s t-test, mean ± SEM). (H) Inflammasome mRNA components, and (I) decreased levels of IL-1b and IL-18 inflammasome related cytokines in the heart at 24 hours post-infarction, (n>3 hearts per group, *p<0.05 by Student’s t-test, mean ± SEM).
Table 1.
Microbiome benefits long-term cardiac structure and function post-MI.
| Disrupted-HF | Normal-HF | Disrupted-Sham | Sham | |
|---|---|---|---|---|
| Echocardiography (n) | 8 | 8 | 8 | 8 |
| Baseline | ||||
| LVIDd (mm) | 3.93±0.04 | 3.95±0.02 | 3.92±0.05 | 3.95±0.02 |
| LVIDs (mm) | 2.34±0.04 | 2.33±0.03 | 2.34±0.04 | 2.33±0.03 |
| EF (%) | 77.48±0.70 | 77.99±0.66 | 77.10±0.77 | 77.94±0.70 |
| FS (%) | 40.51±0.62 | 40.88±0.61 | 40.24±0.72 | 40.86±0.64 |
| HR (bpm) | 470±5 | 463±5 | 468±5 | 475±7 |
| 1-week post-infarction | ||||
| LVIDd (mm) | 5.42±0.15* | 4.97±0.15 | 4.03±0.03 | 4.09±0.03 |
| LVIDs (mm) | 4.33±0.15* | 3.73±0.18 | 2.40±0.04 | 2.43±0.02 |
| EF (%) | 47.12±1.79** | 56.20±2.48 | 77.40±0.71 | 77.56±0.62 |
| FS (%) | 20.26±0.94** | 25.41±1.46 | 40.40±0.64 | 40.54±0.56 |
| HR (bpm) | 451±11 | 462±6 | 461±7 | 460±14 |
| 4-weeks - HF | ||||
| LVIDd (mm) | 5.76±0.12* | 5.26±0.19 | 4.06±0.04 | 4.07±0.05 |
| LVIDs (mm) | 4.63±0.11** | 4.03±0.18 | 2.40±0.04 | 2.44±0.05 |
| EF (%) | 46.04±0.70** | 53.27±1.62 | 78.01±0.65 | 77.11±0.60 |
| FS (%) | 19.68±0.36** | 23.64±0.89 | 40.97±0.61 | 40.16±0.52 |
| HR (bpm) | 456±13 | 473±6 | 475±7 | 470±7 |
| 8-weeks - HF | ||||
| LVIDd (mm) | 5.96±0.17* | 5.39±0.16 | 4.13±0.02 | 4.15±0.04 |
| LVIDs (mm) | 4.81±0.16** | 4.15±0.15 | 2.45±0.03 | 2.46±0.03 |
| EF (%) | 45.18±1.01** | 52.31±1.48 | 77.66±0.54 | 77.67±0.47 |
| FS (%) | 19.27±0.52** | 23.09±0.81 | 40.69±0.48 | 40.63±0.43 |
| HR (bpm) | 473±14 | 460±12 | 479±7 | 478±8 |
| 10-weeks - HF | ||||
| LVIDd (mm) | 6.24±0.23* | 5.55±0.18 | 4.14±0.03 | 4.15±0.05 |
| LVIDs (mm) | 5.13±0.22** | 4.37±0.17 | 2.49±0.02 | 2.50±0.05 |
| EF (%) | 42.20±1.31** | 49.26±1.31 | 76.72±0.52 | 76.72±0.80 |
| FS (%) | 17.78±0.64** | 21.39±0.70 | 39.78±0.45 | 39.80±0.69 |
| HR (bpm) | 483±7 | 468±9 | 474±6 | 469±10 |
| Hemodynamics (HF) | ||||
| LVESP (mmHg) | 80.86±1.53** | 90.46±1.12 | 96.37±1.17 | 98.04±1.91 |
| LVEDP (mmHg) | 2.79±0.81 | 1.21±0.91 | 0.40±0.54 | 0.25±0.40 |
| LVESV (μl) | 43.86±0.80** | 34.39±2.04 | 8.35±0.38 | 8.30±1.45 |
| LVEDV (μl) | 60.20±0.99** | 54.53±1.35 | 34.06±0.63 | 33.02±1.02 |
| +dP/dtmax (mmHg/sec) | 5102±98* | 7176±790 | 10506±352 | 10260±365 |
| -dP/dtmin (mmHg/sec) | -4672±104* | -6317±836 | -9394±562 | -9538±302 |
| SBP (mmHg) | 81.76±1.18** | 89.38±1.01 | 98.11±0.71 | 97.07±1.92 |
| DBP (mmHg) | 54.68±1.78* | 59.57±1.09 | 63.50±1.15 | 63.76±1.18 |
| MAP (mmHg) | 63.07±1.38** | 68.81±0.96 | 74.29±0.71 | 74.11±1.08 |
| HR (bpm) | 544±13 | 557±18 | 506±16 | 549±17 |
| Morphometry (HF) | ||||
| BW (g) | 29.30±0.89 | 31.72±0.98 | 31.27±0.68 | 27.86±0.77 |
| HW (mg) | 210±14.32* | 164±14.09 | 118±2.18 | 122±4.25 |
| HW:BW (mg/g) | 7.06±0.51* | 5.08±0.32 | 3.78±0.05 | 4.18±0.11 |
| HW:TL (mg/mm) | 10.53±0.72* | 7.73±0.45 | 5.87±0.10 | 6.10±0.20 |
LV, left ventricle; LVIDd/s, LV internal dimensions at diastole/systole; %EF, % ejection fraction; %FS, % fractional shortening; HR, heart rate; LVESP, LV end systolic pressure; LVEDP, LV end diastolic pressure; LVESV, LV end systolic volume; LVEDV, LV end diastolic volume; dP/dtmax and dP/dtmin, maximum and minimum first derivative of LV pressure; SBP, systolic blood pressure (BP); DBP, diastolic BP; MAP, mean arterial pressure; BW, body weight; HW, heart weight; TL, tibia length. n=7 mice for morphometrics and hemodynamics. Both HF groups are different from Sham by ANOVA.
p<0.05 Disrupted-HF (3 weeks) vs. all other groups by two-way ANOVA, followed by a Tukey post-hoc test.
p<0.001 Disrupted-HF (3 weeks) vs. all other groups by two-way ANOVA, followed by a Tukey post-hoc test. Values are mean±SEM.
3.4. Gut Microbial Derangement Leads to Exacerbated Cardiac Hemodynamics in HF
We next examined functional changes in our murine model, by measuring in vivo hemodynamics parameters (Fig. 2B, Table 1). We found that as compared to normal-HF mice, the disrupted-HF mice had worse (p<0.001) cardiac profiles including decreased LV end-systolic pressure (80.86±1.53 vs. 90.46±1.12 mmHg), increased LV end systolic volume (43.86±0.80 vs. 34.39±2.04 μl) and LV end diastolic volume (60.20±0.99 vs. 54.53±1.35 μl), and decreased cardiac contractility (p<0.05) dP/dtmax (5102±98 vs. 7176±790 mmHg/sec) and dP/dtmin (−4672±104 vs. −6317±836 mmHg/sec). Thus, an intact gut microbiome appears to benefit healing, while gut microbiota disruption worsens cardiac pathophysiology and exacerbates progression to HF.
3.5. Gut Microbial Derangement and Cardiac Morphometrics
In view of the worsened structural and functional profiles in the hearts of HF mice with a depleted microbiome, we next explored whether there were also differences in cardiac morphometrics. It is important to note that initial infarct size was consistent among normal-HF and disrupted-HF mice (Fig. 2C, left). Thus, despite all infarcts starting the same, disruption of the gut microbiome during critical healing phases post-infarction led to worsened outcome. That is, compared to the normal-HF mice, the disrupted-HF mice had significantly worse cardiac LV dilation, with greater (p<0.05) heart weight (HW, 210±14.32 vs. 164±14.09 mg) and heart weight to body weight ratio (HW:BW, 7.06±0.51 vs. 5.08±0.32 mg/g) (Fig. 2C middle and right, Table 1) by 10 weeks HF. Moreover, histologic analyses of serial LV sections stained with Masson’s Trichrome (Fig. 2D), and the infarct region (Fig. 2E), revealed that the disrupted-HF mice had greater (p<0.05) absolute scar size (58.77±2.21% vs. 40.91±2.43%; Fig. 1F) and thinner infarcts (30.37±1.61 vs. 45.49±1.66 pixels; Fig. 1G), compared to normal-HF. These remodeling data demonstrate a more rapid evolution to LV decompensation in the gut disrupted cohort, and that a healthy gut microbiome is essential for best outcomes in HF.
