Abstract
The high therapeutic potential of psilocybin, a prodrug of the psychotropic psilocin, holds great promise for the treatment of mental disorders such as therapy‐refractory depression, alcohol use disorder and anorexia nervosa. Psilocybin has been designated a ‘Breakthrough Therapy’ by the US Food and Drug Administration, and therefore a sustainable production process must be established to meet future market demands. Here, we present the development of an in vivo psilocybin production chassis based on repression of l‐tryptophan catabolism. We demonstrate the proof of principle in Saccharomyces cerevisiae expressing the psilocybin biosynthetic genes. Deletion of the two aminotransferase genes ARO8/9 and the indoleamine 2,3‐dioxygenase gene BNA2 yielded a fivefold increase of psilocybin titre. We transferred this knowledge to the filamentous fungus Aspergillus nidulans and identified functional ARO8/9 orthologs involved in fungal l‐tryptophan catabolism by genome mining and cross‐complementation. The double deletion mutant of A. nidulans resulted in a 10‐fold increased psilocybin production. Process optimization based on respiratory activity measurements led to a final psilocybin titre of 267 mg/L in batch cultures with a space–time‐yield of 3.7 mg/L/h. These results demonstrate the suitability of our engineered A. nidulans to serve as a production strain for psilocybin and other tryptamine‐derived pharmaceuticals.
Here, we developed an in vivo psilocybin production chassis, based on repression of l‐tryptophan catabolism. By genome mining, we identified two aminotransferase ARO8/9 orthologs in Aspergillus nidulans involved in l‐tryptophan catabolism. Double deletion of the aminotransferase genes aroH1 and aroH2 resulted in a 10‐fold increased psilocybin production.
INTRODUCTION
Psilocybin, a mushroom indolylethylamine alkaloid, represents the chemically stable prodrug of the psychotropic 4‐dephosphoryl analog psilocin (Hofmann et al., 1959). The latter is structurally related to serotonin and acts as a partial agonist of the human serotonin receptor 2B (5‐HT2B) (Fricke et al., 2019). Numerous recent clinical studies convincingly underscored psilocybin's potential to treat patients suffering from therapy‐refractory depression, alcohol use disorder and anorexia nervosa (Goodwin et al., 2022; Jensen et al., 2022; Peck et al., 2023), convincing the US Food and Drug Administration to grant psilocybin with the status of ‘Breakthrough Therapy’, meaning that this candidate (pro)drug may prove substantially superior to current therapies.
The re‐discovered pharmaceutical value of psilocybin has entailed an increased demand for the compound. The molecule has traditionally been produced by total synthesis, for which several protocols have been devised (Fricke et al., 2019). However, the characterization of the biosynthetic enzymes in the ‘magic mushroom’ Psilocybe cubensis set the stage for a biotechnological large‐scale psilocybin production (Fricke et al., 2017). The first approach was a cell‐free biosynthesis through a three‐enzyme in vitro route from the synthetic precursor 4‐hydroxy‐l‐tryptophan to psilocybin. This method harnesses the l‐tryptophan decarboxylase PsiD, the kinase PsiK and the methyltransferase PsiM. These three enzymes, together with the regioselective cytochrome P450‐monooxygenase PsiH, constitute the complete biosynthetic pathway in Psilocybe mushrooms (Blei et al., 2018) (Figure 1A).
FIGURE 1.
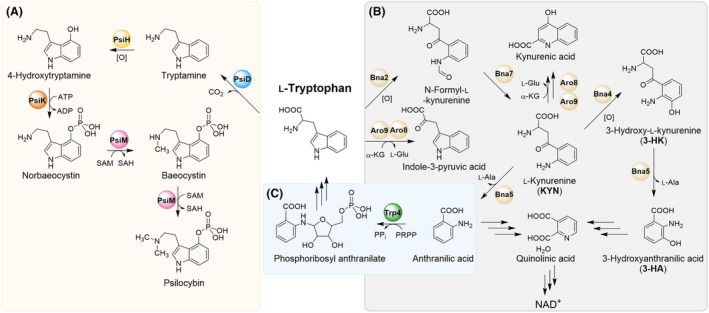
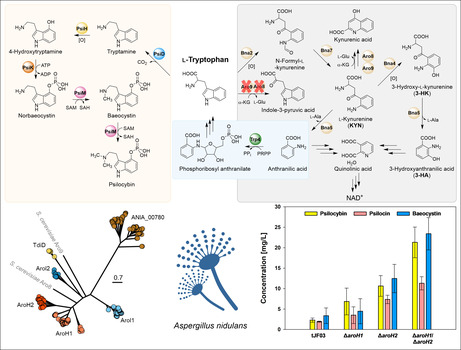
Schematic, simplified overview on l‐tryptophan metabolism. (A) Four‐enzyme psilocybin biosynthetic pathway, reconstituted in S. cerevisiae. (B) In S. cerevisiae, l‐tryptophan is deaminated to indole‐3‐pyruvic acid by the aminotransferases Aro8 and Aro9. A second catabolic route includes oxidative cleavage of the pyrrole ring of l‐tryptophan by the indoleamine‐2,3‐dioxygenase Bna2, to yield N‐formyl‐l‐kynurenine, that is a precursor for the biosynthesis of both kynurenic acid and quinolinic acid. The latter is required for nicotinamide adenine dinucleotide biosynthesis. (C) De novo biosynthesis of l‐tryptophan via the anthranilate phosphoribosyltransferase Trp4. l‐Ala, l‐alanine; l‐Glu, l‐glutamate; NAD+, nicotinamide adenine dinucleotide; PRPP, 5‐phospho‐α‐d‐ribose‐1‐diphosphate; SAH, S‐adenosyl‐l‐homocysteine; SAM, S‐adenosyl‐l‐methionine; α‐KG, α‐ketoglutaric acid.
A second approach was to produce psilocybin heterologously in chassis organisms. To this aim, the filamentous fungus Aspergillus nidulans was previously equipped with a versatile polycistronic expression system for biosynthetic pathways, established for various hosts (Hoefgen et al., 2018; Reimer et al., 2022). Later on, higher titres of 1.16 g/L psilocybin were obtained by fermenting Escherichia coli expressing psiK, psiM and psiD. However, in this case, a targeted feeding with 4‐hydroxyindole was necessary to compensate for the missing P450‐monooxygenase PsiH (Adams et al., 2019). De novo psilocybin synthesis was established in bacteria by producing PsiH and a cytochrome P450 reductase partner in a separate E. coli strain and co‐cultivation of both strains yielded 28.5 mg/L psilocybin (Flower et al., 2023). Finally, a successful high‐scale production of psilocybin in Saccharomyces cerevisiae was obtained by enhancing the native tryptophan biosynthesis and implementing the availability of tryptamine (Milne et al., 2020). However, this approach does not take catabolic routes into account that potentially compete for l‐tryptophan.
Breakdown of tryptophan has been well characterized in S. cerevisiae (Figure 1B) and is mainly initiated by the heme‐dependent indoleamine 2,3‐dioxygenase Bna2 (Yuasa & Ball, 2011). It oxidatively converts l‐tryptophan to N‐formyl‐l‐kynurenine, which is then modified to l‐kynurenine by the kynurenine formamidase Bna7 (Wogulis et al., 2008). A further relevant step, performed by the kynureninase Bna5, is then the cleavage of l‐kynurenine to form l‐alanine and anthranilic acid (Panozzo et al., 2002). Bna5 can also cleave 3‐hydroxy‐l‐kynurenine, releasing 3‐hydroyanthranilic acid (Kim et al., 2018). Subsequently, the two acids produced are precursors in the biosynthesis of quinolinic acid, which after decarboxylation to nicotinic acid serves NAD+ biosynthesis, but also as precursors in the de novo biosynthesis of l‐tryptophan via the anthranilate phosphoribosyltransferase Trp4 (Furter et al., 1986) (Figure 1C). The catabolism of tryptophan can also occur via amino acid transamination by Aro8 and Aro9, the two α‐ketoglutarate‐dependent aminotransferases present in S. cerevisiae (Iraqui et al., 1998).
Several indoleamine 2,3‐dioxygenases have already been characterized in filamentous fungi. In particular, filamentous ascomycetes harbour three BNA2‐like genes, named idoA, idoB and idoC (Yuasa & Ball, 2011). Deletion of idoA has major effects on virulence in both the plant pathogenic fungus Fusarium graminearum and the opportunistic human pathogen Aspergillus fumigatus (Liu et al., 2023; Zelante et al., 2021). Moreover, the aroH gene product has been biochemically characterized in A. fumigatus, confirming its role as a pyridoxal 5′‐phosphate (PLP)‐dependent aminotransferase involved in tryptophan transamination (Dindo et al., 2018).
