Abstract
THO is a protein complex that functions in cotranscriptional mRNP formation. Yeast THO1 and SUB2 (Saccharomyces cerevisiae) were identified as multicopy suppressors of the expression defects of the hpr1Δ mutant of THO. Here we show that multicopy THO1 suppresses the mRNA accumulation and export defects and the hyperrecombination phenotype of THO mutants but not those of sub2Δ, thp1Δ, or spt4Δ. Similarly, Sub2 overexpression suppresses the RNA export defect of hpr1Δ. Tho1 is a conserved RNA binding nuclear protein that specifically binds to transcribed chromatin in a THO- and RNA-dependent manner and genetically interacts with the shuttling hnRNP Nab2. The ability of Tho1 to suppress hpr1Δ resides in its C-terminal half, which contains the RNA binding activity and is located after a SAP/SAF (scaffold-associated protein/scaffold-associated factor) domain. Altogether, these results suggest that Tho1 is an hnRNP that, similarly to Sub2, assembles onto the nascent mRNA during transcription and participates in mRNP biogenesis and export. Overexpression of Tho1 or Sub2 may provide alternative ways for mRNP formation and export in the absence of a functional THO complex.
Transcription is a central cellular function that occurs in the nucleus of eukaryotic cells in coordination with other nuclear processes. RNA polymerase II (RNAPII)-driven transcription takes place in tight association with different mRNA processing steps (46, 59). Indeed, a number of important proteins involved in mRNA processing, such as the capping enzymes, splicing factors, and the cleavage and polyadenylation stimulatory factor responsible for recognizing the poly(A)+ signal, are loaded onto the nascent mRNA by interacting with the C-terminal domain (CTD) of the RNAPII (43). Thus, in cell extracts, a recombinant CTD stimulates poly(A) site cleavage and mRNA splicing (22, 23) and influences 3′-end formation and capping of mRNAs (50, 55). The result of the combined action of molecular processes is a mature mRNA-protein (mRNP) particle proficient for nuclear export. Any failure compromising the formation of an export-proficient mRNP would likely trigger mRNA degradation by the nuclear exosome (28, 54), a process which may also occur cotranscriptionally as shown in Drosophila spp. (6). The nuclear integration of the different processes taking place from transcription to mRNA export was recently observed at the cellular level by the nuclear pore colocalization of actively transcribed GAL genes (8).
An intriguing connection of mRNP biogenesis with other nuclear processes is that observed with genetic stability. Even though there is accumulated evidence for the stimulation of both mutation and recombination by transcription, the functional and physical interconnection between mRNP biogenesis and genetic instability comes from studies with the THO complex of Saccharomyces cerevisiae (2, 3). THO is a four-protein complex composed of stoichiometric amounts of Tho2, Hpr1, Mft1, and Thp2 (10). Null mutations of any component of THO lead to similar phenotypes of transcription impairment, transcription-associated hyperrecombination and RNA export defects (9, 10, 29, 53), and increased levels of exosome-dependent mRNA instability (37, 58). THO has also been purified in humans and Drosophila, and the two major subunits, Tho2 and Hpr1, are present in all of the eukaryotes studied (10, 47, 53). Furthermore, THO forms, together with the RNA export proteins Sub2/UAP56 and Yra1/Aly, a larger complex termed TREX, as identified in yeast (Saccharomyces cerevisiae) and humans (53). In addition to the physical interaction, there is a functional interaction between Sub2, Yra1, and the THO complex. Thus, mutant alleles of SUB2 and YRA1 are synthetic lethal with THO mutations and lead to the same transcription-dependent hyperrecombination and gene expression phenotypes as do THO mutations. In addition, Sub2 overexpression suppresses hpr1Δ (13, 29, 53). THO, Sub2, and Yra1 are recruited to active chromatin in a way in which THO seems to be recruited first to facilitate the subsequent recruitment of Sub2 and Yra1 (36, 53, 58).
The connection between mRNP biogenesis factors and the origin of genetic instability is also provided by the yeast Thp1-Sac3 complex involved in RNA export and located in the proximity of the nuclear pore complex (14), the Mex67-Mtr2 export factor, or the Nab2 hnRNP. Mutations in these factors confer transcription, RNA export, and hyperrecombination phenotypes similar to those of mutants of the THO complex (15, 16, 29).
One major cause for the transcription impairment and hyperrecombination phenotypes of THO mutants is the cotranscriptional accumulation of DNA-RNA hybrids formed behind the elongating RNAPII in the absence of functional THO. This indicates that one role of THO/TREX in mRNP assembly may be to maintain the nascent mRNA apart from the transcribed DNA, impeding undesired interactions of the nascent mRNA with the DNA template (24).
One way of understanding the functional interaction between genetic instability and mRNP biogenesis in general, and the THO complex in particular, has been through the isolation and analysis of multicopy suppressors. Thus, the Sub2 RNA export factor, a putative RNA-dependent ATPase with homology to RNA helicases (32), was found as a multicopy suppressor of hpr1Δ (13, 29). In addition, a new gene of unknown function, THO1, was identified as a multicopy suppressor of hpr1Δ in two different screenings (29, 45). No phenotype has been associated so far with the null mutants of THO1 to provide clues about its in vivo function (45). Here we show that Tho1 is a novel nuclear hnRNP that binds RNA in vitro and is recruited to transcribed chromatin in vivo in a THO-complex- and RNA-dependent manner. Tho1 overexpression suppresses the recombination, RNA export, and gene expression defects of THO− mutants but not those of sub2Δ mutants. Multicopy SUB2 also suppresses the RNA export defect of hpr1Δ strains. The ability of Tho1 to suppress hpr1Δ resides in its RNA binding C-terminal region. Altogether, our study suggests that Tho1, like Sub2, assembles onto the nascent mRNA during transcription and that Tho1 and Sub2 can provide alternative ways for mRNP biogenesis in the absence of a functional THO complex.
MATERIALS AND METHODS
Strains.
Yeast strains used were the previously described W303-1A (wild type) U678-1C (hpr1Δ::HIS3), AYW3-3C (hpr1Δ::HIS3), WRK-1C (tho2Δ::KanMX4), RK2-6C (tho2Δ::KanMX4), AW33-11Bu− (tho1Δ::TRP1) (45), sub2Δ (sub2Δ::HIS3) (27, 29, 38), Sub2-TAP (SUB2-TAP) (53), MFM67-13A (mft1Δ::KanMX4) (10) and WMK-2A (mft1Δ::KanMX4) (10), and the BY4741 (wild type) and its isogenic thp2Δ mutant BY-HR167 (EUROSCARF, Frankfurt, Germany) (10), plus the strains constructed for this study, SUL-1A (MATα ade2 his3 trp1 ura3 sub2Δ HIS3 leu2-k::URA3-ADE2::leu2-k), BSU-S2T-3B (MATα his3 trp1 ura3 leu2-k::URA3-ADE2::leu2-k SUB2-TAP), BSU-S2T-6D (MATa his3 trp1 ura3 hpr1Δ::HIS3 SUB2-TAP), WWT1T and WWT2T (W303 carrying TAP-tagged Tho1 and Tho2, respectively), WHT1T (hpr1Δ::HIS3) carrying TAP-tagged Tho1, WWYL.1D (wild type) and WHYL.2A (hpr1Δ::HIS3) carrying the GAL1pr::YLR454c fusion (53), and NALT-1 and NALT-2 (mutant nab2-1 and double mutant tho1Δ nab2-1), obtained by genetic crosses between AW33-11Bu− and nab2-1 (20).
The chromosomal THO1-TAP fusion was constructed by transforming W303-1A with the 1.17-kb, 5′-end TAP-tagged THO1 sequence obtained by PCR on plasmid pBS1479 (48) using the primers 5′-TCCAGAGTAAGTAAAAACAGGAGAGGCAACCGCTCTGGTTACAGAAGATCCATGGAAAAGAGAAG-3′ and 5′AAAGAACCGAAACTAGAATGAAAAACTCCACCAAAACGGCTTGAGCCTTCGACTCACTATAGGG-3′. Constructions of SUB2-TAP and THO2-TAP were described previously (29, 53).
Plasmids.
