Abstract
Pancreatic cancer has the worst prognosis among all cancers underscoring the need for improved management strategies. Dysregulated mitochondrial function is a common feature in several malignancies, including pancreatic cancer. Although mitochondria have their own genome, most mitochondrial proteins are nuclear-encoded and imported by a multi-subunit translocase of the outer mitochondrial membrane (TOMM). TOMM22 is the central receptor of the TOMM complex and plays a role in complex assembly. Pathobiological roles of TOMM subunits remain largely unexplored. Here we report that TOMM22 protein/mRNA is overexpressed in pancreatic cancer and inversely correlated with disease outcomes. TOMM22 silencing decreased, while its forced overexpression promoted the growth and malignant potential of the pancreatic cancer cells. Increased import of several mitochondrial proteins, including those associated with mitochondrial respiration, was observed upon TOMM22 overexpression which was associated with increased RCI activity, NAD+/NADH ratio, oxygen consumption rate, membrane potential, and ATP production. Inhibition of RCI activity decreased ATP levels and suppressed pancreatic cancer cell growth and malignant behavior confirming that increased TOMM22 expression mediated the phenotypic changes via its modulation of mitochondrial protein import and functions. Altogether, these results suggest that TOMM22 overexpression plays a significant role in pancreatic cancer pathobiology by altering mitochondrial protein import and functions.
Keywords: Pancreatic cancer, mitochondria, protein import, TOMM22, respiratory complex, OXPHOS
Background
Pancreatic cancer is the fourth leading cause of cancer-related death in developed countries and the third in the United States (1). Its incidence and mortality rates are increasing with each coming year and if this trend continues, it will become the second leading cause of cancer-related death in the United States by 2030 or earlier (2). Pancreatic ductal adenocarcinoma (PDAC) is the most common and most aggressive subtype that constitutes more than 90% of all pancreatic malignancies (3). PDAC develops through a series of molecular alterations that promote their tumorigenic and metastatic behavior (3). Despite significant advances in our understanding of the molecular etiology of PDAC, we have made only minimal improvements in the survival of patients during the past decades. Clearly, there is a dire need to develop a more refined understanding of PDAC biology and characterize novel functional targets that could aid in improved diagnosis and therapy.
Mitochondria are important cellular organelles that not only serve as a major source of energy but are also involved in a variety of other biological processes (4). Otto Warburg in 1956 reported that tumor cells sustain their growth by relying upon energy produced via the process of aerobic glycolysis, a phenomenon referred to as the “Warburg effect” (5). Over time, this led to the misconception that mitochondrial respiration is dysfunctional in cancer. This view has been challenged in many studies and it is being increasingly recognized that mitochondrial respiration is functional in cancer cells and required for their proliferation, survival, and progression (6–9). Indeed, some studies have also reported diverse roles of mitochondria in PDAC tumorigenesis (6, 10–14). Mitochondria possess their own genome in multiple copies; however, it encodes for only a fraction of mitochondrial proteins (15). The majority of the mitochondrial proteins (99%) are nuclear-encoded and synthesized as precursors in the cytosol. These proteins are then imported into mitochondria through a well-organized mitochondrial protein import system consisting of seven machineries (15). The Translocase of the outer mitochondrial membrane (TOMM) machinery is comprised of several subunits and is the first to encounter most proteins destined for mitochondria (15).
TOMM22 acts as the central receptor in TOMM machinery and plays a crucial role in the import of nuclear-encoded mitochondrial proteins (4, 15). An earlier study demonstrated the role of TOMM22 in the survival of human umbilical vein endothelial cells (HUVECs) under high glucose conditions (16). However, its role in cancer pathobiology has remained completely unexplored. Here, we demonstrate, for the first time, that TOMM22 is overexpressed in PDAC and associated with poor survival of patients. An increasing level of TOMM22 expression is detectable in progressive pancreatic intraepithelial lesions (PanINs) suggesting its role in early pancreatic carcinogenesis. Utilizing both gain and loss-of-function approaches, we show that abundant TOMM22 expression supports the growth and malignant behavior of pancreatic cancer cells. We also demonstrate the effect of TOMM22 modulation on the mitochondrial protein makeup and define its impact on complex I activity, mitochondrial respiration, mitochondrial membrane potential (MMP), and ATP synthesis. Altogether, our study provides the first evidence for aberrant TOMM22 expression in PDAC development and progression, and suggests its important role in disease pathobiology.
Materials and Methods
Cell lines and culture conditions.
The human pancreatic cancer cell lines used in this study were purchased from ATCC. BxPC3 (RRID: CVCL_0186), HPAF (RRID: CVCL_0313), MIA PaCa (RRID: CVCL_0428), and T3M4 (RRID: CVCL_4056) cells were maintained in culture as adherent monolayer in Roswell Park Memorial Institute Medium (RPMI-1640) (Thermo Fischer Scientific, #11875-119). Panc1 (RRID: CVCL_0480), PaTu 8902 (RRID: CVCL_1845) and SW 1990 (RRID: CVCL_1723) cells were maintained in culture as an adherent monolayer in Dulbecco’s Modified Eagle’s Medium (DMEM) (Thermo Fischer Scientific, #11995-065). Media was supplemented with 10% fetal bovine serum (FBS) (R & D Systems, #S11150H) and 100 μM each of penicillin and streptomycin (VWR, #97063-708). HPDE (RRID: CVCL_4376) cell line was grown in Keratinocyte-SFM media supplemented with Epidermal Growth Factor and Bovine Pituitary Extract (Gibco, #17005042). Cells were cultured in a humidified atmosphere containing 5% CO2 at 37°C. All the cells were tested intermittently and determined to be free of mycoplasma contamination using the Lonza mycoalert mycoplasma detection kit (#LT07-318). Cell authentication was performed using short-tandem repeats genotyping. All experiments were performed with cells cultured in early passages.
In silico analysis of the transcripts of TOMM subunits in PDAC
To examine the differential expression of various TOMM subunits at the transcript level in normal pancreas against PDAC, and their association with patient survival, we screened publicly available GTex and TCGA datasets using web-based applications GEPIA http://gepia.cancer-pku.cn/ (RRID: SCR_018294) and UALCAN http://ualcan.path.uab.edu/ (RRID: SCR_015827).
Immunohistochemistry
Immunohistochemistry (IHC) was performed on formalin-fixed, paraffin-embedded normal pancreas (n=18), PanIN (n=11) and PDAC tissue sections (n=38). Tissue microarrays with adherent demographic information were obtained from US Biomax Inc. Sections were dewaxed in xylene (Azer Scientific) and rehydrated by passing through graded alcohol. Antigen retrieval was done using 0.01M citrate buffer pH 6 (Vector laboratories) and microwave at high heat for 30 minutes. The sections were treated with peroxidase (Vector laboratories) blocking for 5 minutes and then incubated with serum blocking agent for 15 minutes. Afterward, the sections were incubated with the primary antibody (TOMM22) (1:200 dilution) at 4°C overnight and then incubated with a biotinylated secondary antibody (Vector laboratories) for 30 minutes. The antibody was visualized by incubating with 3,3’-diaminobenzidine and counterstaining with Mayer’s hematoxylin (Vector laboratories).
Quantification of TOMM22 staining and clinico-pathological correlation
Immunostaining was assessed using the Aperio digital pathology system (Leica Biosystem, MA, USA; RRID: SCR_020993) with pathologist guidance. The digital pathology system measures the membranous, cytoplasmic and nuclear expression of various proteins on a three-point intensity scale (+1, +2 and +3) combined with the percent positivity of cells (0-100%). The whole tissue section from each subject was evaluated to determine the staining intensity and percent positive cells using the above digital pathology scoring system and Pathologist’s assistance (J.E.C.).
Generation of stable transfectants
To knockdown the expression of TOMM22 in the HPAF cell line, cells were transduced with TOMM22 siRNA lentiviral particles (piLenti-siRNA-GFP-TOMM22) as per the manufacturer’s instructions (Applied Biological Materials #472850910296). In addition, we stably overexpressed TOMM22 in the BxPC3 cell line using lentiviral particles expressing wild-type TOMM22 (pLenti-GIII-CMV-GFP-2A-Puro-TOMM22, #472850610396) (Applied Biological Materials). To generate control derivative cell lines, cells were transduced with lentiviral particles expressing non-targeted scrambled sequence (piLenti-siRNA-GFP-Scrambled) or Empty vector (Lenti-CMV-GFP-2A-Puro-Blank) (Applied Biological Materials #LVP015-G, #LVP690). Stable clones were selected in the presence of puromycin (Sigma Aldrich, #P9620) and examined for TOMM22 expression by western blotting. For our rescue experiments, BxPC3-TOMM22 overexpressing cells were transduced with TOMM22 shRNA particles (psi-LVRU6MH) from Genecopoeia (#LPP-HSH015315-LVRU6MH-100) according to manufacturer’s instructions. The corresponding control cell line was generated by transducing BxPC3-TOMM22 overexpressing cells with shRNA scrambled control-LVRU6MH lentiviral particles (Genecopoeia, #LP521-100). Stable clones were selected in the presence of hygromycin (Invitrogen, #10687010) and examined for TOMM22 expression by Western blotting.
Western blotting
Cells were washed once with cold phosphate-buffered saline (PBS) (Corning, #21-040-CV) and scraped into RIPA lysis buffer (Thermo Fischer Scientific, #89900) containing protease and phosphatase inhibitors (Thermo Fischer Scientific, #78442). Cell lysates were sonicated and subsequently centrifuged at 12,000 r.p.m. for 15 min at 4°C and supernatants were collected. Protein samples were quantified and subsequently resolved by electrophoresis on SDS–PAGE, transferred onto polyvinylidene difluoride (PVDF) membrane (Thermo Fischer Scientific, #88518) and subjected to standard immunodetection procedure using specific antibodies: TOMM22 (1 : 1000) (Abnova, #H00056993-M01J, RRID: AB_519123), MT-CO2 (1 : 1000) (Abcam, #ab79393, RRID: AB_1603751), OXPHOS cocktail (1 : 1000) (Abcam, #ab110411, RRID: AB_2756818), β-actin (1 : 20 000) (Cell Signaling, #12262, RRID: AB_2566811) and PCNA (1 : 1000) (Cell Signaling, #13110, RRID: AB_2636979). Secondary antibodies (Cell Signaling) were used at 1 :2000 dilutions. Blots were processed with SuperSignal West Femto Maximum Sensitivity Substrate Kit (Thermo Fischer Scientific, #34096) and the signal was detected using a ChemiDoc Imaging System (Bio-Rad, CA, USA).
Growth kinetics assay
(1 × 105) HPAF-Scr, HPAF-siTOMM22 cells and (2 × 105) BxPC3-Control, BxPC3-TOMM22, BxPC3-TOMM22-Scr as well as BxPC3-TOMM22-shTOMM22 cells were seeded in 6-well plates (Corning). Media was changed on a daily basis and cell growth was monitored by counting the number of viable cells every day for 7 days using a hemocytometer.
