Abstract
In spliceosome assembly, the 5′ splice site is initially recognized by U1 snRNA. U1 leaves the spliceosome during the assembly process, therefore other factors contribute to the maintenance of 5′ splice site identity as it is loaded into the catalytic site. Recent structural data suggest that human tri-snRNP 27K (SNRP27) M141 and SNU66 H734 interact to stabilize the U4/U6 quasi-pseudo knot at the base of the U6 snRNA ACAGAGA box in pre-B complex. Previously, we found that mutations in Caenorhabditis elegans at SNRP-27 M141 promote changes in alternative 5′ss usage. We tested whether the potential interaction between SNRP-27 M141 and SNU-66 H765 (the C. elegans equivalent position to human SNU66 H734) contributes to maintaining 5′ splice site identity during spliceosome assembly. We find that SNU-66 H765 mutants promote alternative 5′ splice site usage. Many of the alternative 5′ splicing events affected by SNU-66(H765G) overlap with those affected SNRP-27(M141T). Double mutants of snrp-27(M141T) and snu-66(H765G) are homozygous lethal. We hypothesize that mutations at either SNRP-27 M141 or SNU-66 H765 allow the spliceosome to load alternative 5′ splice sites into the active site. Tests with mutant U1 snRNA and swapped 5′ splice sites indicate that the ability of SNRP-27 M141 and SNU-66 H765 mutants to affect a particular 5′ splice alternative splicing event is dependent on both the presence of a weaker consensus 5′ss nearby and potentially nearby splicing factor binding sites. Our findings confirm a new role for the C terminus of SNU-66 in maintenance of 5′ splice site identity during spliceosome assembly.
Keywords: C. elegans, SNRNP27, alternative splicing, snu66, spliceosome
INTRODUCTION
In order for accurate precursor messenger RNA (pre-mRNA) splicing to occur, it is essential that the correct 5′ splice site (5′ss) is identified and maintained during spliceosome assembly and activation (Roca et al. 2008). However, the mechanism of how 5′ss identity is maintained, and the splicing factors involved in this process, are still not well understood. Early in spliceosome assembly, 5′ss recognition occurs via base-pairing with the U1 snRNA, and the branchpoint is recognized by base-pairing with the U2 snRNA. The U4/U6.U5 tri-snRNP is then recruited, forming the pre-B complex (Wilkinson et al. 2020). U4 snRNA base pairs with U6 snRNA to maintain its inactive state; as this interaction is dissociated, the 5′ss is transferred from U1 snRNA to the ACAGAGA box of U6 snRNA, and the 5′ exon associates with loop 1 of U5 snRNA (Staley and Guthrie 1999). Next, U6 snRNA refolds and pairs with U2 snRNA forming the catalytic center, while U4 and U1 are removed from the spliceosome (Fica 2020). This marks the formation of the B complex and the initiation of spliceosome activation (Will and Luhrmann 2011).
Previously, we demonstrated a role for the Caenorhabditis elegans homolog of the tri-SNRP-specific SNRNP27K protein in 5′ss maintenance during spliceosome assembly. Using a genetic screen designed to identify suppressors of cryptic 5′ss usage, we identified a dominant single point mutation, M141T, in the highly conserved C-terminal region of SNRP-27 as a potent suppressor (Dassah et al. 2009). CRISPR-cas9-mediated randomization at amino acid at position 141 revealed that eight amino acid substitutions were viable, three were homozygous lethal, and that viable substitutions at M141 led to changes in alternative 5′ss usage (Zahler et al. 2018). High-throughput mRNA sequencing (mRNA-seq) determined that the M141T mutation specifically changed the alternative 5′ss choice of 26 native genes. Further experiments showed that SNRP-27 is an essential protein (Zahler et al. 2018). Since the tri-snRNP enters the spliceosome after U1 has identified the 5′ss, these results were consistent with the hypothesis that SNRP-27 is involved in maintaining the identity of the 5′ss identified by U1 snRNA. However, the mechanism of action of SNRP-27 and its functional interactions during the maintenance of the 5′ss remained elusive.
In 2019, the Nagai lab published cryo-EM structural models of the human tri-snRNP complex, and the human pre-B complex before U1 snRNP dissociation (Charenton et al. 2019). This was the first study to model SNRNP27K into spliceosomal structures. In these cryo-EM structures, the quasi-pseudoknot composed of nucleotides 63–67 of U4 snRNA anchors one end of the unstructured U6 ACAGAGA sequence loop (Fig. 1). SNRNP27K M141 is sandwiched between H1580 of PRP8 and H734 of SNU66, which in turn stacks on the quasi-pseudoknot. The residues around SNRP-27 M141 appear to be part of a core that ties together multiple parts of the complex and stabilizes the U4/U6 quasi-pseudoknot. The structural model suggests a role for SNRNP27 and SNU66 in helping the U6 snRNA ACAGAGA loop stay in the correct position as it accepts the 5′ss from the U1 snRNA. In the human pre-B structure, the distance from SNU66 H734 to the first base of the intron paired with U1 snRNA is 63.42 Å, indicating that the 5′ss region is distinct from the U6 ACAGAGA region at this stage of assembly (Fig. 1).
FIGURE 1.
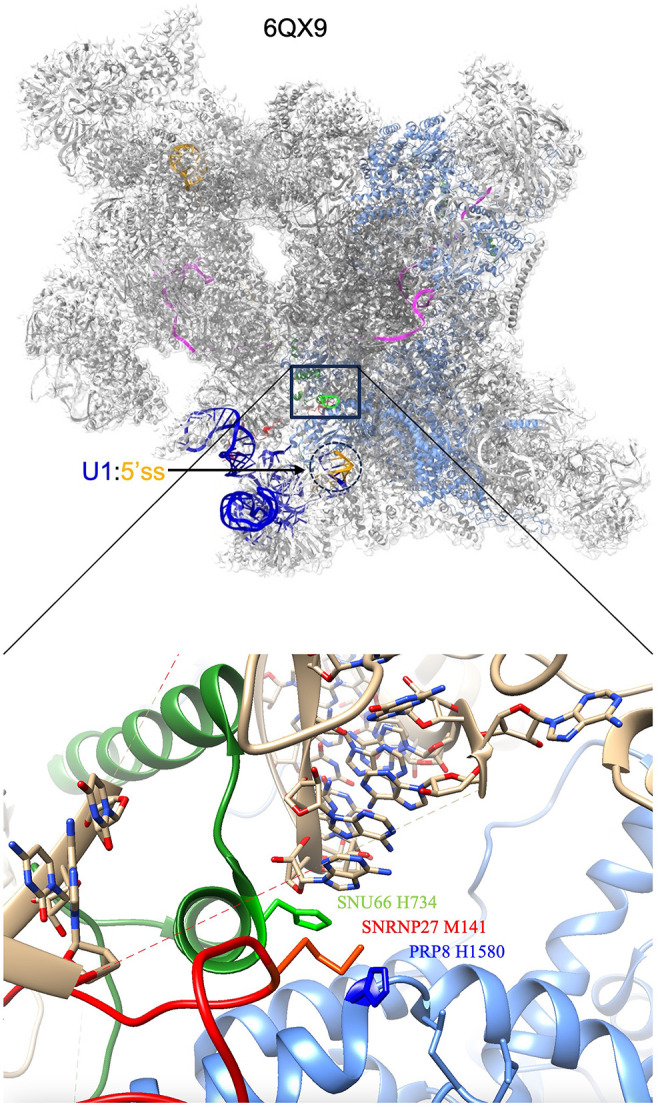
The human pre-B spliceosome and interaction of SNU66 and SNRP27. Coordinates are of the human pre-B complex 6QX9 (Charenton et al. 2019) from the Protein Data Bank (PDB) as rendered in ChimeraX. The top image is the pre-B spliceosome and the bottom is a zoomed-in area in the same orientation showing the SNU66 and SNRP27 interaction. In the top image, PRP8 is in cornflower blue while U1 snRNA is dark blue and the pre-mRNA is orange. The U1 snRNA:5′ss helix is circled with a dotted line. In the bottom image, SNU66 is in green, SNRP27 is in red and PRP8 is in cornflower blue. The key interacting amino acids are labeled. The U4/U6 quasi-pseudoknot is shown stacking above SNU66 H734. The unstructured ACAGAGA region of U6 is indicated by the dashed line in the image.
SNU66 is also known as SART1 and was identified in human cells as an antigen recognized by cytotoxic T lymphocytes and named SART1 (squamous cell carcinoma antigen recognized by T cells 1) (Shichijo et al. 1998). The SART1 gene encodes two proteins of molecular mass 125 and 43 kDa, respectively. Both of the proteins are involved with the regulation of proliferation and have tumor-rejection antigens, but no published role in splicing. The yeast homolog Snu66 (SNUrp associated) was identified as a 66 kDa protein isolated from U4/U6.U5 tri-snRNP (Stevens and Abelson 1999). In the human B complex, the N-terminal half of SNU66 interacts with UBL5, PRP6, and the switch loop and β-finger motifs of Prp8 (Zhan et al. 2018b). Little was known about the role of SNU66's C-terminal half in splicing before its density was mapped in the pre-B complex (Charenton et al. 2019).