Plasmids pCM184-LAUR and p416GAL1lacZ have been described previously (9, 29). Plasmid pT7-HA-THO1, containing an N-terminal His6- hemagglutinin (HA) translational fusion of the complete THO1 coding sequence under the control of the T7 RNA pol promoter, was constructed by inserting into the NheI-ClaI site of pT7-7-His6 a 0.72-kb HA-THO1 fragment obtained by PCR using 5′-GGGTACATGTGCTAGCTATCCCTATGACGTTCCCGACTATGCTATGGCAGATTATTCTTC-3′ and 5′-TACTCCAAGCTTATCGATGCCTTTATCTTCTGTAACCAG-3′ as primers. Plasmids pT7-HA-5′THO1 and pT7-HA-3′THO1 are identical to pT7-HA-THO1 except for containing the 0.21-kb 5′-end and 0.54-kb 3′-end fragments of THO1, respectively. Such copies were obtained by PCR using 5′-GGGTACATGTGCTAGCTATCCCTATGACGTTCCCGACTA-3′ and 5′-GCTAAGGCCCAAGCTTCCTGTGGAGAAACTTCACTTTCAC-3′ as primers for 5′THO1 and 5′-CGGCATGCAGCTAGCGAACAAAACCAGGAACAAGGGTCAG-3′ and 5′-TACTCCAAGCTTATCGATGCCTTTATCTTCTGTAACCAGA-3′ as primers for 3′THO1.
Plasmids pGAL-THO1, pGAL-NTHO1, and pGAL-THO1C were constructed by inserting the respective XbaI-ClaI inserts from the pT7-7His6 plasmids into p416GAL based on pRS416 containing the GAL1 promoter.
Purification of recombinant Tho1 proteins from Escherichia coli.
BL21(DE3) strains transformed with pT7-HA-THO1, pT7-HA-5′THO1, and pT7-HA-3′THO1 were grown in LB medium with 100 μg/ml ampicillin to an optical density at 600 nm (OD600) of 0.8. Expression of the T7 promoter was induced for 4 h by adding IPTG (isopropyl-β-d-thiogalactopyranoside) to a final concentration of 1 mM. Proteins were purified from the lysates by affinity chromatography on a Ni-nitrilotriacetic acid column (QIAGEN) according to the manufacturer's instructions and quantified by Lowry's method.
In situ mRNA export assays.
In situ localization of poly(A)+ RNA was performed as described previously (5) using a digoxigenin-labeled oligo(dT)16 probe.
In vitro RNA binding activity.
RNA binding activity was determined in vitro by RNA band-shifting assays following previously published procedures. As 32P-labeled and cold nucleic acid samples, we used the 90-mer RNA sequence from the polylinker of pBluescript-SK and the 60-mer single-stranded DNA (ssDNA) previously described (29).
Chromatin immunoprecipitation.
For chromatin immunoprecipitation (ChIP) analyses, either the yeast strains WW-T1T and WHT1T, WWT2T, and Sub2-TAP containing the THO1-TAP, THO2-TAP, and SUB2-TAP, respectively, or the yeast strains WWYL.1D and WHYL.2A carrying the GAL1pr::YLR454c fusion were used. For ChIP analyses with PMA1, strains were grown in yeast extract-peptone-dextrose at 30°C up to an OD660 of 0.8. For ChIP with GAL1 and GAL1pr::YLR454c genes, strains were grown in synthetic complete (SC)-2% glycerol-2% lactate to an OD660 of 0.5. The culture was split in two, one half was supplemented with 2% glucose (repressed transcription) and the other with 2% galactose (activated transcription). Samples were taken after 3 h. ChIP assays were performed as described previously (21). RNase treatment was made with 300 U of RNase T1 and 300 U of RNase A for 2 h at 30°C before immunoprecipitation. For experiments with RNase treatment, the time for cross-linking was reduced from 15 to 4 min. Immunoprecipitations were performed by using immunoglobulin-Sepharose for tandem affinity purification (TAP)-tagged proteins and the monoclonal anti-Rpb1-CTD antibody 8WG16 (Berkeley Antibody Company) and protein A-Sepharose for RNAPII. The GFX purification system (Amersham) was used for the last DNA purification step. We used 20- to 27-bp oligonucleotides for the PCR amplification of four fragments of GAL1, five fragments of PMA1, and three fragments of YLR454c, as indicated, and of the bp 9716 to 9863 intergenic region of chromosome V that was used as a negative control. The PCR products obtained were electrophoresed in a 15% acrylamide gel, stained with ethidium bromide, and quantified in a Fuji FLA-3000. For quantification of each ChIP assay, the linear range of each PCR amplification was first established with serial dilutions of the total input (I) and immunoprecipitated (P) DNA. Calculations and normalization of each ChIP were carried out as previously described (4, 30, 33). Briefly, we first performed PCR amplification experiments using different DNA dilutions, so that conditions were established to perform all definitive PCR amplification experiments in the linear range (see Fig. S1 in the supplemental material). The enrichment of the PCR amplification of interest above background levels was calculated by dividing the ratio “gene-specific P signal/intergenic P signal” between the ratio “gene-specific I signal/intergenic I signal.” All experiments were performed at least three times within the linear range with similar results.
Miscellaneous.
Recombination frequencies were obtained from two to three different transformants for each genotype tested. The median frequency of 6 to 12 independent values for each transformant was determined as reported previously (29). Northern analyses were performed according to standard procedures.
RESULTS
Multicopy THO1 and SUB2 suppress all phenotypes of hpr1Δ mutants.
For the isolation of multicopy suppressors of hpr1Δ, we used the LAUR gene expression system containing a 4.15-kb lacZ-URA3 translational fusion under the control of the tet promoter (29). Whereas wild-type cells can express this fusion construct properly, this is not the case for THO mutants. Thus, hpr1Δ cells carrying the LAUR system were unable to form colonies on synthetic medium lacking uracil (SC-Ura-Trp-Leu) and did not express β-galactosidase activity. With this assay, we obtained different overlapping clones that defined only two genes that suppressed hpr1Δ in multicopy, SUB2, whose analysis was previously reported (29), and THO1, a novel gene of unknown function that we have previously found as a multicopy suppressor of the hpr1Δ thermosensitivity and gene expression phenotype (45). In silico analysis revealed that Tho1 has a N-terminal DNA binding domain, termed the SAP domain, that is conserved in eukaryotes and present in a number of scaffold-associated proteins involved in RNA and DNA metabolism (7) (see Fig. S2 in the supplemental material). Interestingly, our in silico analysis revealed that the human ortholog of Tho1 is likely the Hcc-1 protein (see Fig. S2 in the supplemental material), which was identified as a highly expressed protein in human hepatocarcinoma and which has been shown in two-hybrid assays to bind the two DEAD box putative RNA helicases, hSUB2/UAP56/BAT1 and DDX39 (12, 35).
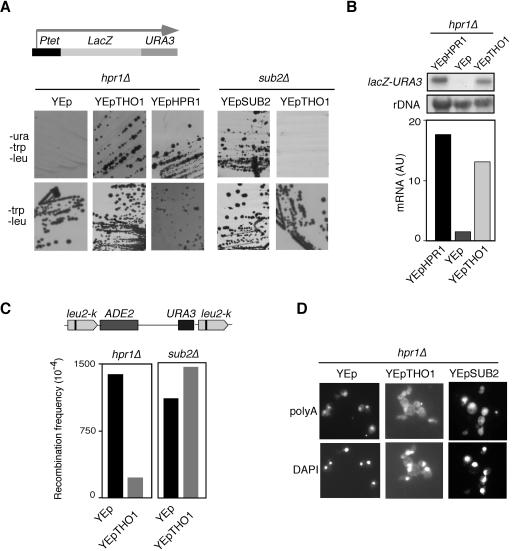
As previously shown for multicopy SUB2, we show that hpr1Δ cells carrying the LAUR system and THO1 in multicopy recovered the abilities to grow on SC-Ura-Trp-Leu and express β-galactosidase activity (Fig. 1A; data not shown). This suppression occurs at the level of mRNA accumulation, as shown by Northern analysis. As can be seen in Fig. 1B, hpr1Δ cells were able to significantly increase the levels of lacZ-URA3 mRNA accumulation when expressing THO1 in multicopy.
FIG. 1.
