Abstract
The identification of genes undergoing genetic or epigenetic alterations and contributing to the development of cancer is critical to our understanding of the molecular mechanisms of carcinogenesis. A new approach in identifying alterations of genes that might be relevant to the process of tumor development was used in this study by examining the gene expression profile in human lung cancer cells exposed to 5-aza-2′-deoxycytidine (5-aza-dC). A cDNA array analysis was carried out on 5-aza-dC-treated and untreated non small cell lung cancer (NSCLC) cell line NCI-H522. Sixteen and 14 genes were upregulated and downregulated, respectively, by 5-aza-dC treatment. Among them, downregulation of tyrosine protein kinase ABL2 (ABL2) gene and upregulation of hint/protein kinase C inhibitor 1 (Hint/PKCI-1), DVL1, TIMP-1, and TRP-1 genes were found in expanded observations in two or three of five 5-aza-dC-treated NSCLC cell lines. Among these genes, we found that cDNA transfer of Hint/PKCI-1 resulted in a significant in vitro growth inhibition in two cell lines exhibiting 5-aza-dC-induced upregulation of Hint/PKCI-1 and significantly reduced in vivo tumorigenicity of one NSCLC cell line. Hint/PKCI-1, which is the only other characterized human histidine triad (HIT) nucleotide-binding protein in addition to tumor-suppressor gene FHIT, might be involved in lung carcinogenesis.
Keywords: Human non small cell lung carcinoma, 5-aza-2′-deoxycytidine, cell growth inhibition, ABL2 gene, Hint/PKCI-1 gene
Introduction
Lung cancer, the majority of which are non small cell lung carcinoma (NSCLC), is the leading cause of cancer death in men and women in the United States [1]. Although most lung cancers are related to tobacco use, it is also ranked second only to bladder cancer in the proportion cases thought to be due to occupational exposures [2]. Increasing evidence demonstrates that the accumulation of epigenetic damage induced by the respiratory epithelium to cigarette smoke and/or occupational carcinogens is one of the major mechanisms responsible for the development of lung cancer. Epigenetic damage, consisting mainly of promoter hypermethylation, disrupts or silences the expression of tumor-suppressor genes, leading to uncontrolled cell proliferation. There are an increasing number of candidate tumor-suppressor genes that are inactivated by promoter hypermethylation in various types of cancer. In human cancer, promoter hypermethylation appears to be involved at least as frequently as point mutations in the disruption of tumor-suppressor genes [3]. Promoter hypermethylation in tumor-suppressor genes, such as p16, death-associated protein kinase (DAPK), FHIT, and Ras effector homologue (RASSF1A), is thought to be an early event in cigarette smoking-related respiratory carcinogenesis [4–6].
One major challenge in lung cancer investigations is the identification of genes that undergo genetic or epigenetic damage during neoplastic development. The basic strategy employed for identifying new candidate genes was the comparison of gene expression between cancer cells and the corresponding noncancerous cells, or between parental tumor cells and anti-tumor agent-treated tumor cells. The comparison of gene expression between 5-aza-2′-deoxycytidine (5-aza-dC)-treated and parental tumor cell lines is a feasible approach for the identification of differentially expressed cancer-related genes [7,8].
5-Aza-dC is the most commonly used DNA demethylation agent and can induce tumor cell differentiation, cell cycle arrest, and apoptosis [9–13]. It is believed that the multiple effects of 5-aza-dC on tumor cells derive from its ability to inhibit DNA methyltransferase during DNA synthesis, leading to DNA demethylation and subsequent activation of the expression of transcriptionally silenced genes [14–16]. The majority of genes activated by 5-aza-dC, such as Rb, p16, E-cadherin, APC, VHL, retinoic acid receptor β, BRCA1, and DLC-1, are tumor-suppressor genes in different types of cancer cells [17–25]. It is known that tumor-suppressor genes can negatively regulate the function of oncogenes or genes promoting tumor cell growth through different signal transduction pathways. Therefore, it is presumed that 5-aza-dC exerts its antitumor function by directly upregulating tumor-suppressor genes through DNA demethylation, and by indirectly downregulating oncogenes through signal transduction pathways. Thus, changes in gene expression, especially the upregulation of tumor-suppressor genes and the downregulation of oncogenes, would be expected in each tumor cell line, which is responsive to 5-aza-dC in cell growth inhibition, cell cycle arrest, or apoptosis, during the transition of DNA methylation status rendered by 5-aza-dC treatment. It can also be understood that the transition of DNA methylation status can provide an opportunity to identify tumor-related genes, either tumor-suppressor gene(s) or oncogene(s).