In this study, we set out to validate the SNU66–SNRNP27K interaction proposed by Charenton et al. (2019) and characterize the role of the C terminus of SNU66 in 5′ss maintenance during spliceosome assembly. We use CRISPR mutagenesis in worms to alter SNU66 at H765, the histidine residue modeled to interact with SNRNP27K M141. The snu-66(H765G) mutation is a suppressor of unc-73(e936) locomotion and splicing defects, similar to the snrp-27(M141T) mutation that we previously found in our forward genetic suppressor screen (Dassah et al. 2009). High-throughput mRNA-seq analysis of SNRP-27 and SNU-66 mutant strains indicates that the snu-66 alleles primarily lead to changes in alternative 5′ss usage for scores of events, displaying significant overlap with the alternative 5′ splicing events observed in SNRP-27 mutants. We show that the alternative splicing events promoted by the mutants rely on additional clues in the pre-mRNA in addition to the sequence content and direction of the alternative splice site. Overall, we have demonstrated the importance of the SNU-66:SNRP-27 interaction in maintaining 5′ss identity after initial recognition by U1 snRNA.
RESULTS
Generating new snu-66 H765 mutants
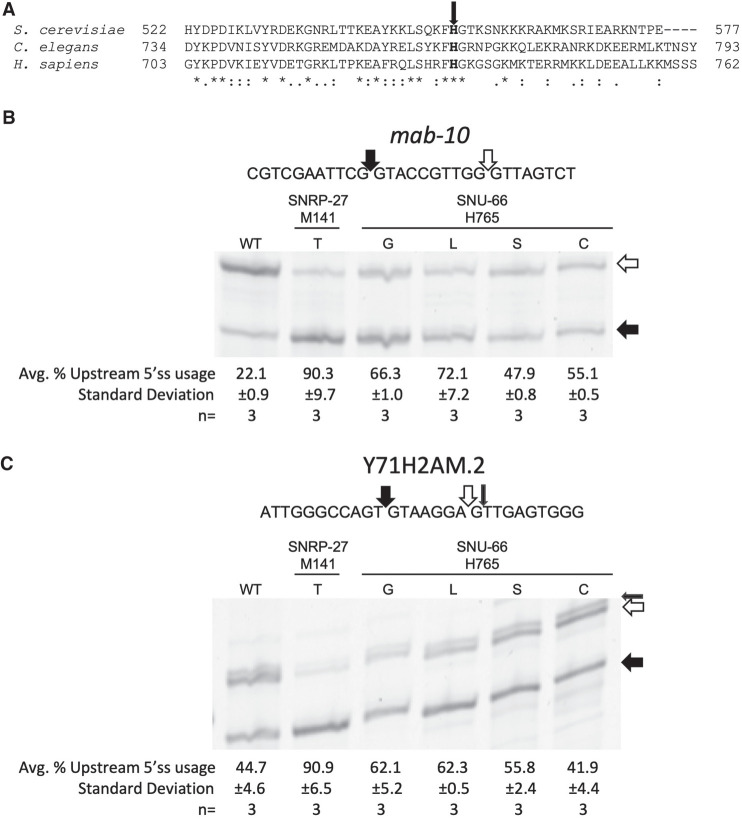
We wanted to test the model that SNU66 H734 (H765 in C. elegans) and PRP8 H1580 (H1573 in C. elegans) functionally interact with M141 of SNRP27 (the M141 position is identical for worms and humans). We hypothesize that interruptions to that interaction would phenocopy the SNRP-27 M141T mutation's impact on 5′ss choice (Zahler et al. 2018). While there is no overall conservation in the C terminus of SNU66, H765 as well as the two flanking amino acids are conserved from Saccharomyces cerevisiae to C. elegans to humans (Fig. 2A), suggesting a functional role for this region of the protein. To interrogate the functionality of the modeled interaction between SNU66, SNRP27, and PRP8, we used a CRISPR–cas9 system coupled with an oligonucleotide-directed recombination repair-based screen to randomize snu-66 at position H765 and prp-8 at position H1573, and screened for viable mutants.
FIGURE 2.
(A) CLUSTAL Omega sequence alignment of the C-terminal region of SNU66 from S. cerevisiae, C. elegans, and human. The bold H with the arrow is the conserved histidine proposed to interact with SNRP-27 M141. (B, C) Cy3 RT-PCR products showing alternative splicing for the alternative 5′ splicing events in mab-10 (B) and Y71H2AM.2 (C) from mixed-stage worm populations. Filled arrows indicate the 5′ss whose usage is known to be promoted by snrp-27(M141T), and the white arrows indicate the 5′ss whose usage is reduced. The small black arrow for Y71H2AM.2 represents the known alternative splice site shown by the doublet on the upper band. The sequences of the alternatively spliced regions are shown above the gels. The percentages that each strain uses the upstream 5′ss with standard deviation (quantified using ImageJ, see Materials and Methods) are shown below the gel images.
We initially recovered nine independent strains, with five different amino acid substitutions at snu-66 position H765; G, L, S, C, and T. These strains grow well and move like wild-type (wt) C. elegans. To test whether these snu-66 H765 mutations affect splicing, we chose to study two of the 26 alternatives 5′ splicing events found to be affected in the snrp-27 M141T mutant; mab-10 and Y71H2AM.2 (Zahler et al. 2018). We performed reverse transcription followed by PCR with cy-3-labeled primers for the five new snu-66 mutant alleles on alternative 5′ splicing events in the mab-10 and Y71H2AM.2 genes, and visualized the PCR products on polyacrylamide gels (Fig. 2B,C). Analysis of the events at mab-10 revealed that the wt strain showed strong usage of the downstream 5′ss, whereas snrp-27(M141T) mutants preferentially used the upstream splice site. The five snu-66 H765 mutant strains all shifted splicing toward the upstream 5′ss, but none as strongly as the snrp-27(M141T). Note that we are not showing the H765T in either experiment in Figure 2B, but it behaved similarly to H765S which is shown. Wild-type worms use the two alternative 5′ss of Y71H2AM.2 with an equal propensity, whereas the snrp-27(M141T) mutant almost exclusively uses the upstream splice site (Fig. 2C). None of the snu-66 H765 mutants preferred the upstream splice site to the extent of snrp-27(M141T); however, snu-66 H765G and H765L promoted the strongest increase in usage of the upstream splice site. The alteration in 5′ss choice due to mutations at snu-66 H765 bolsters the hypothesis that SNU-66 H765 interacting with SNRP-27 M141 is required for proper 5′ss usage.
Mutagenesis of PRP-8 at H1573 yielded five new alleles. Strikingly, four of these H1573 substitution alleles (C, F, S, N) are homozygous lethal yet viable as heterozygotes. The only homozygous viable allele at H1573, H1573R, does not show a change in alternative 5′ splicing for the mab-10 and Y71H2AM.2 events (data not shown). The homozygous lethal disruptions in PRP-8 H1573 may stem from this region of PRP-8 having many roles throughout the splicing cycle (Haselbach et al. 2018; Zhan et al. 2018a, b), as opposed to SNRP-27 and SNU-66 which exit in the transition from pre-B to B complex. The lethal nature of the majority of the substitutions recovered at PRP-8 H1573 impedes our ability to investigate the significance of its interaction with SNRP-27.
To study the combined genetic effects of the snrp-27(M141T) and snu-66 (H765G) we attempted to create a snrp-27(M141T)I; snu-66(H765G)V double mutant strain. Worms were viable if homozygous for one allele and heterozygous for the other. However, we determined that the double homozygous mutation was lethal. This synthetic lethality is consistent with a hypothesis that either mutation weakens the SNRP-27:SNU-66 interaction, and that the double mutant eliminates the interaction. The sensitivity of the disruption of this interaction is consistent with our previous finding that SNRP-27 M141G, F, and E mutant alleles are homozygous lethal, along with our finding that snrp-27 is an essential gene in worms (Zahler et al. 2018).
Alternative splicing analysis of high-throughput mRNA-sequencing of SNRP-27 M141T, SNU-66 H765G, and H765L strains
We wanted to determine if the two strongest alleles of snu-66 (based on Fig. 2B,C), H765G and H765L, lead to global changes in splicing on native genes and compare those results to snrp-27(M141T). We compared high-throughput mRNA-sequencing results of the three mutant strains with wt. Synchronized L3 animals were used in these experiments to control against splicing changes due to different developmental states that might be present in mixed populations (Suzuki et al. 2022). RNA from three biologically independent RNA isolations from each strain was used to prepare libraries for high-throughput sequencing. One hundred and fifty base pairs of paired-end reads from the libraries were mapped to the C. elegans genome using STAR (Dobin et al. 2013). Alternative 5′ss (A5) and alternative 3′ss (A3) events were identified de novo across all the libraries, and the percent spliced in (PSI) for each event in each library was calculated. We also examined Ensembl archived (ensArch65) retained intron, skipped exon, multiple skipped exon, mutually exclusive exon, alternative first exon, and alternative last exon events and determined the PSI for each of these in each library. We then determined ΔPSI in pairwise comparisons for each event in a mutant library versus N2 (wt) strain libraries. Nine pairwise comparisons (three mutant libraries vs. three N2 libraries) were done for each event in each mutant strain. Alternative splicing events that showed ≥15% ΔPSI in all nine pairwise comparisons (pairSum = 9) and ≥20% average ΔPSI were investigated further. Sequencing reads for individual pairSum = 9 events were examined on the UCSC Genome Browser to screen against falsely predicted events.