3.6. Aberrant Inflammatory Responses in the Early Remodeling Period
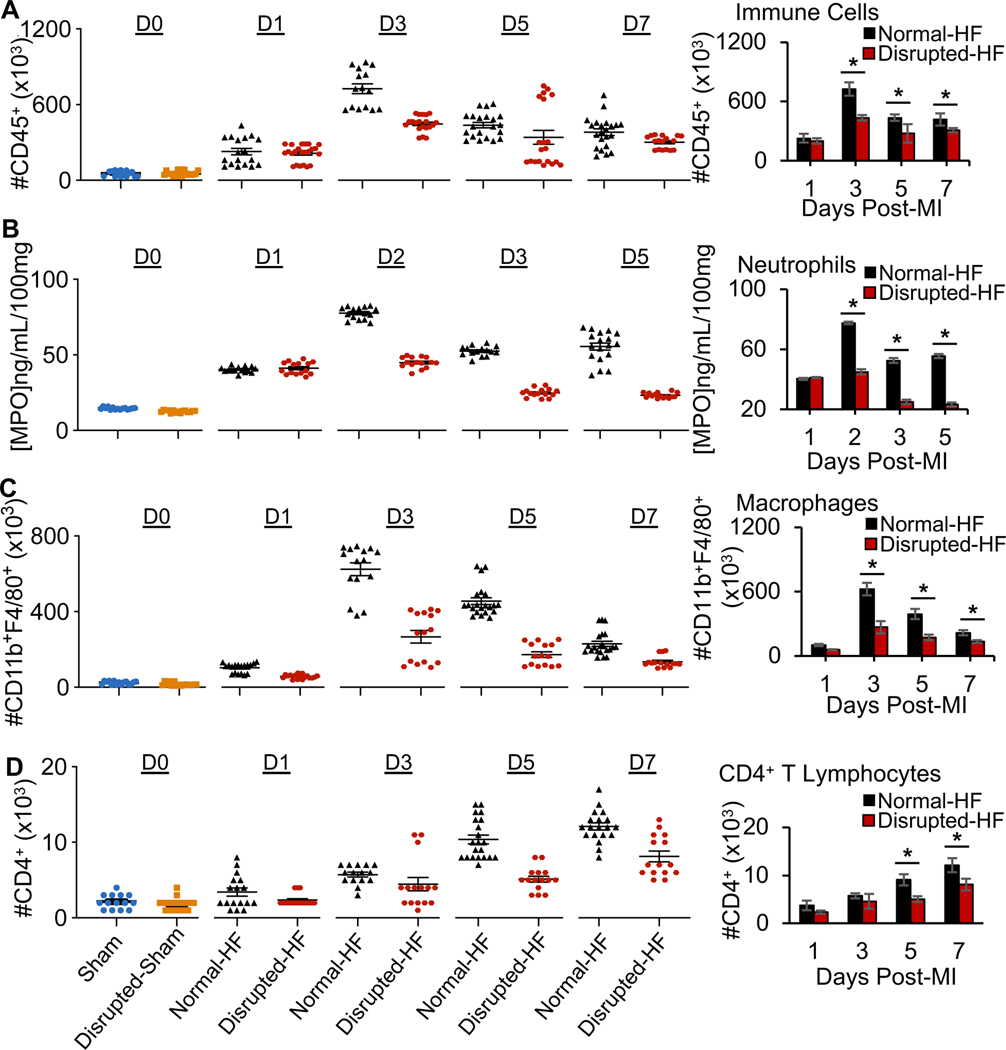
The NLRP3 inflammasome sensor is integral to early cardiomyocyte injury post-infarction. Thus, we explored whether gut microbial derangements affected inflammasome activation in the early remodeling heart, by measuring the inflammasome’s Nlrp3 mRNA and its constituent cytokines. We found similar levels of inflammasome gene expression (Figure 2H) but significantly (p<0.05) reduced levels of IL-1β and IL-18 protein levels in disrupted-HF hearts (Fig. 2I), suggesting that some differences in inflammasome activation could play a role in the worse outcomes in these mice. We also investigated additional measures of inflammatory responses important for cardiac repair. We found significant differences in leukocyte recruitment to infarcted myocardium in the disrupted-HF mice, by flow cytometry analyses (Fig. 3, Supplementary Table 3, see gating strategy in Supplementary Fig. 1). That is, as compared to normal-HF mice, there was decreased (p<0.05) CD45+ immunocyte infiltration in disrupted-HF hearts by day 3 post-infarction (Fig. 3A). Moreover, neutrophil abundance was reduced (p<0.05) in the disrupted-HF hearts over the first week post-infarction (Fig. 3B). In addition, we found that macrophages (CD11b+F4/80+) were also reduced (p<0.05) in the disrupted-HF versus normal-HF hearts (Fig. 3C). Lastly, the adaptive CD4+ T cell infiltration was also reduced (p<0.05) in the disrupted-HF versus normal-HF hearts (Fig. 3D). Collectively, these data suggest that key temporal immune responses involved in the early remodeling period are altered in mice with a disrupted gut microbiome, leading to worse cardiac repair and exacerbated HF outcomes.
Fig. 3.
Gut microbial disruption leads to changes in leukocyte recruitment to infarcted myocardium (scatter plot on left, averages on right). (A) CD45+cells, (B) cardiac myeloperoxidase (MPO, measure of neutrophil infiltration), (C) CD11b+F4/80+ macrophages in the heart, and (D) CD4+ T cells. Hearts were collected at days 0, 1, 2, 3, 5, or 7 post-MI as indicated, (n≥5 hearts per time point per group, with 3 technical replicates, *p<0.05 by Student’s t-test, mean ± SEM, see data values in Supplementary Table 3, and see Supplementary Fig. 1 for gating strategy.
3.7. Altered Gut Microbiome Metabolites and HF Outcomes
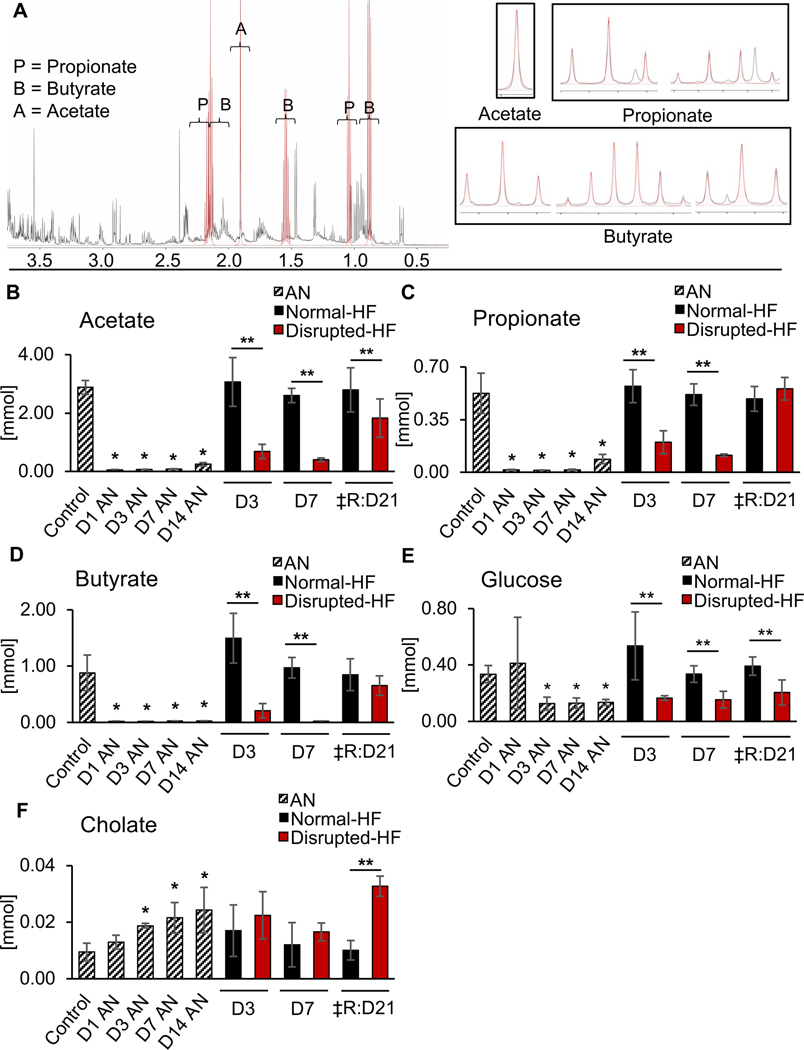
Since the gut microbiome produces short chain fatty acid (SCFA) metabolites that may modulate immune responses, we next examined whether microbiome depletion also altered SCFA levels, which could help to account for the observed changes in cardiac inflammation post-MI (Fig. 4; Supplementary Table 4). The normal 1H nuclear magnetic resonance (NMR) functional profile of cecal SCFA metabolites is shown in Fig. 4A. We found, in our study, that loss of gut microbiota abolished availability of the SCFAs acetate, propionate, and butyrate (Fig. 4B, 4C, 4D, left), resulting in clear differences in profiles in the disrupted-HF versus normal-HF mice (Fig. 4B, 4C, 4D, right). We also found altered luminal glucose levels (Fig. 4E), and cholate (Fig. 4F) in the disrupted-HF versus normal-HF mice. Collectively, these data are consistent with the notion that a healthy gut microbiome produces SCFA metabolites, and that gut microbiota disruption leads to a loss of the metabolites, associated with altered cardiac inflammation and impaired healing in heart disease.
Fig. 4.
Altered gut microbiome metabolites in disrupted-HF mice. (A) Representative WT murine 1H nuclear magnetic resonance (NMR) spectrum from cecal contents. (B) Mice with gut microbiome disruption had reduced concentrations of short-chain fatty acids (SCFA) including acetate, (C) propionate, (D) butyrate, and (E) reduced glucose, and (F) increased cholate. Samples collected at ZT03 in the treatment groups indicated on the x-axes, where D=day of collection, AN=antibiotics, ‡R=recovery, *p<0.05 AN vs. D0 by Student’s t-test. **p<0.05 Disrupted-HF vs. Normal-HF by Student’s t-test, mean ± SD, see data values in Supplementary Table 4.
3.8. Therapeutically Reconstituting the Microbiome Recapitulates Beneficial Healing and Improves HF Outcomes
Given that disrupting the gut microbiome impairs healing after MI, we next investigated whether reconstituting a healthy gut microbiome could be useful after MI to benefit outcome. To do this, we used a cohousing protocol (Fig. 5A) to reconstitute the microbiome, whereby mice with a disrupted microbiome were moved to cages containing the feces and litter from healthy mice after infarction. Because rodents are coprophagic, they readily ingest the feces to re-establish their microbiome. Notably, reconstituting the microbiome benefited cardiac remodeling, as shown by echocardiography and in vivo hemodynamics. Fig. 5B and Table 2 show that the reconstituted-HF mice have significantly better outcomes than disrupted-HF mice, and cardiovascular outcomes are restored to the same levels as seen in the normal-HF mice. This includes improvements in LV dimensions, cardiac contractility, and hemodynamic volumes. Benefits of reconstituting the microbiome on cardiac remodeling were also evident by histological analyses. Serial LV sections stained with Masson’s Trichrome (Fig. 5C), close-up of the infarct region (Fig. 5D) and quantification, showed that the reconstituted-HF mice had smaller absolute scar size (37.34±2.20% versus 54.66±5.05%; Fig. 5E) and less thinning of the infarct region (47.35±3.39 pixels versus 31.41±0.75 pixels; Fig. 5F), compared to the disrupted-HF mice. Fig. 5G shows the gut microbiome in reconstituted-HF mice at the phylum level. Collectively, these data demonstrate that a healthy gut microbiome plays a beneficial role in cardiac remodeling, and moreover that reconstitution of a disrupted gut microbiome is a promising strategy to improve health outcomes (Fig. H).