WST assay
(1× 104) HPAF-Scr and (1.5 × 104) BxPC3-TOMM22 cells were seeded in 96-well plates. After 24 hours, cells were treated with various doses of rotenone (1μM, 5μM, or 10μM) (Sigma-Aldrich, #R8875-5G) for 24 and 48 hours. Thereafter, WST reagent (Biovision, #K304-2500) was added to each well. Cells were incubated for 1 hour and the absorbance was measured using the Tecan plate reader.
Apoptosis assay
Apoptosis assay was conducted using a PE Annexin V Apoptosis Detection kit (BD Biosciences, #559763). (1.5 × 105) HPAF-Scr, HPAF-siTOMM22 cells and (2.0 × 105) BxPC3-Control, BxPC3-TOMM22 cells were seeded in 6-well plates. After 24 hours, serum-free media was added to the cells to induce apoptosis. Serum-containing media was used as a control. After 72 hours, the supernatant was collected in test tubes to preserve floating cells, after which adherent cells were harvested by trypsinization. Subsequently, harvested cells were added to tubes containing corresponding floating cells, centrifuged, washed, and resuspended in binding buffer at a final concentration of 106 cells/ml. 5 μL each of PE Annexin V and 7-AAD was added to 100 μL of cell suspension containing 105 cells followed by 15 min incubation at room temperature protected from light. Afterward, 400 μL of binding buffer was added to each tube and cells were analyzed by flow cytometry.
Colony formation assay
Cells were seeded at low density in 6-well plates; (4 × 102 cells per well) for HPAF-Scr and HPAF siTOMM22 cells and (1.5 × 103 cells per well) for BxPC3-Control and BxPC3-TOMM22 cells. Cells were allowed to form colonies for 2 weeks. Subsequently, colonies were fixed, stained with crystal violet (0.1% w/v), imaged, and counted using the Image analysis software (ImageJ; RRID: SCR_003070).
Migration and Invasion assays
(2.5 × 106) HPAF-Scr, HPAF-siTOMM22 cells and (2.0 × 106) BxPC3-Control, BxPC3-TOMM22 cells were resuspended in serum-free media and seeded in a polyethylene terephthalate membrane insert for migration assay (6-well inserts, pore size 8 μm; Corning, #353093). Inserts were placed into a 6-well plate containing media supplemented with 10% FBS that served as a chemoattractant. For the invasion assay, (7.5 × 105) HPAF-Scr, HPAF-siTOMM22 cells as well as (7 × 105) BxPC3-Control, BxPC3-TOMM22 cells, BxPC3-TOMM22-Scr and BxPC3-TOMM22-shTOMM22 cells were resuspended in serum-free media and seeded in a matrigel-coated polycarbonate membrane insert (24-well inserts, pore size 8 μm, Corning, #354483). In addition, (7.5 × 105) HPAF-Scr cells and (7 × 105) BxPC3-TOMM22 cells were resuspended in serum-free media containing 0μM or 10μM rotenone and 0μM or 5μM rotenone respectively, after which, they were seeded in matrigel-coated polycarbonate membrane inserts for invasion assay. Inserts were placed into a 24-well plate containing media supplemented with 10% FBS that served as a chemoattractant. After 24 hours of incubation, cells on the upper surface of the membrane inserts were removed by cotton swab, and cells that migrated/invaded to the bottom surface were stained using Hema 3™ stat pack (Fisher Scientific, #23-123869). Subsequently, membranes were mounted on a slide, and migrated and invaded cells were counted in 10 random fields of view under a microscope at 10X magnification.
Isolation of mitochondria and mitochondrial protein
Isolation of mitochondria was done using the mitochondrial isolation kit for cultured cells by Thermo Fischer (#89874) according to the manufacturer’s instructions. Thereafter, mitochondrial pellets were lysed using RIPA buffer with protease and phosphatase inhibitors. Mitochondrial protein concentration was determined by Lowry method followed by SDS-PAGE and immunoblotting.
Sample preparation for mass spectrometry
50μg of total mitochondrial protein from 4.7 × 107 (HPAF-Scr and HPAF-siTOMM22 cells) was loaded on 12% Mini-PROTEAN® TGX gels (Bio-Rad, #4561044) and ran at 100V for 1hour 20 min. The gels were stained with Coomassie blue R-250 for 20 min and subsequently destained in a solution containing 45% methanol and 10% acetic acid for 2 hours. Each lane of the gels was cut into smaller pieces (~1mm in size) using a cutting device and transferred into 1.5ml Eppendorf tubes. The gel pieces were washed with 200 μl of 50% 200mM ammonium bicarbonate (ABC)/ 50% methanol three times for 15 min each, after which, they were dehydrated with 200μl acetonitrile (ACN) and dried in a speed vacuum. Subsequently, 50μL urea solution was added to the gel slices. Gel slices were then incubated for 15 min at 37°C in the ThermoMixer. Afterward, 5μL of iodoacetamide (IAA) was added to the gel pieces for the alkylation of the SH groups of the cysteine residues. The gel pieces were then incubated for 15 min at 37°C in a water bath. Afterward, the gel pieces were washed 3 times and again dehydrated with 200μl ACN, after which they were dried in a speed vacuum and rehydrated with 40μl of trypsin solution. Gel slices were then incubated at 37°C in the ThermoMixer overnight. Following overnight digestion, gel pieces were centrifuged for 5 min at 10,000g. After centrifugation, the supernatant (which contains the tryptic peptides) was transferred to MS sample vials. Samples were dried in the speed vacuum and frozen at −80°C until MS analysis. On the day of analysis, the dried samples were dissolved with 35μL of the 2% ACN/ 1% HAc solution and ran on the Thermo Q Exactive Plus mass spectrometer.
LC-MS/MS Analysis
Samples were subjected to 60-minute data-dependent LC-MS/MS analyses using a 2.0 μl injection volume for each. Peptides were loaded onto a Thermo EASY-Spray C18 reversed-phase column (75μm X 150mm, 3μm particle size), and eluted using a linear gradient at 300nl/min. Electrospray ionization was performed in positive mode with the EASY-Spray Ion Source. Mass spectra were acquired from 400-2000 m/z at 70,000 resolution (MS1 data). Data-dependent acquisition was performed on the top 6 most abundant precursor ions using HCD fragmentation (MS2 data) with a normalized collision energy of 27 and an isolation width of 2.0m/z at 17,500 resolution. A 15-second dynamic exclusion was utilized to minimize redundant MS2 scans and to increase the number of peptide identifications. Precursor ions that had a single charge state, or a charge state +5 or greater were excluded. Two blanks were run in between each sample to minimize and monitor for carryover.
Analysis of MS/MS Data
Thermo Proteome Discoverer 2.2 software was used to provide protein identification and relative abundance. The files were searched against the reviewed human sequences from Uniprot (March 2022) (20,292 sequences). Semi-tryptic peptides were allowed, with a precursor mass tolerance of 10 ppm and a fragment tolerance of 0.05Da, permitting a maximum of 2 missed cleavage sites. Cysteine alkylation was set as a static modification (carbamidomethyl, +57.021Da) due to the IAA that was used during preparation. No other post-translational modifications were included. Abundance was based on precursor area, using unique and/or razor peptides only. Peptides were restricted to those with a minimum peptide length of 5 residues. Identified proteins along with their individual abundances per sample, sequest scores and PSMs were compared across both samples/injections.
Complex I enzyme activity assay
Complex I enzyme activity was measured using the Complex I enzyme activity assay kit (Abcam, #ab109721). Total protein was isolated from cells and quantified by Lowry method. After which, an equal amount of protein was loaded in each well of the Complex I microplate. Thereafter, protein samples were incubated for 3 hours. After incubation, protein samples were removed, each well of the microplate was washed and the assay solution was added. Optical density was measured in kinetic mode every 30 seconds for a duration of 30 minutes using a Tecan microplate reader.
Measurement of oxygen consumption rate and extracellular acidification rate
Cellular oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) were measured using a Seahorse Bioscience XF96 Extracellular Flux Analyzer (Seahorse Bioscience). Cells were seeded in 96-well plates at 1× 104 (HPAF Scr, HPAF siTOMM22 cells), 2× 104 (BxPC3-Control, BxPC3-TOMM22 cells) and 1.5× 104 (HPDE, HPAF and BxPC3 cells) per well. After 48 hours, seahorse XF RPMI medium (Seahorse Bioscience, #103576-100) supplemented with 1 mM pyruvate, 2 mM glutamine, and 10 mM glucose was added to the cells for the OCR measurement. After which cells were placed in the non-CO2 incubator for one hour to degas the plate. Subsequently, cells were placed in the seahorse instrument and OCR was measured in response to consecutive addition of 1 μM oligomycin (oligo), 1 μM carbonyl cyanide p-triflouromethoxyphenylhydrazone (FCCP), and 0.5 μM rotenone and antimycin A (Rot/AA). For the ECAR measurement, seahorse XF RPMI medium supplemented with 2 mM glutamine was added to the cells after 48 hours of seeding. Afterward, cells were placed in the non-CO2 incubator for one hour to degas the plate and subsequently placed in the seahorse instrument. ECAR was measured in response to consecutive addition of 10 mM glucose, 1 μM oligomycin, and 50 mM 2-deoxy-glucose (2-DG). Data were normalized to relative cell numbers.
Measurement of mitochondrial membrane potential
1× 104 (HPAF Scr, HPAF siTOMM22 cells) and 2× 104 (BxPC3-Control, BxPC3-TOMM22 cells) were plated in each well of a 96-well plate. At 48 hours, 50μM Carbonyl cyanide m-chlorophenyl hydrazone (CCCP) (Abcam, #ab141229) was added to the media, which was added to the control wells as a positive control. After 15 mins of incubation, media was removed from all wells, cells were rinsed with PBS, 20μM of JC-1 dye (Abcam, #ab113850) was added to PBS, vortexed and the staining solution was added to the cells. Cells were incubated in the staining solution for 20 minutes at 37°C, after which, cells were rinsed once with PBS. Subsequently, PBS was added to each well and JC-1 fluorescence was measured using a Tecan fluorescence plate reader. Data were normalized to relative cell numbers.
ATP measurement
ATP assay was done using the luminescent ATP Detection kit (Abcam, #ab113849). Cells were seeded in 96-well plates at 1× 104 (HPAF Scr, HPAF siTOMM22 cells) and 2× 104 (BxPC3-Control, BxPC3-TOMM22, BxPC3-TOMM22-Scr, BxPC3-TOMM22-shTOMM22 cells) per well. After 48 hours, detergent was added to the cells to lyse them. Subsequently, the substrate was added to the cells and the emitted light was measured using a luminometer. ATP Concentration was calculated from the ATP standard curve and normalized to the respective cell numbers. To determine the effect of rotenone on ATP production, 1× 104 HPAF-Scr and 1.5× 104 BxPC3-TOMM22 cells were seeded in 96-well plates. After 24 hours, HPAF-Scr cells were treated with 0 or 10μM rotenone and BxPC3-TOMM22 cells were treated with 0 or 5μM rotenone for 48 hours, after which, ATP concentration was determined as described above.