In this study, we employed a cDNA array to analyze the changes in gene expression in an in vitro 5-aza-dC-treated human lung adenocarcinoma cell line. Treatment of this cell line with 5-aza-dC resulted in growth inhibition, cell cycle arrest, apoptosis, and changes in mRNA expression of several genes. Among them, the hint/protein kinase C inhibitor 1 (Hint/PKCI-1) gene was upregulated by 5-aza-dC and inhibited lung tumor cells growth.
Materials and Methods
Reagents and Cell Culture
5-Aza-dC was purchased from Sigma (St. Louis, MO). Atlas Human Cancer 1.2 Array, carrying cDNA fragments of a total of 1176 individual genes, was purchased from Clontech (Palo Alto, CA). In Situ Cell Death Detection (TDT-mediated dUTP biotin nick end labeling [TUNEL] assay) Kit was purchased from Roche Molecular Biochemicals (Indianapolis, IN). Human NSCLC cell lines A539, NCI-H23, NCI-H358, NCI-H522, and NCI-H520 were purchased from ATCC (Rockville, MD) and cultured in RPMI 1640 medium (Gibco BRL, Gibco, Carlsbad, CA) containing 10% of fetal bovine serum and 100 U of penicillin and streptomycin.
Cell Proliferation Assay, TUNEL Assay (In Vitro Cell Death Assay), and Cell Cycle Analysis for 5-Aza-dC-Treated NCI-H522 Cells
In cell proliferation assays, 1 x 105 cells were seeded in each T-25 culture flask in triplicate. Cells were either treated or untreated with 1 µM 5-aza-dC, and then trypsinized and collected at 24, 48, 72, 96, and 120 hours of treatment. Viable cells determined by trypan blue (Gibco, Carlsbad, CA) exclusion were counted using a hematocytometer.
In TUNEL assays, 1 day before treatment, tumor cells either treated or untreated with 5-aza-dC were plated in fourwell chamber slides. The cells were fixed at each time point of 24, 72, 96, and 120 hours of treatment by 2% paraformaldehyde solution (in phosphate-buffered saline [PBS], pH 7.4) for 60 minutes at room temperature, permeated in 0.1% Triton X-100/0.1% sodium acetate for 2 minutes on ice, and then labeled with TUNEL reaction mixture containing calf thymus DNA terminal transferase and fluorescein-labeled dNTP at 37°C for 1 hour. After applying antifade and mounting medium on the slide, fluorescein-labeled cells were detected by fluorescence microscopy and the ratio of the number of labeled cells versus the number of total cells was obtained by counting the cells of 10 observation fields.
In the cell cycle analysis, both 1 µM 5-aza-dC-treated and untreated cells were collected at 24, 48, 72, 96, and 120 hours of treatment in PBS buffer containing 10 mM glucose and then fixed in 70% ethanol at 4°C for at least 1 hours. The cells were then stained for 30 minutes in propidium iodide solution containing 7.5 mM propidium iodide, 10 mM glucose, and 100 U/ml RNase A in PBS buffer. Flow cytometric analysis was performed using a FACS Calibur flow cytometer (Becton Dickinson Immunocytometry Systems, San Jose, CA), which was equipped with Cell Quest version 3.1f software (Becton Dickinson Biosciences, San Jose, CA) for cell cycle data collection. Cell cycle distribution was analyzed using ModFit LT Version 2.0 software (Verity Software House, Inc., Topsham, ME).