Fig. 5.
Microbiome reconstitution improves HF outcomes. (A) Experimental outline, mice received antibiotics for 1 week before infarction, and then randomized to either i) no intervention (disrupted-HF) or ii) were co-housed immediately post-MI for 1 week, to reconstitute their microbiome (reconstituted-HF). (B) Reconstituted-HF mice had better outcomes than the disrupted-HF mice, including increased LVIDd, LVIDs, and decreased %EF, %FS, as measured by echocardiography, and better vivo hemodynamic pressure-volume loops (representative image) by 8 weeks HF, (n=6–7 mice per group, *p<0.05 Disrupted-HF(1 week) vs. Reconstituted-HF by Student’s t-test, mean ± SEM, see data values in Table 2). (C) The hearts of the reconstituted-HF mice also had less LV dilation by histology (fewer equidistant sections) and (D) close-up of infarct regions by Masson’s trichrome, and (E) quantification revealing smaller absolute scar size and (F) greater integrity of infarcts, (n=3–4 hearts per group, *p<0.05 by Student’s t-test, mean ± SEM). (G) Relative abundance of cecal microbial phyla normalizing in the reconstituted-HF group. (H) Illustration showing how gut microbial disruption may be altering gut-derived metabolic processes leading to larger infarcts and poorer outcomes, and how microbiome reconstitution shortly after infarction rescues cardiac outcomes.
Table 2.
Co-housing benefits long-term cardiac structure and function after pre-MI microbiome disruption.
| Disrupted-HF | Reconstituted-HF | Normal-HF | |
|---|---|---|---|
| Echocardiography (n) | 7 | 7 | 7 |
| 1-week post-infarction | |||
| LVIDd (mm) | 5.16±0.06 | 4.88±0.18 | 4.89±0.17 |
| LVIDs (mm) | 4.04±0.06* | 3.58±0.21 | 3.57±0.18 |
| EF (%) | 49.93±0.91* | 58.91±2.65 | 59.38±2.02 |
| FS (%) | 21.67±0.50* | 27.04±1.61 | 27.27±1.21 |
| BW (g) | 22.45±0.44 | 23.81±0.32 | 22.48±0.38 |
| HR (bpm) | 463±13 | 459±9 | 467±6 |
| 4-weeks HF | |||
| LVIDd (mm) | 5.73±0.15* | 5.13±0.22 | 5.19±0.18 |
| LVIDs (mm) | 4.52±0.14* | 3.87±0.22 | 3.86±0.19 |
| EF (%) | 48.78±1.04* | 55.47±2.06 | 57.24±2.15 |
| FS (%) | 21.15±0.54* | 24.93±1.18 | 26.00±1.23 |
| BW (g) | 25.12±0.46 | 27.40±0.52 | 24.95±0.53 |
| HR (bpm) | 475±8 | 455±12 | 457±11 |
| 8-weeks HF | |||
| LVIDd (mm) | 6.03±0.14* | 5.30±0.23 | 5.42±0.21 |
| LVIDs (mm) | 4.84±0.14* | 4.02±0.24 | 4.14±0.21 |
| EF (%) | 46.01±0.99** | 54.55±2.13 | 53.41±2.12 |
| FS (%) | 19.73±0.50** | 24.42±1.21 | 23.75±1.18 |
| BW (g) | 28.39±0.74 | 31.11±0.69 | 28.67±0.84 |
| HR (bpm) | 471±9 | 479±10 | 452±10 |
| Hemodynamics (HF) | |||
| LVESP (mmHg) | 83.28±1.74* | 90.55±1.65 | 92.11±1.48 |
| LVEDP (mmHg) | 2.65±0.82 | 2.71±1.94 | 1.08±0.49 |
| LVESV (μl) | 35.84±3.61 | 29.34±1.40 | 32.02±2.08 |
| LVEDV (μl) | 58.02±1.80 | 54.07±1.20 | 53.41±2.48 |
| +dP/dtmax(mmHg/sec) | 5370±163* | 7190±492 | 7304±467 |
| -dP/dtmin (mmHg/sec) | -4562±151* | -6138±486 | -6179±326 |
| SBP (mmHg) | 83.44±0.97* | 92.05±0.94 | 90.39±1.14 |
| DBP (mmHg) | 53.85±1.61 | 57.47±1.64 | 57.50±1.02 |
| MAP (mmHg) | 63.08±1.13* | 68.31±1.30 | 67.78±0.65 |
| HR (bpm) | 508±29 | 474±23 | 504±23 |
LV, left ventricle; LVIDd/s, LV internal dimensions at diastole/systole; %EF, % ejection fraction; %FS, % fractional shortening; BW, body weight; HR, heart rate; LVESP, LV end systolic pressure; LVEDP, LV end diastolic pressure; LVESV, LV end systolic volume; LVEDV, LV end diastolic volume; dP/dtmax and dP/dtmin, maximum and minimum first derivative of LV pressure; SBP, systolic blood pressure (BP); DBP, diastolic BP; MAP, mean arterial pressure; HR, heart rate. n=6 mice for hemodynamics.
p<0.05 Disrupted-HF(1 week) vs. Reconstituted-HF by Student’s t-test.
p<0.01 Disrupted-HF(1 week) vs. Reconstituted-HF by Student’s t-test. Values are mean±SEM.
3.9. Circadian Influence on Gut Physiology to Improve HF Outcomes
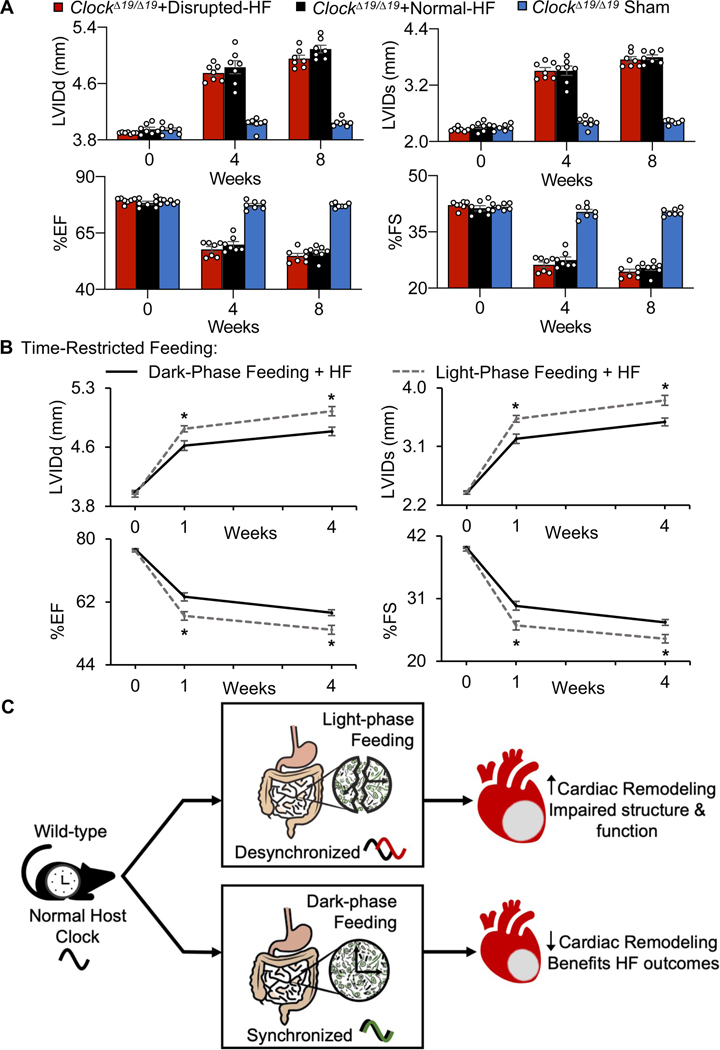
We next examined the circadian influence on gut physiology contributing to cardiac healing after MI. First, we used a genetic model, the canonical circadian mutant ClockΔ19/Δ19 mice (Fig. 6A–C) and observed altered diurnal rhythms in the composition of their gut microbiome, as compared to wildtype (WT) mice (Fig. 6D, Supplementary Table 5). The microbial patterns differed even though the animals were housed under the same conditions, in the same facilities, and were given the same food. Second, we noted that a dysfunctional circadian mechanism in the ClockΔ19/Δ19 mice coincided with a lack of benefits for cardiac remodeling, even in the presence of an intact gut microbiome (Fig. 7A, Supplementary Table 6, Supplementary Fig. 2A–E, Fig. 3, Table 7). Collectively, these findings are consistent with the notion that an intact circadian mechanism in the host is necessary to beneficially influence gut physiology and improve HF outcomes.
Fig. 6.
CLOCK influences the microbiome. (A) Representative genotyping of ClockΔ19/Δ19 mice homozygous for the CLOCK point mutation (Δ19/Δ19), heterozygous littermates (+/Δ19), and wild-type littermates (+/+), (B) running wheel actigraphy of mice under normal LD and in DD, and (C) feeding patterns (left - food intake in the light versus dark period, middle - %food intake light versus dark period, right – total food intake over 24 hours; *p<0.05 by Student’s t-test, mean ± SEM). (D) WT mice exhibit diurnal (light, murine sleep time) (dark, murine wake time) rhythms in their cecal microbiota composition, and these profiles differ in mice lacking a functional CLOCK. Left – WT 24 hour diurnal rhythms (white bars = light period / sleep time, black bars – dark period / wake time) plotted at the family level with JTK cycle values in brackets; middle left - WT sleep vs. wake abundance for adjacent family member; middle right - ZT03 (sleep time) and ZT15 (wake time) representative time points; far right - ClockΔ19/Δ19 at ZT03 and ZT15 time points. RA = Relative Abundance, *p<0.05 by Student’s t-test, mean ± SEM. See also Supplementary Table 5.