In addition to the two major pathways, fungi can use l‐tryptophan as a scaffold to produce indole acetate or secondary metabolites. One example is the aforementioned conversion of tryptophan to tryptamine by tryptophan decarboxylases, as reported in P. cubensis and Rhizoctonia solani (Fricke et al., 2017; Pedras et al., 2005). A combined genetic, biochemical and transcriptomic study within the psilocybin producer Psilocybe mexicana showed a massive downregulation of l‐tryptophan catabolism genes, among them iasA, encoding an indole‐3‐acetaldehyde synthase, when psilocybin biosynthesis set in (Seibold et al., 2024). Additionally, fungi may produce secondary metabolite‐specific enzymes with catalytic similarities to those of primary metabolism, such as the aminotransferase TdiD, which is part of the gene cluster that mediates the biosynthesis of terrequinone A in A. nidulans (Bouhired et al., 2007; Schneider et al., 2007), or the indoleamine 2,3‐dioxygenases involved in the biosynthesis of benzazepine alkaloids (Caesar et al., 2020; Chen et al., 2022; Li et al., 2020).
Here, we report on a strategy to obtain high psilocybin titres in fungal heterologous hosts by suppressing l‐tryptophan catabolism and increasing its intracellular availability. For a proof of concept, we expressed the psilocybin biosynthetic gene cluster (BGC) in an S. cerevisiae triple mutant deficient in genes encoding l‐tryptophan‐degrading enzymes, namely ARO8, ARO9 and BNA2. Batch cultures yielded approximately 50 mg/L of psilocybin. To reach the highest production titres possible, we implemented the concept of suppressing l‐tryptophan catabolism in the mould A. nidulans as well. We successfully identified and deleted two genes coding for functional Aro8/Aro9 orthologs. This approach yielded a strain capable of producing 267 mg/L psilocybin in batch cultures. The herein generated production strain will enable the development of a sustainable, reproducible and scalable biotechnological production process for the pharmaceutically relevant prodrug psilocybin.
EXPERIMENTAL PROCEDURE
Strains, media and growth conditions
The wild‐type strain S. cerevisiae BY4741, Δaro8 and Δaro9 single mutant derivatives and Δbna2 were retrieved from Euroscarf (Scientific Research and Development GmbH, Oberursel, Germany). In preparation for yeast transformation, strains were grown in YPD complete medium. Selection and cultivation of transformants were performed using SD minimal media with the appropriate dropout amino acid solution. For the production of psilocybin, l‐kynurenine and anthranilic acid, an SD ‐Uracil/Uridine (‐Ura) pre‐culture was prepared overnight. The main culture consisted of 20 mL SD ‐Ura +1 mM l‐tryptophan in 100 mL triple‐baffled Erlenmeyer flasks and was inoculated with the pre‐culture to give an OD600 of 0.5. All strains were grown in triplicates at 30°C and 180 rpm for 2 days. For plate assays, the SD ‐Ura overnight culture was diluted with water to OD600 = 0.1–1 × 10−4 and 10 μL of each dilution was incubated on selective plates (SD ‐Ura +/− Trp) for 3 days.
The cultivations of A. nidulans were performed in Aspergillus minimal medium (AMM) consisting of 6 g/L NaNO3, 0.52 g/L KCl, 1.52 g/L KH2PO4, 0.52 g/L MgSO4 × 7 H2O and 1 mL/L Hutner's trace elements. The Hutner's trace elements were prepared as 1000× stock solution and contained 50 g/L EDTA, 22 g/L ZnSO4 × 7 H2O, 11 g/L H3BO3, 5 g/L MnCl2 × 4 H2O, 1.6 g/L CoCl2 × 6 H2O, 1.6 g/L CuSO4 × 5 H2O, 1.1 g/L (NH4)6Mo7O24 × 4 H2O and 5 g/L FeSO4 × 7 H2O. For expression analysis of the A. nidulans wild‐type strain RMSO11, 50 mL AMM (supplemented with 3 mg/L p‐aminobenzoic acid and 5 mM l‐arginine) was inoculated with 1 × 107 conidia/mL in a 300 mL Erlenmeyer flask and shaken for 1 day at 37°C and 180 rpm. To induce gene expression, the mycelium was shifted to new flasks with or without 10 mM l‐tryptophan for 1 h. Expression analysis via qRT‐PCR was performed as previously described (Janevska et al., 2020) with primers listed in Table S2. For A. nidulans plate assays, AMM (with p‐aminobenzoic acid and l‐arginine) was supplemented with 5/10 mM l‐tryptophan or 5 mM nicotinamide and serial 1:10 spore dilutions started with a 5 μL drop of 1 × 105 conidia/mL. Single drops in the centre of the plate were 5 μL of 1 × 103 conidia/mL.
For psilocybin production in A. nidulans, 1 × 107 conidia/mL were inoculated in 50 mL AMM in a 300 mL Erlenmeyer flask with supplementation of 3 mg/L p‐aminobenzoic acid, 5 mM l‐arginine, 1 mM l‐tryptophan, 0.1 mM nicotinamide and 10% (w/v) glucose, cultivated for 18 h in shaking condition at 37°C, 180 rpm. Afterwards, 20 μg/mL doxycycline was added to induce the expression of the psi gene cluster. Then, incubation continued for another 24 h at 37°C and 180 rpm.
Plasmid construction and generation of transformants
To construct the yeast psi expression plasmid pYes‐psi, the previously created promoterless plasmid pJF33 harbouring all psilocybin biosynthetic genes in the order psiH, psiD, psiK and psiM and the tev protease‐coding sequence was used (Hoefgen et al., 2018). A PCR fragment obtained from the empty overexpression plasmid pYes2‐TEF1 (Hoefgen et al., 2018), amplified with primers [1]//[2] (Table S2), was used together with the PacI‐treated pJF33 to obtain pJF34 based on transformation‐associated recombination (TAR) cloning using the protocol reported by Hoefgen et al. (2018). Then, venusN and venusC were removed by amplification based on pJF34 with primer pairs [3]//[4] and [5]//[6]. These two fragments were assembled by TAR cloning to give pYes‐psi. This plasmid harbours the psilocybin polycistronic cluster under the control of the TEF1 promoter, 2 μ as origin of replication and URA3 as auxotrophic marker and was subsequently used to transform S. cerevisiae strains.
The generation of yeast double and triple mutants was achieved by homologous recombination. ARO8 was exchanged for LEU2 in the Δaro9 background using flanks of ca. 500 bp. The ARO8 deletion construct was generated by fusion PCR. Therefore, ARO8 upstream and downstream flanks including the required overhangs were amplified with oligonucleotide primer pairs [7]//[8] and [11]//[12] (Table S2), respectively, using genomic DNA of S. cerevisiae BY4741 as template. LEU2 including overhangs was amplified with primer pair [9]//[10], using genomic DNA of a prototrophic yeast as template (Jena Microbial Research Collection, JMRC: STI25222). The fusion PCR was performed by applying the three PCR products and the outer primers [7]//[12].
Furthermore, S. cerevisiae BNA2 was exchanged for HIS3 in the Δaro8/Δaro9 background. The BNA2 deletion construct was generated by TAR cloning. Therefore, BNA2 upstream and downstream flanks of ca. 900 bp including the required overhangs were amplified with primer pairs [13]//[14] and [15]//[16] (Table S2), respectively, based on genomic DNA of S. cerevisiae BY4741. HIS3 including promoter and terminator sequences was amplified with the primer pair [17]//[18], based on genomic DNA of the prototrophic yeast STI25222. S. cerevisiae BY4741 was transformed with the three PCR products as well as with the HindIII/XbaI‐digested shuttle vector pYes2 (Life Technologies, Darmstadt, Germany), to create the deletion vector pΔbna2. For the transformation of Δaro8/Δaro9, the BNA2 deletion construct was amplified with primers [13]//[16] from the deletion vector and the PCR reaction was DpnI‐digested to remove the template.
For complementation of S. cerevisiae Δaro8/Δaro9, A. nidulans potential aro‐like aminotransferase genes were amplified from cDNA: aroH1 [19]//[20], aroH2 [21]//[22], aroI1 [23]//[24], aroI2 [25]//[26], tdiD [27]//[28] and ANIA_00780 [29]//[30] (Table S2). As controls, S. cerevisiae ARO8 [31]//[32] and ARO9 [33]//[34] were amplified from gDNA of BY4741. The inserts were cloned into HindIII/BamHI‐digested pYes2‐TEF1 via primer‐introduced overhangs and TAR cloning.
Similarly, to complement S. cerevisiae Δbna2, A. nidulans potential ido‐like indoleamine 2,3‐dioxygenase genes were amplified from cDNA: idoA [35]//[36], idoB [37]//[38] and idoC [39]//[40] (Table S2). For complementation of S. cerevisiae Δbna5, A. nidulans kynU2 [41]//[42] was amplified. As controls, S. cerevisiae BNA2 [43]//[44] and BNA5 [45]//[46] were amplified from gDNA of BY4741.