Suppression of the gene expression, hyperrecombination, and RNA export phenotypes of hpr1Δ by multicopy THO1 and SUB2. (A) Analysis of the capacity of hpr1Δ (U678-4C) and sub2Δ (sub2Δ) cells carrying the Ptet::lacZ-URA3 construct to form colonies in SC lacking Ura (sub2Δ cells are slow-growing but viable null sub2 Ura− mutants (29) derived from W303 isogenic strain DLY23 (27, 38). The cells analyzed were transformants with the empty multicopy vector YEp351 (YEp, negative control) or YEp351-HPR1 (YEp-HPR1, positive control), YEp351-THO1 (YEp-THO1) or YEp351-SUB2 (YEp-SUB2) carrying the HPR1, THO1, and SUB2 genes, respectively. The top panel shows a scheme of the Ptet::lacZ-URA3 translational fusion used to study expression of lacZ and URA3. (B) Northern analyses of the same hpr1Δ transformant strains shown in panel A. RNA was isolated from mid-log-phase cultures grown in SC lacking Trp and Leu. As a 32P-labeled DNA probe we used the 3-kb BamHI lacZ fragment and an internal 589-bp 25S rRNA fragment obtained by PCR. mRNA quantifications were performed in a Fuji FLA3000 and normalized with respect to the rRNA levels of each sample, and are given in arbitrary units (AU) (C) Recombination frequency of the intrachromosomal direct-repeat system leu2-k::URA3-ADE2::leu2-k in hpr1Δ (U678-4C) and sub2Δ (SUL-1A) mutants transformed with multicopy vector YEp351 and YEp351-THO1. The top panel shows a scheme of the recombination assay used. Recombination frequencies are the median value of a total of 12 independent colonies obtained from SC lacking Leu. Ura− recombinants were selected in SC plus 5-fluoroorotic acid plates. (D) Subcellular localization of poly(A)+ RNAs detected in situ with digoxigenin-labeled oligo(dT)16. Samples were taken from mid-log-phase cultures of hpr1Δ (U678-4C) cells transformed with either YEp351, YEp351-THO1 or YEp351-SUB2 and shifted to 37° for 4 h in SC lacking Leu. Nuclei (DNA) were visualized by staining with 10 μg/ml DAPI (4′,6′-diamidino-2-phenylindole). Analyses were performed with an Olympus AHBT3 microscope.
In addition to the gene expression defect, two other hallmark phenotypes of THO mutants are the hyperrecombination and mRNA export defect phenotypes. If multicopy THO1 suppressed the absence of the THO complex in the cell directly and not just the gene expression defect of hpr1Δ cells in an indirect manner, the recombination and mRNA export phenotypes of hpr1Δ might also be suppressed. To test this, we determined the frequency of direct-repeat recombination in the chromosomal leu2-k::URA3-ADE2::leu2-k system and the subcellular localization of bulk poly(A)+ RNA in mutant cells transformed with the YEp351-THO1 multicopy plasmid. As can be seen in Fig. 1C, recombination levels in hpr1Δ cells carrying THO1 in multicopy were sevenfold lower than those in hpr1Δ cells transformed with the empty multicopy vector YEp351. In a previous study (45), we did not detect a significant suppression of hyperrecombination of the first THO1 clone obtained. Apparently, as we now found a sixfold reduction in the recombination of hpr1Δ using a different E. coli-amplified clone from that study (data not shown), the E. coli-amplified clone used in that previous study accumulated some mutation that limited its capacity to reduce hyperrecombination. In contrast, multicopy THO1 was able to suppress neither the gene expression defect (Fig. 1A) nor the high levels of recombination (Fig. 1C) of sub2Δ cells. It is important to note that we used slow-growing but viable sub2Δ strains isogenic to W303-1A that were previously shown to be viable (38). The ability of multicopy SUB2 to suppress the hyperrecombination of hpr1Δ cells was reported previously (13).
In situ localization of poly(A)+ RNA by digoxigenin-labeled oligo(dT) revealed that hpr1Δ cells accumulated poly(A)+ RNA in the nucleus after 4 h of shifting to 37° if transformed with YEp351 and in both the nucleus and the cytoplasm if transformed with multicopy YEp351-THO1 (Fig. 1D). Similar results were obtained with multicopy YEp351-SUB2 carrying SUB2 (Fig. 1D). Therefore, the RNA export defect of hpr1Δ cells was also suppressed by multicopy THO1 and SUB2.
Multicopy THO1 suppresses all mutants of the THO complex.
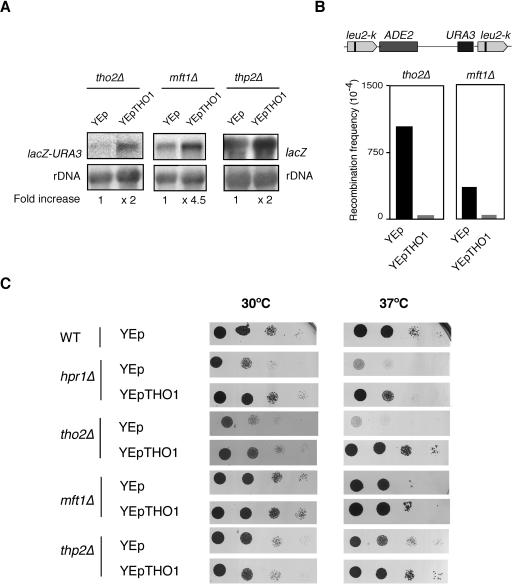
In order to determine whether THO1 suppression was specific to hpr1Δ or general to other mutants of the THO complex, we tested the ability of multicopy THO1 to suppress different phenotypes of the other three THO-null mutants, tho2Δ, mft1Δ, and thp2Δ. As can be seen in Fig. 2, multicopy THO1 partially suppressed the incapacity of tho2Δ and mft1Δ to express the lacZ-URA3 construct to different degrees as determined by uracil prototrophy (data not shown) and Northern analysis (Fig. 2A). The suppression of gene expression phenotypes was also observed in thp2Δ strains (Fig. 2A), in which case Northern analyses were performed with the GAL1-lacZ system because the genetic markers of the thp2Δ strains were incompatible with those of the lacZ-URA3 construct. Hyperrecombination was reduced 19.4- and 16.6-fold in tho2Δ and mft1Δ, respectively (Fig. 2B). Finally, the ability of multicopy THO1 to suppress all THO mutants was tested with the thermosensitivity (ts) phenotype. As can be seen in Fig. 2C, the ts phenotype of all THO-null mutants (hpr1Δ, tho2Δ, mft1Δ, and thp2Δ) was suppressed by multicopy THO1. These results are in agreement with the observation that THO is a structural and functional unit (10) as suppression is observed for all phenotypes. Nevertheless, the variability observed in the intensity of suppression may reflect a difference in the in vivo relevance of each subunit for each phenotype tested, in particular in the presence of multicopy THO1.
FIG. 2.
Suppression of different phenotypes of THO mutants by multicopy THO1. (A) Northern analyses of tho2Δ (RK2-6C), mft1Δ (WMK-2A) cells carrying the Ptet::lacZ-URA3 construct, and thp2Δ (BY-HR167) carrying the GAL1pr::lacZ construct under transcription activation conditions. (B) Recombination frequency of the intrachromosomal direct-repeat system leu2-k::URA3-ADE2::leu2-k mft1Δ (MFM67-13A) and tho2Δ (WRK-1C) mutants transformed with multicopy vector YEp351 and YEp351-THO1. Details are as described for Fig. 1. (C) Suppression of the thermosensitivity (ts) phenotype of hpr1Δ, tho2Δ, mft1Δ and thp2Δ by multicopy THO1. Viability of hpr1Δ (U678-4C) tho2Δ (WRK-1C), mft1Δ (MFM67-13A), and thp2Δ (BY-HR167) cells transformed with either YEp351 or YEp351-THO1. As controls, we used wild-type (WT) W303-1A and BY4741. As they gave identical results, only one is shown. Transformants were spotted as 10-fold serial dilutions on selective medium. Photographs were taken after 3 or 5 days at 30 or 37°C, respectively, as indicated.
The suppression effect of multicopy THO1 was specific to the THO complex mutants, since null mutations of THP1 and SPT4, involved in RNA export and/or transcription and with effects on mRNA accumulation similar to those of THO mutants, were not suppressed by multicopy THO1 (data not shown). The fact that multicopy THO1 suppresses the different phenotypes of THO mutants and not those of sub2Δ suggests a functional relationship of Tho1 with THO rather than with TREX. Nevertheless, in contrast to THO mutations and to sub2Δ cells, tho1Δ did not have any effect on direct-repeat recombination and gene expression or on mRNA export (45; data not shown).
hpr1Δ mutants show reduced processivity of transcription elongation that is suppressed by multicopy THO1 or SUB2.
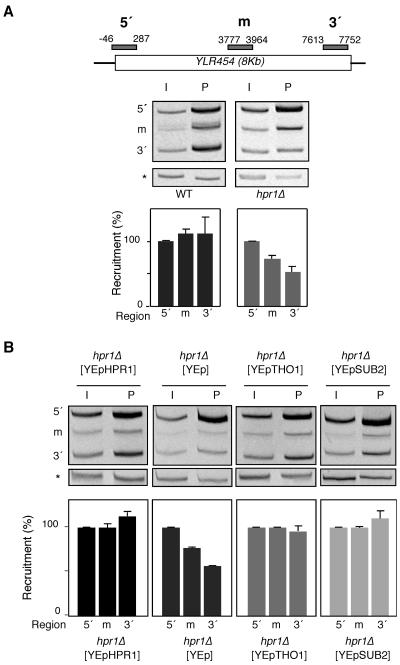
Mutants of the THO complex are impaired in transcription elongation as determined in vivo and in vitro (9, 41, 49). To specifically analyze transcription impairment in THO mutants in vivo independently of any possible effect in RNA stability, we determined RNAPII recruitment by ChIP analysis. This was performed at the 8-kb-long YLR454c gene fused to the GAL1 promoter in three different regions (5′ end, middle, and 3′ end). Consistent with recent results (41), Fig. 3A shows that, whereas more than 95% of RNAPIIs reached the 3′ end of the gene in wild-type cells, this value was reduced to 50% in hpr1Δ mutants, indicating that the processivity of transcription elongation is decreased in THO mutants. Importantly, Tho1 and Sub2 overexpression suppressed this transcription processivity defect of hpr1Δ cells (Fig. 3B). Altogether, the ability of Tho1 to suppress all phenotypes of THO− mutants indicates that the Tho1 and the THO complex must function at the same molecular level.