Fig. 7.
Circadian influence on gut physiology improves cardiac repair. (A) Mice with a disrupted circadian mechanism (ClockΔ19/Δ19) show no benefits on cardiac remodeling, even in the presence of an intact microbiome, by echocardiography to 8 weeks HF, (n=7 mice per group, *p<0.05 ClockΔ19/Δ19 mice vs. all other groups by two-way ANOVA, followed by a Tukey post-hoc test, mean ± SEM). See data values in Supplementary Table 6, Supplementary Fig. 2 for histopathology data and infarct region analysis, flow cytometry leukocyte recruitment to infarcted myocardium, and Supplementary Fig. 1 for flow cytometry gating strategy. n=8 hearts per time point per group, three technical repeats for each, p>0.05 by Student’s t-test, mean ± SEM. For NMR data for cecal SCFAs acetate, propionate, and butyrate, n≥6 mice per group, *p<0.001 by Student’s t-test, mean ± SEM. See Supplementary Fig. 3 for microbiome reconstitution experiments. (B) WT mice with time restricted feeding (TRF) during the dark phase (wake time) had significantly improved outcomes as compared to TRF mice during the light phase (sleep time), by echocardiography to 4 weeks HF, (n≥8 mice per group, *p<0.01 by Student’s t-test, mean ± SEM, see data values in Table 3. (C) Illustration highlighting the circadian influence on gut physiology to benefit cardiac repair.
Next, a model of behavior-induced peripheral tissue circadian disruption was employed. We used time-restricted feeding to specifically investigate the importance of circadian regulation on the gut for healing post-MI. We found that mice randomized to the dark phase-restricted feeding (wake time) prior to MI surgery had improved outcomes on cardiac structure and function, as compared to mice fed only in the light phase (sleep time) (Fig. 7B, Table 3). Thus, disrupting circadian rhythms in gut physiology, without affecting the central host circadian clock, impairs cardiac remodeling. Collectively, these findings demonstrate that the circadian system influences gut physiology and this is important for cardiac repair and can improve post-MI and HF outcomes (Fig. 7C).
Table 3.
Time Restricted Feeding significantly improves post-MI cardiac structure and function.
| Light-Restricted Feeding | Dark-Restricted Feeding | |
|---|---|---|
| Echocardiography (n) | 7 | 9 |
| Baseline | ||
| LVIDd (mm) | 3.95±0.03 | 3.98±0.02 |
| LVIDs (mm) | 2.40±0.03 | 2.39±0.02 |
| EF (%) | 76.64±0.33 | 77.06±0.19 |
| FS (%) | 39.64±0.29 | 39.99±0.16 |
| BW (g) | 22.25±0.33 | 22.16±0.32 |
| HR (bpm) | 461±9 | 475±5 |
| 1-week post-infarction | ||
| LVIDd (mm) | 4.78±0.04* | 4.57±0.06 |
| LVIDs (mm) | 3.53±0.05** | 3.22±0.07 |
| EF (%) | 58.00±1.22** | 63.45±1.16 |
| FS (%) | 26.27±0.77** | 29.75±0.77 |
| BW (g) | 23.72±0.37 | 23.26±0.40 |
| HR (bpm) | 463±9 | 468±9 |
| 4-weeks post-infarction | ||
| LVIDd (mm) | 5.01±0.06** | 4.75±0.05 |
| LVIDs (mm) | 3.81±0.08** | 3.48±0.06 |
| EF (%) | 54.04±1.27** | 58.93±0.84 |
| FS (%) | 23.95±0.73** | 26.82±0.52 |
| BW (g) | 26.16±0.60 | 25.40±0.49 |
| HR (bpm) | 469±7 | 481±7 |
LV, left ventricle; LVIDd/s, LV internal dimensions at diastole/systole; %EF, % ejection fraction; %FS, % fractional shortening; BW, body weight; HR, heart rate.
p<0.05 Dark-Restricted Feeding (DF-HF) vs. Light-Restricted Feeding (LF-HF) by Student’s t-test. *p<0.01 Dark-Restricted Feeding (DF-HF) vs. Light-Restricted Feeding (LF-HF) by Student’s t-test. Mean ± SEM.
4. Discussion
In this study, we investigate the circadian influence on gut physiology and how it can benefit cardiac repair in a well-established murine model of myocardial infarction (MI) leading to heart failure (HF). First, we show that the gut microbiome displays diurnal rhythms in composition. Second, we determine that gut microbial disruption impairs cardiac repair post-MI, exacerbating pathophysiology leading to HF. Gut disruption impairs healing at multiple levels, including enhancing gene expression of cardiac biomarkers of heart disease, and worsening structural and functional parameters in the remodeling heart. Gut microbial derangement also coincides with the production of an aberrant inflammatory response in the early cardiac remodeling period, including altered inflammasome cytokine abundance, and immunocyte recruitment to infarcted myocardium, possibly underlying the worse cardiac remodeling in these mice. Conversely, reconstituting the gut microbiome after infarction recapitulates a beneficial healing phenotype, consistent with the notion that gut physiology contributes to cardiac repair. A functional circadian clock mechanism in the host is important for coordinating the gut microbial responses to improve healing, as no benefits accrued in ClockΔ19/Δ19 mice. Finally, we demonstrate that time-restricted feeding is a feasible therapeutic approach to confer beneficial circadian influences on gut physiology to improve cardiac repair and HF outcomes.
One of the key findings of this study is that the gut microbiome contributes to cardiac repair post-MI and reduces pathophysiology to limit HF outcomes in WT mice. Our findings are consistent with reports that the gut microbiome is important for health and disease, including cardiac remodeling [80–82], metabolic conditions [63, 83] and recovery after spinal cord injury [64]. These preclinical studies are revealing that there is significant translational potential, and highlight the importance of a healthy gut microbiota composition for healing from disease in humans [84, 85]. In our studies, we used a simple cohousing strategy to promote the re-uptake of a healthy gut microbiome in disrupted mice. We recognize that microbiome constitution is as exists in our animal housing environment, that it was beneficial where these experiments were performed; however, microbiome composition may differ at other facilities, and this could have other or additional effects on outcomes. Nevertheless, the gut microbiome exhibits day and night rhythms in abundance and metabolite production, and the downstream targets of these gut microbial pathways likely function in a diurnal manner to impact cardiac repair and outcomes. The analogous approach in humans is fecal microbiota transfer, as is currently used to treat Clostridioides difficile infection, and other novel approaches that are also being developed to promote the transfer of a multi-species community to recipients [86, 87].
In the future, similar approaches to gut microbial reconstitution may also be applied to the management and treatment of other cardiovascular conditions. For example, 1) many people experience gut microbial disruption as a consequence of antibiotic treatments, as they are frequently prescribed for a wide variety of conditions. Antibiotic use is especially prevalent among our aging population in long-term care facilities (e.g. [88, 89]); notably, these elderly individuals are also at increased risk of heart disease (reviews, [1, 2] ). Reconstituting a healthy microbiome in our aging population could readily reduce cardiovascular morbidities, helping these individuals to live longer and healthier lives. 2) Gut microbial disruption can modify SCFA levels. This is important because SCFAs are critical for healthy human physiology [90] and modulate host immune responses through G-protein coupled receptors (GPR) GPR41 and GPR43 [91], which are commonly found on immune cells and are involved in cellular differentiation, activation, and other activities (reviewed in [92]). In terms of cardiovascular diseases, modulating gut microbiome mediated SCFA levels could benefit treatment of hypertension [93], and atherosclerosis [92]. Our findings further suggest that modulating these SCFAs may also modify patterns of infiltrating inflammatory cells involved in cardiac repair post-MI and in HF. However, it is important to note that conclusions regarding diurnal abundance of SCFAs, or the temporal effects on downstream cellular targets including macrophages and T-cells, requires further investigation. Indeed, in the context of MI or HF, many recent studies suggest a complex interplay of pro-inflammatory and reparative macrophages (e.g. reviews, [94–100]), and deleterious CD4 positive (e.g. reviews [101–103]) and beneficial CD4 positive (e.g. reviews [104, 105]) T cell populations are involved in cardiac remodeling and outcomes. In summary, there is a circadian influence on gut physiology, and approaches to translate these findings can benefit cardiovascular health and repair. Clearly, over the next few decades, gut microbiome therapies will influence clinical medicine, including giving rise to new approaches to treat cardiovascular disease in humans. It is worth noting that our studies investigated what happens in male mice, but future studies in female mice could also prove interesting, and lead to new insights into cardiac repair in both biological sexes.
An important and novel finding of this study is that there is a circadian influence on gut physiology that can benefit cardiac repair. We used several different approaches to investigate the circadian influence on gut physiology and HF outcomes. First, profiling of the murine cecal microbiome over 24 hours by 16S rRNA gene sequencing and bioinformatics analyses revealed day and night rhythms in the healthy gut microbiome. Second, we noted that these diurnal microbial rhythms were not affected by MI. However, antibiotics disrupted the composition and functionality of the gut microbiome. Next, we investigated microbial rhythms in the ClockΔ19/Δ19 mice, a canonical genetic model of circadian disruption [47, 68], and found that gut microbial rhythms were altered in these mice. Fifth, we found that a functional circadian clock mechanism in the host was necessary for the benefits conferred by the gut microbiome on cardiac repair. That is, in contrast to the WT mice, an intact gut microbiome did not appear to benefit HF outcomes in the ClockΔ19/Δ19 mice. The divergent outcomes may be related to findings that ClockΔ19/Δ19 mice have altered immune cell recruitment to infarcted myocardium post-MI [39], and impaired cardiac hypertrophy responses [36]. Thus, benefits for cardiac repair in the ClockΔ19/Δ19 mice may be attenuated by downstream circadian dependent inflammatory and/or hypertrophy responses. Although beyond the scope of this study, it would be interesting to investigate if these findings are specific to CLOCK, or also apply to other core mechanism factors. This is especially interesting as pharmacology is being developed to target components of the circadian mechanism, and early studies suggest that it can benefit HF outcomes [40]. To further investigate the circadian influence on gut physiology, we used a model where circadian rhythms in the gut were altered, without disrupting the host. Thus, the final approach that we used to investigate the circadian influence on gut physiology and HF outcomes involved a time-of-day restricted feeding (TRF) approach. Mice are nocturnal and preferentially eat during the dark phase, so light-phase restricted feeding has been shown to cause a 12-hour phase shift in the microbiome, without affecting the central pacemaker [54, 58]. TRF has also been used therapeutically to improve benefits in experimental [106, 107], and clinical [108–110] metabolic studies. Moreover, studies have demonstrated that the benefits of TRF can be attributed to improving the day/night profile of the gut microbiome, and consequently the diurnal functions of the gut [78, 111]. However, this has not yet been demonstrated to benefit cardiac repair. In this study, we found that feeding restricted to the animal’s wake-time, but not sleep-time, improved cardiac healing post-MI, demonstrating the importance of the circadian influence on the gut microbiome. This opens the door for feasible circadian-based translational approaches, such as TRF, to influence gut biology and improve cardiac post-MI and HF outcomes in patients.