To delete A. nidulans aroH1, ca. 1 kb flanks were amplified with primers [47]//[48] and [49]//[50] using gDNA of A. nidulans RMSO11 wild type (Table S2). The pabA resistance gene (An03g03130), including its promoter and terminator, was amplified with [51]//[52] from Aspergillus niger gDNA. The fusion PCR was performed by applying the three PCR products and the outer primers [47]//[50]. Similarly, to delete A. nidulans aroH2, ca. 1 kb flanks were amplified with [53]//[54] and [55]//[56] from RMSO11 gDNA. The ptrA resistance cassette was amplified with primers [57]//[58] and plasmid pSK275 as template (Krappmann et al., 2006). Then, the fusion PCR was performed with the outer primers [53]//[56]. Finally, to delete A. nidulans idoA, ca. 1 kb flanks were amplified with primers [59]//[60] and [61]//[62] from A. nidulans gDNA. The ptrA cassette and the fusion PCR [59]//[62] were amplified as described above.
Phylogenetic analysis
RefSeq Fungi Database files were downloaded from the National Center for Biotechnology Information (NCBI). The search for conserved amino acids in fungal genomes was carried out using the BLAST® Command Line Applications (Camacho et al., 2009). The search was performed using standard default parameters. Hits were extracted and double‐checked using the CDD Database.
Amino acid sequences for the ARO‐ and IDO‐like proteins (Table S1) were aligned using MUSCLE (Edgar, 2004) v3.8 with a maximum of 16 iterations. Phylogenetic inference was performed using IQ‐TREE2 (Minh et al., 2020) v2.2.0.3 and 1000 ultrafast bootstraps. The ModelFinder (Kalyaanamoorthy et al., 2017) module of IQ‐TREE2 was used to identify JTT+I+G4 (Jones et al., 1992) as the best‐fitting substitution model for both ortholog sets. Phylogenetic trees were visualized using ggtree (Yu et al., 2017).
Chemical analysis
After the cultivation of yeast, 300 μL of the culture was mixed with 300 μL MeOH prior to bead milling. Then, samples were filtered for LC‐HRMS analysis (samples were 1:2 dilutions).
After the cultivation of A. nidulans, 20 mL culture together with 10 mL H2O and 10 mL MeOH was homogenized. Then, 100 μL of crude extract was pipetted with a cut tip and diluted with 900 μL H2O, filtered and analysed by LC‐HRMS (samples were diluted 1:20).
LC‐HRMS was performed on a Q‐Exactive Plus Hybrid Quadrupole Orbitrap mass spectrometer using electrospray ionization and a Dionex UltiMate 3000 UHPLC system (Thermo Fisher Scientific) equipped with a Kinetex C18 column (2.1 × 150 mm, 2.5 μm, 100 Å, Phenomenex). The analytical method is a gradient elution of solvents A (water, 0.1%, v/v, formic acid) and B (acetonitrile, 0.1%, v/v, formic acid) at a flow rate of 0.3 mL/min: 5% B for 0.5 min, a linear gradient to 97% B for 11.5 min, then 97% B for 3 min and 5% B for 3 min. Calibration curves for psilocybin, baeocystin and psilocin were determined using peak areas of HPLC‐HRMS measurements of samples with concentration ranges from 0 to 10, 0 to 68 and 0 to 0.625 μM, respectively. All samples were analysed in triplicates and error bars represent the standard error.
HPLC method organic acids
Organic acids and ethanol were analysed by HPLC. The X‐LC® system (JASCO International Co., Tokyo, Japan) was equipped with an Aminex HPX‐87H Ion Exclusion Column (300 × 7.8 mm, 9 μm, Bio‐Rad Laboratories Inc., Hercules CA, USA), maintained at 50°C, a refractive index detector and an UV detector (210 nm). The substances were separated under isocratic conditions with 0.5 mL/min of 0.005 M H2SO4 as the mobile phase. For injection, 50 μL of diluted culture filtrate (1:10 with 0.005 M H2SO4) was used.
Growth and metabolic activity monitoring
Cultivations were conducted in 250 mL unbaffled shake flasks filled with 30 mL AMM containing 100 g/L glucose, buffered to pH 7.0 with 100 mM 3‐(N‐morpholino)propanesulfonic acid (MOPS) and supplemented with 3 mg/L p‐aminobenzoic acid, 0.1 mM nicotinamide and 1 mM l‐tryptophan. A. nidulans strains tJF03 (Hoefgen et al., 2018) and ΔaroH1/ΔaroH2_psi were inoculated to 1 × 107 conidia/mL and grown at 300 rpm with 50 mm shaker throw for 192 h. To determine the OTR and CTR, the incubators were equipped with the Kühner Transfer‐Rate Online Measurement (TOM) system, enabling online exhaust gas analysis. Aeration with compressed air was set to 11 mL/min and one measuring cycle of 20 min included 6 min measuring time. Psilocybin formation was induced with 50 μg/mL doxycycline at 31 h after the OTR peaked. For better production of biosynthetic enzymes, the incubation temperature was reduced from 37 to 30°C. For offline analysis during cultivation, duplicate samples were taken in the form of entire shake flask cultures that were run in parallel.
Offline analytics
To determine the cell dry weight, 10 mL culture broth was vacuum filtrated through a pre‐dried and pre‐weighed Whatman® 589/2 filter, washed with 100 mL Milli‐Q® water and dried and weighed using an MA160 moisture analyser. Glucose concentration was measured in the supernatant using a YSI 2950 Biochemistry Analyser. The pH measurement was conducted with a Schott pH meter. For psilocybin, psilocin and baeocystin extraction, 0.5 mL culture broth was diluted 1:2 with Milli‐Q® water. Subsequently, 0.5 mL diluted culture broth was diluted 1:2 with methanol, added to 0.5 g glass beads (0.75–1.0 mm) and milled three times for 60 s at 6.5 m/s and 4°C (FastPrep ‐24™ Sample Preparation System) for cell disruption. The cell lysate was centrifuged at 17,000 g, 4°C for 10 min and the supernatant was diluted 1:5 with Milli‐Q® water before filtering through 0.22 μm cellulose acetate syringe filters and storing at −20°C until analysis. For each sample, triplicate extraction was performed.
RESULTS AND DISCUSSION
Psilocybin production in l‐tryptophan catabolism‐repressed S. cerevisiae
The removal of competing pathways is one of the most commonly used strategies in metabolic engineering to enhance precursor supply (Lee et al., 2012; Qian et al., 2011). Therefore, we hypothesized that disabling l‐tryptophan degradation in the production host would route the available cellular l‐tryptophan pool preferentially towards psilocybin biosynthesis. To test this hypothesis, the psi cluster was expressed in S. cerevisiae strains deficient for either the ARO8, the ARO9 (aminotransferases) or BNA2 (indoleamine 2,3‐dioxygenase) genes. Deletion of ARO9 resulted in mutants that were more sensitive to elevated l‐tryptophan concentrations (5 mM). The concomitant deletion of ARO8 in the Δaro9 background increased this sensitivity, which rendered double mutants unable to grow in the presence of 5 mM l‐tryptophan and confirmed previous results (Figure 2A) (Ohashi et al., 2017). However, the additional deletion of BNA2, resulting in the triple mutant ∆3, did not show increased sensitivity towards l‐tryptophan in comparison to the double mutant, indicating that the production of kynurenic acid does not contribute to the detoxification of excess l‐tryptophan (Figure 2A). Based on these results, S. cerevisiae production cultures contained 1 mM l‐tryptophan, which allowed for robust growth of all strains.
FIGURE 2.
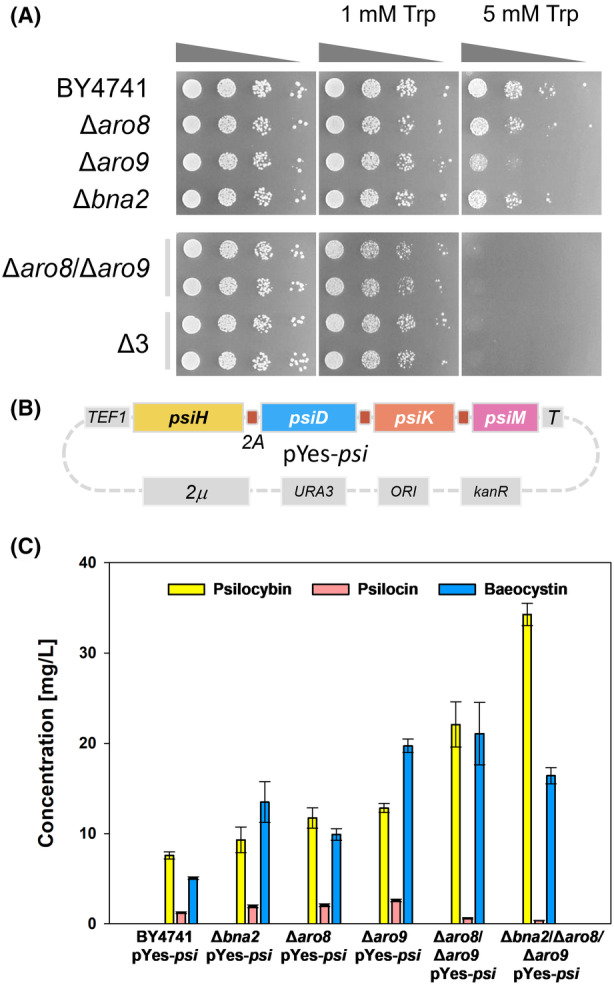
Psilocybin production in S. cerevisiae mutant strains. (A) The phenotypic analysis of l‐tryptophan catabolism‐repressed S. cerevisiae confirmed their sensitivity to high l‐tryptophan (Trp) concentrations. (B) Genetic map of the expression plasmid pYes‐psi carrying the psi cluster expressed as a polycistron under the control of the constitutive TEF1 promoter. (C) Production of psilocybin, psilocin and baeocystin in S. cerevisiae wild type and tryptophan catabolism‐repressed mutants after 48 h of cultivation.