FIG. 3.
Analysis of the distribution of RNAPII along the GAL1pr::YLR454c fusion construct located at the endogenous YLR454c chromosomal locus. (A) ChIP analysis in wild-type (WT) and hpr1Δ isogenic strains. (B) ChIP analysis in hpr1Δ cells transformed with YEp351-HPR1, YEp351, YEp351-THO1, or YEp351-SUB2. Strains used were WWYL-1D (WT) and WHYL-2A (hpr1Δ). The scheme of the gene analyzed and the PCR-amplified fragments are shown. All experiments were performed with two different transformants. ChIPs were repeated three times for each sample. One representative acrylamide electrophoresis for each experiment is shown. The other fragments correspond to the DNA regions studied. The relative abundance of each DNA fragment was calculated as the ratio between each DNA fragment and the intergenic region quantification results of the precipitated fractions (P) normalized with respect to the corresponding ratios of the input fractions (I). The recruitment data shown are referred to the value of the 5′ region, taken as 100%. Error bars indicate standard deviations. The PCR fragment corresponding to the bp 9716 to 9863 intergenic region of chromosome V used as a control is indicated by asterisks. m, middle.
Tho1 is a conserved nuclear protein that binds to actively transcribed chromatin in a THO- and RNA-dependent manner.
As expected from the nuclear localization of the THO complex (10), Tho1 is predominantly nuclear, as we have shown with an N-terminal Tho1-GFP fusion (data not shown), and as has been observed in a high-throughput analysis (18).
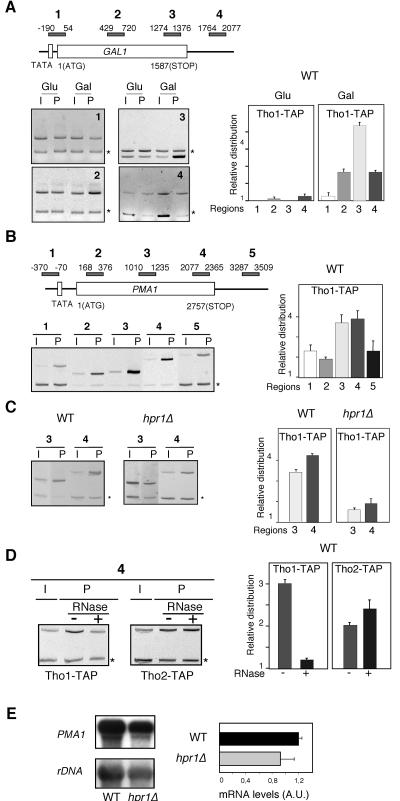
We next wondered if Tho1, as previously shown for THO/TREX, was present in actively transcribed chromatin. This was determined by ChIP assays using yeast strains containing the TAP-tagged version of THO1 at the chromosomal locus. We studied the distribution of Tho1 at four different positions of the GAL1 locus: the promoter 5′ end of the coding region, the middle part of the open reading frame, the 3′ end of the open reading frame, and the untranslated 3′-end region (UTR) of the gene. As can be seen in Fig. 4A, there was no amplification of the DNA sequences analyzed when GAL1 was repressed (2% glucose), indicating no recruitment of Tho1 to inactive chromatin. However, the recruitment of Tho1 to GAL1 chromatin was observed when transcription was active. Recruitment was poor at the promoter 5′-end region and high in the rest of the regions tested. The largest recruitment of Tho1 was observed at the 3′ end of the coding region, with its presence being reduced at the UTR.
FIG. 4.
Tho1 binds to actively transcribed chromatin in a THO- and RNA-dependent manner. The scheme of the genes analyzed and the PCR fragments amplified by PCR are shown. One representative acrylamide electrophoresis for each experiment is shown, with the PCR fragment corresponding to the bp 9716 to 9863 intergenic region of chromosome V used as a control indicated with asterisks. (A) ChIP analysis of Tho1 at the endogenous GAL1 gene in the wild-type strain WWT1T. (B) ChIP analysis of Tho1 at the endogenous PMA1 gene in the wild-type (WT) WWT1T strains carrying the TAP-tagged Tho1. (C) Importance of the THO complex for the recruitment of Tho1 to chromatin at the PMA1 gene, as determined by comparative ChIP analysis in the wild type (WWT1T) and hpr1Δ mutant (WHT1T) (D) Effect of RNase treatment on Tho1 and Tho2 recruitment to active chromatin at the PMA1 gene, as determined by modified ChIP analyses. These were performed in the wild-type WWT1T and WWT2T strains, carrying TAP-tagged Tho1 and Tho2, respectively, treated with (+) or without (−) RNase before immunoprecipitation. Cross-linking times were reduced to 4 min in either treated or nontreated samples. (E) Northern analyses of the expression of PMA1 in WT and hpr1Δ congenic strains RNA was isolated from mid-log-phase cultures grown in rich media. As a 32P-labeled DNA probe, we used the 2.7-kb PMA1 fragment and an internal 589-bp 25S rRNA fragment, both obtained by PCR. Other details are as described for Fig. 1 and 2. AU, arbitrary units. Error bars indicate standard deviations.
To test whether the recruitment of Tho1 was specific for the regulated GAL genes, we extended the ChIP analysis to the constitutively transcribed PMA1 gene. Five different regions of PMA1 were analyzed: the promoter, the 5′ end, the middle part, and the 3′ end of the coding sequence, and the 3′ UTR. As can be seen in Fig. 4B, poor but detectable recruitment of Tho1 was observed at the promoter, the 5′ end of the coding sequence, and the UTR, and as in the GAL1 gene, recruitment of Tho1 was higher toward the 3′ end of the coding sequence. Therefore, we can conclude that Tho1 is recruited to active chromatin regardless of whether it is regulated (GAL1) or constitutive (PMA1).
As has been shown recently for Sub2 (1), we wondered whether Tho1 recruitment to transcribed chromatin was dependent on the THO complex and on the nascent RNA. To assay whether the THO complex was required or whether it enhanced recruitment of Tho1 to active chromatin, we compared the results of the ChIP analysis of Tho1-TAP in wild-type and hpr1Δ strains. As can be seen in Fig. 4C, Tho1 recruitment was clearly reduced in hpr1Δ cells, suggesting that the THO complex is required for Tho1 recruitment. To rule out the possibility that the THO dependency of Tho1 recruitment to chromatin was due to an incapacity of hpr1Δ to transcribe PMA1, we analyzed the levels of PMA1 transcription by Northern analysis and found that they were only slightly reduced in hpr1Δ versus wild-type cells (Fig. 4E). This was expected, as we had previously shown that the effect of hpr1Δ on transcription is significant in highly transcribed long or GC-rich DNA sequences (11), whereas PMA1 does not belong to this class of genes, and is consistent with previous Northern analyses of PMA1 in hpr1Δ strains (58).
To determine whether the Tho1 recruitment is RNA dependent, a ChIP analysis was performed with or without RNases A and T1 before immunoprecipitation. Given that Tho2 recruitment was reported to be independent of RNA (1), we used a TAP-tagged Tho2 protein as a negative control. As can be seen in Fig. 4D, in RNase-treated samples, Tho1 recruitment to the PMA1 gene was abolished, whereas Tho2 was strongly recruited. As expected in these experiments, Sub2 recruitment was also reduced after RNase treatment (data not shown). Tho1 recruitment to transcribed chromatin is therefore dependent on THO and RNA.
In vitro RNA binding activity of Tho1.
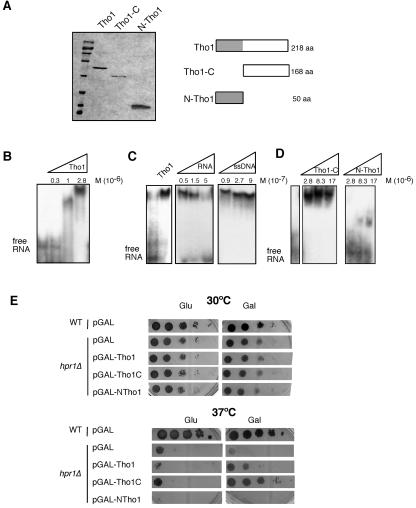
Given all of the previous results, we wondered if Tho1 had the ability to bind RNA in vitro. Since Tho1 has two structurally different halves, the N-terminal region that contains the SAP/SAF domain and the C-terminal half, with no known domain, we constructed His6-tagged versions of the full 218-amino-acid (aa) Tho1 protein, the 50-aa N-terminal region containing the SAP domain (N-Tho1), and the 168-aa C-terminal part lacking the SAP domain (Tho1-C). Such proteins were expressed in E. coli from the T7 promoter and purified with Ni-nitrilotriacetic acid columns in order to determine their nucleic acid binding properties in vitro.