NAD+/NADH measurement
NAD+/NADH ratio was determined using the NAD+/NADH quantification kit (Abcam #ab176723) according to the protocol provided by the manufacturer. Cells were lysed in lysis buffer, after which, cell lysates and NADH standards were added to 96-well plates. Subsequently, NAD+/NADH extraction solution and NAD+/NADH control solution were added to the sample wells and standard wells respectively. After 15 minutes of incubation at 37°C, a neutralization solution was added to neutralize the NAD+ and NADH extracts respectively, after which, the reaction mixture was added to each well. Thereafter, the plate was incubated at room temperature in the dark for 2 hours and the fluorescence was measured using a Tecan plate reader. NAD+/NADH ratio was calculated by dividing the NAD+ values by NADH and normalized according to protein concentration.
Measurement of reactive oxygen species (ROS)
3× 105 (HPAF Scr, HPAF siTOMM22 cells) and 4× 105 (BxPC3-Control, BxPC3-TOMM22 cells) were seeded in 6-well plates. After 48 hours of incubation, 5μM MitoSox Red dye (Invitrogen) was added to serum-free media, mixed and added to the cells. Cells were incubated for 30 mins at 37°C. Thereafter, cells were rinsed once with PBS, trpsinized and centrifuged. After removing the supernatant, cells were resuspended in PBS and MitoSox Red fluorescence was measured by flow cytometry.
Statistical analysis
All experiments described above were performed at least three times and data obtained are presented as mean ± SEM. Unpaired Student’s t-test was used to calculate the statistical significance between two groups. One-way analysis of variance (ANOVA) was used to calculate statistical significance among three groups. When two independent variables and a dependent variable were analyzed, a two-way ANOVA was performed. All analyses were performed using GraphPad Prism version 9 (GraphPad, Inc.; RRID: SCR_002798). p ≤ 0.05 was considered significant.
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (17) partner repository with the dataset identifier PXD040198. All other data can be made available upon reasonable request.
Results
TOMM22 is overexpressed in PDAC patients and associated with poor survival.
To examine the protein expression pattern of TOMM22 in human PDAC tissues, PanIN lesions and normal pancreas tissues, we procured tissue microarrays with adherent demographic information and performed immunohistochemistry. We confirmed TOMM22-positive staining in the cytoplasm of both PanIN and PDAC tumors (Figure 1A, Supplementary figure 1). Further, we observed progressively increased expression of TOMM22 in PanIN lesions (n=11) compared to normal tissues (n=18) and was even higher in PDAC tissues (n=38) (p<0.0001) (Figure 1A, bar graph). TOMM22 expression did not correlate with advancing cancer grades (Figure 1B). However, TOMM22 expression was detectable in early-grade tumors. Notably, five out of the nine grade 1 tumors were clinically stage 4 tumors, and 2 each were stage 2 and stage 1 tumors respectively (Figure 1C). All five stage 4 tumors exhibit progressively higher levels of TOMM22 expression compared to the stage 1 and stage 2 tumors (Figure 1D). We also observed an association between increased TOMM22 expression and progressive cancer stages (p<0.0001) (Figure 1E). In parallel, we also determined the expression pattern of TOMM22 and various other TOMM subunits at the mRNA level in PDAC and their association with patients’ survival utilizing the GTex and TCGA datasets. We observed a significantly elevated level of TOMM22 mRNA expression in PDAC subjects (n=179) compared to the normal (n=171) (p<0.01) (Figure 1F). High TOMM22 mRNA expression predicted the worst patient survival of the PDAC patients (p=0.0051) (Figure 1G). Among the other TOMM subunits, we observed significantly increased mRNA expression of TOMM40, TOMM5 and TOMM6 in PDAC (n=179) compared to normal cases (n=171) (p<0.01) (Supplementary figure 2A–F). However, there was no association between TOMM40, TOMM5 and TOMM6 mRNA expression and patients’ survival (p=0.31-0.99) (Supplementary figure 2G–I). Taken together, these findings suggest an important role of TOMM22 in PDAC development and progression.
Figure 1. TOMM22 expression is increased in PDAC and negatively correlates with patients’ survival.
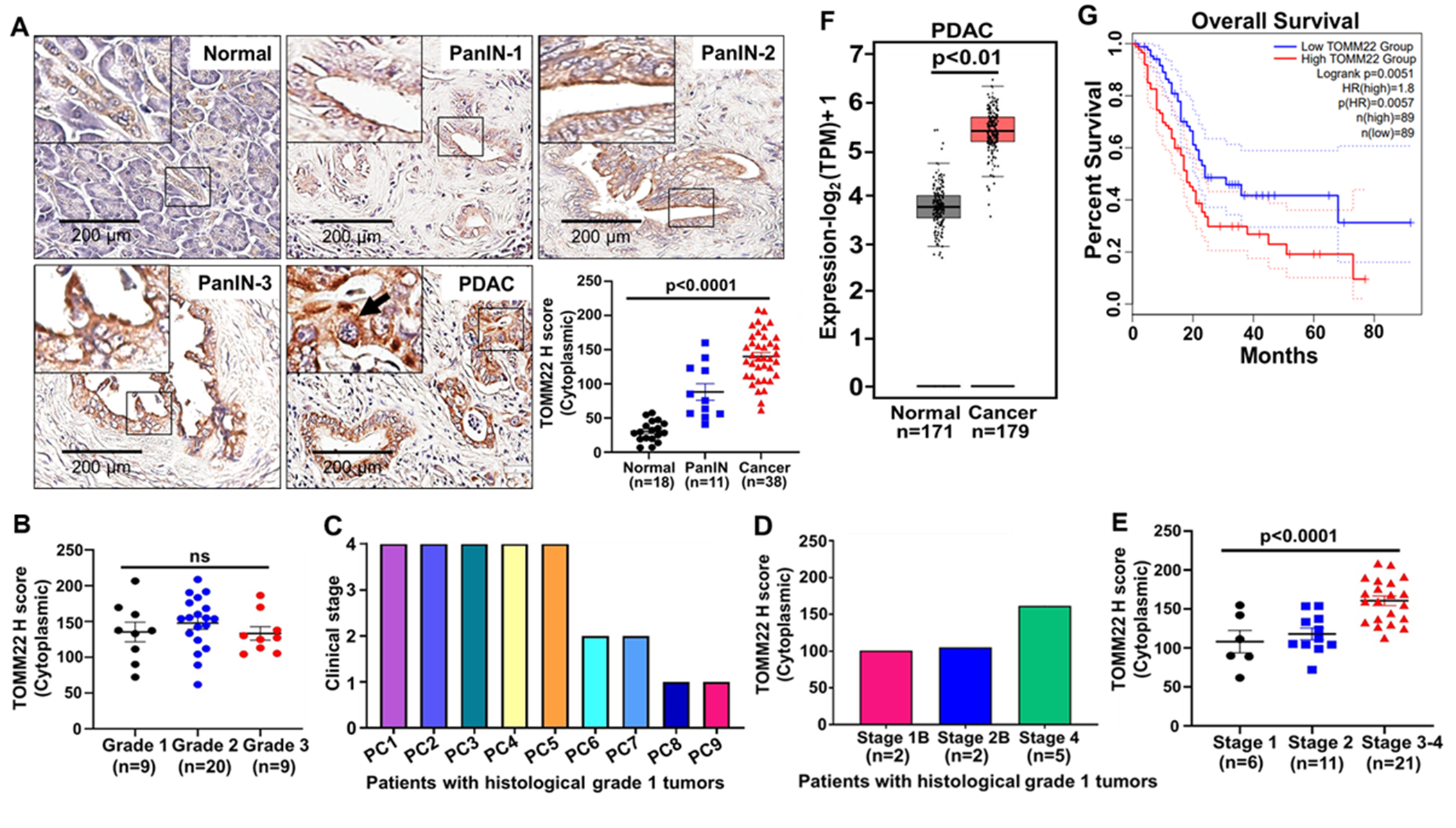
TOMM22 expression in the normal pancreas (n=18), PanIN (n=11) and PDAC tissues (n=38) were determined by immunohistochemistry (A). TOMM22 staining was found in the cytoplasm (arrow). The bar graph indicates the relative abundance of TOMM22 in various groups. Assessment of immunostaining was carried out by the Aperio digital pathology system (Leica Biosystem) with the pathologist’s guidance. Association of TOMM22 expression with grades (B), clinical stages of the grade 1 pancreatic cancer (PC) patients (C), TOMM22 expression pattern in the nine grade 1 tumors from various clinical stages (D). Association of TOMM22 expression with stages (E). Data are presented as mean ± SEM. Statistical analysis by one-way ANOVA. mRNA expression level of TOMM22 in pancreatic cancer (n=179) and normal (n=171) samples (F) and its association with patient survival (G) was screened using datasets from GTex and TCGA. GTex-Genotype-tissue expression, TCGA-The cancer genome atlas, PanIN-Pancreatic intraepithelial neoplasia, PDAC-pancreatic ductal adenocarcinoma.
TOMM22 overexpression is associated with PDAC cell growth, clonogenicity, and malignant behavior.
To evaluate the biological relevance of TOMM22 in PDAC, first, we analyzed the protein expression of TOMM22 in a panel of seven pancreatic cancer cell lines. Among these lines, HPAF cells established from a metastatic PDAC patient exhibited the highest expression of TOMM22. On the other hand, BxPC3 cells established from a primary PDAC tumor had the lowest expression of TOMM22 (Figure 2A). To further investigate the association of high TOMM22 expression with PDAC cell proliferation, we knocked down TOMM22 expression in the HPAF cell line using lentiviral particles harboring a pool of TOMM22 targeting siRNAs. Conversely, we established a stable BxPC3 cell line forcedly overexpressing TOMM22 using a similar lentiviral transduction system. After the selection of stable clones, both cell lines were examined for TOMM22 expression by Western blotting (Figure 2B–C). Following that, we performed growth and clonogenic assays in the TOMM22 modulated cell lines and their respective controls. We observed that HPAF-siTOMM22 (TOMM22 knockdown) cells grew slower than control HPAF-Scr cells with approximately 31% (p<0.0001) difference in their growth on the 7th day (Figure 2D). On the contrary, BxPC3-TOMM22 (TOMM22 overexpressing) cells grew at a faster rate compared to BxPC3-Control cells with approximately 53% (p<0.0001) increased growth on the 7th day (Figure 2E). Furthermore, our data demonstrate a 4.3-fold decrease in the clonogenic ability (p<0.0001) of the HPAF-siTOMM22 cells compared to the control HPAF-Scr cells (Figure 2F), whereas the colony-forming ability of BxPC3-TOMM22 cells was recorded as 7-fold higher (p=0.0008) compared to the BxPC3-Control cells (Figure 2G). To further confirm these phenotypic effects, we knocked down the expression of TOMM22 in the TOMM22-overexpressing BxPC3-TOMM22 cell line using targeted shRNAs and examined for TOMM22 expression by Western blotting after the selection of stable clones (Supplementary figure 3A). We next performed a growth assay and observed that BxPC3-TOMM22-shTOMM22 cells grew slower compared to BxPC3-TOMM22-Scr cells with approximately 42% difference (p<0.0001) in their growth on the 7th day (Supplementary figure 3B). Together, our data establish a growth promoting role of TOMM22 in PDAC cells. Since the growth rate can be influenced by the rate of apoptosis, we performed an apoptosis assay to examine the impact of TOMM22 overexpression on apoptosis. Interestingly, TOMM22 silencing had no effect on the apoptotic rate in HPAF cells (Supplementary figure 4A), however, TOMM22 overexpression led to reduced apoptosis rate in BxPC3 cells (Supplementary figure 4B), both under serum-containing (p=0.03) and serum-starved conditions (p=0.0006).