The plasmid harbouring the psilocybin biosynthetic genes (Figure 2B) was used to transform the Δaro8, Δaro9, and Δbna2 single mutants, the Δaro8/Δaro9 double mutant and a mutant lacking all three genes (Δ3). Quantitative analysis revealed that psilocybin production had increased in the single mutants, compared to the wild‐type control expressing the psi cluster (Figure 2C). In particular, the production of baeocystin, that is, the monomethylated immediate precursor to psilocybin, was significantly higher in the Δaro9 mutant strain, suggesting a more active pathway as consequence of disabled l‐tryptophan catabolism. For full conversion of baeocystin to psilocybin, additional copies of the psiM methyltransferase gene could be integrated into the psi cluster as previously shown (Milne et al., 2020).
The above results suggested that the more severely l‐tryptophan catabolism is blocked, the better the heterologous psilocybin pathway would function as the sole sink for this amino acid (Lee et al., 2012; Qian et al., 2011). To test this hypothesis and strongly inhibit l‐tryptophan catabolism, the Δaro8/Δaro9 double mutant and Δ3 triple mutant with the plasmid harbouring the psi cluster were analysed (Figure 2C). For the Δaro8/Δaro9 double mutant background, we observed a significantly increased psilocybin production while the baeocystin level was similar to that of the single Δaro9 background. This trend was also observed for the Δ3 mutant background, where psilocybin titres were further increased to 34.3 mg/L, but not baeocystin titres. Curiously, the latter mutants, which produced higher titres of psilocybin and baeocystin, showed strongly reduced levels of psilocin, pointing towards more stable production of psilocybin and reduced dephosphorylation compared to Milne et al. (2020). Overall, the expression of the polycistronic psi cluster yielded ~7.5 mg/L of psilocybin and ~5 mg/L of baeocystin in the wild‐type background while the repression of the genes involved in l‐tryptophan catabolism led to up to fivefold higher psilocybin titres. Nevertheless, these titres were still fivefold lower than those produced by a strain from Milne et al. (2020). The reason for that is likely the lack of an appropriate cytochrome P450 reductase responsible for the transfer of electrons between NADPH and the cytochrome P450 enzyme PsiH.
Tryptophan catabolism in A. nidulans
The results obtained in S. cerevisiae batch cultures clearly showed the potential of repression of l‐tryptophan catabolism to implement psilocybin production. Furthermore, the product spectrum was less diverse and much more focused on the actual pharmaceutically relevant compound psilocybin, compared to a previous yeast‐based approach (Milne et al., 2020). However, the performance of the obtained strains did not seem to quantitatively advance this prior endeavour. We therefore decided to transfer this knowledge to filamentous fungi and study the genes linked to tryptophan catabolism in Aspergillus. We chose the model fungus A. nidulans, which was also the first chassis used for the heterologous production of psilocybin (Hoefgen et al., 2018).
BLASTp analysis revealed that the A. nidulans genome encodes six homologues of potential ARO‐like aminotransferases (Table 1). Among them, only the tdiD gene has been previously identified as part of the terrequinone A gene cluster (Bok et al., 2006; Schneider et al., 2007). Using the six ARO‐like proteins from A. nidulans as queries, we performed a BLASTp search on the NCBI RefSeq database (Sayers et al., 2021) and identified highly similar proteins in all represented Aspergilli (Table S1). Analysis of the selected sequences using the Conserved Domains Database (CDD) (Wang et al., 2023) confirmed that all of these are putative PLP‐dependent aminotransferases and that, in general, each Aspergillus genome encodes between four and six ARO‐like proteins (Table S1).
TABLE 1.
BLAST analysis of A. nidulans genome for aro‐like, ido‐like and kynureninase genes.
| Acc. number | Gene name | Function of gene product |
|---|---|---|
| ANIA_00780 | ANIA_00780 | Putative aminotransferase |
| ANIA_01857 | kynU2 | Kynureninase |
| ANIA_01858 | idoB | Indoleamine 2,3‐dioxygenase |
| ANIA_02509 | idoA | Indoleamine 2,3‐dioxygenase |
| ANIA_09108 | idoC | Indoleamine 2,3‐dioxygenase |
| ANIA_04156 | aroI2 | Putative aminotransferase |
| ANIA_05041 | aroH2 | α‐Ketoglutarate‐dependent aminotransferase |
| ANIA_06338 | aroH1 | α‐Ketoglutarate‐dependent aminotransferase |
| ANIA_08172 | aroI1 | Putative aminotransferase |
| ANIA_08516 | tdiD | α‐Ketoglutarate‐dependent aminotransferase |
Phylogenetic analysis revealed the presence of six clades (Figure 3A). We identified two groups of proteins similar to AroH from A. fumigatus (Spizzichino et al., 2022). However, within these clades, we observed a few Aspergilli with two AroH‐like enzymes, including A. nidulans. A second, less represented clade included putative aminotransferases/transaminases similar to AroI from A. fumigatus (Choera et al., 2017). Next, the least represented clade (Figure 3A, yellow) refers to proteins similar to TdiD, which were only identified in species containing the respective tdi BGC (Figure S1). Finally, a further group included putative and yet uncharacterized transaminases similar to Aro8 from yeast.
FIGURE 3.
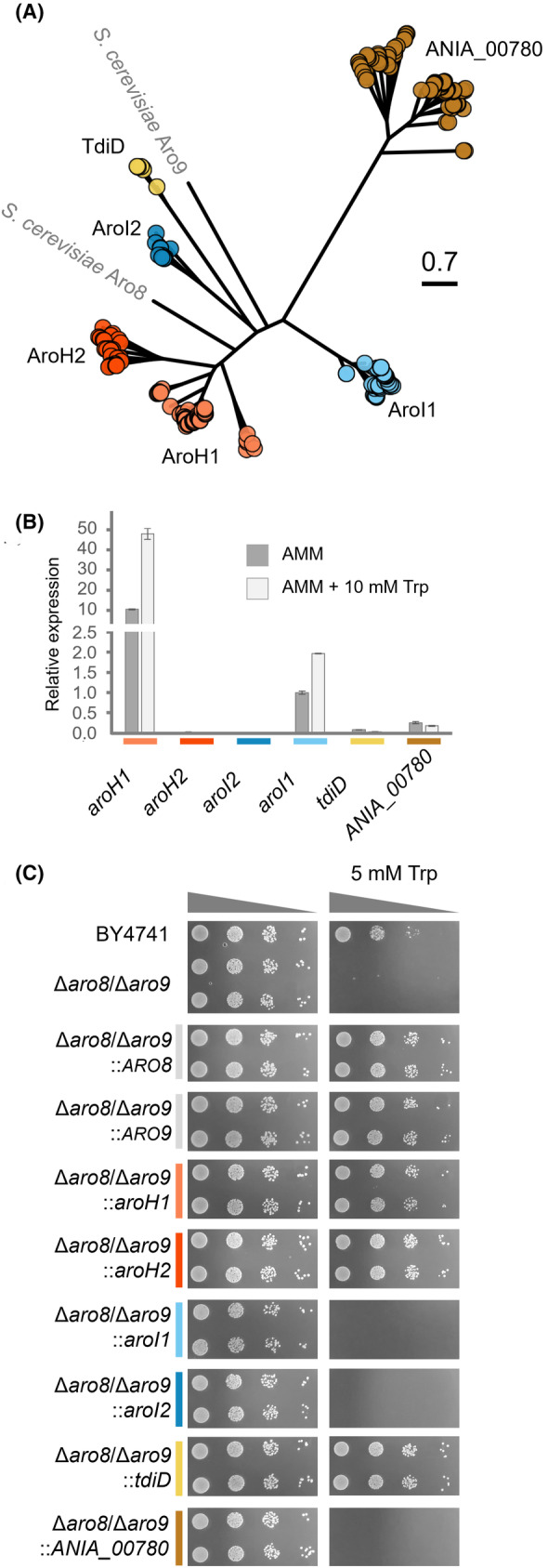
Identification and analysis of ARO‐like aminotransferases from A. nidulans. (A) Evolutionary distance among ARO‐like aminotransferases in Aspergilli. Clades are named based on the genes identified in A. fumigatus (AroH, AroI) and A. nidulans (TdiD). (B) qRT‐PCR analysis of the A. nidulans aro‐like genes in response to tryptophan (Trp). (C) Functional complementation analysis of the identified genes from A. nidulans in the S. cerevisiae Δaro8/Δaro9 mutant strain. BY4741 and Δaro8/Δaro9 contained the empty plasmid pYes2‐TEF1.