The three polypeptides were purified (Fig. 5A), and RNA binding was determined by gel-shifting assays using a 32P-labeled 90-nucleotide RNA fragment synthesized in vitro. As can be seen in Fig. 5B, the full Tho1 protein binds to RNA. Band shifting was directly proportional to the amount of Tho1 protein used. This binding could be competed with cold RNA in a manner proportional to the amount of cold RNA added, whereas it was not competed by higher amounts of cold ssDNA (Fig. 5C). Therefore, binding to RNA was specific rather than a manifestation of unspecific binding to any type of single-stranded nucleic acid. We therefore performed a more detailed analysis to determine which region was conferring the main ability of the protein to bind to RNA using the N-Tho1- and Tho1-C-truncated versions. Figure 5D shows that although the N-Tho1 version containing only the SAP domain had a weak ability to bind RNA, the major RNA binding activity resided in the C-terminal part of the protein outside of the SAP domain, although no known RNA binding motif appears in this region. As expected from its N-terminal SAP domain, Tho1 also binds double-stranded DNA (see Fig. S3 in the supplemental material).
FIG. 5.
In vitro RNA binding activity of Tho1. (A) Twelve percent sodium dodecyl sulfate-polyacrylamide gel electrophoresis of a His6-tagged version of the complete Tho1 protein and its truncated N-Tho1 and Tho1-C forms obtained from E. coli and stained with Coomassie blue. Proteins were purified from the BL21 E. coli strain transformed with pT7-HA-THO1, pT7-HA-5′THO1, and pT7-HA-3′THO1. A scheme of each protein purified is shown on the right, with the SAP domain shown in gray. (B) Band shift assays of the RNA binding activity of Tho1 using 5 ng of the 90-bp, 32P-labeled RNA from the polylinker of pBluescript-SK (0.05 M 10−7) and increasing amounts of purified Tho1. (C) Competition assays of RNA binding activity using increasing amounts of the same 90-mer RNA molecule used before and a 60-mer ssDNA as cold competitor. (D) RNA binding assays with increasing equimolecular amounts of the N-Tho1 and Tho1-C truncated forms of Tho1. Other details are as described for panel B. (E) Comparison of the ability to suppress the thermosensitivity (ts) phenotype of hpr1Δ by overexpression of truncated Tho1 versions. Viability of hpr1Δ (U678-4C) cells transformed with either pGAL-THO1, pGAL-NTHO1, and pGAL-THO1C or the empty vector p416GAL (pGAL). Transformants were spotted on selective medium with glucose to control for cell number and on galactose-containing medium to induce gene expression. Other details are as described for Fig. 2. WT, wild type.
Given these results, we tested the capacities of both truncated Tho1 proteins to suppress the hpr1Δ mutation. For this, we constructed new plasmids in which the N-Tho1- and Tho1-C-encoding DNA sequences were fused to the regulated GAL1 promoter. As can be seen in Fig. 5E, analysis of yeast hpr1Δ transformed with these plasmids revealed that, whereas N-Tho1 overexpression was unable to suppress the thermosensitivity (ts) of hpr1Δ, Tho1-C overexpression suppressed this ts phenotype with a much better efficiency than did the whole Tho1 protein.
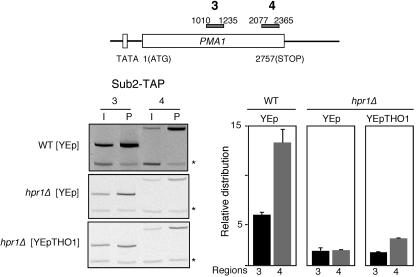
Multicopy THO1 does not enhance Sub2 recruitment to active chromatin.
It has been shown that THO enhances the ability of Sub2 to bind active chromatin, suggesting that THO helps recruit Sub2 to the site of transcription (58). We therefore wondered whether the suppression of THO mutations by multicopy THO1 was due to the capacity of Tho1 to help recruit Sub2 to active chromatin. To test this possibility, we assayed, by ChIP analysis, the ability of TAP-Sub2 to be recruited to the PMA1 gene in hpr1Δ cells transformed with the empty YEp351 vector or with the multicopy YEp351-THO1 carrying THO1. As can be seen in Fig. 6, the recruitment of Sub2 to the two different PMA1 regions tested was reduced to 40 and 20% of the wild-type levels, regardless of whether the hpr1Δ strains expressed THO1 in multicopy. Therefore, Tho1 does not stabilize or help to recruit Sub2 to the site of transcription.
FIG. 6.
Effect of multicopy THO1 on Sub2 recruitment to active chromatin at the PMA1 gene in hpr1Δ and wild-type strains. ChIP analyses were performed with strains BSU-S2T.3B (WT) and BSU-S2T.6D (hpr1Δ) transformed with plasmid YEp351-THO1, carrying THO1 in multicopy or the empty vector. Other details are as described for Fig. 3. Error bars indicate standard deviations. The PCR fragment corresponding to the bp 9716 to 9863 intergenic region of chromosome V used as a control is indicated by asterisks.
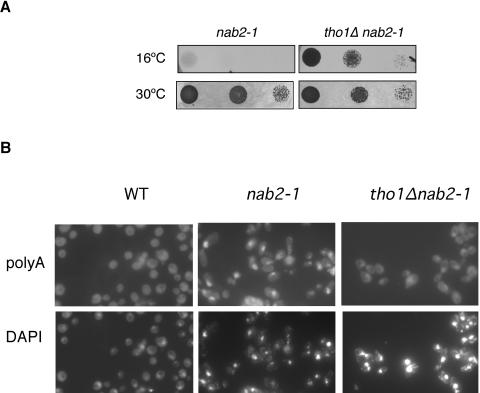
Genetic interaction of Tho1 with the Nab2 RNA binding protein.
Thus far, we were not able to identify a phenotype associated with tho1Δ mutants and our identification of a role of Tho1 in mRNP biogenesis derives from the effects caused by Tho1 overexpression. Consequently, by directly studying the effect of tho1Δ in double mutant combinations, we next determined whether THO1 could genetically interact with other functions of mRNP biogenesis and export. We analyzed the viability of double mutants of tho1Δ and mutations in genes encoding different protein complexes that may act subsequently in the mRNP biogenesis pathway in which THO participates, such as sub2Δ, mex67-5, nab2-1, and thp1Δ. These single mutants show similar gene expression and hyperrecombination phenotypes to those of THO mutants (16, 29). All double mutants constructed were viable at 30°C (data not shown), as was also shown for tho1Δ hpr1Δ mutants (45). Interestingly, although tho1Δ nab2-1 had no phenotype at the permissive temperature (30°C), tho1Δ suppressed the cryosensitivity (cs) phenotype of nab2-1 cells. Thus, nab2-1 tho1Δ cells formed colonies at 16°C, whereas nab2-1 did not (Fig. 7A), and the nuclear poly(A)+ RNA accumulation phenotype observed in nab2-1 mutants at 16°C was reduced in tho1Δ nab2-1 double mutants (Fig. 7B). This result provides a demonstration that the function of wild-type levels of Tho1 is related to RNP biogenesis and export and establishes a functional link between Tho1 and the Nab2 poly(A)+-RNA binding protein.
FIG. 7.
Deletion of THO1 rescues the cs phenotype and the mRNA export defects of nab2-1. (A) Tenfold serial dilutions of nab2-1 (NALT-1) and tho1Δ nab2-1 (NALT-2) were spotted on selective plates and incubated at 16°C and 30°C for 5 days. (B) Subcellular localization of poly(A)+ RNA detected in situ with digoxigenin-labeled oligo(dT)16. Other details are as described for Fig. 1.
DISCUSSION
This study reveals that Tho1, a SAF-box/SAP domain protein, is a nuclear hnRNP with a strong capacity to bind RNA at its C-terminal half and that has a functional relationship to THO and Sub2 in cotranscriptional mRNP biogenesis. Multicopy THO1, like SUB2, suppresses all phenotypes of THO-complex mutants. As is the case for Sub2, Tho1 is specifically recruited to active chromatin in a RNA- and THO-dependent manner.