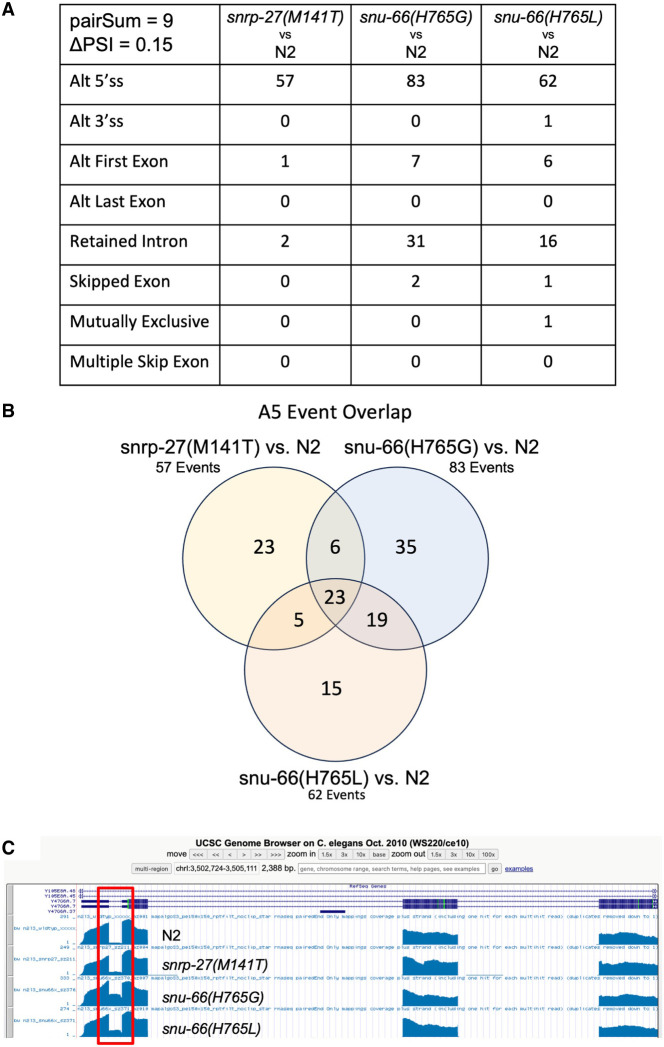
The predominant change in alternative splicing that we found in the three mutant strains was in alternative 5′ss usage. We found 57 alternative 5′ss events for snrp-27(M141T), 83 alternative 5′ss events for snu-66(H765G), and 62 alternative 5′ss events for snu-66(H765L) that met these stringent criteria (Fig. 3A). We compared these 5′ss events and found 23 were common all three mutant strain experiments (Fig. 3B). We performed a Fisher's exact test from the R software package for statistical computing, to determine the significance of the overlap in A5 splicing events identified in the three different mutant strains. We identified a total of 4375 A5 events de novo from these libraries, and these A5 events were used to search for alternative splicing changes in the mutant strains (see Materials and Methods). A P-value < 2.2×10−16 was obtained for the A5 event overlap in all three pairwise comparisons between the mutant strains, indicating that the overlap in splicing events shown in Figure 3B is indeed significant. The alternative events from Figure 3 are listed in Supplemental Table S1. It should be noted that this experiment increases the number of previously identified snrp-27(M141T) native alternative splicing targets from 26 to 57, likely due to a much greater depth of sequencing than the previous study (Zahler et al. 2018).
FIGURE 3.
(A) Table showing the number of alternative splicing events of each class found for each strain relative to wt in larval stage L3 synchronized animals. Each event must have a minimum of 15 junction spanning reads in each library, a ΔPSI > 0.15 for all nine pairwise comparisons, and a mean ΔPSI > 0.20 for the nine pairwise comparisons. (B) Venn diagram showing the overlapping and unique alternative 5′ splicing events identified for the three mutant strains. (C) UCSC Genome Browser screenshot showing the first four exons and first three introns of gene Y47G6A.7. Coverage tracks for one library for each of the indicated strains are shown. The first intron, subject to intron retention (IR) alternative splicing, is boxed in red.
We applied de novo alternative 5′ss discovery for our mRNA-seq analysis, enabling us to determine how many of the A5 events that change in the mutant strains represent cryptic splicing versus alternative splicing. We looked for junctions that were found specifically in mutant but not wt libraries, which would be evidence of activation of cryptic splicing as opposed to a change in the ratio of an expected alternative splicing event. The number of events representing cryptic splicing was small, with only 3/57 events for snrp-27(M141T), 8/83 for snu-66(H765G), and 4/62 for snu-66(H765L) in which the alternative 5′ss promoted by the mutant was not detected in wt L3 animals (11 unique events). Therefore, >90% of the A5 events affected by the mutants are native alternative 5′ splicing events.
Another prominent category of alternative splicing events revealed by our mRNA seq analysis was IR, an alternative splicing event where introns can be retained in mature mRNAs instead of being spliced out. Interestingly, retained introns are known to play a role in the regulation of gene expression, and IR is associated with Alzheimer's disease and cancer (Zheng et al. 2020). The snu-66(H765G) and snu-66(H765L) alleles have 35 and 16 IR events, respectively, that met our stringent criteria; snrp-27(M141T) only led to two examples of retained introns (Fig. 3A). In looking at the alignment of RNA-seq data to the C. elegans genome on the UCSC Genome Browser (Kent et al. 2002), it was clear that IR in these genes is specific to the known IR event; the other introns in the same gene showed complete and efficient removal in the wt and mutant strains. An example of this is shown in Figure 3C for the gene Y47G6A.7. The first intron (red box) has 5% IR in wt but rises to 40% IR in the snu-66(H765G) mutant strain (average ΔPSI = 34.4%). All other introns in Y47G6A.7 (introns 2 and 3 are also shown in Fig. 3C) show highly efficient intron removal. Therefore, the snu-66 H765G and H765L alleles are triggering a known IR event, as opposed to an overall decrease in splicing efficiency, indicating that SNU-66 H765 may normally have a specific role in promoting the splicing of these introns.
We confirmed several of the new alternative 5′ splicing events from the snu-66 mutant sequencing data experimentally. The events for validation were chosen based on a high ΔPSI for snu-66 mutants, and included alternative 5′ss that were either upstream or downstream from the splice site used predominantly in the wt strain. Three 5′ splicing events were chosen for analysis by reverse transcription followed by PCR with cy-3 5′-labeled primers, and visualized by gel electrophoresis. These assays showed high concordance of the alternative 5′ splicing changes found through the high-throughput sequencing analysis (Fig. 4).
FIGURE 4.
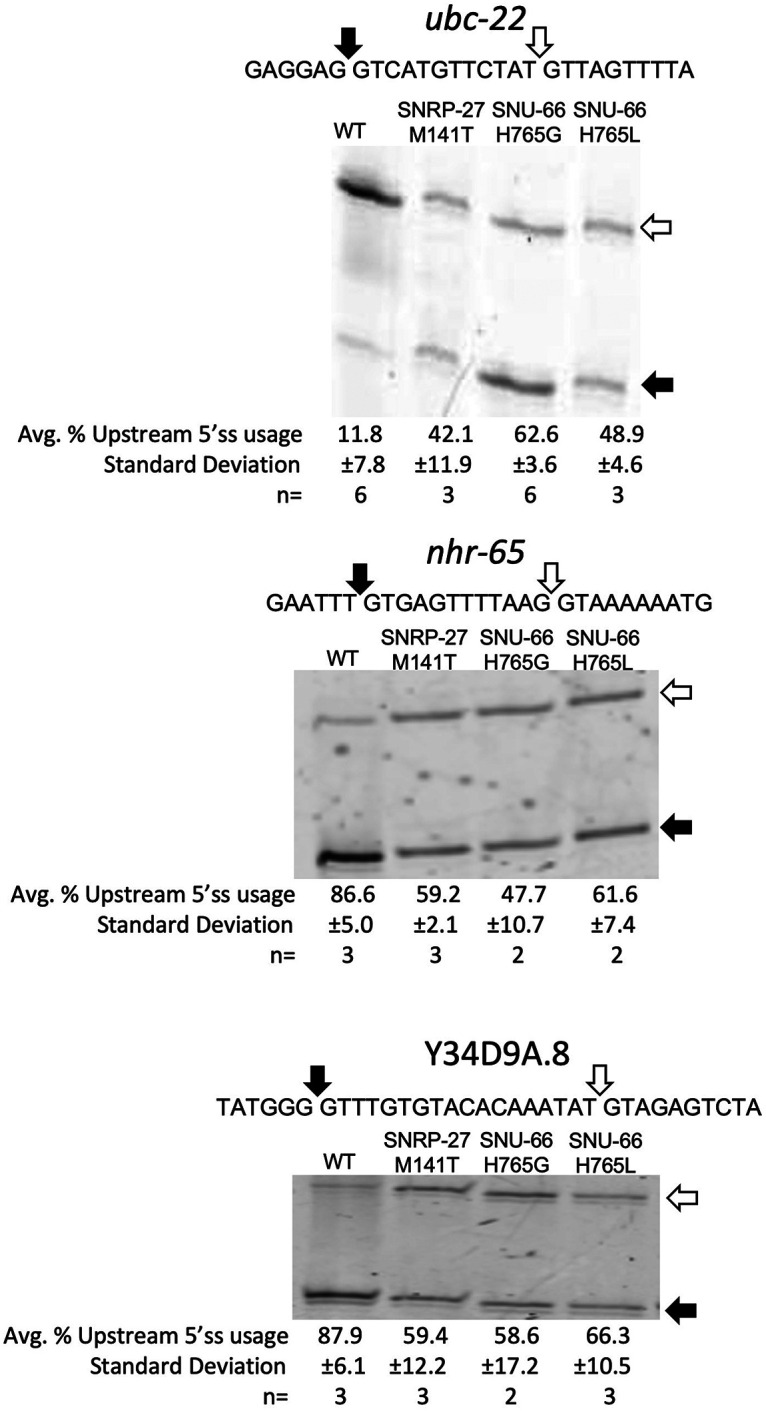
Confirmation of selected alternative 5′ss splicing events identified through mRNA-seq analysis. Events were detected by Cy3 RT-PCR followed by separation on 6% polyacrylamide denaturing gels. The sequences of each alternative splicing event and the location of splice sites are indicated above the gels. Quantification of gel band intensity to determine relative alternative splice site usage is indicated below the gels.