To better understand the role of these genes during tryptophan catabolism, we investigated their expression upon induction by l‐tryptophan (Figure 3B). qRT‐PCR analysis revealed that aroH1 and arol1 were the only genes with measurable basal expression, which further increased in the presence of higher l‐tryptophan concentrations. Next, each corresponding cDNA was cloned into a yeast expression vector and transferred into the S. cerevisiae Δaro8/Δaro9 mutant background (Figure 3C). The over‐expression of aroH1 and aroH2 completely restored the tryptophan sensitivity of the yeast strain, confirming their function as transaminases responsible for indole pyruvic acid production. Moreover, the assay also confirmed the function of TdiD as transaminase, however, the gene is specific to the biosynthesis of terrequinone A and not expressed under laboratory conditions (Figure 3B). Therefore, it was excluded as target for our repression strategy of general tryptophan catabolism. The other expressed genes aroI1, aroI2 and ANIA_00780 did not complement S. cerevisiae Δaro8/Δaro9, suggesting an alternative role for the encoded enzymes.
A second step in the analysis was the search for indoleamine 2,3‐dioxygenases (IDOs) that are functionally similar to yeast Bna2. We identified numerous ido‐like genes in Aspergilli. Overall, each Aspergillus species harbours three genes encoding potential IDO‐like proteins and, as expected, this number may be variable in case of enzymes involved in secondary metabolism (Table 1, Table S1). This situation resembles previous observations for F. graminearum and A. fumigatus (Liu et al., 2023; Zelante et al., 2021). The phylogenetic analysis resulted in a more scattered tree but, on closer inspection, we could group the potential orthologs not only based on their amino acid sequences but also on gene synteny (Figure 4A). We noticed that idoB is encoded adjacent to a putative kynureninase gene, annotated as kynU2, which is also potentially involved in tryptophan catabolism (Figure 4B). The two genes share a bidirectional promoter and this locus is present in all Aspergilli from the RefSeq Database. Additionally, we noticed this two‐gene cluster is commonly present in other filamentous fungi of the Ascomycete division as well, including the Alternaria, Botrytis, Fusarium, Penicillium, Sclerotinia, Sordaria and Trichoderma genera, just to mention the most studied ones.
FIGURE 4.
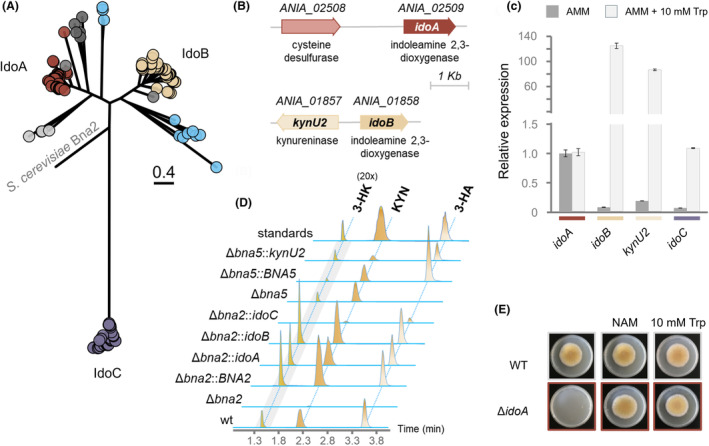
Identification and analysis of indoleamine 2,3‐dioxygenases from A. nidulans. (A) Phylogenetic tree of IDO‐like proteins identified in Aspergilli. The three main clades are named based on previous analyses made in A. fumigatus. Reported IdoA‐ and IdoB‐related proteins are shown in maroon and sand, respectively, which also show a conserved gene synteny (shown in panel B). IdoC‐like proteins are shown in violet, while proteins shown in light blue likely participate in secondary metabolism. The remaining IDO‐like proteins are depicted in grey. Scale bar indicates amino acid substitutions/site. (B) Genetic loci for the idoA and idoB genes in A. nidulans. (C) qRT‐PCR analysis of the A. nidulans idoA, idoB, kynU2 and idoC genes in response to tryptophan (Trp). (D) Functional complementation of S. cerevisiae Δbna2 and Δbna5 with idoA, idoB, idoC and kynU2, respectively. Corresponding masses are: 3‐HK [+m/z] 225.0870; KYN [+m/z] 209.0921; 3‐HA [+m/z] 138.0550. (E) Phenotypic analysis of A. nidulans ΔidoA on minimal media and in presence of nicotinamide (NAM) and tryptophan (Trp).
A second prominent clade was the one including all IdoA‐related enzymes (Figure 4A). In this case as well, we noticed that, in Aspergilli, this gene is generally adjacent to a gene encoding a putative cysteine desulfurase, similar to yeast Nfs1, which catalyses the removal of sulphur from cysteine to gain alanine (Figure 4B) (Kispal et al., 1999). However, this synteny does not seem to be as conserved as that of idoB and was mainly found in Aspergillus species. Lastly, a third putative indoleamine 2,3‐dioxygenase, IdoC, was shown to be highly conserved in Aspergilli, being part of a third large clade in the analysis (Figure 4A).
By performing the phylogenetic analysis, we found that some Aspergilli may contain additional ido‐like genes, whose functions are difficult to predict. Some of them (Figure 4A, in grey) may be the result of gene duplication. Nonetheless, we observed the formation of clades (Figure 4A, in sky blue), comprising a number of putative indoleamine 2,3‐dioxygenases most likely connected to secondary metabolism. All of the genes coding for these enzymes are located in close proximity to natural product genes, for example, AcdA from Aspergillus candidus, for the alkaloid pyrrolobenzazepine aspcandine (Chen et al., 2022) and TzpB, related to the biosynthesis of terreazepine (Caesar et al., 2020).
In order to confirm the link between the ido genes and tryptophan catabolism, we investigated their gene expression in response to 10 mM l‐tryptophan (Figure 4C). While idoA expression was not affected by the presence of tryptophan, expression levels of idoB and idoC were strongly increased. Moreover, the kynU2 gene was also induced by tryptophan, confirming that the shared promoter regulates both genes. The obtained results confirmed previous observations for A. fumigatus (Zelante et al., 2021). The function of the identified genes was further confirmed by complementation of the S. cerevisiae loss of function mutants (Figure 4D). As reported, the deletion of BNA2 blocks the production of l‐kynurenine derivatives (via blocking N‐formyl‐l‐kynurenine production), while by removing BNA5 (kynureninase), only the biosynthesis of anthranilic acid is abolished (Figure 1) (Panozzo et al., 2002). We analysed the S. cerevisiae Δbna2 mutant and confirmed that l‐kynurenine (KYN), 3‐hydroxy‐l‐kynurenine (3‐HK) and 3‐hydroyanthranilic acid (3‐HA) were not produced (Figure 4D). In the Δbna2 mutant strain, we noticed a small peak with the retention time of KYN, but with a different mass. Functional complementation analysis performed by the expression of A. nidulans idoA, idoB and idoC cDNA in the S. cerevisiae Δbna2 background restored the l‐tryptophan catabolic pathway. Similarly, the cDNA of A. nidulans kynU2 was able to rescue the loss of 3‐HA production in the S. cerevisiae Δbna5 mutant (Figure 4D).
As final proof for the functional conservation of IDO‐like proteins in Aspergilli, the deletion of A. nidulans idoA resulted in a lethal phenotype, when grown on minimal media, which could be rescued by the addition of either nicotinamide or tryptophan (Figure 4E). The latter compound most likely rescues the lethal phenotype by activating the repressed and functionally redundant genes idoB and idoC.