Figure 2. Increased TOMM22 expression promotes PDAC cell growth, clonogenic ability and malignant potential.
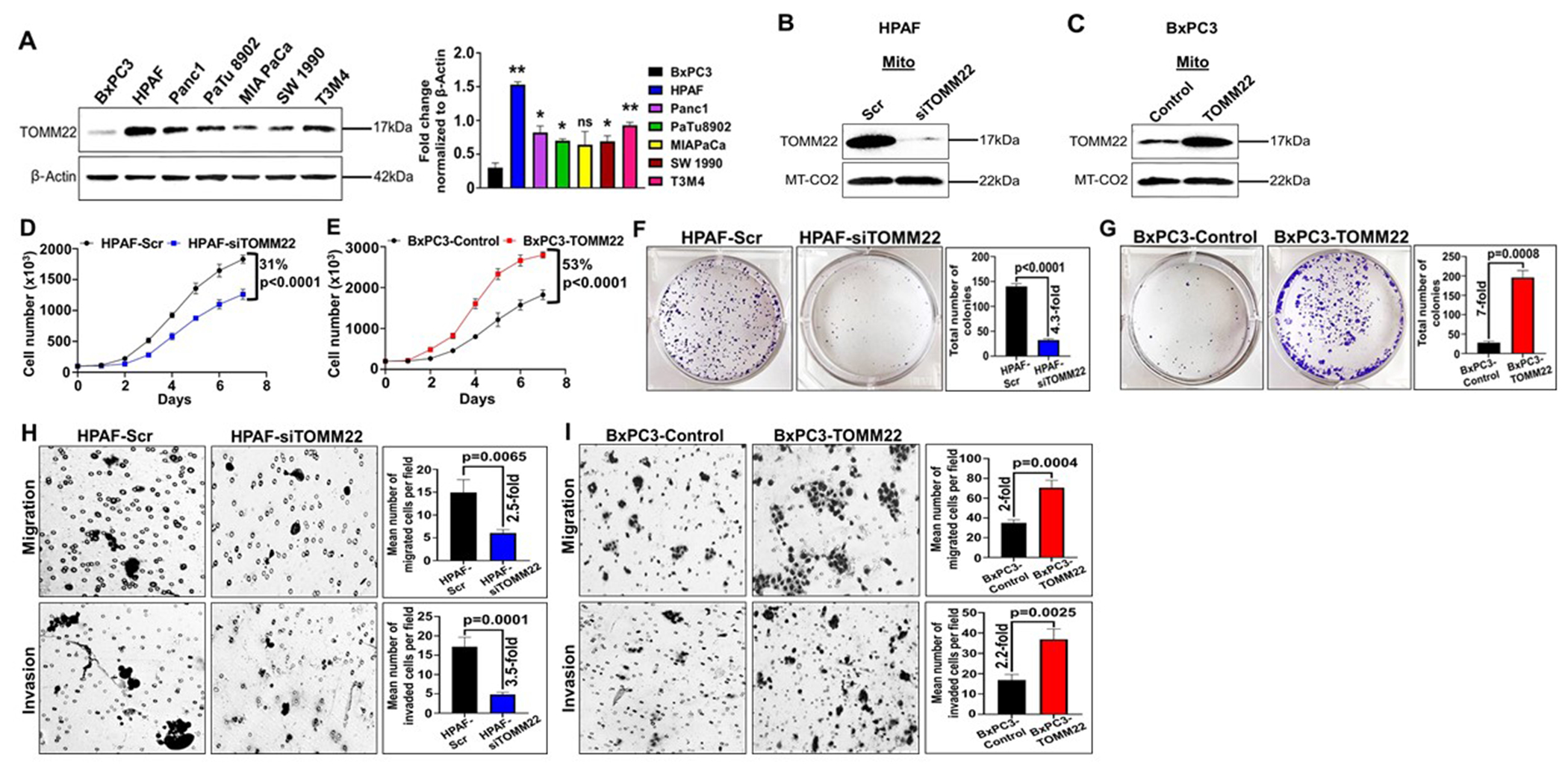
Protein expression of TOMM22 in the whole cell lysate of seven pancreatic cancer cell lines and densitometric evaluation of TOMM22/ β-Actin ratio (A). β-Actin was used as a loading control. Data represent mean ± SEM, (n=3). TOMM22/ β-Actin ratio of BxPC3 was compared to the TOMM22/ β-Actin ratio of other pancreatic cancer cell lines. Statistical analysis by unpaired Student’s t-test. (*p < 0.05, **p < 0.01). Protein expression of TOMM22 in the mitochondrial lysate of TOMM22 silenced cells [HPAF-siTOMM22] (B), TOMM22 overexpressing cells [BxPC3-TOMM22] (C) and their respective control cells. MT-CO2 was used as a loading control. Growth kinetics measurement of PDAC cell lines exhibiting silencing [HPAF-siTOMM22] (D) or forced overexpression [BxPC3-TOMM22] (E) of TOMM22 relative to their respective controls. Data represent mean ± SEM, (n=3). Statistical analysis by two-way ANOVA. Colony formation assay of TOMM22 silenced cells (F), TOMM22 overexpressing cells (G) and their controls. Representative images of pancreatic cancer colonies and their quantification are shown. Data is presented as mean ± SEM, (n=3). Statistical analysis by unpaired Student’s t-test. HPAF-Scr, HPAF-siTOMM22 cells (H) and BxPC3-Control, BxPC3-TOMM22 cells (I) were resuspended in serum free media and seeded on non-coated or matrigel-coated membranes for migration and invasion assays respectively. Media supplemented with 10% FBS was used as a chemoattractant in lower wells and cells were allowed to migrate or invade for 24 hours. Data are presented as the mean number of cells in 10 random fields ± SEM, (n=3). Statistical analysis by unpaired Student’s t-test. Scr: Scrambled control; siTOMM22: TOMM22 targeted siRNA pool (a set of 4 targets); Control: empty vector; TOMM22: lentivirus with wild type TOMM22.
To determine the impact of TOMM22 expression on the aggressive behavior of PDAC cells, we performed migration and invasion assays. A 2.5-fold decrease (p=0.0065) was observed in the migration of HPAF-siTOMM22 cells compared to HPAF-Scr cells. Further, the number of invaded cells from the HPAF-siTOMM22 group decreased by 3.5-fold (p=0.0001) compared to control HPAF-Scr cells (Figure 2H). Conversely, the migration and invasion of BxPC3-TOMM22 cells were enhanced by 2-fold (p=0.0004) and 2.2-fold (p=0.0025) respectively compared to the control cells (Figure 2I). Additionally, the knockdown of TOMM22 in the BxPC3-TOMM22 cells reversed this phenotype, which demonstrated a 3-fold (p=0.0004) decrease in their invasion compared to BxPC3-TOMM22-Scr cells (Supplementary figure 3C).
TOMM22 regulates the import of cytosolic pre-proteins in the mitochondria.
Since TOMM22 is crucial for the import of mitochondrial proteins, we examined the effect of its silencing on mitochondrial protein import. Mitochondria were isolated from HPAF-Scr and HPAF-siTOMM22 cells followed by total mitochondrial protein preparation. Then, we loaded the protein samples on polyacrylamide gel for electrophoretic separation to remove chemical impurities from the mitochondrial isolation process. Gel was digested to extract proteins followed by MS analysis (Supplementary figure 5A). The purity of mitochondrial protein preparation was examined by immunoblotting for PCNA (nuclear marker) and MT-CO2 (mitochondrial marker) prior to MS analysis (Figure 3A). Chromatograms generated during MS analysis were analyzed using the Thermo Proteome Discoverer 2.2 software (Supplementary figure 5B, C), which provided label-free quantification of proteins using the peak areas of identified peptides. Subsequently, the abundance of each protein was normalized to the abundance of MT-CO2, a mitochondrial DNA-encoded protein whose expression remains unchanged upon TOMM22 modulation.
Figure 3. Mass spectrometry analysis reveals differential import of mitochondrial proteins.