The functional relationship of Tho1 with the THO complex suggests a function of Tho1 in mRNP biogenesis/processing that is supported by the ability of Tho1 to bind RNA in vitro. Despite the functional interconnection of Sub2 and Tho1, the primary functions of both proteins may be different. Thus, sub2Δ cells are either sick and thermosensitive or inviable, depending on genetic background (19, 38), and show gene expression and RNA export defects and a strong hyperrecombination phenotype (13, 29). tho1Δ mutants are viable and show no phenotypes and, whereas hpr1 sub2 mutants are inviable, hpr1 tho1 and sub2 tho1 are viable (45; data not shown). Either the function of Tho1 in mRNP metabolism is redundant or dispensable. Nonetheless, other genes with a putative role in mRNP biogenesis and export for which only their overexpression and not their null mutation have a phenotype have been defined. An example of that is Gbp2, which interacts physically with the TREX complex (25, 53, 57).
Sub2 may be an RNA-dependent ATPase, as shown for the human ortholog UAP56 (51) that forms, with the major RNA binding protein Yra1, a heterodimer that physically associates with THO in a larger complex termed TREX. The requirement of Sub2-Yra1 in RNA export has been well established (52), and the associated phenotypes of gene expression defect and transcription-dependent hyperrecombination conferred by their mutations suggest that Sub2 function is essential for the cotranscriptional formation of RNA export-competent mRNPs. The association of Sub2 and THO with active chromatin is consistent with this idea. Although THO facilitates the recruitment of Sub2 to active chromatin, multicopy THO1 does not suppress THO− mutants by helping to recruit Sub2 nor does it stabilize the TREX complex at the site of transcription (Fig. 6). Nevertheless, we cannot rule out the possibility that cotranscriptional Sub2 recruitment takes longer than usual in the THO1-rescued hpr1Δ strains and is not detected cotranscriptionally. It is likely that Tho1 and Sub2 as well as the THO complex function in vivo in a multiribonucleoprotein structure at the site of transcription. In the absence of THO, overexpression of Tho1 or Sub2 may partially provide either a THO-like function or an alternative pathway of mRNP biogenesis and assembly. Overexpressed Tho1 or Sub2 may prevent the undesired reactivity of nascent mRNA with template DNA that has been observed in hpr1 mutants (24). The higher accumulation of Tho1 and Sub2 at the 3′ ends of transcribed coding regions (Fig. 4) (58) opens the possibility that Tho1 and Sub2 might also be important for transcription termination or 3′-end processing. Interestingly, we have recently found a genetic and functional interaction between THO and the RNA cleavage and polyadenylation factors (40).
The observation that multicopy THO1 does not suppress sub2 but only THO− mutations suggests that the THO complex constitutes a functional unit independent of Sub2-Yra1 RNA export factors. This is consistent with the facts that THO is a stable complex in the absence of Sub2 (29), whereas THO becomes unstable in hpr1Δ mutants (our unpublished observation). Tho1 cannot substitute for Sub2 in the cell, only for THO. Indeed THO has been shown to copurify with Sub2 in yeast and humans, but this has not been possible yet for Drosophila THO (47) and recent evidence suggests that human THO components can be purified free of UAP56/Sub2 and Yra1/ALY (42). These results are in agreement with the observations that only Sub2 but not THO complex recruitment to chromatin depends on RNA (1), suggesting the possibility that the association of THO with Sub2-Yra1 in the TREX complex might be mediated by the mRNA molecule.
Tho1 may belong to a SAP/SAF-box class of hnRNPs with a function related to mRNP biogenesis and export. Indeed, the SAP domain was identified in scaffold-associated proteins (7) such as SAF-A/hnRNPU, which is an RNA binding protein that copurifies with RNAPII and cooperates with actin for productive transcription by RNAPII (31, 34), and SAF-B, which interacts with SR proteins and affects splicing in vivo (44). Nevertheless, the observation that overexpression of the RNA binding C-terminal half of Tho1 suppresses hpr1Δ more efficiently than the whole Tho1 protein, whereas overexpression of the SAF-box has no effect (Fig. 5), indicates that the RNA binding C-terminal half of Tho1, rather than the SAP domain, is primarily responsible for its mRNP biogenesis/export-related function.
Two additional observations support that Tho1 is an hnRNP with a role in mRNP biogenesis and export. The putative structural homolog of Tho1 in humans is a nuclear-matrix protein, Hcc-1 (see Fig. S2 in the supplemental material), which has been shown in two-hybrid assays to bind the two DEAD box putative RNA helicases, hSUB2/UAP56/BAT1 and DDX39 (12, 35). Whether Tho1 might interact with RNA-dependent ATPases important in the assembly of export-competent mRNPs is a possibility to be explored. It has been reported in a high-throughput two-hybrid analysis that Tho1 and Sub2 interact (26), and we have observed that His6-tagged Tho1 purified from E. coli coimmunoprecipitates with Sub2 (unpublished data). The physiological relevance of this interaction is not clear and needs further investigation since Tho1 overexpression does not enhance Sub2 recruitment (Fig. 6) and we cannot discard the possibility that such Sub2-Tho1 interaction could be mediated by RNA. It is also interesting to note that Tho1 coprecipitates with many other factors involved in mRNA metabolism with a Flag-tagged Spt5 transcription factor (39).
Thp1-Sac3 is a nuclear protein complex located at nuclear pores, and the mutation of any of its components has been shown to lead to similar phenotypes in gene expression and genomic instability to those of THO mutants (14, 16). The overexpression of Nab2, a poly(A)+ binding hnRNP, suppresses thp1Δ mutations (16), and interestingly, tho1Δ suppresses the cryosensitivity cs phenotype of nab2-1. Like tho1Δ, null mutations of the two tetratricopeptide repeat-like proteins Mlp1/Mlp2 attached to the nuclear pores suppress the ts phenotype of the N-terminal deletion of NAB2 (56). One possible explanation for this result is that Tho1 could facilitate Nab2 loading onto the mRNP particle. In this scenario, Nab2-1 mutant protein could form a suboptimal mRNP particle that would not be exported from the nucleus and would be degraded. A role for Mlp1/Mlp2 in the control of export and degradation of unprocessed mRNAs has been proposed (17). The genetic interaction between Tho1 and Nab2 strengthens the possibility that Tho1 functions along the mRNP biogenesis process from the DNA to the nuclear pore, establishing a further link between transcription and RNA export as well as between THO and Thp1-Sac3.
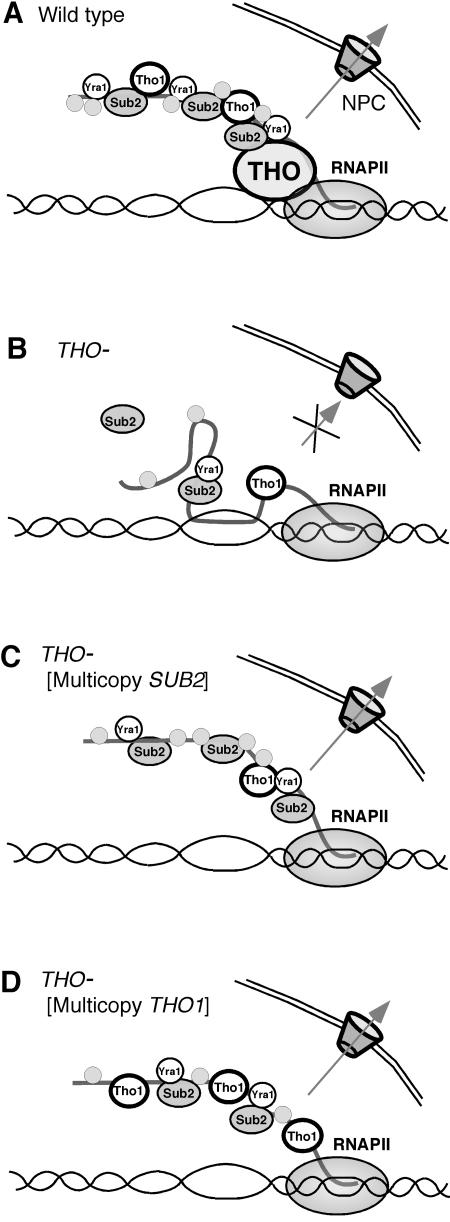
Considering all results, we suggest a working hypothesis in which the THO complex would facilitate loading of Tho1 and Sub2-Yra1 onto the nascent mRNA (Fig. 8). Consistent with the high abundance of Sub2 per cell (18) numerous Sub2 molecules could be recruited onto the nascent mRNA molecule as it is extended, allowing formation of an export-competent mRNP (Fig. 8). In the absence of a functional THO complex, only a few Sub2 subunits would gain access to the RNA by a THO-independent, low-affinity mechanism leading to an export-incompetent suboptimal mRNP that could form DNA-RNA hybrids behind the elongating RNAPII. Efficient recruitment of Tho1 onto the nascent mRNA would also require THO. In THO− mutants, there would be two alternatives. Sub2 overexpression would increase its own probability to bind to the nascent mRNA, bypassing the requirement of THO to assemble an export-competent mRNP. Tho1 overexpression would allow low-affinity recruitment of Tho1 to yield an export-competent mRNP with the essential Sub2-Yra1 heterodimers loaded in a THO- and Tho1-independent manner. The model is consistent with the idea that Yra1 would be the adapter to the Mex67/Mtr2 export factor responsible for the export of the mRNP in all scenarios considered. Whether Tho1 has a structural role in mRNP formation and export or has a biochemical activity required to catalyze a particular step of mRNP biogenesis and export will have to be deciphered in the future.