Psilocybin production in tryptophan catabolism‐repressed A. nidulans
Analysis of the A. nidulans genes potentially involved in tryptophan catabolism suggested the presence of two main catabolic branches formed by the AroH‐like and Ido‐like enzymes, respectively. As for the tdi BGC, its role in intracellular tryptophan consumption does not seem to be relevant, as tdiD was not expressed (Figure 3B) and terrequinone A was not produced under the conditions tested. Complete depletion of idoA, idoB and idoC gene expression has already been shown for F. graminearum and A. fumigatus, resulting in mutants able to grow only in the presence of nicotinamide, similar to what we are reporting for A. nidulans idoA deletion here (Figure 4E). For this reason, we focused on the two aroH genes characterized here. Growth of the single (ΔaroH1, ΔaroH2) and double mutants (ΔaroH1/ΔaroH2) was not affected by the addition of up to 10 mM l‐tryptophan (Figure S2), suggesting that the A. nidulans mutants were more resistant than the respective S. cerevisiae mutants (Figure 2A).
The psi biosynthetic cluster was expressed as a polycistron without codon optimization under the control of the TetON inducible promoter and the analysis of the resulting mutant strains was performed using fluorescent microscopy, exploiting the previously reported split green fluorescent protein (GFP) system (Hoefgen et al., 2018). Briefly, the psilocybin biosynthetic genes are flanked by two GFP gene subunits and a functional GFP is produced only when the entire mRNA is correctly translated. Consequently, transformants that display a nuclear localization of the GFP correctly express all genes of interest.
The generated single and double deletion mutants of aroH1 and aroH2 in A. nidulans were then transformed with the psi BGC. Product formation was initially investigated in batch cultures for 48 h, comparing the strain expressing the psi cluster in a wild type (tJF03) (Hoefgen et al., 2018) versus the ΔaroH single mutants as background, which revealed a higher production of psilocybin (Figure 5). This effect was even more pronounced when both aroH genes were deleted, resulting in a 10 times higher production.
FIGURE 5.
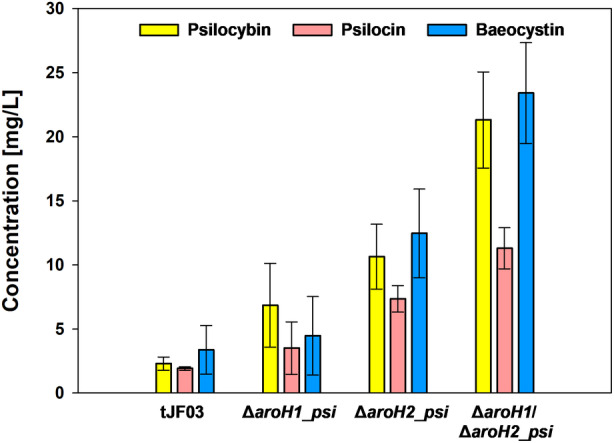
Psilocybin, psilocin and baeocystin production in A. nidulans mutant strains. Production of psilocybin, psilocin and baeocystin during shake flask cultivations of A. nidulans aroH single and double mutant strains expressing the psi cluster as a polycistron after 48 h (24 h post‐induction). Data are presented as averages and error bars indicate the standard error.
The best‐producing strain, ΔaroH1/ΔaroH2_psi, was chosen for further characterization and optimization of growth and product formation based on the online monitoring of the oxygen transfer rate (OTR) as well as carbon dioxide formation. A. nidulans ΔaroH1/ΔaroH2_psi was compared to tJF03 and monitored for 192 h (Figure 6). Until 21 h, both strains grew exponentially to their maximal oxygen transfer rates of 23 mmol/L/h (ΔaroH1/ΔaroH2_psi) and 24 mmol/L/h (tJF03) (Figure 6A). Although glucose was available at 21 h (Figure 6B), the OTR declined slowly, indicating a secondary substrate limitation (Anderlei & Büchs, 2001; Vrabl et al., 2019). Interestingly, the OTR and carbon dioxide transfer rate (CTR) of ΔaroH1/ΔaroH2_psi peaked again shortly after induction, whereas a plateau was observed for tJF03. From 48 h on, the respiratory activity decreased for both strains, also caused by glucose exhaustion around 72 h (ΔaroH1/ΔaroH2_psi) and until 96 h (tJF03). Due to the limitation of a second substrate, biomass formation was limited at maximum cell dry weights (CDW) of 30 g/L (tJF03) and 28 g/L (ΔaroH1/ΔaroH2_psi) after 96 h and 72 h. There was a considerable difference in pH between the two strains, with the ΔaroH1/ΔaroH2_psi cultures becoming more acidic at pH 6.1 (48 h), compared to pH 6.5 for the tJF03 cultures. By HPLC analysis, 2 g/L succinic acid and 1.4 g/L fumaric acid were detected after 48 h, however, simulating these amounts in media were not leading to the same pH shift, leaving the causality for pH differences elusive. Generally, a growth deficit was not apparent, even though tryptophan catabolism is repressed in ΔaroH1/ΔaroH2_psi.
FIGURE 6.
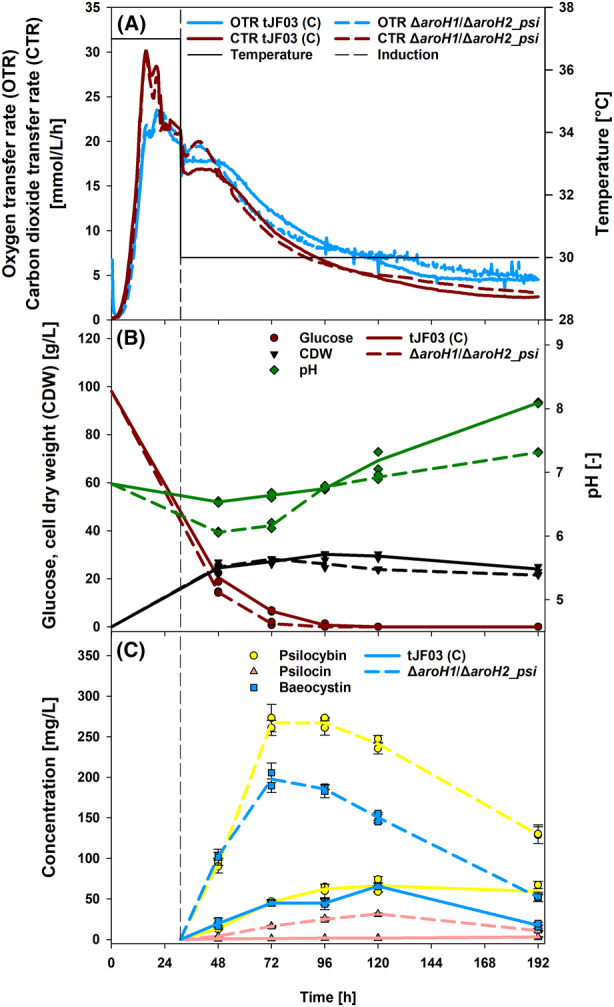
Process characterization and respiratory activity monitoring. (A) Online data of oxygen transfer rate (OTR) and carbon dioxide transfer rate (CTR) is shown for the A. nidulans double mutant ΔaroH1/ΔaroH2 and the control strain tJF03, both expressing the psi cluster as a polycistron. Induction was performed at 31 h upon decrease of metabolic activity observed by the decline in OTR signal. For improved enzyme production, the temperature was shifted to 30°C as indicated. (B) Offline data of glucose concentration, cell dry weight (CDW) and pH progression is shown over the cultivation time for both strains. (C) Production of psilocybin, psilocin and baeocystin over the time of cultivation for the wild type (tJF03) and double mutant backgrounds. Data are presented as averages from duplicate shake flask cultivations and triplicate product quantification with standard errors presented as error bars.
Regarding psilocybin formation, a maximum of 267 mg/L was formed in ΔaroH1/ΔaroH2_psi until 72 h (Figure 6C). This is fourfold higher and 1.7 times faster than the maximum of tJF03 with 66 mg/L at 120 h. Increased tryptophan, 4‐hydroxytryptamine and norbaeocystin concentrations observed in the double mutant further prove the hypothesis that the cellular l‐tryptophan pool is increased and preferentially routed towards psilocybin biosynthesis (Figure S3). Interestingly, no tryptamine was detected suggesting sufficient efficiency of PsiD. Compared to the initial expression test (Figure 5), the psilocybin concentration was increased 12‐fold by increasing cultivation time and modifying cultivation conditions (induction time, temperature regime and pH buffering). Similar to the previous results, a considerable amount of the precursor baeocystin was formed (Figure 5). This is probably due to the inadequate amount of psiM present in the polycistronic psi cluster. A strategy to further optimize the product formation could be achieved by introduction of additional copies of the gene in the psi cluster similar to the reported S. cerevisiae strain by Milne et al. (2020). In contrast to the yeast strain, psilocin production comprised minor amounts for both strains in this study, showing a less diverse product spectrum and more specific psilocybin production. Finally, in this shake flask cultivation, 267 mg/L of psilocybin was produced in 72 h in A. nidulans ΔaroH1/ΔaroH2_psi, resulting in a space–time‐yield (STY) of 3.7 mg/L/h. Although this was only a batch process, the STY is even higher than the 2.9 mg/L/h (627 mg/L in 213.7 h) achieved by the engineered S. cerevisiae during the fed‐batch bioprocess (Milne et al., 2020). A combination of both, the repression of catabolic pathways as well as the optimization of the biosynthetic pathway will probably lead to the highest psilocybin formation and will be matter of future investigations.