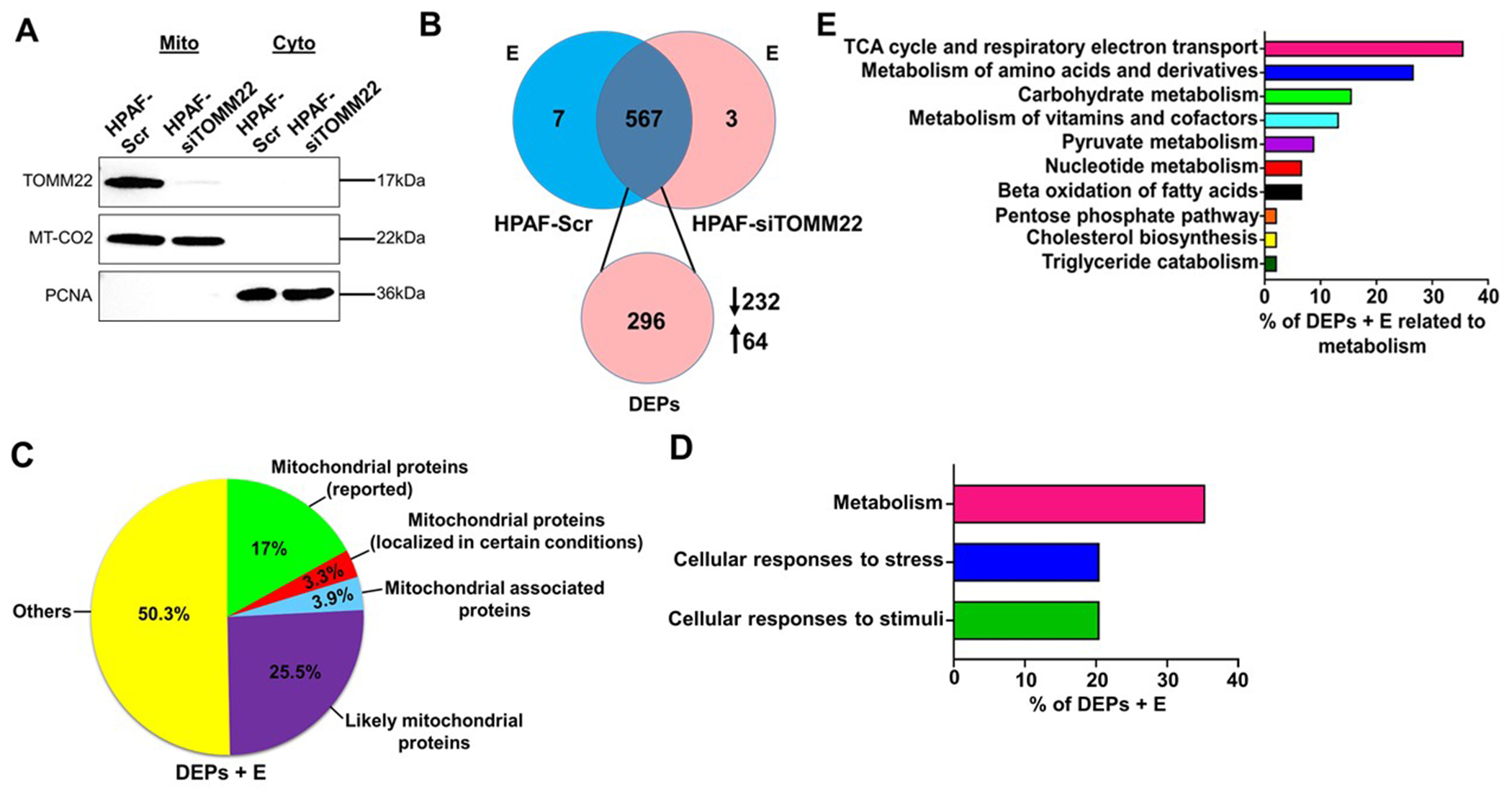
Expression of TOMM22 protein in mitochondrial and cytosolic lysates of HPAF-Scr and HPAF-siTOMM22 cells. MT-CO2 was used as a mitochondrial loading control and PCNA was used to ascertain the purity of the mitochondrial fraction (A). The Venn diagram represents the total number of proteins and DEPs identified by mass spectrometry. The downward arrow indicates the number of proteins whose abundance was decreased in the HPAF-siTOMM22 samples compared to HPAF-Scr samples while the upward arrow shows the number of proteins whose abundance was increased in the HPAF-siTOMM22 samples compared to HPAF-Scr samples (B). Pie chart represents the distribution of (DEPs + E) into various groups (C). Pathway analysis of (DEPs + E) known to localize to or associate with mitochondria was done using Reactome and KEGG databases (D, E). DEPs: Differentially expressed proteins; E: proteins exclusive to either sample.
A total of 577 proteins were detected, of which 7 and 3 were exclusively present in the HPAF-Scr and HPAF-siTOMM22 samples, respectively, and 296 were differentially present (Figure 3B). Out of 296 differentially expressed proteins (DEPs), 232 exhibited ≥ 1.5-fold decrease in the HPAF-siTOMM22 relative to the HPAF-Scr, whereas 64 showed ≥ 1.5-fold increased levels in the HPAF-siTOMM22 samples as opposed to the HPAF-Scr (Figure 3B). DEPs and proteins exclusive to either sample (DEPs + E) were compared using the Uniprot and Mitocarta databases. 17% of the (DEPs +E) in our study were those that are reported as mitochondrial proteins, while 3.3% are known to localize to the mitochondria in certain diseased conditions (Figure 3C). 3.9% of the (DEPs + E) were categorized as mitochondria-associated proteins and 25.5% as likely mitochondrial proteins (proteins that have not yet been reported to localize to mitochondria but whose isoforms have been reported to localize therein). 50.3% of the (DEPs + E) were classified as others since they have not yet been identified to localize or associate with mitochondria (Figure 3C). Pathway analysis of the 152 (DEPs + E) known to localize to or associate with mitochondria (Supplementary table 1) using Reactome and KEGG databases identified metabolism as the most enriched pathway in the HPAF-Scr cells beside cellular responses to stress and cellular responses to stimuli pathways (Figure 3D). Amongst the enriched metabolism related pathways, we observed that most of the (DEPs + E) were involved in the citric acid cycle and respiratory electron transport pathways (Figure 3E).
TOMM22 enhances the import of nuclear-encoded subunits of the electron transport chain and complex I activity.
Since the citric acid cycle is responsible for generating reducing equivalents that will be oxidized at the electron transport chain (ETC) and both of these pathways are enriched in the HPAF-Scr cell lines; we confirmed the effect of TOMM22 modulation on the import of subunits of the various respiratory complexes (RC) by immunoblotting. The expression of the nuclear encoded ATP5A (RCV), UQCRC2 (RCIII), SDHB (RCII) and NDUFB8 (RCI) was markedly decreased (p< 0.05) in HPAF-siTOMM22 cells compared to the control cells (Figure 4A). We also recorded a 50% reduction (p=0.0302) of the RCI activity in the HPAF-siTOMM22 cell lines compared to HPAF-Scr cells (Figure 4B). On the other hand, restoration of TOMM22 in the BxPC3 cells (BxPC3-TOMM22), reversed the expression of the above RC proteins (Figure 4C) with a concomitant increase in RCI activity by 38% (p=0.0285) (Figure 4D). Since RCI is responsible for oxidizing NADH to NAD+, we measured NAD+/NADH ratio to further confirm the increased activity of RCI in the TOMM22 overexpressing cell lines. Our data show that NAD+/NADH ratio is higher in TOMM22 overexpressing cells compared to cells with a low level of TOMM22 expression (Figure 4E–F). Altogether, these data show that TOMM22 regulates the import of nuclear encoded mitochondrial proteins, thereby influencing RCI activity.
Figure 4. TOMM22 overexpression is associated with increased import of nuclear-encoded subunits of the electron transport chain, Complex I activity and NAD+/NADH ratio.
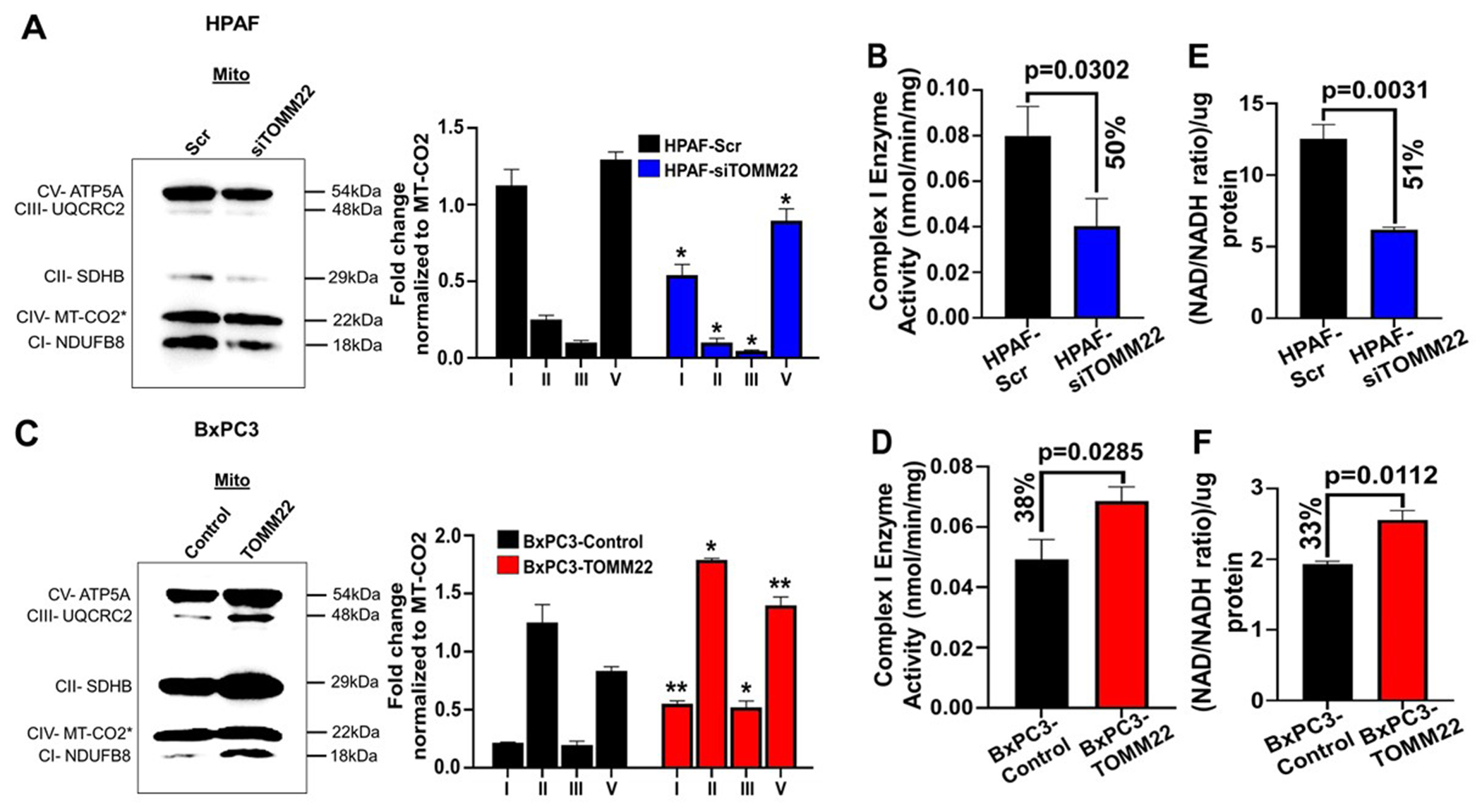
Protein expression of ATP5A (RCV), UQCRC2 (RCIII), SDHB (RCII), MT-CO2 (RCIV) and NDUFB8 (RCI) in the mitochondrial lysate of TOMM22 silenced cells [HPAF-siTOMM22] (A), TOMM22 overexpressing cells [BxPC3-TOMM22] (C) and their respective control cells, and densitometric evaluation of each nuclear encoded subunit/ MT-CO2 ratio. Protein with asterisk (*) (MT-CO2) is a mitochondrial DNA encoded protein from RCIV. Data represent mean ± SEM, (n=3). Statistical analysis by unpaired Student’s t-test. (*p < 0.05, **p < 0.01). Complex I enzyme activity of HPAF-siTOMM22 cells (B), BxPC3-TOMM22 cells (D) and their controls. Data represent mean ± SEM, (n=3). Statistical analysis by unpaired Student’s t-test. NAD+/NADH ratio of TOMM22 depleted cells [HPAF-siTOMM22] (E) and TOMM22 overexpressing cells [BxPC3-TOMM22] (F), relative to their respective controls. NAD+/NADH ratio was calculated by dividing the NAD+ values by NADH and normalized according to protein concentration. Data represent mean ± SEM, (n=3). Statistical analysis by unpaired Student’s t-test.