Analyzing the alternative 5′ splicing events identified by mRNA-seq
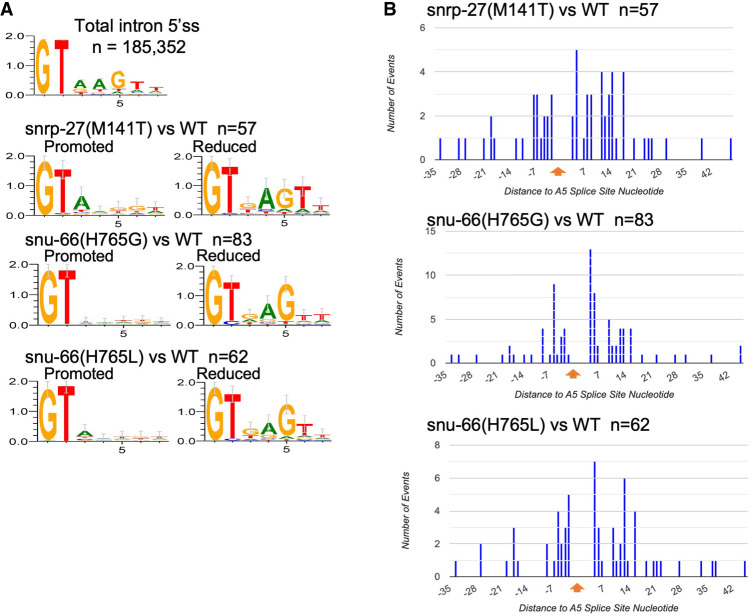
The consensus sequence for the first seven nucleotides of the 5′ss intronic regions for C. elegans is GTAAGTT (Fig. 5A). This sequence interacts by base-pairing with the U1 snRNA during the E complex. The consensus sequence is then handed off to the U6 snRNA in the formation of the B complex. We analyzed the sequence context of the 5′ss for the alternative isoforms (Fig. 5A). In the pairs of alternative 5′ss, we refer to the one whose usage is increased in the presence of the mutant allele relative to wt as “promoted” in the mutant strain, and the other member of the pair whose usage is reduced in the mutant strain relative to wt as “reduced” in the mutant strain. Aside from the GT at the +1 and +2 positions of the intron, the 5′ss sequences promoted in the snrp-27 and snu-66 mutants have no consensus, with the exception of the A at the +3 position for snrp-27(M141T), which we observed previously (Zahler et al. 2018). The 5′ss promoted in all three mutant strains are less conserved relative to total introns and relative to the 5′ss whose usage is reduced (Fig. 5A). This indicates that, outside of the GT dinucleotide to start the intron, the mutations in SNRP-27 and SNU-66 that we tested allow the spliceosome to be less stringent when selecting an alternative 5′ss sequence. 5′ss sequences whose usage is reduced in these strains tended to exclude A residues at the +3 position, with the other positions in agreement with the consensus 5′ss (AGT at positions +4, +5, and +6, respectively). Our previous studies also saw the exclusion of A residues at the +3 position of alternative 5′ss reduced in SNRP-27 M141T (Zahler et al. 2018).
FIGURE 5.
(A) Pictograms of the consensus sequence for the first seven nucleotides of the introns. The top is for 185,352 wt C. elegans introns. Below that, arranged in pairs, are the intron starts for the alternative 5′ss whose usage is promoted or reduced for the SNU-66 and SNRP-27 mutant strains. Strains and number of events analyzed are listed. Sequence logos were generated using WebLogo (Crooks et al. 2004). (B) Histogram showing the distance of alternative 5′ss whose usage is increased in each mutant strain relative to the position of the alternative 5′ss whose usage is decreased.
We next examined the distance in nucleotides from the alternative 5′ss whose usage is reduced in the mutant strains to the alternative splice site promoted in the mutant strains. These are displayed as histograms in Figure 5B. The site whose usage is reduced in the mutants is set at 0 nt. Alternative 5′ss promoted by the mutants are then recorded in the histogram based on distance; mutant-promoted splice sites upstream are to the left of 0 and mutant-promoted splice sites downstream are shown to the right of 0. The number of nucleotides from the wt splice site ranged from −34 to +45, with a roughly even distribution of upstream and downstream sites for all three mutants. These results suggest that the promotion of alternative 5′ss by the snrp-27 and snu-66 alleles is not directional, but that these changes are dependent on the presence of a suboptimal alternative 5′ss within a reasonable distance, ≤33 nt upstream or ≤45 nt downstream.
Exploring the potential mechanism for the SNRP-27 M141T, SNU-66 H765G, and H765L mutants on 5′ splice site choice
In studying the SNU-66 H765 mutants’ effects on global splicing and its similarity to the SNRP-27(M141T) effects, it became clear that SNU-66 H765 is important for 5′ss maintenance. SNRP-27 and SNU-66 enter the spliceosome with the tri-snRNP, after U1 snRNA has bound to the 5′ss. The cryo-EM structure shows interactions between SNRP27 M141 and SNU66 H765 that stabilize the U4/U6 quasi-pseudo knot (Charenton et al. 2019). One hypothesis to explain the alternative 5′ss usage in the presence of these mutations is that these SNRP-27:SNU-66:quasi-pseudoknot interactions are keeping the U6 ACAGAGA box sequestered from interacting with an alternative 5′ss while U1 snRNA is bound to the initial 5′ss. Disrupting the stacking interactions through mutating these amino acids could potentially allow the ACAGAGA box loop to prematurely interact with the pre-mRNA while the U1 snRNA is still bound to the 5′ss. This could allow the U6 ACAGAGA box to interact with an alternative 5′ss while the correct one is occupied by U1 snRNA.
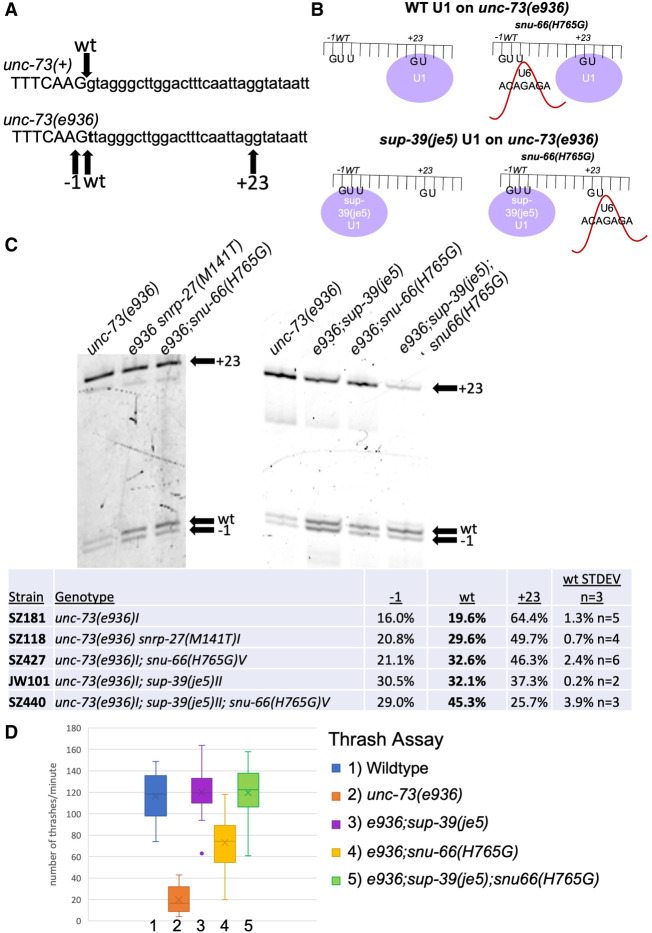
Previously, we identified one unc-73(e936) suppressor, sup-39(je5), which is one of the 12 U1 snRNA genes. This suppressor U1 snRNA has a compensatory mutation that allows for U1 base-pairing with introns beginning with UU instead of the canonical GU (Zahler et al. 2004). It causes the spliceosome to prefer the unc-73(e936) wt cryptic site beginning with UU, leading to increases in splicing at the −1 and wt sites, and a decrease in +23 splice site usage (Fig. 6A). If snu-66 mutant alleles are promoting alternative 5′ss usage by promoting a different 5′ss than is occupied by U1 snRNA independent of directionality, we hypothesized that changing U1 snRNA 5′ss occupation with the mutant sup-39 allele would lead to the snu-66 mutant allele promoting splicing in the opposite direction. This hypothesis is represented in cartoon form in Figure 6B.
FIGURE 6.