cDNA Expression Array Analysis and Reverse Transcription-Polymerase Chain Reaction (RT-PCR) Confirmation for Gene Expression
Polyadenylated RNA from 5-aza-dC-treated NCI-H522 and untreated control cells was extracted with TriZol reagent (Life Technologies, Inc., Grand Island, NY) and purified with magnetic oligo(dT) beads (DYNAL, Inc., Lake Success, NY). In each cDNA array analysis, 0.6 mg of mRNA was radiolabeled with a mixture of gene-specific primers during the reverse transcription procedure. The labeled probe of a total of 7.5 x 106 cpm activity was hybridized with each array membrane. Membrane washing was performed by following the procedure described in the user's manual. The hybridized membrane was exposed to a PhosphoImage plate and processed using Software ImageQuant Version 3.3 (Molecular Dynamics, Sunnyval, CA). The quantitative data analysis was performed using AtlasImage 2.0 Software (cat no. V1212-1; Clontech). For verifying the alterations of gene expression detected by cDNA array analysis, total full-length cDNA of both treated and untreated cells were synthesized by reverse transcription and amplified by the SMART PCR cDNA Synthesis Kit in a low cycle number (BD Biosciences, Palo Alto, CA). All cDNA samples were diluted into equal concentration according to the PCR production of GAPDH before use in the comparison of gene expression.
Antisense Oligo Transfection and Full-Length cDNA Transfection
For antisense oligo transfection, a stretch of 23-bp-long DNA sequences of tyrosine protein kinase ABL2 (ABL2) cDNA from Genebank (accession no. M35296) was chosen for synthesizing both sense and antisense oligos (sense, agagcagggatggggcagcaggt; antisense, acctgctgccccatccctgctct), which cover the ATG start codon of the gene. Both oligos are incorporated with phosphorothioate to increase their intracellular stability. In each transfection, 0.55 µM oligo and 10 µg of lipofectamine in 1 ml of serum-free 1640 medium were applied to the growing cells in a T-25 flask containing 0.5 x 106 to 1 x 106 cells. Triplicate flasks for both sense and antisense oligo transfection were used. After incubation at 37°C for 8 hours, 4 ml of complete RPMI 1640 medium was added and cells were incubated for another 48 hours, and then trypsinized and counted. In cDNA transfection studies, the full-length cDNA of the Hint/PKCI-1 or TRP-1 gene, amplified by RT-PCR from cell line NCI-H522, were cloned into the XhoI recognition site of the pLXSN vector (Clontech). Primer sequences used for cloning the full-length cDNA of the Hint/PKCI-1 gene were: upstream, acgtctcgaggcgagatggcagatgagattg; downstream, atgtctcgagacgtgcttaaccaggaggccaatg. Primer sequences for cloning full-length cDNA of TRP-1 gene were: upstream, gaccctcgagcagaatgagtgctcctaaactcc; downstream, gctactcgagagtagggcatttgttagaccacagac. The insert and its orientation from recombinant clones were confirmed by restriction analysis and DNA sequencing. Five micrograms of plasmid DNA containing either forward-oriented or reverse-oriented full-length cDNA in the expression vector was used in each lipotransfection. After 48 hours of incubation, cells were selected by 200 µg/ml G418 for about 20 days. The mixture of G418-resistant clones was then collected.
Nude Mice Tumorigenicity Assay
Exponentially growing cells collected from stable transfection were harvested and resuspended in serum-free medium. After washing twice with serum-free medium, 2 x 106 cells were inoculated subcutaneously at the proximal dorsal midline of 4- to 6-week-old female Balb/cathymic nude mice (Jackson Laboratories, Inc., Bar Harbor, ME). Tumor size was measured in two dimensions twice a week. Tumor tissue generated at the injection site was dissected at the end of the assay, fixed in 4% paraformaldehyde buffer (pH 7.0), and embedded in paraffin for pathologic examination. The weight of tumor dissected was measured and a t-test was used to compare the numbers obtained from different injection groups. Tumor growth was typically observed for 90 days. Following the institute's guidance on animal care, nude mice with steadily growing tumors reaching a size of 10-15 x 10-15 mm2 were sacrificed before the end of observation period. Histopathologic examination was performed on the specimens stained with hematoxylin and eosin.