TOMM22 overexpression enhances mitochondrial respiratory function in PDAC cells
Since aberrant TOMM22 expression altered RC expression and activity, we were interested to see how its modulation affected cellular metabolism. Considering that OXPHOS and glycolysis are the two processes involved in cellular energy production, we assessed the oxygen consumption rate (OCR) of the cells, which reflects a measure of mitochondrial respiration, and extracellular acidification rate (ECAR) which reflects glycolysis. We first examined the effect of TOMM22 silencing on OCR in HPAF cells. Our data revealed a lower basal (p=0.0007), ATP-linked (p=0.0018) and maximal OCR (p<0.0001) in the TOMM22 silenced HPAF-siTOMM22 cells compared to the control HPAF-Scr cells (Figure 5A). An elevated level of basal (p=0.0008), ATP-linked (p=0.0087) and maximal OCR (p=0.0020) was observed in TOMM22-overexpressing BxPC3-TOMM22 cells compared to the control BxPC3 cells (Figure 5A). Overall, this finding reflects a higher mitochondrial respiration rate in TOMM22 overexpressing cells (HPAF-Scr, BxPC3-TOMM22) as opposed to the cells with a lower level of TOMM22 expression (HPAF-siTOMM22, BxPC3-Control). To determine the impact of TOMM22 overexpression on extracellular acidification rate (ECAR), we performed the Seahorse XF Glycolysis Stress test and discovered that glycolysis-linked ECAR (p=0.0021) and maximal ECAR (p<0.0001) were lower in the HPAF-siTOMM22 cells as opposed to the HPAF-Scr cells (Supplementary figure 6A). Correspondingly, BxPC3-TOMM22 cells had higher glycolysis-linked ECAR (p=0.0050) and maximal ECAR (p=0.0006) compared to BxPC3-Control cells (Supplementary figure 6A). Interestingly we also found that overall, wild type HPAF cells were mostly OXPHOS dependent while wild type BXPC3 cells displayed high glycolysis compared to non-cancerous HPDE cells, as suggested by a measure of their OCR and ECAR (Supplementary figure 7A–G). Maintenance of mitochondrial membrane potential (MMP) is a key indicator of mitochondrial function and cell health (18, 19). As a reason, we determined the impact of TOMM22 overexpression on the MMP of pancreatic cancer cells. We observed downregulation of MMP in HPAF-siTOMM22 cells compared to HPAF-Scr cells (p=0.0022) (Figure 5B). In parallel, BxPC3-TOMM22 cells have an augmented MMP as opposed to their control (p=0.0003) (Figure 5C). Further, to investigate the impact of TOMM22 modulation on ATP production, we performed an ATP assay and recorded a lower level of ATP concentration in TOMM22-depleted HPAF cells compared to HPAF-Scr cells (p=0.0038). (Figure 5D). On the other hand, total ATP was higher in the BXPC3-TOMM22 cells compared to BxPC3-Control cells (p=0.0081) (Figure 5E). In addition, ATP concentration was reduced in the BxPC3-TOMM22 cells upon TOMM22 knockdown (p= 0.001) (Supplementary figure 3D). These results, therefore, demonstrate a functional association between high TOMM22 expression and enhanced mitochondrial respiration in PDAC cells. Mitochondria are the primary site of ROS production in the cells as ROS is mainly produced as a by-product of mitochondrial respiration at the ETC. We determined the influence of TOMM22 modulation on ROS levels through the assessment of mitochondrial ROS in both TOMM22 overexpressing cells and cells with reduced TOMM22 expression. We observed that mitochondrial ROS levels were higher in the HPAF-siTOMM22 cells compared to HPAF-Scr cells (p=0.0054) (Supplementary figure 7H). Conversely, mitochondrial ROS was elevated in the BxPC3-TOMM22 cells compared to BxPC3-Control cells (p=0.0354) (Supplementary figure 7I). These results therefore demonstrate that the effect of TOMM22 overexpression on ROS production is context dependent.
Figure 5. TOMM22 modulates the mitochondrial function of pancreatic cancer cells.
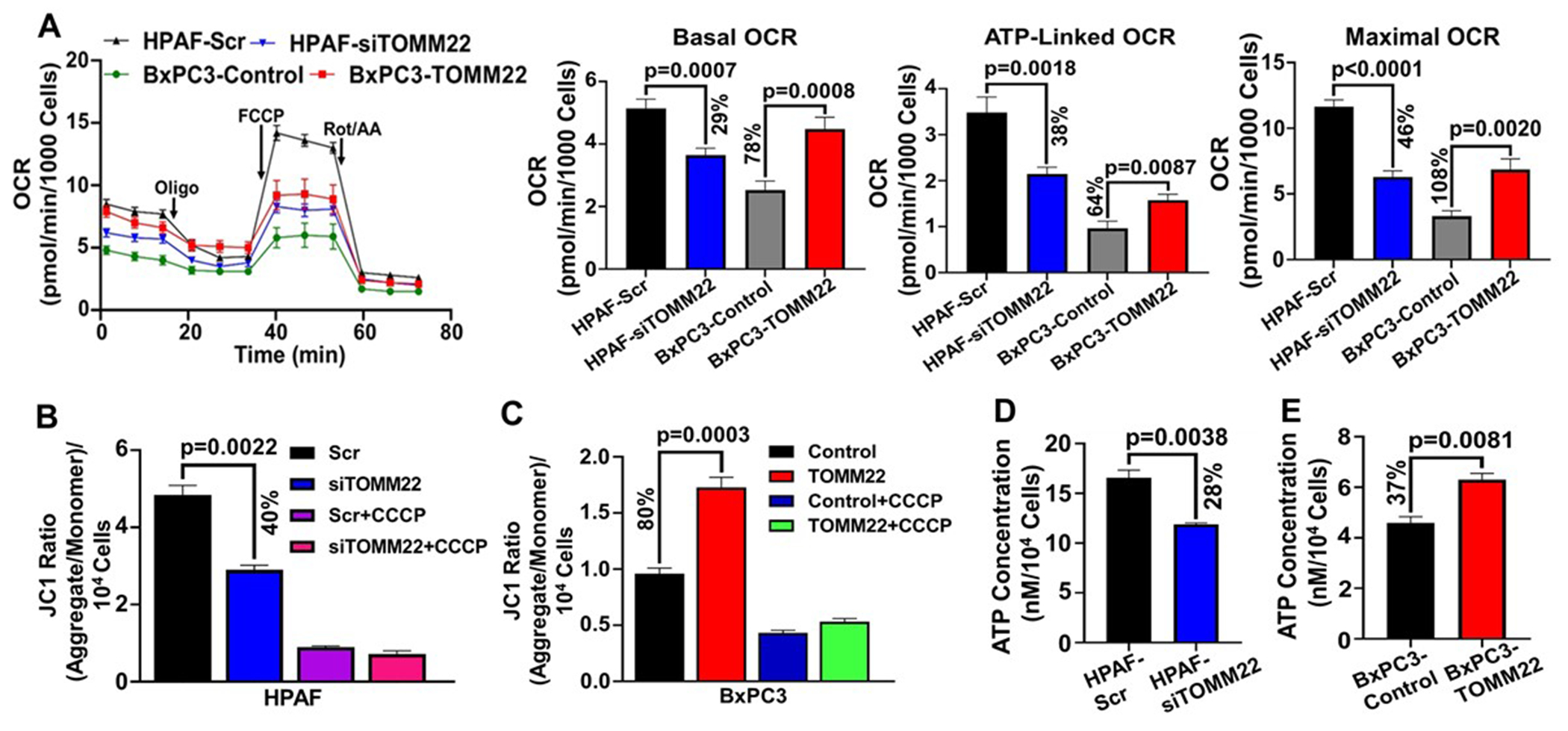
OCR of TOMM22 silenced cells [HPAF-siTOMM22] and TOMM22 overexpressing cells [BxPC3-TOMM22], relative to their respective controls (A). Data were normalized to relative cell numbers. Data represent mean ± SEM, (n=3). Statistical analysis by unpaired Student’s t-test. MMP of HPAF-siTOMM22 cells (B), BxPC3-TOMM22 cells (C) along with their respective control cells. JC-1 ratio was calculated by dividing the aggregate reading by the monomer reading and normalized to the respective cell numbers. CCCP was added to the cells as a positive control. Data represent mean ± SEM, (n=3). Statistical analysis by unpaired Student’s t-test. ATP Concentration of TOMM22 depleted cells (D) and TOMM22 overexpressing cells (E), relative to their respective controls. ATP Concentration was calculated from the ATP standard curve and normalized to the respective cell numbers. Data represent mean ± SEM, (n=3). Statistical analysis by unpaired Student’s t-test.
Inhibition of RCI reduces the growth, invasion, and ATP production of pancreatic cancer cells
To confirm that TOMM22 mediated promotion of PDAC progression is due to alterations in mitochondrial protein import and functions, we treated cells with rotenone, a known inhibitor of RCI activity. Following the rotenone treatment, we determined its impact on the growth, invasion capabilities and ATP production of the PDAC cells with higher TOMM22 expression (HPAF-Scr, BxPC3-TOMM22). Our results show a dose dependent decrease in the growth of HPAF-Scr and BxPC3-TOMM22 cells. Treatment of the cells with 5μM of rotenone for 48 hours led to 34.3% (p<0.0001) and 56% (p<0.0001) reduction in the growth of HPAF-Scr and BxPC3-TOMM22 cells respectively (Figure 6A–B). Similarly, treatment of the cells with 10μM of rotenone for 48 hours resulted in 46.5% (p<0.001) and 70% (p<0.001) decrease in the growth of HPAF-Scr and BxPC3-TOMM22 cells respectively (Figure 6A–B). Since treatment of HPAF-Scr cells with 10μM of rotenone led to 46.5% decrease in growth after 48 hours and treatment of BxPC3-TOMM22 cells with 5μM rotenone led to 56% decrease in growth after 48 hours, as well as 74% (p=0.0485) and 76% (p=0.0492) reduction in complex I activity of HPAF-Scr and BxPC3-TOMM22 cells respectively (Figure 6C–D), these doses were chosen for subsequent experiments.
Figure 6. Complex I inhibition decreases pancreatic cancer cell growth, invasion, and ATP production.