(A) Sequences of the wt (top) and e936 mutant allele (bottom) at the start of intron 16 of the unc-73 gene. The g → t mutation is indicated for e936 in bold. The arrow above points to the wt 5′ss used in unc-73(+), and the arrows below point to the −1, wt, and +23 cryptic 5′ss activated in the unc-73(e936) mutant. (B) Cartoon representation of the hypothesis we are testing of premature U6 interaction with a weak 5′ss while U1 snRNA is still bound to a nearby 5′ss. The top part shows binding of U1 snRNA preferentially to the unc-73(e936) + 23 cryptic 5′ss on the left, and on the right side, we propose premature binding of U6 ACAGAGA region to the upstream cryptic splice sites in the presence of the snu-66(H765G) mutant. The bottom shows the U1 mutant sup-39(je5) which promotes splicing at introns that begin with UU occupying the upstream cryptic 5′ss. On the bottom right is a hypothetical outcome of splicing under these conditions, of U6 ACAGAGA interacting with the downstream 5′ss when mutant U1 occupies the upstream 5′ss. (C) Cryptic splicing of the unc-73 gene was measured using cy3 RT-PCR and separation on 6% denaturing polyacrylamide gels. The strains tested are indicated at the top and the +23, wt, and −1 splice sites are indicated on the right side of each part. Quantitation of the relative usage of the three cryptic splice sites is shown below the gel images. (D) Box and whisker chart representing the thrash test data to demonstrate phenotypic suppression of unc-73(e936) uncoordination. The y-axis represents the average number of times a worm bent across their body axis in 1 min; 20 L4 worms were assayed for each strain.
To test this hypothesis, we did crosses to generate new strains so that we could test unc-73(e936) cryptic splicing and movement changes for snu-66(H765G). The e936 allele of unc-73 has a G → T point mutation at the beginning of intron 16. This leads to the activation of three cryptic splice sites, +23 into the intron, −1 upstream into the exon (the G → T mutation leads to a new GU dinucleotide). Importantly, splicing is detected at the wt splice site, even though that intron now begins with UU. Usage of the +23 and −1 cryptic sites lead to out-of-frame messages, and we found identified extragenic suppressors that increase the relative usage of the wt splice site (even though the mutant intron begins with UU), lead to improved locomotion (Roller et al. 2000; Dassah et al. 2009; Mayerle et al. 2019; Cartwright-Acar et al. 2022; Suzuki et al. 2022). Even though use of the +23 and −1 cryptic sites leads to messages that are out-of-frame, transcripts that use these sites are not subject to nonsense-mediated decay. This indicates that the relative splice site usage seen in this assay is consistent with the output of the spliceosome (Roller et al. 2000). To test whether snu-66(H765G) changes cryptic splicing of e936, we performed reverse transcription followed by cy-3 labeled PCR and ran the products on a denaturing polyacrylamide gel with single nucleotide resolution (Fig. 6C). In an almost identical way to snrp-27(M141T), the snu-66(H765G) allele drove splicing away from the +23 and toward the −1 and wt splice site. The increase from 19.6% in-frame splicing in the unsuppressed strain to 32.6% splicing in the snu-66(H765G) suppressor strain is consistent with snu-66(H765G) being a suppressor of the locomotion defect of e936. This was confirmed in a thrash assay for locomotion defect suppression (Fig. 6D). These results indicate that snu-66(H765G) is indeed a locomotion defect suppressor of unc-73(e936) at the level of splicing, phenocopying the mutation in its interacting partner residue snrp-27(M141T). Given that so many of the suppressor alleles have been dominant, we have yet to saturate the unc-73(e936) suppressor screen (Cartwright-Acar et al. 2022). Therefore, it is satisfying that a specific point mutation at a position identified as an SNRP-27(M141T)-interacting residue in cryo-EM studies is a previously undiscovered unc-73(e936) suppressor.
Next we tested strains with sup-39(je5) and a double mutant of sup-39(je5);snu-66(H765G) for e936 suppression. The sup-39(je5) U1 snRNA suppressor increases wt splice site usage in e936 to a level similar to snu-66(H765G), with ∼32% of transcripts being in the frame in both suppressors. The sup-39(je5);snu-66(H765G) double suppressor mutant strain shows an additive effect on unc-73(e936) cryptic splicing to 45.3% of the messages using the in-frame wt site, a stronger change than either suppressor allele alone (Fig. 6C). We also tested the sup-39(je5);snu-66(H765G) double mutant strain for its effect on unc-73(e936) locomotion defect suppression using the thrash assay. The double mutant suppresses unc-73(e936) to the same level as sup-39(je5) alone, with both equivalent to wt (Fig. 6D). If our initial hypothesis was correct that snu-66(H765G) promotes splicing of the alternative 5′ss not occupied by U1 snRNA, with the sup-39 mutant U1 snRNA occupying the wt splice site, we would have expected snu-66(H765G) to promote splicing in the opposite direction of sup-39(je5) instead of the additive effect we observe. This indicates that something in addition to a lack of U1 occupancy at the alternative 5′ss determines the site promoted by the snu-66(H765G) allele.
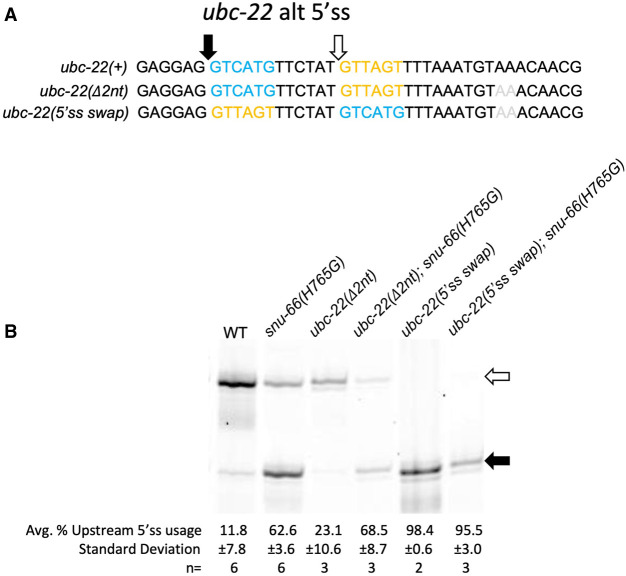
We next asked whether the alternative 5′ss promoted by the snu-66(H765G) allele was based on the sequence of the alternative 5′ss themselves or the relative locations of the splice sites. In the gene ubc-22, there is an alternative 5′ splicing event in which snrp-27(M141T) and snu-66(H765G and H765L) alleles promote an upstream 5′ss (Fig. 4). We designed a new allele of ubc-22 in which the first six intronic bases of the pair of alternative splice sites, separated by 12 nt, were swapped (Fig. 7A). We combined this new ubc-22 allele (along with a control allele with a 2 nt deletion 16 nt downstream from the downstream 5′ss) with the snu-66(H765G) allele and determined the effect on splicing (Fig. 7B). As shown previously, the wt strains almost exclusively chose the downstream splice site in wt ubc-22, and the snu-66(H765G) mutation promoted the upstream splice site. This same effect held true in the 2 nt intronic deletion control allele of ubc-22. In the ubc-22 5′ swap allele in a wt background, the upstream 5′ss was used almost exclusively, and in the presence of snu-66(H765G), there was no switching to the weaker downstream 5′ss. If U1 snRNA occupancy at the stronger consensus alternative 5′ss is what led to promotion of usage of the weaker alternative consensus 5′ss in the presence of the snrp-27 and snu-66 mutants, then we might have predicted the opposite splice site usage in the ubc-22 5′ss swap event in the presence of snu-66(H765G), where U1 is being directed to the stronger consensus upstream splice site. This would have been consistent with our observation that snu-66(H765G) promotes alternative 5′ss usage of native substrates in both directions, depending on the splicing event. However, our results on both unc-73(e936) in the presence of sup-39 (Fig. 6) and the 5′ss swap of ubc-22 (Fig. 7) indicate that there must be a component outside of the 5′ss interaction sequence that also contributes to the identity and direction of the snu-66(H765G)-promoted alternative splicing event.
FIGURE 7.
(A) Sequences for the ubc-22 alternative 5′ splicing event. The top sequence is the wt sequence for ubc-22. The middle sequence shows ubc-22 with the AA dinucleotide deletion, which is a control for the 5′ swap experiment below. The bottom sequence is ubc-22 with the 5′ss sequences swapped and the AA dinucleotide deletion. The blue sequence and black arrow represent the wt upstream splice site, while the yellow sequence and white arrow show the wt downstream sequence. The gray AA are the two nucleotides that were deleted. (B) Cy3 RT-PCR products separated on a 6% denaturing polyacrylamide gel for the alternatively spliced region of ubc-22. The genotype [ubc-22 mutant status and snu-66(H765G) mutant status] is shown above each lane. Quantitation is shown below the gel image.
DISCUSSION
SNRNP27K was initially identified as a component of purified tri-snRNPs that could undergo phosphorylation in its N-terminal RS domain (Fetzer et al. 1997). Our genetic study identified the dominant mutation M141T that suppresses cryptic splicing and changes 5′ss use in C. elegans for a number of native alternative 5′ splicing events (Dassah et al. 2009; Zahler et al. 2018). Our results indicate an essential role of the highly conserved C terminus of SNRP-27, but it was still a poorly understood splicing factor. Subsequently, the cryo-EM structure of the human tri-snRNP and pre-B complex gave a hint to possible SNRP-27-protein interactions, and to when in the splicing cycle it performs its role (Charenton et al. 2019). Human SNRP-27 M141 was modeled to interact with H734 of SNU66 and H1580 of PRP8 and to stabilize the U4/U6 quasi-pseudoknot, positioning the ACAGAGA loop correctly before accepting the 5′splice site from the U1 snRNA. SNU66 was known in splicing as a component of U4/U6.U5 tri-snRNP, with its N-terminal half having a role during B complex formation by stabilizing PRP8 (Zhan et al. 2018b). The potential of a SNU66 residue interacting with SNRP-27 M141 and the quasi-pseudoknot indicated that the previously unstudied SNU66 C-terminal half could have a role with SNRP-27 in maintaining 5′ss identity during the handoff from U1 snRNA to U6 snRNA.