Results
5-Aza-dC Induces Cell Apoptosis and Cell Cycle Arrest in NCI-H522 Cells
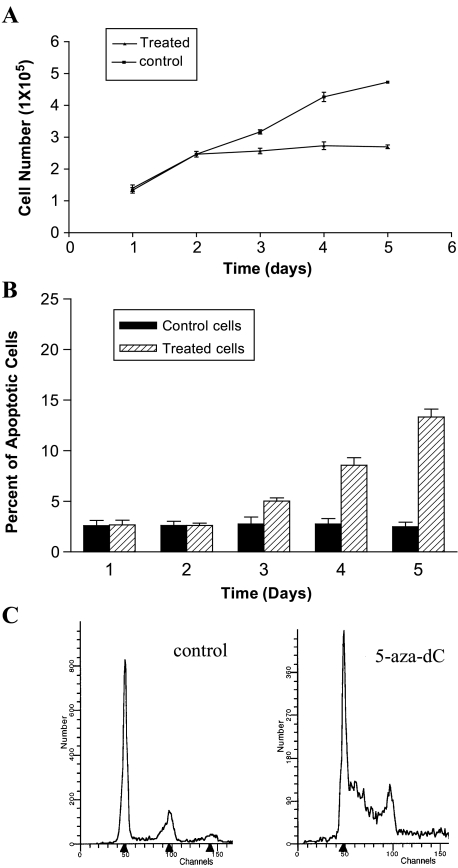
To find an appropriate cell line whose response to 5-aza-dC is derived mainly from drug-induced DNA demethylation, five NSCLC cell lines were screened by 1 µM 5-aza-dC treatment for 4 days. In all lines, 5-aza-dC caused a significant inhibition of cell growth compared to the untreated cell lines. However, all but the NCI-H522 cell line exhibited profound cytotoxicity as demonstrated by dramatic cell death and detachment from culturing flask after 5-aza-dC treatment. 5-Aza-dC-induced cell growth inhibition in NCI-H522 cells was further characterized by cell proliferation assay, TUNEL assay, and cell cycle analysis. The cell proliferation assay showed that a slowdown on cell growth started at 48 hours after 1 µM 5-aza-dC treatment, followed by nearly a complete cessation of cell proliferation (Figure 1A). The TUNEL assay showed that apoptotic cells started to increase on the third day after 5-aza-dC treatment and reached 13% on the fifth day postdemethylation treatment compared with 2% in untreated cells (Figure 1B). The flow cytometry assay indicated that the percentage of cells in S-phase was dramatically increased on the fifth day of 1 µM 5-aza-dC treatment (Figure 1C). These data indicated that the 5-aza-dC-induced cell growth inhibition was partially attributable to cell cycle arrest and apoptosis.
Figure 1.
Cell proliferation assay, TUNEL assay, and cell cycle analysis for NCI-H522 cells treated with 1 µM 5-aza-dC. Cell proliferation assay (A) demonstrated that cells started slower growth at the second day of 5-aza-dC treatment. TUNEL assay (B) showed that about 13% of cells becomes apoptotic in drug-treated cells compared with 2% of apoptotic cells in untreated cells. Cell cycle analysis (C) indicated that 5-aza-dC treatment can cause a significant S-phase arrest in the cell cycle.
Alterations of mRNA Expression Were Identified in 5-Aza-dC-Treated NCI-H522 Cells
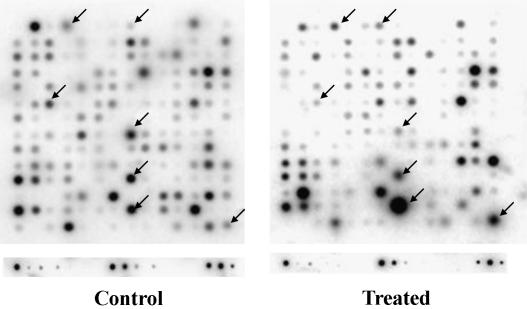
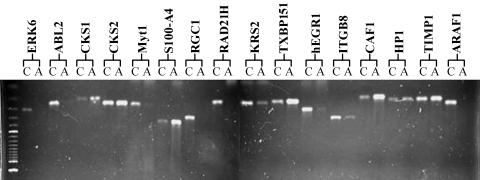
To avoid ambiguity caused by potential 5-aza-dC-induced cytotoxicity during gene expression analysis, only the NCI-H522 cell line was chosen for cDNA array analysis. To identify genes undergoing changes in mRNA expression after 5-aza-dC treatment, a membrane cDNA array analysis covering 1176 genes was employed. The cDNA array analysis was performed by using a comparison between 4 days of 1 µM 5-aza-dC-treated and untreated cells. Quantitative cDNA array data were determined by the ratio of the adjusted intensity of each gene with that of the average of all housekeeping genes provided in the same cDNA array, including ubiquitin and GAPDH. Under our experimental conditions, the average intensity values of all housekeeping genes were consistent with that of ubiquitine and GAPDH between treated and untreated cells. The genes with an absolute adjusted intensity over 500 and with a ratio difference over five-fold between treated and untreated samples were chosen for further analysis. A representative cDNA array analysis is shown in Figure 2. Alteration of mRNA expression of a total of 73 genes was determined by these criteria from repeated experiments. After further confirmation by RT-PCR, 30 genes, either downregulated or upregulated in 5-aza-dC-treated cells, were considered as candidate genes for further characterization (Table 1). Examples such as downregulation of ABL2, membrane-associated protein (Myt-1), Rho-Gap hematopoietic protein C1 (RGC1), and early growth response protein 1 (hEGR1) genes; and upregulation of Hint/PKCI-1, dishevelled 1 (DVL1), chromatin assembly factor 1 (CAF1), tissue metalloproteinase inhibitor 1 (TIMP-1), and tyrosinase-related protein 1 (TRP-1) genes are shown in Figure 3.