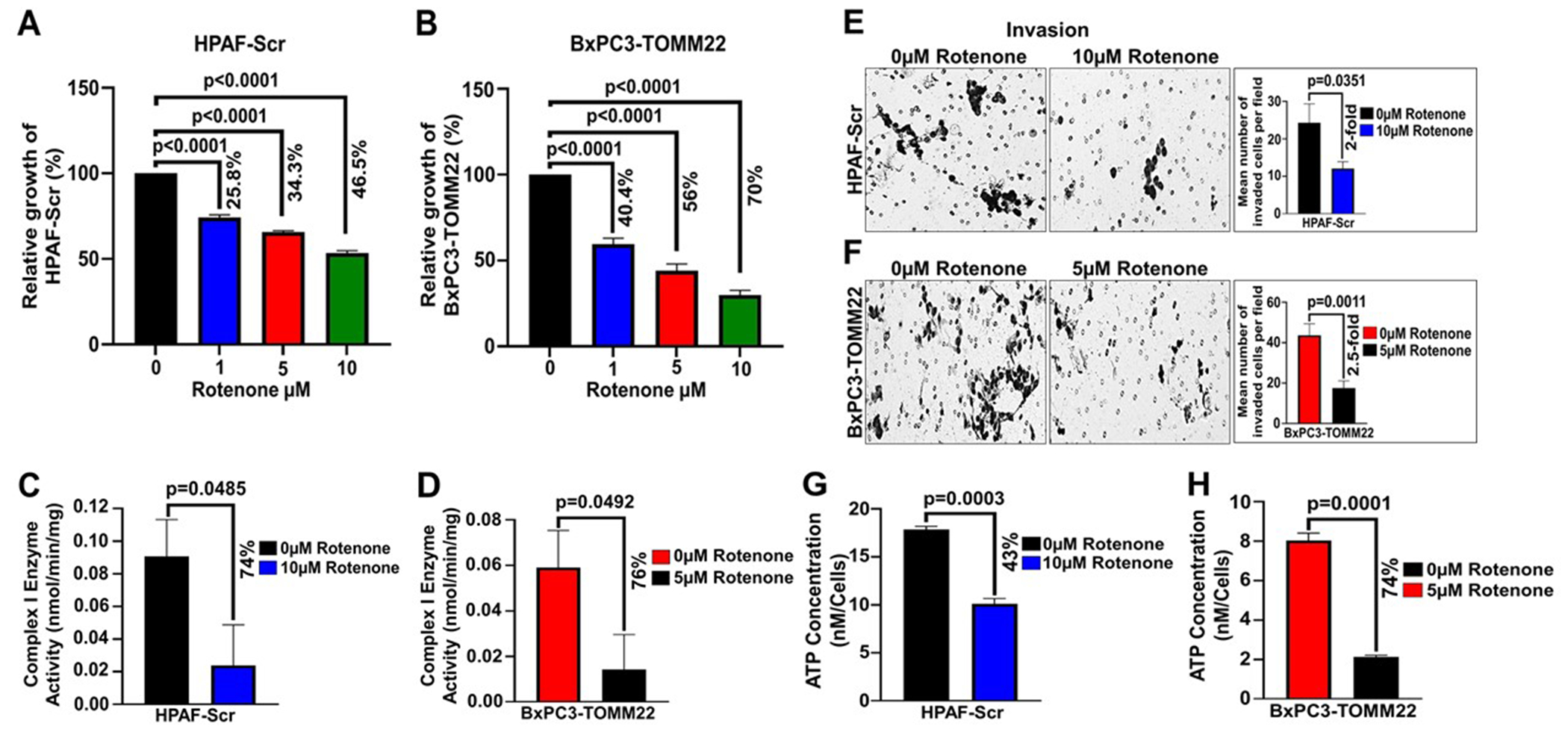
HPAF-Scr (A) and BxPC3-TOMM22 (B) cells were seeded in 96 well plates. After 24 hours, cells were treated with 0μM, 1μM, 5μM or 10μM rotenone and WST assay was performed after 48 hours. Data represent mean ± SEM, (n=3). Statistical analysis by unpaired Student’s t-test. HPAF-Scr (C) and BxPC3-TOMM22 (D) cells were treated with 0μM or 10μM rotenone and 0μM or 5μM rotenone respectively for 48 hours, after which, complex I enzyme activity was done. Data represent mean ± SEM, (n=3). Statistical analysis by unpaired Student’s t-test. HPAF-Scr (E) and BxPC3-TOMM22 (F) cells were resuspended in serum free media containing 0μM or 10μM rotenone and 0μM or 5μM rotenone respectively. Thereafter, cells were seeded on matrigel-coated polycarbonate membrane inserts for invasion assay. Media supplemented with 10% FBS was used as a chemoattractant in lower wells and cells were allowed to invade for 24 hours. Data is presented as mean number of cells in 10 random fields ± SEM, (n=3). Statistical analysis was done using an unpaired Student’s t-test. HPAF-Scr (G) and BxPC3-TOMM22 (H) cells were seeded in 96 well plates. After 24 hours, HPAF-Scr cells were treated with 0μM or 10μM rotenone, BxPC3-TOMM22 cells were treated with 0μM or 5μM rotenone, and ATP assay was performed after 48 hours. ATP Concentration was calculated from the ATP standard curve and normalized to the respective cell numbers. Data represent mean ± SEM, (n=3). Statistical analysis by unpaired Student’s t-test.
Next, we performed an invasion assay and observed that treatment of HPAF-Scr cells with 10μM rotenone for 24 hours led to a 2-fold (p=0.0351) reduction in the invasion of HPAF-Scr cells compared to cells without rotenone treatment (Figure 6E). Similarly, the invasion of BxPC3-TOMM22 cells decreased by 2.5-fold (p=0.0011) upon treatment with 5μM rotenone for 24 hours (Figure 6F). To determine the impact of rotenone treatment on ATP production, we performed an ATP assay and observed that rotenone treatment led to 43% (p=0.0003) and 74% (p=0.0001) decrease in the ATP concentration of HPAF-Scr and BxPC3-TOMM22 cells respectively (Figure 6G–H). These results indicate that the inhibition of RCI activity by rotenone impairs mitochondrial function (ATP production), which in turn leads to reduced growth and invasion of TOMM22 overexpressing PDAC cells (HPAF-Scr, BxPC3-TOMM22). Thus, TOMM22 overexpression promotes the growth and malignant behavior of PDAC cells via its regulation of nuclear encoded mitochondrial protein import and functions.
Discussion
Despite recent advancements unraveling the molecular basis of pancreatic tumorigenesis, timely management and long-term survival of the patients is dismal. Early detection of this lethal malignancy and appropriate interventions for curative intent are thus dire needs. Rewiring of mitochondrial metabolism, apoptosis, and redox balance play important roles in human tumorigenesis and therapy resistance including PDAC, a mitochondria enriched cancer (10–14). In this light, mitochondria could be exploited as attractive targets for the efficient management of PDAC. Cancer cells wisely utilize mitochondria to fulfill their constant energy requirement and avoid intrinsic apoptosis. Remarkably, the proteins involved in tumor metabolism (except for the 13 mtDNA-encoded proteins), regulation of apoptosis and other mitochondrial pathways must be imported into mitochondria through the unique mitochondrial protein import system (15). The OMM translocase subunit TOMM22 acts as the central receptor of the TOMM complex, which is the most important counterpart of all the machineries in the mitochondrial protein import system since it is the first one to come in contact with the majority of the nuclear-encoded mitochondrial proteins to allow their entry into the intermembrane space (15). At present, the expression pattern and functional role of TOMM22 are largely unknown in PDAC. Overexpression of TOMM22 at both mRNA and protein levels and its association with poor disease outcome suggest a potential role of TOMM22 in fueling PDAC tumorigenesis by the sustained import of the key metabolic and redox regulators into mitochondria. Elevated TOMM22 expression in the preneoplastic lesions suggests its early activation in driving PDAC tumorigenesis. Interestingly, a progressive abundance of TOMM22 expression detected in advanced staged tumors with low histopathological grades, reveals TOMM22 as a better prognostic indicator than tumor grade. This observation was further supported by the correlation of TOMM22 expression with stage-wise progression of pancreatic cancer. Thus, TOMM22 appears to be promising as an early diagnostic and prognostic biomarker for PDAC. Analysis of malignant tumors and early lesions from a larger cohort of patients would be necessary to further validate our findings.
Using two different approaches (gain and loss of function), we demonstrated the functional significance of TOMM22 in PDAC. Our data show that TOMM22 overexpression plays a crucial role in promoting the growth and clonogenicity of PDAC cells. Apart from the growth-promoting role of TOMM22 overexpression, our data showed that increased TOMM22 expression promotes the malignant potential (migration and invasion) of PDAC cells. Although there is no information regarding the role of TOMM22 in cancer, a recent study reported that knockdown of TOMM40, led to decreased proliferation of epithelial ovarian cancer cells and xenografts (20). Furthermore, another study demonstrated that TOMM20 overexpression led to increased proliferation, migration and invasion of colorectal cancer cells (21). Although TOMM22 silencing did not affect the apoptotic rate in HPAF cells, its overexpression reduced the rate of apoptosis in BxPC3 cells, suggesting that the impact of TOMM22 overexpression on apoptosis is context dependent.
In the OXPHOS system, RCI acts as the entry gate and docking site to initiate the electron transport process through the transfer of electrons from NADH to ubiquinone. Enhanced import of RCI subunit/activity of RCI, high NAD+/NADH ratio accompanied with the increased import of RCII, RCIII and RCV components as well as increased OCR, MMP and ATP production indicate the dependency of the PDAC cells on TOMM22 for progressive growth. Remarkable loss of growth and malignant potential following the blockade of RCI in TOMM22 proficient and rescued PDAC cells confirmed our findings. Enhanced electron transfer as a result of high OXPHOS rate favors ROS production, likewise, destabilization of electron transfer through the electron transport chain can also increase ROS production (22). Our data is also in compliance with these facts as high mitochondrial ROS in the BxPC3-TOMM22 cells could be explained by these cells displaying a higher OXPHOS rate. On the other hand, increased ROS levels in the TOMM22 depleted HPAF cells could be due to the reduced import of the subunits of the ETC, leading to its reduced functionality and inefficient electron transfer, thus resulting in increased electron leak and ROS generation.
In addition to the OXPHOS, the MS analysis identified other relevant mitochondrial pathways associated with TOMM22 mediated import. Furthermore, MS analysis uncovers a significant amount of proteins, which have not yet been identified to localize to or associate with mitochondria. This could be due to the fact that the available bioinformatics tools for detecting mitochondrial targeting sequences have low sensitivity and specificity. The development of more efficient and highly sensitive bioinformatics tools is needed for the accurate prediction of cellular localization of proteins, including the mitochondrial compartment. Of note, manipulation of TOMM22 did not affect the expression of mitochondria encoded MT-CO2, a subunit of the respiratory complex IV (RCIV) encoded by the mitochondrial DNA. This finding reiterates the significance of TOMM22 as a moderator of nuclear encoded mitochondrial protein import and thus, mitochondrial function. Enhanced ECAR in TOMM22-overexpressing pancreatic cancer cells, which were primarily glycolytic in nature, suggest a potential role of TOMM22 in maintaining both OXPHOS and Glycolytic influx to promote aggressive growth. However, the precise molecular mechanism remains to be determined. We recognize the limitation of being unable to overexpress TOMM22 in the HPAF-siTOMM22 cell line as an additional control for our experiments, because the knockdown of TOMM22 in the HPAF-siTOMM22 cell line is stable. To address this limitation, we knocked down the expression of TOMM22 in the BxPC3-TOMM22 cell line as additional control and observed that TOMM22 knockdown reversed the phenotypic effects caused by TOMM22 overexpression as well as resulted in a reduction in ATP production.