The activation of the pre-B to B complex is initiated by the BRR2 helicase binding to U4 snRNA to unwind it from U6. SNU66 has been modeled to have extensive contact with BRR2 in S. cerevisiae tri-snRNP spliceosomes, wrapping around the BRR2 C-terminal cassette (Nguyen et al. 2016). This interaction has been proposed to negatively regulate BRR2 unwinding of U4/U6 (Ulrich et al. 2016). In contrast, for human tri-snRNP and pre-B complexes, only a few central amino acids of SNU66 interact with BRR2, and these cannot explain BRR2 regulation (Charenton et al. 2019). Instead, BRR2 activation is proposed to be downstream from 5′ss binding to U6, which would destabilize SNU66 and SNRP27 and liberate the PRP8 RNaseH and Endo domains, allowing BRR2 to be activated and bind to U4 for U6 unwinding (Charenton et al. 2019). Our studies have looked at the role of SNRP-27 and SNU66 interaction in maintaining the identity of the 5′ss after U1 snRNA recognition. Given that its metazoan interacting partner SNRP-27 is not found in S. cerevisiae, it is perhaps not surprising that SNU66 may have a different role in regulating splicing steps in higher organisms.
Based on modeling from the structural data of the human pre-B complex (Charenton et al. 2019), we further studied SNU66 and determined that H734 has a role in splicing. Analysis of the C. elegans SNU-66(H765G) and (H765L) mutants by mRNA-seq showed that alternative 5′ss promoted by the mutation are a weaker match to the 5′ss consensus sequence than the splice sites in the alternative pair whose usage is reduced (Fig. 5A). This suggests that mutations at this position in SNU-66 disrupt the transfer of the optimal 5′ss from U1 snRNA to U6 snRNA, and promotes usage of weaker nearby sites, similar to the SNRP-27(M141T) mutation. Mutating this SNU-66 position also suppresses the unc-73(e936) phenotype (Fig. 6), meaning that the mutant spliceosome is more likely to splice at the noncanonical UU 5′ss. Our data suggest that snu-66(H765) mutations allow the spliceosome to splice at positions with less stringent 5′ss consensus sequences.
The results also suggest that SNU-66 and SNRP-27 work together and in additive ways to maintain the 5′ss. The alternative 5′ splicing events found through mRNA-seq for the snu-66 and snrp-27 mutants saw significant overlaps of the events shared between them (Fig. 3B). Although snu-66(H765G) and snrp-27(M141T) C. elegans strains are viable with no significant phenotypical changes, the double mutant is lethal. Perhaps each mutant destabilizes the protein–protein interaction between SNRP-27 and SNU-66 slightly, leading to similar alternative splicing changes, while the double mutant abolishes the interaction leading to lethality, consistent with certain alleles of snrp-27(M141) mutations being homozygous lethal and the essential nature of snrp-27 for viability (Zahler et al. 2018).
The alternative 5′ss promoted in the mutant strains could be upstream or downstream from the 5′ss whose usage is reduced, depending on the splicing event (Fig. 5B). This relatively even distribution demonstrates that the mutations do not have a specific directional effect on alternative 5′ss choice across all events. When we swapped the sequence of the ubc-22 alternative 5′ss, we switched splicing specificity in a wt SNU-66 strain, consistent with wt splicing going to the stronger consensus sequence 5′ss. However, with the 5′ss sequence switch, we no longer saw a change in splicing to the other 5′ss in the presence of snu-66(H765G) (Fig. 7). This seemed to contradict the possibility that 5′ss preference is due to activation of the alternative site while U1 was still bound to the stronger 5′ss. This hypothesis was further explored using a U1 snRNA mutant that promoted a 5′ss starting with UU (Fig. 6). In that case, the unc-73(e936);sup-39(je5);snu-66(H765G) triple mutant saw the strongest change from the +23 splice site to the overlapping −1 and wt 5′ss in unc-73(e936) cryptic splicing, even though the sup-39(je5) allele led U1 snRNA to prefer 5′ss beginning with UU. Ultimately, our experiments suggest that the ability of SNU-66 and SNRP-27 mutants to alter 5′ss choice depends on additional components of the pre-mRNA than just the presence of a 5′ss not occupied by U1 snRNA in the region of the initially assembled 5′ss.
When U6 snRNA binds the 5′ss, a Watson–Crick base pair is made between the 5′ss +4 U and the central adenosine of the U6 ACAGAGA box (in bold). If this adenosine is methylated at the N6 position (m6A) by METTL16, 5′ss preferences for the +4 position of the intron become more flexible (Parker et al. 2023). A recent comparative genomics study found SNRP-27 may be necessary in genomes where U6 snRNA is m6A-modified, because its presence is evolutionarily correlated to more variance of the 5′ss +4 position when the U6 snRNA m6A modification is present (note that SNRP-27 is not found in the S. cerevisiae genome) (Parker et al. 2023). In addition, they hypothesize that the SNRP-27 M141 location, close to the U6 snRNA m6A, implies that SNRP-27 may stabilize the U6/5′ss interaction by chaperoning the central A in the U6 ACAGAGA sequence for 5′ss docking. This is consistent with our findings that SNRP-27(M141T) and SNU-66(H765G/L) cause a shift in 5′ splice usage toward 5′ss with less stringency at the +4 position and away from the +4A (Fig. 5A).
SNRP-27 has an N-terminal region strikingly rich in alternating phosphoserines and arginines, known as an RS domain (Fetzer et al. 1997). These phosphoserine/arginine domains are found at the C termini of the SR protein family splicing factors, and proteins with these domains have a significant role in the recruitment of spliceosomal components throughout the splicing cycle, especially in the early stages of initial spliceosome recruitment of U1 snRNA to the pre-mRNA (Zahler et al. 1992; Zahler and Roth 1995). The U1-70K protein component of U1 snRNP also has an RS domain. These RS domains may serve as phospho-regulated interaction domains (Shepard and Hertel 2009; Howard and Sanford 2015). Given that SNRP-27 has an RS domain, it is possible that it may be recruited to spliceosomes through SR protein interactions independent of its binding to snu-66 in the tri-snRNP. This may explain the directionality independence of 5′ss sequence and U1 snRNA occupation that we see in the ubc-22 5′ss swap and unc-73(e936);sup-39 experiments. The ubc-22 alternative splicing event contains the sequence GAGGAG directly upstream of the 5′ss, and G-A-R sequences, where R is a purine, are known to bind to a subset of SR proteins (Nagel et al. 1998). It is possible that this sequence GAR sequence recruits an SR protein, and its presence at the upstream 5′ss in either version of the ubc-22 swap experiment could contribute to the upstream 5′ss preference of both of the snu-66 mutants. Perhaps the snrp-27(M141T) and snu-66(H765G) alleles allow for alternative 5′ss usage and U6 interaction with alternative splice sites when there are other elements of the pre-mRNA directing components of the spliceosome toward the alternative sites. Studying the role of the RS domain of SNRP-27, and the position of SR protein binding sites near 5′ss at events whose splicing changes in snrp-27 M141 and snu-66 H765 mutants could lead to more insights into the mechanism of the role of these proteins in 5′ss identity maintenance during active site formation. Given the high degree of amino acid identity between human and C. elegans SNRP-27 (amino acids 137–153 that are modeled into Fig. 1 are identical between the species), it is highly likely that the function of the factor in the two species is identical. It is tempting to consider that regulating SNRP-27 activity between cell types may be a way of controlling alternative 5′ splicing, but there is no evidence for that at this time. The phosphorylated RS domain of SNRP-27 activity might potentially be a target of posttranslational regulation of its activity.
MATERIALS AND METHODS
C. elegans strains
C. elegans strains used in this study are indicated in Table 1.
TABLE 1.
Strains used in this study
| Strain name | Genotype | Notes |
|---|---|---|
| JW101 | unc-73(e936)I; sup-39(je5)II | je5 is U1 snRNA e936 suppressor |
| SZ118 | unc-73(e936) snrp-27(az26)I | snrp-27(M141T) e936 suppressor strain |
| SZ181 | unc-73(e936)I | |
| SZ211 | snrp-27(az56)I | snrp-27(M141T) |
| SZ326 | snu-66(az138)V | snu-66(H765S) |
| SZ370 | snu-66(az160)V | snu-66(H765G) |
| SZ371 | snu-66(az161)V | snu-66(H765L) |
| SZ372 | snu-66(az162)V | snu-66(H765C) |
| SZ427 | unc-73(e936)I; snu-66(az160)V | snu-66(H765G) e936 suppressor strain |
| SZ444 | ubc-22(az187)X | ubc-22 alt 5′ss swap |
| SZ445 | unc-73(e936)I; sup-39(je5)II; snu-66(az160)V | sup-39; snu-66(H765G) double e936 suppressor |
| SZ447 | ubc-22(az188)X | ubc-22 2 nt intron deletion control |
| SZ448 | snu-66(az160)V; ubc-22(az187)X | ubc-22 alt 5′ss swap with snu-66(H765G) |
| SZ457 | snu-66(az160)V; ubc-22(az188)X | ubc-22 2 nt intron del. control with snu-66(H765G) |
Thrash assay
A thrash test was performed to measure how well the suppressed worms move in comparison to the uncoordinated mutant unc-73(e936) worms (Run et al. 1996). Live L4 worms were transferred to a drop of M9 solution on an NGM agar plate and observed at 20°C for 60 sec. The number of times that they bent across their body axis was recorded. Twenty L4 animals were assayed for each strain.