CONCLUSION
Currently, the demand for psilocybin for clinical studies and pharmaceutical purposes is met by chemical synthesis. Engineered microorganisms represent a viable in vivo alternative to reconstitute metabolic pathways for biotechnological production. Fungal hosts offer proper posttranslational processing of enzymes as well as cofactors. Specific to pathways including P450 enzymes, such as for psilocybin, compatible cytochrome P450 reductases are intrinsically provided by the host cell. In this study, we successfully demonstrated the positive effect of tryptophan catabolism repression on biosynthesis of the tryptamine‐derived prodrug psilocybin in S. cerevisiae and A. nidulans. Deletion of the aminotransferase genes ARO8 and ARO9 as well as the indoleamine 2,3‐dioxygenase gene BNA2 significantly increased psilocybin titres in S. cerevisiae. Genome mining of the A. nidulans genome revealed six genes coding for potential ARO‐like aminotransferases, two of which were verified to complement the S. cerevisiae Δaro8/Δaro9 aminotransferase‐deficient mutant. Simultaneous deletion of aroH1 and aroH2 in A. nidulans yielded a 10‐fold increased psilocybin production. Characterization and initial optimization of the process with the engineered A. nidulans strain based on online respiratory activity measurement resulted in a titre of 267 mg/L psilocybin. These findings contribute to a better understanding of tryptophan catabolism in fungi and enable improved biosyntheses of tryptophan‐derived alkaloids of pharmaceutical relevance in fungal hosts.
AUTHOR CONTRIBUTIONS
Slavica Janevska: Conceptualization; data curation; investigation; formal analysis; supervision; writing – original draft; funding acquisition. Sophie Weiser: Writing – review and editing; investigation; formal analysis; data curation; visualization. Ying Huang: Methodology; formal analysis; visualization. Jun Lin: Methodology; formal analysis; visualization. Sandra Hoefgen: Methodology; formal analysis; visualization. Katarina Jojić: Visualization; formal analysis. Amelia E. Barber: Methodology; formal analysis; visualization. Tim Schäfer: Formal analysis; visualization. Janis Fricke: Formal analysis; visualization. Dirk Hoffmeister: Supervision; funding acquisition; writing – review and editing. Lars Regestein: Formal analysis; supervision; funding acquisition; project administration. Vito Valiante: Writing – original draft; visualization; funding acquisition; supervision. Johann E. Kufs: Conceptualization; writing – review and editing; project administration; visualization; supervision.
CONFLICT OF INTEREST STATEMENT
The authors declare no competing interests.
Supporting information
Data S1.
ACKNOWLEDGEMENTS
We thank Daniela Hildebrandt for her excellent technical support. This work was supported by the Leibniz Research Cluster in the frame of the BMBF Strategic Process Biotechnology 2020+ (V.V.) and by a grant from the Free State of Thuringia and the European Social Fund (project HoWi, 2019FGR0079, L.R., S.W.). Work in S.J.'s laboratory is currently supported by a grant from the Free State of Thuringia and the European Social Fund Plus (project FusInfect, 2022FGR0007). Work in D.H.'s laboratory is funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation)—SFB 1127, Project‐ID 239748522 and under Germany's Excellence Strategy—EXC 2051, Project‐ID 390713860. S.W. and D.H. are authorized to handle controlled compounds according to the Narcotics Act of the Federal Republic of Germany.
Janevska, S. , Weiser, S. , Huang, Y. , Lin, J. , Hoefgen, S. , Jojić, K. et al. (2024) Optimized psilocybin production in tryptophan catabolism‐repressed fungi. Microbial Biotechnology, 17, e70039. Available from: 10.1111/1751-7915.70039
Slavica Janevska and Sophie Weiser contributed equally to this work.
Contributor Information
Slavica Janevska, Email: slavica.janevska@leibniz-hki.de.
Johann E. Kufs, Email: johann.kufs@uni-bielefeld.de.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Adams, A.M. , Kaplan, N.A. , Wei, Z. , Brinton, J.D. , Monnier, C.S. , Enacopol, A.L. et al. (2019) In vivo production of psilocybin in E. coli . Metabolic Engineering, 56, 111–119. [DOI] [PubMed] [Google Scholar]
- Anderlei, T. & Büchs, J. (2001) Device for sterile online measurement of the oxygen transfer rate in shaking flasks. Biochemical Engineering Journal, 7, 157–162. [DOI] [PubMed] [Google Scholar]
- Blei, F. , Baldeweg, F. , Fricke, J. & Hoffmeister, D. (2018) Biocatalytic production of psilocybin and derivatives in tryptophan synthase‐enhanced reactions. Chemistry—a European Journal, 24, 10028–10031. [DOI] [PubMed] [Google Scholar]
- Bok, J.W. , Hoffmeister, D. , Maggio‐Hall, L.A. , Murillo, R. , Glasner, J.D. & Keller, N.P. (2006) Genomic mining for aspergillus natural products. Chemistry & Biology, 13, 31–37. [DOI] [PubMed] [Google Scholar]
- Bouhired, S. , Weber, M. , Kempf‐Sontag, A. , Keller, N.P. & Hoffmeister, D. (2007) Accurate prediction of the Aspergillus nidulans terrequinone gene cluster boundaries using the transcriptional regulator LaeA. Fungal Genetics and Biology, 44, 1134–1145. [DOI] [PubMed] [Google Scholar]
- Caesar, L.K. , Robey, M.T. , Swyers, M. , Islam, M.N. , Ye, R. , Vagadia, P.P. et al. (2020) Heterologous expression of the unusual terreazepine biosynthetic gene cluster reveals a promising approach for identifying new chemical scaffolds. MBio, 11, e01691‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho, C. , Coulouris, G. , Avagyan, V. , Ma, N. , Papadopoulos, J. , Bealer, K. et al. (2009) BLAST+: architecture and applications. BMC Bioinformatics, 10, 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L. , Tang, J.W. , Liu, Y.Y. & Matsuda, Y. (2022) Aspcandine: a pyrrolobenzazepine alkaloid synthesized by a fungal nonribosomal peptide synthetase‐polyketide synthase hybrid. Organic Letters, 24, 4816–4819. [DOI] [PubMed] [Google Scholar]
- Choera, T. , Zelante, T. , Romani, L. & Keller, N.P. (2017) A multifaceted role of tryptophan metabolism and indoleamine 2,3‐dioxygenase activity in Aspergillus fumigatus‐host interactions. Frontiers in Immunology, 8, 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dindo, M. , Costanzi, E. , Pieroni, M. , Costantini, C. , Annunziato, G. , Bruno, A. et al. (2018) Biochemical characterization of Aspergillus fumigatus AroH, a putative aromatic amino acid aminotransferase. Frontiers in Molecular Biosciences, 5, 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar, R.C. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research, 32, 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flower, J.E. , Gibbons, W.J., Jr. , Adams, A.M. , Wang, X. , Broude, C.N. & Jones, J.A. (2023) Biosynthesis of psilocybin and its nonnatural derivatives by a promiscuous psilocybin synthesis pathway in Escherichia coli . Biotechnology and Bioengineering, 120, 2214–2229. [DOI] [PubMed] [Google Scholar]
- Fricke, J. , Blei, F. & Hoffmeister, D. (2017) Enzymatic synthesis of psilocybin. Angewandte Chemie, International Edition, 56, 12352–12355. [DOI] [PubMed] [Google Scholar]
- Fricke, J. , Lenz, C. , Wick, J. , Blei, F. & Hoffmeister, D. (2019) Production options for psilocybin: making of the magic. Chemistry—a European Journal, 25, 897–903. [DOI] [PubMed] [Google Scholar]
- Furter, R. , Paravicini, G. , Aebi, M. , Braus, G. , Prantl, F. , Niederberger, P. et al. (1986) The TRP4 gene of Saccharomyces cerevisiae: isolation and structural analysis. Nucleic Acids Research, 14, 6357–6373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin, G.M. , Aaronson, S.T. , Alvarez, O. , Arden, P.C. , Baker, A. , Bennett, J.C. et al. (2022) Single‐dose psilocybin for a treatment‐resistant episode of major depression. New England Journal of Medicine, 387, 1637–1648. [DOI] [PubMed] [Google Scholar]
- Hoefgen, S. , Lin, J. , Fricke, J. , Stroe, M.C. , Mattern, D.J. , Kufs, J.E. et al. (2018) Facile assembly and fluorescence‐based screening method for heterologous expression of biosynthetic pathways in fungi. Metabolic Engineering, 48, 44–51. [DOI] [PubMed] [Google Scholar]