These circadian approaches are especially translationally relevant to individuals in our society who experience disrupted circadian biology. Some common examples of how we experience circadian biology disruption include night-time shift work [112], circadian rhythm sleep-wake disorders (e.g. reviewed in [113]), social jet lag [114], or nocturnal sleep disorders like obstructive sleep apnea (e.g. reviewed in [115]). Importantly, individuals with disturbed circadian rhythms are more susceptible to cardiovascular diseases, and have worse outcomes [116–121]; also reviewed [45, 46]. Similarly, recent experimental studies also suggest that circadian misalignment may be relevant to intensive care settings [39], impairing healing in our most vulnerable patients. These and other studies have led to the development of a new field of medicine, called circadian medicine, and it may be especially relevant to the treatment of cardiovascular diseases [122]. Promising translational applications to add to this toolbox then, are strategies to recapitulate the normal diurnal gut physiology and the benefits that accrue to patients with cardiovascular disease.
5. Conclusions
The contributions of gut physiology to how we heal from disease are only beginning to come to light; this study clearly demonstrates that it is an important factor to consider for reducing post-MI adverse remodeling and progression to HF in mice. In this study, we also demonstrate that the circadian influence on gut physiology plays an important role in benefiting cardiac repair. Our findings are certainly not conclusive for human patients, but at a minimum they focus attention on the pervasive and possibly detrimental effects of disturbing our circadian gut physiology on cardiovascular repair processes. We hope our preclinical study will stimulate translation of basic circadian biology concepts to clinical cardiovascular health, such as maintaining a normal diurnal environment, promoting diurnal gut physiology, and by applying circadian strategies to benefit patients with cardiovascular disease, leading to longer and healthier lives.
Supplementary Material
Acknowledgements
The authors thank Dr. Sameer Al-Abdul-Wahid for his technical assistance and expertise with the NMR experiments, and Jeff Gross at the University of Guelph Genomics Facility for running our samples on the Illumina Miseq.
Funding Sources
This work was supported by grants from the Canadian Institutes of Health Research (CIHR) to T.A.M. P.M. is supported by a CIHR MSc award. T.A.M. is a career investigator of the Heart and Stroke Foundation of Canada.
Footnotes
Disclosures
The authors declare that there are no competing interests.
Appendix A. Supplementary Information
References
- [1].Mozaffarian D, Benjamin EJ, Go AS, Arnett DK, Blaha MJ, Cushman M, de Ferranti S, Despres JP, Fullerton HJ, Howard VJ, Huffman MD, Judd SE, Kissela BM, Lackland DT, Lichtman JH, Lisabeth LD, Liu S, Mackey RH, Matchar DB, McGuire DK, Mohler ER 3rd, Moy CS, Muntner P, Mussolino ME, Nasir K, Neumar RW, Nichol G, Palaniappan L, Pandey DK, Reeves MJ, Rodriguez CJ, Sorlie PD, Stein J, Towfighi A, Turan TN, Virani SS, Willey JZ, Woo D, Yeh RW, Turner MB, American C. Heart Association Statistics, S. Stroke Statistics, Heart disease and stroke statistics−-2015 update: a report from the American Heart Association, Circulation 131(4) (2015) e29–322. [DOI] [PubMed] [Google Scholar]
- [2].Anderson L, Thompson DR, Oldridge N, Zwisler AD, Rees K, Martin N, Taylor RS, Exercise-based cardiac rehabilitation for coronary heart disease, Cochrane Database Syst Rev (1) (2016) CD001800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].World Health Organization (WHO), New initiative launched to tackle cardiovascular disease, the world’s number one killer, 2016. http://www.who.int/cardiovascular_diseases/global-hearts/Global_hearts_initiative/en/. (Accessed December 10, 2019.
- [4].World Health Organization (WHO), Cardiovascular diseases (CVDs), 2017. https://www.who.int/cardiovascular_diseases/world-heart-day-2017/en/ (Accessed December 10, 2019.
- [5].Clarke JM, Hamer J, Shelton JR, Taylor S, Venning GR, The rhythm of the normal human heart, Lancet 1(7984) (1976) 508–12. [DOI] [PubMed] [Google Scholar]
- [6].Millar-Craig MW, Bishop CN, Raftery EB, Circadian variation of blood-pressure, Lancet 1(8068) (1978) 795–7. [DOI] [PubMed] [Google Scholar]
- [7].Hu K, Ivanov P, Hilton MF, Chen Z, Ayers RT, Stanley HE, Shea SA, Endogenous circadian rhythm in an index of cardiac vulnerability independent of changes in behavior, Proc Natl Acad Sci U S A 101(52) (2004) 18223–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Muller JE, Stone PH, Turi ZG, Rutherford JD, Czeisler CA, Parker C, Poole WK, Passamani E, Roberts R, Robertson T, et al. , Circadian variation in the frequency of onset of acute myocardial infarction, N Engl J Med 313(21) (1985) 1315–22. [DOI] [PubMed] [Google Scholar]
- [9].Muller JE, Tofler GH, Stone PH, Circadian variation and triggers of onset of acute cardiovascular disease, Circulation 79(4) (1989) 733–43. [DOI] [PubMed] [Google Scholar]
- [10].Montaigne D, Marechal X, Modine T, Coisne A, Mouton S, Fayad G, Ninni S, Klein C, Ortmans S, Seunes C, Potelle C, Berthier A, Gheeraert C, Piveteau C, Deprez R, Eeckhoute J, Duez H, Lacroix D, Deprez B, Jegou B, Koussa M, Edme JL, Lefebvre P, Staels B, Daytime variation of perioperative myocardial injury in cardiac surgery and its prevention by Rev-Erbalpha antagonism: a single-centre propensity-matched cohort study and a randomised study, Lancet 391(10115) (2018) 59–69. [DOI] [PubMed] [Google Scholar]
- [11].Mistry P, Duong A, Kirshenbaum L, Martino TA, Cardiac Clocks and Preclinical Translation, Heart Fail Clin 13(4) (2017) 657–672. [DOI] [PubMed] [Google Scholar]
- [12].Martino T, Arab S, Straume M, Belsham DD, Tata N, Cai F, Liu P, Trivieri M, Ralph M, Sole MJ, Day/night rhythms in gene expression of the normal murine heart, J Mol Med (Berl) 82(4) (2004) 256–64. [DOI] [PubMed] [Google Scholar]
- [13].Martino TA, Tata N, Belsham DD, Chalmers J, Straume M, Lee P, Pribiag H, Khaper N, Liu PP, Dawood F, Backx PH, Ralph MR, Sole MJ, Disturbed diurnal rhythm alters gene expression and exacerbates cardiovascular disease with rescue by resynchronization, Hypertension 49(5) (2007) 1104–13. [DOI] [PubMed] [Google Scholar]
- [14].Martino TA, Oudit GY, Herzenberg AM, Tata N, Koletar MM, Kabir GM, Belsham DD, Backx PH, Ralph MR, Sole MJ, Circadian rhythm disorganization produces profound cardiovascular and renal disease in hamsters, Am J Physiol Regul Integr Comp Physiol 294(5) (2008) R1675–83. [DOI] [PubMed] [Google Scholar]
- [15].Podobed P, Pyle WG, Ackloo S, Alibhai FJ, Tsimakouridze EV, Ratcliffe WF, Mackay A, Simpson J, Wright DC, Kirby GM, Young ME, Martino TA, The day/night proteome in the murine heart, Am J Physiol Regul Integr Comp Physiol 307(2) (2014) R121–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Podobed PS, Alibhai FJ, Chow CW, Martino TA, Circadian regulation of myocardial sarcomeric Titin-cap (Tcap, telethonin): identification of cardiac clock-controlled genes using open access bioinformatics data, PLoS One 9(8) (2014) e104907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, Weitz CJ, Extensive and divergent circadian gene expression in liver and heart, Nature 417(6884) (2002) 78–83. [DOI] [PubMed] [Google Scholar]
- [18].Durgan DJ, Pulinilkunnil T, Villegas-Montoya C, Garvey ME, Frangogiannis NG, Michael LH, Chow CW, Dyck JR, Young ME, Short communication: ischemia/reperfusion tolerance is time-of-day-dependent: mediation by the cardiomyocyte circadian clock, Circ Res 106(3) (2010) 546–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Rudic RD, McNamara P, Reilly D, Grosser T, Curtis AM, Price TS, Panda S, Hogenesch JB, FitzGerald GA, Bioinformatic analysis of circadian gene oscillation in mouse aorta, Circulation 112(17) (2005) 2716–24. [DOI] [PubMed] [Google Scholar]
- [20].Oyama Y, Bartman CM, Bonney S, Lee JS, Walker LA, Han J, Borchers CH, Buttrick PM, Aherne CM, Clendenen N, Colgan SP, Eckle T, Intense Light-Mediated Circadian Cardioprotection via Transcriptional Reprogramming of the Endothelium, Cell Rep 28(6) (2019) 1471–1484 e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Tsimakouridze EV, Straume M, Podobed PS, Chin H, LaMarre J, Johnson R, Antenos M, Kirby GM, Mackay A, Huether P, Simpson JA, Sole M, Gadal G, Martino TA, Chronomics of pressure overload-induced cardiac hypertrophy in mice reveals altered day/night gene expression and biomarkers of heart disease, Chronobiol Int 29(7) (2012) 810–21. [DOI] [PubMed] [Google Scholar]