CRISPR/Cas9 genomic editing
Cas9 guides were chosen from the CRISPR guide track (Haeussler et al. 2016) on the UCSC Genome Browser (Kent et al. 2002) C. elegans reference assembly (WS220/ce10). crRNAs were synthesized by Integrated DNA Technologies (www.idtdna.com) (Table 2). Cas9/CRISPR RNA guides were assembled with a standard tracrRNA (IDT); these RNAs were heated to 95°C and incubated at room temperature to allow for annealing. The full guides were then incubated with Cas9 protein (UC Berkeley QB3 MacroLab) to allow for the assembly of the Cas9 RNA complex (Paix et al. 2017). That mix, along with a single-stranded homology-directed repair guide oligonucleotide (Table 3) was then micro-injected into the syncytial gonad of young adult hermaphrodite animals. A dpy-10(cn64) co-CRISPR strategy was used to identify F1 animals showing homology-directed CRISPR repair in their genomes (Arribere et al. 2014). Silent restriction sites were incorporated into or deleted from the repair oligo design so that mutations could be easily tracked by restriction digestion of PCR products from DNA extracted from single worms. Injected animals were moved to plates using a recovery buffer, allowed to recover for 4 h, and surviving worms were plated individually. F1 offspring were screened for the dpy-10(cn64) dominant roller (Rol) coinjection marker phenotype. F1 nondumpy roller animals (heterozygous for cn64) were plated individually, allowed to lay eggs, and then the adult was removed and checked for allele of interest by PCR and restriction enzyme digestion followed by gel electrophoresis. PCR primers used for mutational screening are indicated in Table 4. If an F1 worm showed the presence of a heterozygous DNA fragment matching the programmed restriction site, nonroll animals in the F2 progeny (indicating loss of the co-CRISPR marker) were screened by electrophoresis of digested PCR products. Individuals that had lost the coinjection marker but were homozygous for the allele of interest were retained and sequenced at the gene of interest to verify error-free insertion of sequences guided by the repair oligo.
TABLE 2.
CRISPR crRNA guide RNA sequences (entered as DNA into the IDTdna.com crRNA order form)
| Strain | Sequence |
|---|---|
| snu-66 H765 randomized | CAATCCGGGTAAGAAACAGT |
| Prp8 H1573 randomized | TCTTTTGCCACAAGTGAGCA |
| ubc-22 5′ss swap ΔAA and ubc-22 ΔAA | AGTTTTAAATGTAAACAACG |
TABLE 3.
CRISPR repair oligonucleotides
| Strain | Sequence |
|---|---|
| snu-66 H765 randomized | TGAGTGTGTCTAGTGAATTTTGAAATTTACAACTTTTTTTTTTCAGGTTCNNNGGTCGCAATCCGGGTAAGAAACAGTTcGAaAAACGAGCTAATCGTAAGGA |
| Prp8 H1573 randomized | ACGGAAAGATCCCGACGCTCAAGATTTCTCTCATTCAAATaTTtCGTGCTNNNTTGTGGCAAAAGATTCACGAGTCAGTAGTTATGGATCTGTGTCAAGTTTT |
| ubc-22 5′ss swap ΔAA | TATTGAAGATTTATTTCGATCAATCGAGGTAGACGAGGAGGTTAGTTTCTATGTCATGTTTAAATGT∧ACAACGTGGCATTTTCTTTCTAAAAACTTAAGCATTTGACTATTAAAACTGTT |
| ubc-22 ΔAA | TATTGAAGATTTATTTCGATCAATCGAGGTAGACGAGGAGGTCATGTTCTATGTTAGTTTTAAATGT∧ACAACGTGGCATTTTCTTTCTAAAAACTTAAGCATTTGACTATTAAAACTGTT |
NNN indicates that a codon was replaced with randomly incorporated nucleotides at that position. Lower case nucleotides are silent mutations (differ from wt). Underlined are the nucleotides that are mutated. Bold sequences are the restriction enzyme site used. For ubc-22 repair oligos, ∧ indicates the site of a 2 nt (AA) deletion, 15 nt into the downstream intron, leading to the formation of a BsrGI restriction enzyme cleavage site (TGTACA).
TABLE 4.
PCR primers used in screening for CRISPR mutations and tracking alleles
| Strain | Forward primer | Reverse primer |
|---|---|---|
| snrp-27(M141T) | GAGTCGTTACAAAGTGGAGC | TTCGCCATGGTCAAATTCCC |
| snu-66 H765 randomized | GTGGACCAGTTATGCCATTC | CCACTGAGCACAAGATACGG |
| Prp8 H1573 randomized | CTCTGGTGGTCTCCAACTATC | CATAATCTCCCCAACGAAGC |
| ubc-22 5′ss swap | CTCACGGTACGCTGTCATTT | GATGTTCAGTATGTGGGCTTCA |
RNA extraction, cDNA production, and PCR amplification
RNA from indicated strains was extracted from mixed staged or L3 populations of worms using TRIzol (Invitrogen), before phase separation with CHCl3 and isopropanol precipitation. RNA was reverse transcribed with specific primers for each gene using AMV reverse transcriptase (Promega). cDNA was PCR-amplified for 25 cycles with 5′-Cy3-labeled reverse primers (IDT) and unlabeled forward primers using Phusion high-fidelity polymerase (NEB). Table 5 has a list of primers used for the Cy3 RT-PCR experiments. PCR products were separated on 40 cm tall 6% polyacrylamide urea denaturing gels and visualized using a Molecular Dynamics Typhoon Scanner. Band intensity quantitation was performed using ImageJ software (https://imagej.nih.gov/ij/). For quantitation, a box of the same size was drawn around each alternative splicing product for a .gel image in ImageJ, and a control background box of the same size was drawn between products in each lane. The background volume value was subtracted from each band's value within a lane, and then the relative usage of the splice sites was calculated.
TABLE 5.
Oligonucleotide primers for RT-PCR Cy3 experiments
| Event | Forward primer | Cy3 reverse primer |
|---|---|---|
| mab-10 | TTTGAGTCAGCTGTCGTCAG | CAGATGAGGTGGATGATGAC |
| Y71H2AM.2 | ACAAGATCTACATGGTGCCG | ATTTGGATTTCAGCCGGGTG |
| ubc-22 | TTCTTCTGCCAGTCATCGTCT | AATGTTTCGGAGGCACTGTC |
| nhr-65 | TTTCGTGCTCCAGTGTGACC | TACGTGAACCATTTGGTGGC |
| Y34D9A.8 | CAGCGGTTATCGTCGTTGTC | AATGGCGGATTCGCTTCTCTC |
| unc-73 | TCAACCAGAAGCTGTTGGTG | TCCCTTAAAGTAGGCTCGTG |
High-throughput RNA-seq
Total RNA isolations from three biological replicates were done for strains SZ211 [snrp-27(M141T)], SZ370 [snu-66(H765G)], SZ371 [snu-66(H765L)], and N2 (wt). Poly(A) mRNA isolation and sequencing library preparation for each RNA sample was performed by Azenta; 150 nt × 150 nt paired-end reads were obtained. After filtering of repeat and noncoding RNA sequences and duplicate reads, unique reads were two-pass aligned to UCSC Genome Browser C. elegans reference assembly (WS220/ce10) (this earlier assembly release was used to facilitate comparison to previous RNA-seq data sets obtained by our laboratory) using STAR (Dobin et al. 2013). An average of 10.87M distinct 150 × 150 bp reads were mapped to ce10 for each library (range from 8.69 to 11.97M per library).
High stringency ΔPSI analysis
Alternative 5′ (A5) and alternative 3′ (A3) splicing events were identified de novo from the STAR mappings of all the libraries. We identified mapped introns which shared one end in common and one end different, with at least five reads of support (total across all samples) for each alternative variant and a maximum of 50 nt between the alternative ends (either 5′ or 3′, respectively). In addition, alternative first exon (AF), alternative last exon (AL), skipped exon (SE), retained intron (RI), mutually exclusive exon (MX), and multiple skipped exon (MS) events were derived from the Ensembl gene predictions Archive 65 of WS220/ce10 (EnsArch65) using junctionCounts “infer pairwise events” function (https://github.com/ajw2329/junctionCounts). The PSI for each event in each sample was derived using junctionCounts with the option suppress_eij_use for A3 and A5 events. Each strain had three biological replicates; therefore, between any two strains, there are a total of nine possible pairwise comparisons for each event. For each suppressor strain, only alternative splicing events with a minimum of 10 junction counts per library that showed a change in the same direction >15% ΔPSI compared to the N2 wt control in all nine pairwise comparisons (pairSum = 9) were considered further. Those events with a mean ΔPSI > 20% across the nine comparisons were chosen for examination. The reads supporting each alternative splice site choice event were examined individually on the UCSC Genome Browser (Kent et al. 2002) C. elegans reference assembly (WS220/ce10), to ensure that the algorithmically flagged events were real examples of alternative splice site choice.