In summary, we have identified TOMM22 as a novel driver of PDAC tumor growth and aggressiveness via its modulation of mitochondrial protein import and functions. Since TOMM22 overexpression alters the metabolism of PDAC cells, future work aimed at identifying the oncometabolites generated as a result of its overexpression and its potential impact on retrograde signaling would be helpful in shedding light on the diverse mechanisms through which TOMM22 overexpression promotes PDAC growth and progression, thereby, establishing TOMM22 as a useful therapeutic target. Based on the expression pattern in clinical samples, TOMM22 could potentially be useful for developing early detection, monitoring and surveillance strategies to improve the overall survival of patients.
Supplementary Material
Visual Overview. TOMM22 is overexpressed in pancreatic cancer and promotes pancreatic cancer progression via regulation of nuclear encoded mitochondrial protein import and functions.
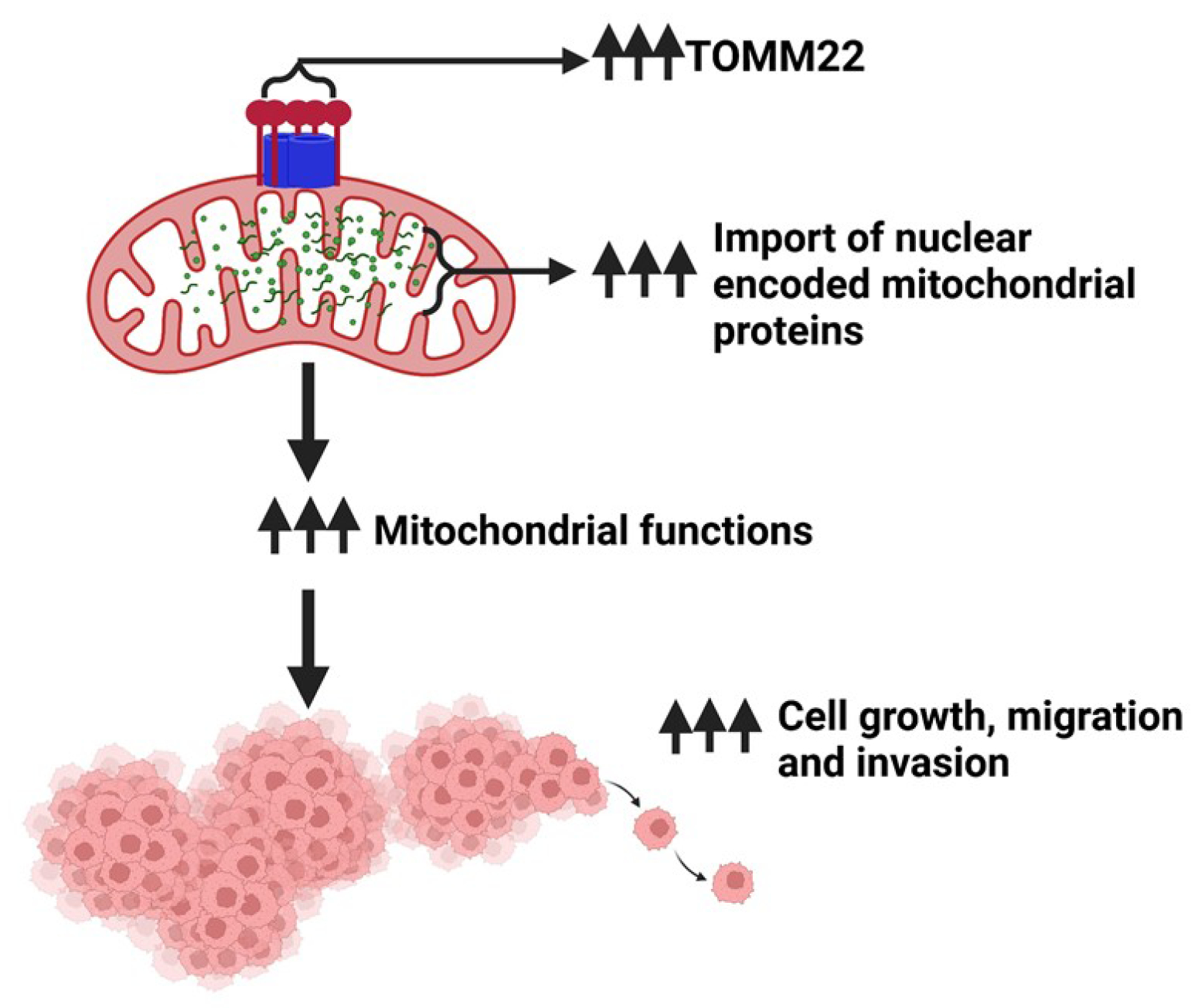
Visual overview was created with Biorender.com.
Implications:
TOMM22 bears potential for early diagnostic/prognostic biomarker development and therapeutic targeting for better management of pancreatic cancer patients.
Acknowledgments
The authors would like to thank Ms. Lindsay Schambeau and Dr. Marie Migaud, University of South Alabama Mass Spectrometry Core Facility for their assistance with the generation of the Mass Spectrometry data. The authors would also like to acknowledge the funding and resource support from the University of South Alabama – (Mitchell Cancer Institute), NCI/NIH (R01CA224306-04, A. Singh) and DOD funding (DURIP W911NF-20-1-0155).
Abbreviations
- PDAC
Pancreatic ductal adenocarcinoma
- PanIN
Pancreatic intraepithelial neoplasia
- TOMM
Translocase of the outer mitochondrial membrane
- MT
Mitochondrial
- IMS
Intermembrane space
- GTex
Genotype-Tissue Expression
- TCGA
The Cancer Genome Atlas
- ETC
Electron transport chain
- RC
Respiratory complex
Footnotes
Competing interests
The authors declare no competing interests.
Ethical Statement: Not applicable
References
- 1.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin 2022;72(1):7–33 doi 10.3322/caac.21708. [DOI] [PubMed] [Google Scholar]
- 2.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res 2014;74(11):2913–21 doi 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 3.Grant TJ, Hua K, Singh A. Molecular Pathogenesis of Pancreatic Cancer. Prog Mol Biol Transl Sci 2016;144:241–75 doi 10.1016/bs.pmbts.2016.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wiedemann N, Pfanner N. Mitochondrial Machineries for Protein Import and Assembly. Annu Rev Biochem 2017;86:685–714 doi 10.1146/annurev-biochem-060815-014352. [DOI] [PubMed] [Google Scholar]
- 5.WARBURG O. On the origin of cancer cells. Science 1956;123(3191):309–14 doi 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 6.Viale A, Pettazzoni P, Lyssiotis CA, Ying H, Sánchez N, Marchesini M, et al. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature 2014;514(7524):628–32 doi 10.1038/nature13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Porporato PE, Filigheddu N, Pedro JMB, Kroemer G, Galluzzi L. Mitochondrial metabolism and cancer. Cell Res 2018;28(3):265–80 doi 10.1038/cr.2017.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dong LF, Kovarova J, Bajzikova M, Bezawork-Geleta A, Svec D, Endaya B, et al. Horizontal transfer of whole mitochondria restores tumorigenic potential in mitochondrial DNA-deficient cancer cells. Elife 2017;6 doi 10.7554/eLife.22187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng G, Hardy M, Topchyan P, Zander R, Volberding P, Cui W, et al. Potent inhibition of tumour cell proliferation and immunoregulatory function by mitochondria-targeted atovaquone. Sci Rep 2020;10(1):17872 doi 10.1038/s41598-020-74808-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu Y, Ricciardiello F, Yang G, Qiu J, Huang H, Xiao J, et al. The Role of Mitochondria in the Chemoresistance of Pancreatic Cancer Cells. Cells 2021;10(3) doi 10.3390/cells10030497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qin C, Yang G, Yang J, Ren B, Wang H, Chen G, et al. Metabolism of pancreatic cancer: paving the way to better anticancer strategies. Mol Cancer 2020;19(1):50 doi 10.1186/s12943-020-01169-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fernald K, Kurokawa M. Evading apoptosis in cancer. Trends Cell Biol 2013;23(12):620–33 doi 10.1016/j.tcb.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martinez-Useros J, Li W, Cabeza-Morales M, Garcia-Foncillas J. Oxidative Stress: A New Target for Pancreatic Cancer Prognosis and Treatment. J Clin Med 2017;6(3) doi 10.3390/jcm6030029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aggarwal V, Tuli HS, Varol A, Thakral F, Yerer MB, Sak K, et al. Role of Reactive Oxygen Species in Cancer Progression: Molecular Mechanisms and Recent Advancements. Biomolecules 2019;9(11) doi 10.3390/biom9110735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haastrup MO, Vikramdeo KS, Singh S, Singh AP, Dasgupta S. The Journey of Mitochondrial Protein Import and the Roadmap to Follow. Int J Mol Sci 2023;24(3) doi 10.3390/ijms24032479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zeng Y, Pan Q, Wang X, Li D, Lin Y, Man F, et al. Impaired Mitochondrial Fusion and Oxidative Phosphorylation Triggered by High Glucose Is Mediated by Tom22 in Endothelial Cells. Oxid Med Cell Longev 2019;2019:4508762 doi 10.1155/2019/4508762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perez-Riverol Y, Bai J, Bandla C, García-Seisdedos D, Hewapathirana S, Kamatchinathan S, et al. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res 2022;50(D1):D543–D52 doi 10.1093/nar/gkab1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rovini A, Heslop K, Hunt EG, Morris ME, Fang D, Gooz M, et al. Quantitative analysis of mitochondrial membrane potential heterogeneity in unsynchronized and synchronized cancer cells. FASEB J 2021;35(1):e21148 doi 10.1096/fj.202001693R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perry SW, Norman JP, Barbieri J, Brown EB, Gelbard HA. Mitochondrial membrane potential probes and the proton gradient: a practical usage guide. Biotechniques 2011;50(2):98–115 doi 10.2144/000113610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang W, Shin HY, Cho H, Chung JY, Lee EJ, Kim JH, et al. TOM40 Inhibits Ovarian Cancer Cell Growth by Modulating Mitochondrial Function Including Intracellular ATP and ROS Levels. Cancers (Basel) 2020;12(5) doi 10.3390/cancers12051329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park SH, Lee AR, Choi K, Joung S, Yoon JB, Kim S. TOMM20 as a potential therapeutic target of colorectal cancer. BMB Rep 2019;52(12):712–7 doi 10.5483/BMBRep.2019.52.12.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raimondi V, Ciccarese F, Ciminale V. Oncogenic pathways and the electron transport chain: a dangeROS liaison. Br J Cancer 2020;122(2):168–81 doi 10.1038/s41416-019-0651-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (17) partner repository with the dataset identifier PXD040198. All other data can be made available upon reasonable request.