- [22].Chalmers JA, Martino TA, Tata N, Ralph MR, Sole MJ, Belsham DD, Vascular circadian rhythms in a mouse vascular smooth muscle cell line (Movas-1), Am J Physiol Regul Integr Comp Physiol 295(5) (2008) R1529–38. [DOI] [PubMed] [Google Scholar]
- [23].Martino TA, Tata N, Bjarnason GA, Straume M, Sole MJ, Diurnal protein expression in blood revealed by high throughput mass spectrometry proteomics and implications for translational medicine and body time of day, Am J Physiol Regul Integr Comp Physiol 293(3) (2007) R1430–7. [DOI] [PubMed] [Google Scholar]
- [24].Martino TA, Tata N, Simpson JA, Vanderlaan R, Dawood F, Kabir MG, Khaper N, Cifelli C, Podobed P, Liu PP, Husain M, Heximer S, Backx PH, Sole MJ, The primary benefits of angiotensin-converting enzyme inhibition on cardiac remodeling occur during sleep time in murine pressure overload hypertrophy, J Am Coll Cardiol 57(20) (2011) 2020–8. [DOI] [PubMed] [Google Scholar]
- [25].Khaper N, Bailey CDC, Ghugre NR, Reitz C, Awosanmi Z, Waines R, Martino TA, Implications of disturbances in circadian rhythms for cardiovascular health: A new frontier in free radical biology, Free Radic Biol Med 119 (2018) 85–92. [DOI] [PubMed] [Google Scholar]
- [26].Eckle T, Hartmann K, Bonney S, Reithel S, Mittelbronn M, Walker LA, Lowes BD, Han J, Borchers CH, Buttrick PM, Kominsky DJ, Colgan SP, Eltzschig HK, Adora2b-elicited Per2 stabilization promotes a HIF-dependent metabolic switch crucial for myocardial adaptation to ischemia, Nat Med 18(5) (2012) 774–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Rabinovich-Nikitin I, Lieberman B, Martino TA, Kirshenbaum LA, Circadian-Regulated Cell Death in Cardiovascular Diseases, Circulation 139(7) (2019) 965–980. [DOI] [PubMed] [Google Scholar]
- [28].Bray MS, Shaw CA, Moore MW, Garcia RA, Zanquetta MM, Durgan DJ, Jeong WJ, Tsai JY, Bugger H, Zhang D, Rohrwasser A, Rennison JH, Dyck JR, Litwin SE, Hardin PE, Chow CW, Chandler MP, Abel ED, Young ME, Disruption of the circadian clock within the cardiomyocyte influences myocardial contractile function, metabolism, and gene expression, Am J Physiol Heart Circ Physiol 294(2) (2008) H1036–47. [DOI] [PubMed] [Google Scholar]
- [29].Durgan DJ, Hotze MA, Tomlin TM, Egbejimi O, Graveleau C, Abel ED, Shaw CA, Bray MS, Hardin PE, Young ME, The intrinsic circadian clock within the cardiomyocyte, Am J Physiol Heart Circ Physiol 289(4) (2005) H1530–41. [DOI] [PubMed] [Google Scholar]
- [30].Jeyaraj D, Haldar SM, Wan X, McCauley MD, Ripperger JA, Hu K, Lu Y, Eapen BL, Sharma N, Ficker E, Cutler MJ, Gulick J, Sanbe A, Robbins J, Demolombe S, Kondratov RV, Shea SA, Albrecht U, Wehrens XH, Rosenbaum DS, Jain MK, Circadian rhythms govern cardiac repolarization and arrhythmogenesis, Nature 483(7387) (2012) 96–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Schroder EA, Burgess DE, Zhang X, Lefta M, Smith JL, Patwardhan A, Bartos DC, Elayi CS, Esser KA, Delisle BP, The cardiomyocyte molecular clock regulates the circadian expression of Kcnh2 and contributes to ventricular repolarization, Heart Rhythm 12(6) (2015) 1306–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Schroder EA, Lefta M, Zhang X, Bartos DC, Feng HZ, Zhao Y, Patwardhan A, Jin JP, Esser KA, Delisle BP, The cardiomyocyte molecular clock, regulation of Scn5a, and arrhythmia susceptibility, Am J Physiol Cell Physiol 304(10) (2013) C954–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Wang Z, Tapa S, Francis Stuart SD, Wang L, Bossuyt J, Delisle BP, Ripplinger CM, Aging Disrupts Normal Time-of-day Variation in Cardiac Electrophysiology, Circ Arrhythm Electrophysiol (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Duong ATH, Reitz CJ, Louth EL, Creighton SD, Rasouli M, Zwaiman A, Kroetsch JT, Bolz SS, Winters BD, Bailey CDC, Martino TA, The Clock Mechanism Influences Neurobiology and Adaptations to Heart Failure in Clock(19/19) Mice With Implications for Circadian Medicine, Sci Rep 9(1) (2019) 4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Chalmers JA, Lin SY, Martino TA, Arab S, Liu P, Husain M, Sole MJ, Belsham DD, Diurnal profiling of neuroendocrine genes in murine heart, and shift in proopiomelanocortin gene expression with pressure-overload cardiac hypertrophy, J Mol Endocrinol 41(3) (2008) 117–24. [DOI] [PubMed] [Google Scholar]
- [36].Alibhai FJ, LaMarre J, Reitz CJ, Tsimakouridze EV, Kroetsch JT, Bolz SS, Shulman A, Steinberg S, Burris TP, Oudit GY, Martino TA, Disrupting the key circadian regulator CLOCK leads to age-dependent cardiovascular disease, J Mol Cell Cardiol 105 (2017) 24–37. [DOI] [PubMed] [Google Scholar]
- [37].Alibhai FJ, Reitz CJ, Peppler WT, Basu P, Sheppard P, Choleris E, Bakovic M, Martino TA, Female ClockDelta19/Delta19 mice are protected from the development of age-dependent cardiomyopathy, Cardiovasc Res 114(2) (2018) 259–271. [DOI] [PubMed] [Google Scholar]
- [38].Glen Pyle W, Martino TA, Circadian rhythms influence cardiovascular disease differently in males and females: role of sex and gender, Current Opinion in Physiology 5 (2018) 30–37. [Google Scholar]
- [39].Alibhai FJ, Tsimakouridze EV, Chinnappareddy N, Wright DC, Billia F, O’Sullivan ML, Pyle WG, Sole MJ, Martino TA, Short-term disruption of diurnal rhythms after murine myocardial infarction adversely affects long-term myocardial structure and function, Circ Res 114(11) (2014) 1713–22. [DOI] [PubMed] [Google Scholar]
- [40].Reitz CJ, Alibhai FJ, Khatua TN, Rasouli M, Bridle BW, Burris TP, Martino TA, SR9009 administered for one day after myocardial ischemia-reperfusion prevents heart failure in mice by targeting the cardiac inflammasome, Commun Biol 2 (2019) 353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Bennardo M, Alibhai F, Tsimakouridze E, Chinnappareddy N, Podobed P, Reitz C, Pyle WG, Simpson J, Martino TA, Day-night dependence of gene expression and inflammatory responses in the remodeling murine heart post-myocardial infarction, Am J Physiol Regul Integr Comp Physiol 311(6) (2016) R1243–R1254. [DOI] [PubMed] [Google Scholar]
- [42].Reitz CJ, Martino TA, Disruption of Circadian Rhythms and Sleep on Critical Illness and the Impact on Cardiovascular Events, Curr Pharm Des 21(24) (2015) 3505–11. [DOI] [PubMed] [Google Scholar]
- [43].Martino TA, Sole MJ, Molecular time: an often overlooked dimension to cardiovascular disease, Circ Res 105(11) (2009) 1047–61. [DOI] [PubMed] [Google Scholar]
- [44].Sole MJ, Martino TA, Diurnal physiology: core principles with application to the pathogenesis, diagnosis, prevention, and treatment of myocardial hypertrophy and failure, J Appl Physiol (1985) 107(4) (2009) 1318–27. [DOI] [PubMed] [Google Scholar]
- [45].Alibhai FJ, Tsimakouridze EV, Reitz CJ, Pyle WG, Martino TA, Consequences of Circadian and Sleep Disturbances for the Cardiovascular System, Can J Cardiol 31(7) (2015) 860–72. [DOI] [PubMed] [Google Scholar]
- [46].Martino TA, Young ME, Influence of the cardiomyocyte circadian clock on cardiac physiology and pathophysiology, J Biol Rhythms 30(3) (2015) 183–205. [DOI] [PubMed] [Google Scholar]
- [47].Vitaterna MH, King DP, Chang AM, Kornhauser JM, Lowrey PL, McDonald JD, Dove WF, Pinto LH, Turek FW, Takahashi JS, Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior, Science 264(5159) (1994) 719–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, King DP, Takahashi JS, Weitz CJ, Role of the CLOCK protein in the mammalian circadian mechanism, Science 280(5369) (1998) 1564–9. [DOI] [PubMed] [Google Scholar]
- [49].Hogenesch JB, Gu YZ, Jain S, Bradfield CA, The basic-helix-loop-helix-PAS orphan MOP3 forms transcriptionally active complexes with circadian and hypoxia factors, Proc Natl Acad Sci U S A 95(10) (1998) 5474–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Reddy P, Zehring WA, Wheeler DA, Pirrotta V, Hadfield C, Hall JC, Rosbash M, Molecular analysis of the period locus in Drosophila melanogaster and identification of a transcript involved in biological rhythms, Cell 38(3) (1984) 701–10. [DOI] [PubMed] [Google Scholar]
- [51].Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, Maywood ES, Hastings MH, Reppert SM, mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop, Cell 98(2) (1999) 193–205. [DOI] [PubMed] [Google Scholar]
- [52].Griffin EA Jr., Staknis D, Weitz CJ, Light-independent role of CRY1 and CRY2 in the mammalian circadian clock, Science 286(5440) (1999) 768–71. [DOI] [PubMed] [Google Scholar]
- [53].Young ME, Razeghi P, Taegtmeyer H, Clock genes in the heart - Characterization and attenuation with hypertrophy, Circulation Research 88(11) (2001) 1142–1150. [DOI] [PubMed] [Google Scholar]