FIG. 8.
Model explaining the roles of the THO complex, Sub2, and Tho1 in mRNP cotranscriptional mRNP biogenesis in different genetic backgrounds. (A) In wild-type cells, the THO complex would facilitate loading of the Sub2-Yra1 heterodimers onto the nascent mRNA. Numerous Sub2 molecules are recruited onto the mRNA molecule as it is extended, allowing the formation of an export-competent mRNP, which is exported by Mex67-Mtr2. (B) In the absence of a functional THO complex, Sub2 is not loaded properly onto the nascent mRNA and only a few molecules gain access to the RNA by a THO-independent low-affinity mechanism. As a consequence, an improperly assembled export-incompetent mRNP is generated, which could form DNA-RNA hybrids behind the elongating RNAPII. (C) In the absence of a functional THO complex, overexpression of Sub2 increases the probability of Sub2 or Sub2-Yra1 to directly bind to the nascent mRNA, bypassing the requirement of THO to assemble an export-competent mRNP. (D) Overexpression of the Tho1 hnRNP will allow loading of Tho1 onto the nascent mRNA in the absence of THO complex. An alternative export-competent mRNP molecule would be formed with the essential Sub2-Yra1 subunits loaded in a THO-independent manner. This would explain why sub2Δ, in contrast to THO null mutations, cannot be suppressed by multicopy THO1.
Supplementary Material
Acknowledgments
We thank K. Struhl, E. Hurt, and A. H. Corbett for kindly providing reagents, F. Prado and R. E. Wellinger for reading the manuscript, and D. Haun for style supervision.
This work was supported by grants from the Ministry of Science of Education of Spain (SAF2003-00204) and the Junta de Andalucia (CVI-102). S.J. was the recipient of a training grant from the Spanish Ministry of Health.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Abruzzi, K. C., S. Lacadie, and M. Rosbash. 2004. Biochemical analysis of TREX complex recruitment to intronless and intron-containing yeast genes. EMBO J. 23:2620-2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aguilera, A. 2002. The connection between transcription and genomic instability. EMBO J. 21:195-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aguilera, A. 2005. Cotranscriptional mRNP assembly: from the DNA to the nuclear pore. Curr. Opin. Cell Biol. 17:242-250. [DOI] [PubMed] [Google Scholar]
- 4.Ahn, S. H., M. Kim, and S. Buratowski. 2004. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol. Cell 13:67-76. [DOI] [PubMed] [Google Scholar]
- 5.Amberg, D. C., A. L. Goldstein, and C. N. Cole. 1992. Isolation and characterization of RAT1: an essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes Dev. 6:1173-1189. [DOI] [PubMed] [Google Scholar]
- 6.Andrulis, E. D., J. Werner, A. Nazarian, H. Erdjument-Bromage, P. Tempst, and J. T. Lis. 2002. The RNA processing exosome is linked to elongating RNA polymerase II in Drosophila. Nature 420:837-841. [DOI] [PubMed] [Google Scholar]
- 7.Aravind, L., and E. V. Koonin. 2000. SAP—a putative DNA-binding motif involved in chromosomal organization. Trends Biochem. Sci. 25:112-114. [DOI] [PubMed] [Google Scholar]
- 8.Casolari, J. M., C. R. Brown, S. Komili, J. West, H. Hieronymus, and P. A. Silver. 2004. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell 117:427-439. [DOI] [PubMed] [Google Scholar]
- 9.Chávez, S., and A. Aguilera. 1997. The yeast HPR1 gene has a functional role in transcriptional elongation that uncovers a novel source of genome instability. Genes Dev. 11:3459-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chávez, S., T. Beilharz, A. G. Rondón, H. Erdjument-Bromage, P. Tempst, J. Q. Svejstrup, T. Lithgow, and A. Aguilera. 2000. A protein complex containing Tho2, Hpr1, Mft1 and a novel protein, Thp2, connects transcription elongation with mitotic recombination in Saccharomyces cerevisiae. EMBO J. 19:5824-5834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chávez, S., M. García-Rubio, F. Prado, and A. Aguilera. 2001. Hpr1 is preferentially required for transcription of either long or G+C-rich DNA sequences in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:7054-7064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choong, M. L., L. K. Tan, S. L. Lo, E. C. Ren, K. Ou, S. E. Ong, R. C. Liang, T. K. Seow, and M. C. Chung. 2001. An integrated approach in the discovery and characterization of a novel nuclear protein over-expressed in liver and pancreatic tumors. FEBS Lett. 496:109-116. [DOI] [PubMed] [Google Scholar]
- 13.Fan, H. Y., R. J. Merker, and H. L. Klein. 2001. High-copy-number expression of Sub2p, a member of the RNA helicase superfamily, suppresses hpr1-mediated genomic instability. Mol. Cell. Biol. 21:5459-5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischer, T., K. Strasser, A. Racz, S. Rodriguez-Navarro, M. Oppizzi, P. Ihrig, J. Lechner, and E. Hurt. 2002. The mRNA export machinery requires the novel Sac3p-Thp1p complex to dock at the nucleoplasmic entrance of the nuclear pores. EMBO J. 21:5843-5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gallardo, M., and A. Aguilera. 2001. A new hyperrecombination mutation identifies a novel yeast gene, THP1, connecting transcription elongation with mitotic recombination. Genetics 157:79-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gallardo, M., R. Luna, H. Erdjument-Bromage, P. Tempst, and A. Aguilera. 2003. Nab2p and the Thp1p-Sac3p complex functionally interact at the interface between transcription and mRNA metabolism. J. Biol. Chem. 278:24225-24232. [DOI] [PubMed] [Google Scholar]
- 17.Galy, V., O. Gadal, M. Fromont-Racine, A. Romano, A. Jacquier, and U. Nehrbass. 2004. Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell 116:63-73. [DOI] [PubMed] [Google Scholar]
- 18.Ghaemmaghami, S., W. K. Huh, K. Bower, R. W. Howson, A. Belle, N. Dephoure, E. K. O'Shea, and J. S. Weissman. 2003. Global analysis of protein expression in yeast. Nature 425:737-741. [DOI] [PubMed] [Google Scholar]
- 19.Giaever, G., A. M. Chu, L. Ni, C. Connelly, L. Riles, S. Veronneau, S. Dow, A. Lucau-Danila, K. Anderson, B. Andre, A. P. Arkin, A. Astromoff, M. El-Bakkoury, R. Bangham, R. Benito, S. Brachat, S. Campanaro, M. Curtiss, K. Davis, A. Deutschbauer, K. D. Entian, P. Flaherty, F. Foury, D. J. Garfinkel, M. Gerstein, D. Gotte, U. Guldener, J. H. Hegemann, S. Hempel, Z. Herman, D. F. Jaramillo, D. E. Kelly, S. L. Kelly, P. Kotter, D. LaBonte, D. C. Lamb, N. Lan, H. Liang, H. Liao, L. Liu, C. Luo, M. Lussier, R. Mao, P. Menard, S. L. Ooi, J. L. Revuelta, C. J. Roberts, M. Rose, P. Ross-Macdonald, B. Scherens, G. Schimmack, B. Shafer, D. D. Shoemaker, S. Sookhai-Mahadeo, R. K. Storms, J. N. Strathern, G. Valle, M. Voet, G. Volckaert, C. Y. Wang, T. R. Ward, J. Wilhelmy, E. A. Winzeler, Y. Yang, G. Yen, E. Youngman, K. Yu, H. Bussey, J. D. Boeke, M. Snyder, P. Philippsen, R. W. Davis, and M. Johnston. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387-391. [DOI] [PubMed] [Google Scholar]
- 20.Green, D. M., K. A. Marfatia, E. B. Crafton, X. Zhang, X. Cheng, and A. H. Corbett. 2002. Nab2p is required for poly(A) RNA export in Saccharomyces cerevisiae and is regulated by arginine methylation via Hmt1p. J. Biol. Chem. 277:7752-7760. [DOI] [PubMed] [Google Scholar]
- 21.Hecht, A., and M. Grunstein. 1999. Mapping DNA interaction sites of chromosomal proteins using immunoprecipitation and polymerase chain reaction. Methods Enzymol. 304:399-414. [DOI] [PubMed] [Google Scholar]
- 22.Hirose, Y., and J. L. Manley. 1998. RNA polymerase II is an essential mRNA polyadenylation factor. Nature 395:93-96. [DOI] [PubMed] [Google Scholar]
- 23.Hirose, Y., R. Tacke, and J. L. Manley. 1999. Phosphorylated RNA polymerase II stimulates pre-mRNA splicing. Genes Dev. 13:1234-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huertas, P., and A. Aguilera. 2003. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol. Cell 12:711-721. [DOI] [PubMed] [Google Scholar]