Consensus motifs
Consensus motifs were created using WebLogo (Crooks et al. 2004); https://weblogo.berkeley.edu/logo.cgi.
Multiple sequence alignments
Multiple sequence alignments were generated using CLUSTAL Omega at the online web interface; https://www.ebi.ac.uk/Tools/msa/clustalo/.
DATA ACCESS
Raw mRNA sequencing data for 12 libraries in fastq format, along with .gtf files for all analyzed alternative splicing events, are available at the NCBI Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/) accession GSE252787.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
The authors would like to thank Kenneth Osterhoudt for many helpful discussions, Catiana Cartwright-Acar for technical assistance, and Martin Kinsu for help with WebLogo analysis and for critical reading of the manuscript. We are grateful to our colleagues Melissa Jurica, Joshua Arribere, and Manny Ares for helpful discussions and comments on the manuscript. This research was funded by a grant from the National Institute of General Medical Sciences, R01GM135221, to A.M.Z. K.S. was a fellow of the UCSC IMSD Training Program, R25GM058903. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.079971.124.
Freely available online through the RNA Open Access option.
REFERENCES
- Arribere JA, Bell RT, Fu BX, Artiles KL, Hartman PS, Fire AZ. 2014. Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics 198: 837–846. 10.1534/genetics.114.169730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright-Acar CH, Osterhoudt K, Suzuki J, Gomez DR, Katzman S, Zahler AM. 2022. A forward genetic screen in C. elegans identifies conserved residues of spliceosomal proteins PRP8 and SNRNP200/BRR2 with a role in maintaining 5′ splice site identity. Nucleic Acids Res 50: 11834–11857. 10.1093/nar/gkac991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charenton C, Wilkinson ME, Nagai K. 2019. Mechanism of 5′ splice site transfer for human spliceosome activation. Science 364: 362–367. 10.1126/science.aax3289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res 14: 1188–1190. 10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassah M, Patzek S, Hunt VM, Medina PE, Zahler AM. 2009. A genetic screen for suppressors of a mutated 5′ splice site identifies factors associated with later steps of spliceosome assembly. Genetics 182: 725–734. 10.1534/genetics.109.103473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fetzer S, Lauber J, Will CL, Luhrmann R. 1997. The U4/U6.U5 tri-snRNP-specific 27K protein is a novel SR protein that can be phosphorylated by the snRNP-associated protein kinase. RNA 3: 344–355. [PMC free article] [PubMed] [Google Scholar]
- Fica SM. 2020. Cryo-EM snapshots of the human spliceosome reveal structural adaptions for splicing regulation. Curr Opin Struct Biol 65: 139–148. 10.1016/j.sbi.2020.06.018 [DOI] [PubMed] [Google Scholar]
- Haeussler M, Schönig K, Eckert H, Eschstruth A, Mianné J, Renaud JB, Schneider-Maunoury S, Shkumatava A, Teboul L, Kent J, et al. 2016. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol 17: 148. 10.1186/s13059-016-1012-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haselbach D, Komarov I, Agafonov DE, Hartmuth K, Graf B, Dybkov O, Urlaub H, Kastner B, Lührmann R, Stark H. 2018. Structure and conformational dynamics of the human spliceosomal Bact complex. Cell 172: 454–464.e11. 10.1016/j.cell.2018.01.010 [DOI] [PubMed] [Google Scholar]
- Howard JM, Sanford JR. 2015. The RNAissance family: SR proteins as multifaceted regulators of gene expression. Wiley Interdiscip Rev RNA 6: 93–110. 10.1002/wrna.1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. 2002. The human genome browser at UCSC. Genome Res 12: 996–1006. 10.1101/gr.229102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayerle M, Yitiz S, Soulette C, Rogel LE, Ramirez A, Ragle JM, Katzman S, Guthrie C, Zahler AM. 2019. Prp8 impacts cryptic but not alternative splicing frequency. Proc Natl Acad Sci 116: 2193–2199. 10.1073/pnas.1819020116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel RJ, Lancaster AM, Zahler AM. 1998. Specific binding of an exonic splicing enhancer by the pre-mRNA splicing factor SRp55. RNA 4: 11–23. [PMC free article] [PubMed] [Google Scholar]
- Nguyen THD, Galej WP, Bai XC, Oubridge C, Newman AJ, Scheres SHW, Nagai K. 2016. Cryo-EM structure of the yeast U4/U6.U5 tri-snRNP at 3.7 Å resolution. Nature 530: 298–302. 10.1038/nature16940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paix A, Folkmann A, Seydoux G. 2017. Precision genome editing using CRISPR-Cas9 and linear repair templates in C. elegans. Methods 121–122: 86–93. 10.1016/j.ymeth.2017.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker MT, Fica SM, Barton GJ, Simpson GG. 2023. Inter-species association mapping links splice site evolution to METTL16 and SNRNP27K. Elife 12: e91997. 10.7554/eLife.91997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca X, Olson AJ, Rao AR, Enerly E, Kristensen VN, Børresen-Dale AL, Andresen BS, Krainer AR, Sachidanandam R. 2008. Features of 5′-splice-site efficiency derived from disease-causing mutations and comparative genomics. Genome Res 18: 77–87. 10.1101/gr.6859308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roller AB, Hoffman DC, Zahler AM. 2000. The allele-specific suppressor sup-39 alters use of cryptic splice sites in Caenorhabditis elegans. Genetics 154: 1169–1179. 10.1093/genetics/154.3.1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Run JQ, Steven R, Hung MS, van Weeghel R, Culotti JG, Way JC. 1996. Suppressors of the unc-73 gene of Caenorhabditis elegans. Genetics 143: 225–236. 10.1093/genetics/143.1.225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard PJ, Hertel KJ. 2009. The SR protein family. Genome Biol 10: 242. 10.1186/gb-2009-10-10-242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shichijo S, Nakao M, Imai Y, Takasu H, Kawamoto M, Niiya F, Yang D, Toh Y, Yamana H, Itoh K. 1998. A gene encoding antigenic peptides of human squamous cell carcinoma recognized by cytotoxic T lymphocytes. J Exp Med 187: 277–288. 10.1084/jem.187.3.277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley JP, Guthrie C. 1999. An RNA switch at the 5′ splice site requires ATP and the DEAD box protein Prp28p. Mol Cell 3: 55–64. 10.1016/S1097-2765(00)80174-4 [DOI] [PubMed] [Google Scholar]
- Stevens SW, Abelson J. 1999. Purification of the yeast U4/U6⋅U5 small nuclear ribonucleoprotein particle and identification of its proteins. Proc Natl Acad Sci 96: 7226–7231. 10.1073/pnas.96.13.7226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki J, Osterhoudt K, Cartwright-Acar CH, Gomez DR, Katzman S, Zahler AM. 2022. A genetic screen in C. elegans reveals roles for KIN17 and PRCC in maintaining 5′ splice site identity. PLoS Genet 18: e1010028. 10.1371/journal.pgen.1010028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich AKC, Seeger M, Schütze T, Bartlick N, Wahl MC. 2016. Scaffolding in the spliceosome via single α helices. Structure 24: 1972–1983. 10.1016/j.str.2016.09.007 [DOI] [PubMed] [Google Scholar]
- Wilkinson ME, Charenton C, Nagai K. 2020. RNA splicing by the spliceosome. Annu Rev Biochem 89: 359–388. 10.1146/annurev-biochem-091719-064225 [DOI] [PubMed] [Google Scholar]
- Will CL, Luhrmann R. 2011. Spliceosome structure and function. Cold Spring Harb Perspect Biol 3: a003707. 10.1101/cshperspect.a003707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahler AM, Roth MB. 1995. Distinct functions of SR proteins in recruitment of U1 small nuclear ribonucleoprotein to alternative 5′ splice sites. Proc Natl Acad Sci 92: 2642–2646. 10.1073/pnas.92.7.2642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahler AM, Lane WS, Stolk JA, Roth MB. 1992. SR proteins: a conserved family of pre-mRNA splicing factors. Genes Dev 6: 837–847. 10.1101/gad.6.5.837 [DOI] [PubMed] [Google Scholar]
- Zahler AM, Tuttle JD, Chisholm AD. 2004. Genetic suppression of intronic +1G mutations by compensatory U1 snRNA changes in Caenorhabditis elegans. Genetics 167: 1689–1696. 10.1534/genetics.104.028746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahler AM, Rogel LE, Glover ML, Yitiz S, Ragle JM, Katzman S. 2018. SNRP-27, the C. elegans homolog of the tri-snRNP 27K protein, has a role in 5′ splice site positioning in the spliceosome. RNA 24: 1314–1325. 10.1261/rna.066878.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan X, Yan C, Zhang X, Lei J, Shi Y. 2018a. Structure of a human catalytic step I spliceosome. Science 359: 537–545. 10.1126/science.aar6401 [DOI] [PubMed] [Google Scholar]
- Zhan X, Yan C, Zhang X, Lei J, Shi Y. 2018b. Structures of the human pre-catalytic spliceosome and its precursor spliceosome. Cell Res 28: 1129–1140. 10.1038/s41422-018-0094-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng JT, Lin CX, Fang ZY, Li HD. 2020. Intron retention as a mode for RNA-seq data analysis. Front Genet 11: 586. 10.3389/fgene.2020.00586 [DOI] [PMC free article] [PubMed] [Google Scholar]