- Hofmann, A. , Heim, R. , Brack, A. , Kobel, H. , Frey, A. , Ott, H. et al. (1959) Psilocybin und Psilocin, zwei psychotrope Wirkstoffe aus mexikanischen Rauschpilzen. Helvetica Chimica Acta, 42, 1557–1572. [Google Scholar]
- Iraqui, I. , Vissers, S. , Cartiaux, M. & Urrestarazu, A. (1998) Characterisation of Saccharomyces cerevisiae ARO8 and ARO9 genes encoding aromatic aminotransferases I and II reveals a new aminotransferase subfamily. Molecular & General Genetics, 257, 238–248. [DOI] [PubMed] [Google Scholar]
- Janevska, S. , Ferling, I. , Jojić, K. , Rautschek, J. , Hoefgen, S. , Proctor, R.H. et al. (2020) Self‐protection against the sphingolipid biosynthesis inhibitor fumonisin B1 is conferred by a FUM cluster‐encoded ceramide synthase. MBio, 11, e00455‐20. Available from: 10.1128/mbio.00455-00420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, M.E. , Stenbæk, D.S. , Juul, T.S. , Fisher, P.M. , Ekstrøm, C.T. , Knudsen, G.M. et al. (2022) Psilocybin‐assisted therapy for reducing alcohol intake in patients with alcohol use disorder: protocol for a randomised, double‐blinded, placebo‐controlled 12‐week clinical trial (the QUANTUM trip trial). BMJ Open, 12, e066019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, D.T. , Taylor, W.R. & Thornton, J.M. (1992) The rapid generation of mutation data matrices from protein sequences. Bioinformatics, 8, 275–282. [DOI] [PubMed] [Google Scholar]
- Kalyaanamoorthy, S. , Minh, B.Q. , Wong, T.K.F. , von Haeseler, A. & Jermiin, L.S. (2017) ModelFinder: fast model selection for accurate phylogenetic estimates. Nature Methods, 14, 587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H.T. , Na, B.K. , Chung, J. , Kim, S. , Kwon, S.K. , Cha, H. et al. (2018) Structural basis for inhibitor‐induced hydrogen peroxide production by kynurenine 3‐monooxygenase. Cell Chemical Biology, 25, 426–438.e424. [DOI] [PubMed] [Google Scholar]
- Kispal, G. , Csere, P. , Prohl, C. & Lill, R. (1999) The mitochondrial proteins Atm1p and Nfs1p are essential for biogenesis of cytosolic Fe/S proteins. The EMBO Journal, 18, 3981–3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krappmann, S. , Jung, N. , Medic, B. , Busch, S. , Prade, R.A. & Braus, G.H. (2006) The Aspergillus nidulans F‐box protein GrrA links SCF activity to meiosis. Molecular Microbiology, 61, 76–88. [DOI] [PubMed] [Google Scholar]
- Lee, J.W. , Na, D. , Park, J.M. , Lee, J. , Choi, S. & Lee, S.Y. (2012) Systems metabolic engineering of microorganisms for natural and non‐natural chemicals. Nature Chemical Biology, 8, 536–546. [DOI] [PubMed] [Google Scholar]
- Li, H. , Gilchrist, C.L.M. , Phan, C.S. , Lacey, H.J. , Vuong, D. , Moggach, S.A. et al. (2020) Biosynthesis of a new benzazepine alkaloid nanangelenin a from Aspergillus nanangensis involves an unusual l‐kynurenine‐incorporating NRPS catalyzing regioselective lactamization. Journal of the American Chemical Society, 142, 7145–7152. [DOI] [PubMed] [Google Scholar]
- Liu, X. , Wang, L. , Choera, T. , Fang, X. , Wang, G. , Chen, W. et al. (2023) Paralogous FgIDO genes with differential roles in tryptophan catabolism, fungal development and virulence in Fusarium graminearum . Microbiological Research, 272, 127382. [DOI] [PubMed] [Google Scholar]
- Milne, N. , Thomsen, P. , Mølgaard Knudsen, N. , Rubaszka, P. , Kristensen, M. & Borodina, I. (2020) Metabolic engineering of Saccharomyces cerevisiae for the de novo production of psilocybin and related tryptamine derivatives. Metabolic Engineering, 60, 25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh, B.Q. , Schmidt, H.A. , Chernomor, O. , Schrempf, D. , Woodhams, M.D. , von Haeseler, A. et al. (2020) IQ‐TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Molecular Biology and Evolution, 37, 1530–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi, K. , Chaleckis, R. , Takaine, M. , Wheelock, C.E. & Yoshida, S. (2017) Kynurenine aminotransferase activity of Aro8/Aro9 engage tryptophan degradation by producing kynurenic acid in Saccharomyces cerevisiae . Scientific Reports, 7, 12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panozzo, C. , Nawara, M. , Suski, C. , Kucharczyka, R. , Skoneczny, M. , Bécam, A.M. et al. (2002) Aerobic and anaerobic NAD+ metabolism in Saccharomyces cerevisiae . FEBS Letters, 517, 97–102. [DOI] [PubMed] [Google Scholar]
- Peck, S.K. , Shao, S. , Gruen, T. , Yang, K. , Babakanian, A. , Trim, J. et al. (2023) Psilocybin therapy for females with anorexia nervosa: a phase 1, open‐label feasibility study. Nature Medicine, 29, 1947–1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedras, M.S. , Yu, Y. , Liu, J. & Tandron‐Moya, Y.A. (2005) Metabolites produced by the phytopathogenic fungus Rhizoctonia solani: isolation, chemical structure determination, syntheses and bioactivity. Zeitschrift für Naturforschung Section C, 60, 717–722. [DOI] [PubMed] [Google Scholar]
- Qian, Z.G. , Xia, X.X. & Lee, S.Y. (2011) Metabolic engineering of Escherichia coli for the production of cadaverine: a five carbon diamine. Biotechnology and Bioengineering, 108, 93–103. [DOI] [PubMed] [Google Scholar]
- Reimer, C. , Kufs, J.E. , Rautschek, J. , Regestein, L. , Valiante, V. & Hillmann, F. (2022) Engineering the amoeba Dictyostelium discoideum for biosynthesis of a cannabinoid precursor and other polyketides. Nature Biotechnology, 40, 751–758. [DOI] [PubMed] [Google Scholar]
- Sayers, E.W. , Beck, J. , Bolton, E.E. , Bourexis, D. , Brister, J.R. , Canese, K. et al. (2021) Database resources of the national center for biotechnology information. Nucleic Acids Research, 49, D10–D17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, P. , Weber, M. , Rosenberger, K. & Hoffmeister, D. (2007) A one‐pot chemoenzymatic synthesis for the universal precursor of antidiabetes and antiviral bis‐indolylquinones. Chemistry & Biology, 14, 635–644. [DOI] [PubMed] [Google Scholar]
- Seibold, P.S. , Dörner, S. , Fricke, J. , Schäfer, T. , Beemelmanns, C. & Hoffmeister, D. (2024) Genetic regulation of l‐tryptophan metabolism in Psilocybe mexicana supports psilocybin biosynthesis. Fungal Biology and Biotechnology, 11, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spizzichino, S. , Pampalone, G. , Dindo, M. , Bruno, A. , Romani, L. , Cutruzzolà, F. et al. (2022) Crystal structure of Aspergillus fumigatus AroH, an aromatic amino acid aminotransferase. Proteins, 90, 435–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrabl, P. , Schinagl, C.W. , Artmann, D.J. , Heiss, B. & Burgstaller, W. (2019) Fungal growth in batch culture—what we could benefit if we start looking closer. Frontiers in Microbiology, 10, 2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J. , Chitsaz, F. , Derbyshire, M.K. , Gonzales, N.R. , Gwadz, M. , Lu, S. et al. (2023) The conserved domain database in 2023. Nucleic Acids Research, 51, D384–D388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wogulis, M. , Chew, E.R. , Donohoue, P.D. & Wilson, D.K. (2008) Identification of formyl kynurenine formamidase and kynurenine aminotransferase from Saccharomyces cerevisiae using crystallographic, bioinformatic and biochemical evidence. Biochemistry, 47, 1608–1621. [DOI] [PubMed] [Google Scholar]
- Yu, G. , Smith, D.K. , Zhu, H. , Guan, Y. & Lam, T.T.‐Y. (2017) GGTREE: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods in Ecology and Evolution, 8, 28–36. [Google Scholar]
- Yuasa, H.J. & Ball, H.J. (2011) Molecular evolution and characterization of fungal indoleamine 2,3‐dioxygenases. Journal of Molecular Evolution, 72, 160–168. [DOI] [PubMed] [Google Scholar]
- Zelante, T. , Choera, T. , Beauvais, A. , Fallarino, F. , Paolicelli, G. , Pieraccini, G. et al. (2021) Aspergillus fumigatus tryptophan metabolic route differently affects host immunity. Cell Reports, 34, 108673. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