- 25.Hurt, E., M. J. Luo, S. Rother, R. Reed, and K. Strasser. 2004. Cotranscriptional recruitment of the serine-arginine-rich (SR)-like proteins Gbp2 and Hrb1 to nascent mRNA via the TREX complex. Proc. Natl. Acad. Sci. USA 101:1858-1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ito, T., K. Tashiro, and T. Kuhara. 2001. Systematic analysis of Saccharomyces cerevisiae genome: gene network and protein-protein interaction network. Tanpakushitsu Kakusan Koso. 46:2407-2413. (In Japanese.) [PubMed] [Google Scholar]
- 27.Jensen, T. H., J. Boulay, M. Rosbash, and D. Libri. 2001. The DECD box putative ATPase Sub2p is an early mRNA export factor. Curr. Biol. 11:1711-1715. [DOI] [PubMed] [Google Scholar]
- 28.Jensen, T. H., K. Dower, D. Libri, and M. Rosbash. 2003. Early formation of mRNP: license for export or quality control? Mol. Cell 11:1129-1138. [DOI] [PubMed] [Google Scholar]
- 29.Jimeno, S., A. G. Rondón, R. Luna, and A. Aguilera. 2002. The yeast THO complex and mRNA export factors link RNA metabolism with transcription and genome instability. EMBO J. 21:3526-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim, M., S. H. Ahn, N. J. Krogan, J. F. Greenblatt, and S. Buratowski. 2004. Transitions in RNA polymerase II elongation complexes at the 3′ ends of genes. EMBO J. 23:354-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim, M. K., and V. M. Nikodem. 1999. hnRNP U inhibits carboxy-terminal domain phosphorylation by TFIIH and represses RNA polymerase II elongation. Mol. Cell. Biol. 19:6833-6844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kistler, A. L., and C. Guthrie. 2001. Deletion of MUD2, the yeast homolog of U2AF65, can bypass the requirement for sub2, an essential spliceosomal ATPase. Genes Dev. 15:42-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Komarnitsky, P., E. J. Cho, and S. Buratowski. 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 14:2452-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kukalev, A., Y. Nord, C. Palmberg, T. Bergman, and P. Percipalle. 2005. Actin and hnRNP U cooperate for productive transcription by RNA polymerase II. Nat. Struct. Mol. Biol. 12:238-244. [DOI] [PubMed] [Google Scholar]
- 35.Leaw, C. L., E. C. Ren, and M. L. Choong. 2004. Hcc-1 is a novel component of the nuclear matrix with growth inhibitory function. Cell. Mol. Life Sci. 61:2264-2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lei, E. P., H. Krebber, and P. A. Silver. 2001. Messenger RNAs are recruited for nuclear export during transcription. Genes Dev. 15:1771-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Libri, D., K. Dower, J. Boulay, R. Thomsen, M. Rosbash, and T. H. Jensen. 2002. Interactions between mRNA export commitment, 3′-end quality control, and nuclear degradation. Mol. Cell. Biol. 22:8254-8266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Libri, D., N. Graziani, C. Saguez, and J. Boulay. 2001. Multiple roles for the yeast SUB2/yUAP56 gene in splicing. Genes Dev. 15:36-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lindstrom, D. L., S. L. Squazzo, N. Muster, T. A. Burckin, K. C. Wachter, C. A. Emigh, J. A. McCleery, J. R. Yates III, and G. A. Hartzog. 2003. Dual roles for Spt5 in pre-mRNA processing and transcription elongation revealed by identification of Spt5-associated proteins. Mol. Cell. Biol. 23:1368-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luna, R., S. Jimeno, M. Marin, P. Huertas, M. García-Rubio, and A. Aguilera. 2005. Interdependence between transcription and mRNP processing and export, and its impact on genetic stability. Mol. Cell 18:711-722. [DOI] [PubMed] [Google Scholar]
- 41.Mason, P. B., and K. Struhl. 2005. Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol. Cell 17:831-840. [DOI] [PubMed] [Google Scholar]
- 42.Masuda, S., R. Das, H. Cheng, E. Hurt, N. Dorman, and R. Reed. 2005. Recruitment of the human TREX complex to mRNA during splicing. Genes Dev. 19:1512-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McCracken, S., N. Fong, E. Rosonina, K. Yankulov, G. Brothers, D. Siderovski, A. Hessel, S. Foster, S. Shuman, and D. L. Bentley. 1997. 5′-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev. 11:3306-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nayler, O., W. Stratling, J. P. Bourquin, I. Stagljar, L. Lindemann, H. Jasper, A. M. Hartmann, F. O. Fackelmayer, A. Ullrich, and S. Stamm. 1998. SAF-B protein couples transcription and pre-mRNA splicing to SAR/MAR elements. Nucleic Acids Res. 26:3542-3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Piruat, J. I., and A. Aguilera. 1998. A novel yeast gene, THO2, is involved in RNA pol II transcription and provides new evidence for transcriptional elongation-associated recombination. EMBO J. 17:4859-4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Proudfoot, N. J., A. Furger, and M. J. Dye. 2002. Integrating mRNA processing with transcription. Cell 108:501-512. [DOI] [PubMed] [Google Scholar]
- 47.Rehwinkel, J., A. Herold, K. Gari, T. Kocher, M. Rode, F. L. Ciccarelli, M. Wilm, and E. Izaurralde. 2004. Genome-wide analysis of mRNAs regulated by the THO complex in Drosophila melanogaster. Nat. Struct. Mol. Biol. 11:558-566. [DOI] [PubMed] [Google Scholar]
- 48.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030-1032. [DOI] [PubMed] [Google Scholar]
- 49.Rondón, A. G., S. Jimeno, M. García-Rubio, and A. Aguilera. 2003. Molecular evidence that the eukaryotic THO/TREX complex is required for efficient transcription elongation. J. Biol. Chem. 278:39037-39043. [DOI] [PubMed] [Google Scholar]
- 50.Schroeder, S. C., B. Schwer, S. Shuman, and D. Bentley. 2000. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev. 14:2435-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shi, H., O. Cordin, C. M. Minder, P. Linder, and R. M. Xu. 2004. Crystal structure of the human ATP-dependent splicing and export factor UAP56. Proc. Natl. Acad. Sci. USA 101:17628-17633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Strasser, K., and E. Hurt. 2001. Splicing factor Sub2p is required for nuclear mRNA export through its interaction with Yra1p. Nature 413:648-652. [DOI] [PubMed] [Google Scholar]
- 53.Strasser, K., S. Masuda, P. Mason, J. Pfannstiel, M. Oppizzi, S. Rodriguez-Navarro, A. G. Rondón, A. Aguilera, K. Struhl, R. Reed, and E. Hurt. 2002. TREX is a conserved complex coupling transcription with messenger RNA export. Nature 417:304-308. [DOI] [PubMed] [Google Scholar]
- 54.Stutz, F., and E. Izaurralde. 2003. The interplay of nuclear mRNP assembly, mRNA surveillance and export. Trends Cell Biol. 13:319-327. [DOI] [PubMed] [Google Scholar]
- 55.Uguen, P., and S. Murphy. 2003. The 3′ ends of human pre-snRNAs are produced by RNA polymerase II CTD-dependent RNA processing. EMBO J. 22:4544-4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vinciguerra, P., N. Iglesias, J. Camblong, D. Zenklusen, and F. Stutz. 2005. Perinuclear Mlp proteins downregulate gene expression in response to a defect in mRNA export. EMBO J. 24:813-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Windgassen, M., and H. Krebber. 2003. Identification of Gbp2 as a novel poly(A)+ RNA-binding protein involved in the cytoplasmic delivery of messenger RNAs in yeast. EMBO Rep. 4:278-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zenklusen, D., P. Vinciguerra, J. C. Wyss, and F. Stutz. 2002. Stable mRNP formation and export require cotranscriptional recruitment of the mRNA export factors Yra1p and Sub2p by Hpr1p. Mol. Cell. Biol. 22:8241-8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zorio, D. A., and D. L. Bentley. 2004. The link between mRNA processing and transcription: communication works both ways. Exp. Cell Res. 296:91-97. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.