- [54].Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela F, Schibler U, Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus, Genes Dev 14(23) (2000) 2950–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Stokkan KA, Yamazaki S, Tei H, Sakaki Y, Menaker M, Entrainment of the circadian clock in the liver by feeding, Science 291(5503) (2001) 490–3. [DOI] [PubMed] [Google Scholar]
- [56].Vollmers C, Gill S, DiTacchio L, Pulivarthy SR, Le HD, Panda S, Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression, Proc Natl Acad Sci U S A 106(50) (2009) 21453–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Mukherji A, Kobiita A, Ye T, Chambon P, Homeostasis in intestinal epithelium is orchestrated by the circadian clock and microbiota cues transduced by TLRs, Cell 153(4) (2013) 812–27. [DOI] [PubMed] [Google Scholar]
- [58].Thaiss CA, Zeevi D, Levy M, Zilberman-Schapira G, Suez J, Tengeler AC, Abramson L, Katz MN, Korem T, Zmora N, Kuperman Y, Biton I, Gilad S, Harmelin A, Shapiro H, Halpern Z, Segal E, Elinav E, Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis, Cell 159(3) (2014) 514–29. [DOI] [PubMed] [Google Scholar]
- [59].Liang X, Bushman FD, FitzGerald GA, Rhythmicity of the intestinal microbiota is regulated by gender and the host circadian clock, Proc Natl Acad Sci U S A 112(33) (2015) 10479–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Leone V, Gibbons SM, Martinez K, Hutchison AL, Huang EY, Cham CM, Pierre JF, Heneghan AF, Nadimpalli A, Hubert N, Zale E, Wang Y, Huang Y, Theriault B, Dinner AR, Musch MW, Kudsk KA, Prendergast BJ, Gilbert JA, Chang EB, Effects of diurnal variation of gut microbes and high-fat feeding on host circadian clock function and metabolism, Cell Host Microbe 17(5) (2015) 681–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Thaiss CA, Levy M, Korem T, Dohnalova L, Shapiro H, Jaitin DA, David E, Winter DR, Gury-BenAri M, Tatirovsky E, Tuganbaev T, Federici S, Zmora N, Zeevi D, Dori-Bachash M, Pevsner-Fischer M, Kartvelishvily E, Brandis A, Harmelin A, Shibolet O, Halpern Z, Honda K, Amit I, Segal E, Elinav E, Microbiota Diurnal Rhythmicity Programs Host Transcriptome Oscillations, Cell 167(6) (2016) 1495–1510 e12. [DOI] [PubMed] [Google Scholar]
- [62].Tahara Y, Yamazaki M, Sukigara H, Motohashi H, Sasaki H, Miyakawa H, Haraguchi A, Ikeda Y, Fukuda S, Shibata S, Gut Microbiota-Derived Short Chain Fatty Acids Induce Circadian Clock Entrainment in Mouse Peripheral Tissue, Sci Rep 8(1) (2018) 1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI, An obesity-associated gut microbiome with increased capacity for energy harvest, Nature 444(7122) (2006) 1027–31. [DOI] [PubMed] [Google Scholar]
- [64].Kigerl KA, Hall JC, Wang L, Mo X, Yu Z, Popovich PG, Gut dysbiosis impairs recovery after spinal cord injury, J Exp Med 213(12) (2016) 2603–2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Zhang C, Zhang W, Zhang J, Jing Y, Yang M, Du L, Gao F, Gong H, Chen L, Li J, Liu H, Qin C, Jia Y, Qiao J, Wei B, Yu Y, Zhou H, Liu Z, Yang D, Li J, Gut microbiota dysbiosis in male patients with chronic traumatic complete spinal cord injury, J Transl Med 16(1) (2018) 353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Chen GY, Shaw MH, Redondo G, Nunez G, The innate immune receptor Nod1 protects the intestine from inflammation-induced tumorigenesis, Cancer Res 68(24) (2008) 10060–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Li G, Xie C, Lu S, Nichols RG, Tian Y, Li L, Patel D, Ma Y, Brocker CN, Yan T, Krausz KW, Xiang R, Gavrilova O, Patterson AD, Gonzalez FJ, Intermittent Fasting Promotes White Adipose Browning and Decreases Obesity by Shaping the Gut Microbiota, Cell Metab 26(4) (2017) 672–685 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].King DP, Zhao Y, Sangoram AM, Wilsbacher LD, Tanaka M, Antoch MP, Steeves TD, Vitaterna MH, Kornhauser JM, Lowrey PL, Turek FW, Takahashi JS, Positional cloning of the mouse circadian clock gene, Cell 89(4) (1997) 641–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF, Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities, Appl Environ Microbiol 75(23) (2009) 7537–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD, Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform, Appl Environ Microbiol 79(17) (2013) 5112–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y, Brown CT, Porras-Alfaro A, Kuske CR, Tiedje JM, Ribosomal Database Project: data and tools for high throughput rRNA analysis, Nucleic Acids Res 42(Database issue) (2014) D633–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].du Pre B, Van Veen T, Crnko S, Vos M, Deddens J, Doevendans P, Van Laake L, Variation within Variation: Comparison of 24-h Rhythm in Rodent Infarct Size between Ischemia Reperfusion and Permanent Ligation, Int J Mol Sci 18(8) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Chomczynski P, Sacchi N, Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction, Anal Biochem 162(1) (1987) 156–9. [DOI] [PubMed] [Google Scholar]
- [74].Labrecque N, Cermakian N, Circadian Clocks in the Immune System, J Biol Rhythms 30(4) (2015) 277–90. [DOI] [PubMed] [Google Scholar]
- [75].Shi X, Xiao C, Wang Y, Tang H, Gallic acid intake induces alterations to systems metabolism in rats, J Proteome Res 12(2) (2013) 991–1006. [DOI] [PubMed] [Google Scholar]
- [76].Wu J, An Y, Yao J, Wang Y, Tang H, An optimised sample preparation method for NMR-based faecal metabonomic analysis, Analyst 135(5) (2010) 1023–30. [DOI] [PubMed] [Google Scholar]
- [77].Yen S, McDonald JA, Schroeter K, Oliphant K, Sokolenko S, Blondeel EJ, Allen-Vercoe E, Aucoin MG, Metabolomic analysis of human fecal microbiota: a comparison of feces-derived communities and defined mixed communities, J Proteome Res 14(3) (2015) 1472–82. [DOI] [PubMed] [Google Scholar]
- [78].Zarrinpar A, Chaix A, Yooseph S, Panda S, Diet and feeding pattern affect the diurnal dynamics of the gut microbiome, Cell Metab 20(6) (2014) 1006–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Hughes ME, Hogenesch JB, Kornacker K, JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets, J Biol Rhythms 25(5) (2010) 372–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Tang TWH, Chen HC, Chen CY, Yen CYT, Lin CJ, Prajnamitra RP, Chen LL, Ruan SC, Lin JH, Lin PJ, Lu HH, Kuo CW, Chang CM, Hall AD, Vivas EI, Shui JW, Chen P, Hacker TA, Rey FE, Kamp TJ, Hsieh PCH, Loss of Gut Microbiota Alters Immune System Composition and Cripples Postinfarction Cardiac Repair, Circulation 139(5) (2019) 647–659. [DOI] [PubMed] [Google Scholar]
- [81].Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, Feldstein AE, Britt EB, Fu X, Chung YM, Wu Y, Schauer P, Smith JD, Allayee H, Tang WH, DiDonato JA, Lusis AJ, Hazen SL, Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease, Nature 472(7341) (2011) 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Yang W, Zhang S, Zhu J, Jiang H, Jia D, Ou T, Qi Z, Zou Y, Qian J, Sun A, Ge J, Gut microbe-derived metabolite trimethylamine N-oxide accelerates fibroblast-myofibroblast differentiation and induces cardiac fibrosis, J Mol Cell Cardiol 134 (2019) 119–130. [DOI] [PubMed] [Google Scholar]
- [83].Backhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, Gordon JI, The gut microbiota as an environmental factor that regulates fat storage, Proc Natl Acad Sci U S A 101(44) (2004) 15718–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Shanahan F, van Sinderen D, O’Toole PW, Stanton C, Feeding the microbiota: transducer of nutrient signals for the host, Gut 66(9) (2017) 1709–1717. [DOI] [PubMed] [Google Scholar]
- [85].Cani PD, Jordan BF, Gut microbiota-mediated inflammation in obesity: a link with gastrointestinal cancer, Nat Rev Gastroenterol Hepatol 15(11) (2018) 671–682. [DOI] [PubMed] [Google Scholar]
- [86].Petrof EO, Gloor GB, Vanner SJ, Weese SJ, Carter D, Daigneault MC, Brown EM, Schroeter K, Allen-Vercoe E, Stool substitute transplant therapy for the eradication of Clostridium difficile infection: ‘RePOOPulating’ the gut, Microbiome 1(1) (2013) 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Cammarota G, Ianiro G, Gasbarrini A, Fecal microbiota transplantation for the treatment of Clostridium difficile infection: a systematic review, J Clin Gastroenterol 48(8) (2014) 693–702. [DOI] [PubMed] [Google Scholar]
- [88].van Buul LW, van der Steen JT, Veenhuizen RB, Achterberg WP, Schellevis FG, Essink RT, van Benthem BH, Natsch S, Hertogh CM, Antibiotic use and resistance in long term care facilities, J Am Med Dir Assoc 13(6) (2012) 568 e1–13. [DOI] [PubMed] [Google Scholar]
- [89].Daneman N, Gruneir A, Bronskill SE, Newman A, Fischer HD, Rochon PA, Anderson GM, Bell CM, Prolonged antibiotic treatment in long-term care: role of the prescriber, JAMA Intern Med 173(8) (2013) 673–82. [DOI] [PubMed] [Google Scholar]
- [90].Oliphant K, Allen-Vercoe E, Macronutrient metabolism by the human gut microbiome: major fermentation by-products and their impact on host health, Microbiome 7(1) (2019) 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Brown AJ, Goldsworthy SM, Barnes AA, Eilert MM, Tcheang L, Daniels D, Muir AI, Wigglesworth MJ, Kinghorn I, Fraser NJ, Pike NB, Strum JC, Steplewski KM, Murdock PR, Holder JC, Marshall FH, Szekeres PG, Wilson S, Ignar DM, Foord SM, Wise A, Dowell SJ, The Orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids, J Biol Chem 278(13) (2003) 11312–9. [DOI] [PubMed] [Google Scholar]
- [92].Ohira H, Tsutsui W, Fujioka Y, Are Short Chain Fatty Acids in Gut Microbiota Defensive Players for Inflammation and Atherosclerosis?, J Atheroscler Thromb 24(7) (2017) 660–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Bartolomaeus H, Balogh A, Yakoub M, Homann S, Marko L, Hoges S, Tsvetkov D, Krannich A, Wundersitz S, Avery EG, Haase N, Kraker K, Hering L, Maase M, Kusche-Vihrog K, Grandoch M, Fielitz J, Kempa S, Gollasch M, Zhumadilov Z, Kozhakhmetov S, Kushugulova A, Eckardt KU, Dechend R, Rump LC, Forslund SK, Muller DN, Stegbauer J, Wilck N, Short-Chain Fatty Acid Propionate Protects From Hypertensive Cardiovascular Damage, Circulation 139(11) (2019) 1407–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Williams JW, Giannarelli C, Rahman A, Randolph GJ, Kovacic JC, Macrophage Biology, Classification, and Phenotype in Cardiovascular Disease: JACC Macrophage in CVD Series (Part 1), J Am Coll Cardiol 72(18) (2018) 2166–2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Moore KJ, Koplev S, Fisher EA, Tabas I, Bjorkegren JLM, Doran AC, Kovacic JC, Macrophage Trafficking, Inflammatory Resolution, and Genomics in Atherosclerosis: JACC Macrophage in CVD Series (Part 2), J Am Coll Cardiol 72(18) (2018) 2181–2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Fayad ZA, Swirski FK, Calcagno C, Robbins CS, Mulder W, Kovacic JC, Monocyte and Macrophage Dynamics in the Cardiovascular System: JACC Macrophage in CVD Series (Part 3), J Am Coll Cardiol 72(18) (2018) 2198–2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Lavine KJ, Pinto AR, Epelman S, Kopecky BJ, Clemente-Casares X, Godwin J, Rosenthal N, Kovacic JC, The Macrophage in Cardiac Homeostasis and Disease: JACC Macrophage in CVD Series (Part 4), J Am Coll Cardiol 72(18) (2018) 2213–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Swirski FK, Nahrendorf M, Cardioimmunology: the immune system in cardiac homeostasis and disease, Nat Rev Immunol 18(12) (2018) 733–744. [DOI] [PubMed] [Google Scholar]
- [99].Prabhu SD, The Cardiosplenic Axis Is Essential for the Pathogenesis of Ischemic Heart Failure, Trans Am Clin Climatol Assoc 129 (2018) 202–214. [PMC free article] [PubMed] [Google Scholar]
- [100].Wang Y, Epelman S, Cardiac Macrophages, Reactive Oxygen Species, and Development of Left Ventricular Dysfunction, JACC Basic Transl Sci 2(6) (2017) 699–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Mathes D, Weirather J, Nordbeck P, Arias-Loza AP, Burkard M, Pachel C, Kerkau T, Beyersdorf N, Frantz S, Hofmann U, CD4+ Foxp3+ T-cells contribute to myocardial ischemia-reperfusion injury, J Mol Cell Cardiol 101 (2016) 99–105. [DOI] [PubMed] [Google Scholar]
- [102].Tang TT, Zhu YC, Dong NG, Zhang S, Cai J, Zhang LX, Han Y, Xia N, Nie SF, Zhang M, Lv BJ, Jiao J, Yang XP, Hu Y, Liao YH, Cheng X, Pathologic T-cell response in ischaemic failing hearts elucidated by T-cell receptor sequencing and phenotypic characterization, Eur Heart J 40(48) (2019) 3924–3933. [DOI] [PubMed] [Google Scholar]
- [103].Bansal SS, Ismahil MA, Goel M, Patel B, Hamid T, Rokosh G, Prabhu SD, Activated T Lymphocytes are Essential Drivers of Pathological Remodeling in Ischemic Heart Failure, Circ Heart Fail 10(3) (2017) e003688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Rieckmann M, Delgobo M, Gaal C, Buchner L, Steinau P, Reshef D, Gil-Cruz C, Horst ENT, Kircher M, Reiter T, Heinze KG, Niessen HW, Krijnen PA, van der Laan AM, Piek JJ, Koch C, Wester HJ, Lapa C, Bauer WR, Ludewig B, Friedman N, Frantz S, Hofmann U, Ramos GC, Myocardial infarction triggers cardioprotective antigen-specific T helper cell responses, J Clin Invest 130 (2019) 4922–4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Bansal SS, Ismahil MA, Goel M, Zhou G, Rokosh G, Hamid T, Prabhu SD, Dysfunctional and Proinflammatory Regulatory T-Lymphocytes Are Essential for Adverse Cardiac Remodeling in Ischemic Cardiomyopathy, Circulation 139(2) (2019) 206–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Chaix A, Lin T, Le HD, Chang MW, Panda S, Time-Restricted Feeding Prevents Obesity and Metabolic Syndrome in Mice Lacking a Circadian Clock, Cell Metab (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Hatori M, Vollmers C, Zarrinpar A, DiTacchio L, Bushong EA, Gill S, Leblanc M, Chaix A, Joens M, Fitzpatrick JA, Ellisman MH, Panda S, Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet, Cell Metab 15(6) (2012) 848–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Sutton EF, Beyl R, Early KS, Cefalu WT, Ravussin E, Peterson CM, Early Time-Restricted Feeding Improves Insulin Sensitivity, Blood Pressure, and Oxidative Stress Even without Weight Loss in Men with Prediabetes, Cell Metab 27(6) (2018) 1212–1221 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Gabel K, Hoddy KK, Haggerty N, Song J, Kroeger CM, Trepanowski JF, Panda S, Varady KA, Effects of 8-hour time restricted feeding on body weight and metabolic disease risk factors in obese adults: A pilot study, Nutr Healthy Aging 4(4) (2018) 345–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Wilkinson MJ, Manoogian ENC, Zadourian A, Lo H, Fakhouri S, Shoghi A, Wang X, Fleischer JG, Navlakha S, Panda S, Taub PR, Ten-Hour Time-Restricted Eating Reduces Weight, Blood Pressure, and Atherogenic Lipids in Patients with Metabolic Syndrome, Cell Metab 31(1) (2020) 92–104 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Ren J, Hu D, Mao Y, Yang H, Liao W, Xu W, Ge P, Zhang H, Sang X, Lu X, Zhong S, Alteration in gut microbiota caused by time-restricted feeding alleviate hepatic ischaemia reperfusion injury in mice, J Cell Mol Med 23(3) (2019) 1714–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].World Health Organization (WHO), World Health Report 2002: Reducing Risks, Promoting Healthy Life, 2002. http://www.who.int/whr/2002/en/ (Accessed March 28, 2020.
- [113].Ashbrook LH, Krystal AD, Fu YH, Ptacek LJ, Genetics of the human circadian clock and sleep homeostat, Neuropsychopharmacology 45(1) (2020) 45–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Wittmann M, Dinich J, Merrow M, Roenneberg T, Social jetlag: misalignment of biological and social time, Chronobiol Int 23(1–2) (2006) 497–509. [DOI] [PubMed] [Google Scholar]
- [115].Bradley TD, Floras JS, Sleep apnea and heart failure: Part I: obstructive sleep apnea, Circulation 107(12) (2003) 1671–8. [DOI] [PubMed] [Google Scholar]
- [116].Bradley TD, Floras JS, Obstructive sleep apnoea and its cardiovascular consequences, Lancet 373(9657) (2009) 82–93. [DOI] [PubMed] [Google Scholar]
- [117].Floras JS, Sleep apnea in heart failure: implications of sympathetic nervous system activation for disease progression and treatment, Curr Heart Fail Rep 2(4) (2005) 212–7. [DOI] [PubMed] [Google Scholar]
- [118].Marin JM, Carrizo SJ, Vicente E, Agusti AG, Long-term cardiovascular outcomes in men with obstructive sleep apnoea-hypopnoea with or without treatment with continuous positive airway pressure: an observational study, Lancet 365(9464) (2005) 1046–53. [DOI] [PubMed] [Google Scholar]
- [119].Vyas MV, Garg AX, Iansavichus AV, Costella J, Donner A, Laugsand LE, Janszky I, Mrkobrada M, Parraga G, Hackam DG, Shift work and vascular events: systematic review and meta-analysis, BMJ 345 (2012) e4800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Kawachi I, Colditz GA, Stampfer MJ, Willett WC, Manson JE, Speizer FE, Hennekens CH, Prospective study of shift work and risk of coronary heart disease in women, Circulation 92(11) (1995) 3178–82. [DOI] [PubMed] [Google Scholar]
- [121].Knutsson A, Akerstedt T, Jonsson BG, Orth-Gomer K, Increased risk of ischaemic heart disease in shift workers, Lancet 2(8498) (1986) 89–92. [DOI] [PubMed] [Google Scholar]
- [122].Martino TA, Young ME, RE: Introduction to Special Issue. On the clock. Ray and Travis 986–987, Science (E-letter February 12, 2017). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.