Figure 2.
A representative cDNA array analysis of NCI-H522 cells. The changes in gene expression, either downregulation or upregulation, after 4 days of 1 µM 5-aza-dC treatment are indicated by arrows.
Table 1.
Alterations in Gene Expression in NCI-H522 Cells Treated by 5-Aza-dC.
| Accession | Gene or Protein Name (Symbol) | Primers User for RT-PCR |
| Genes downregulated by 5-aza-dC | ||
| L07868 | ERBB4 receptor tyrosine kinase | cttcaagcattggataatoccgaatatcac/agcttacaccacagtattocggtgtctgta |
| L24038 | A-raf protooncogene homolog 1 (ARAF1) | tcaaagtatacctgcocaacaagcaac/cttcaaggaoctcgacaatgagctc |
| AF010310 | p53-induced protein | gtgcgcagatcggctatgaggacc/gggtgoccttcatgaggctgctg |
| X79483 | Extracellular signal-regulated kinase 6 (ERK6) | agctgaagatcctggacttcggoc/gggaggcccttcatgtagttcttgg |
| M35296 | Tyrosine protein kinase ABL2 (ABL2) | aggtagctgaggagcttgggagag/tttgctttcgaggcagtgctgggg |
| AF014118 | Membrane-associated kinase 1 (MYT1) | gggocatggctcctacggagag/ccaggcttcacagtgttgctgcag |
| D87119 | Cancellous bone osteoblast | ccagctggtgccggacgtcaac/ccatgctacgtggtcagtcagctc |
| U48296 | pTPCAAX1 nuclear tyrosine phosphatase | cctggttgttgtattgctgttcattgc/tgaccgttggaatctttgaaacgcag |
| X78817 | rho-GAP hematopoietic protein C 1 (RGC1) | ctgcttagcctggctagtgtcaacg/gtctcaatggtctgtcggtocagg |
| U60207 | Serine/threonine protein kinase KRS2 (KRS2) | ggcttgcctcatgtttgttagccag/ctcaactaggagtctctgttcctgg |
| D38551 | RAD21 (S. pombe) homolog (RAD21H) | gtatcaatgggtgggcctgatagtc/gggctctaattgtcttgctatccaac |
| U43431 | DNA topoisomerase III α | gagaccacagtggagatcgacatcg/gcatocgtaccaatgccatgcttct |
| X52541 | Early growth response protein 1 (hEGR1) | tggcttccaggttoccatgatoccc/ggcaagcgtaagggcgttcgtggg |
| M73780 | Integrin precursor (ITGB8) | gtgcccaatgacggaaactgtcatctg/cattgctcgtcactttctgcatocttc |
| Genes upregulated by 5-aza-dC in NCI-H522 cells | ||
| M63618 | Bullous pemphigoid antigen 1 | tgttgcagggtattggctgactgctag/gagtgaacctgtggctctatcaacoct |
| M73980 | Notch protein homolog 1 | cgcaggcttcagcgggatocac/gtactgggtgtgggtctgccagc |
| M14505 | Cyclin-dependent kinase 4 (CDK4) | cttcocatcagcacagttcgtgagg/cttgactgttccaocacttgtcacc |
| X54941 | Cyclin-dependent kinase regulatory subunit 1 (CKS1) | catgtcgcacaaacaaatttactattcgg/agatgtgaggttctggttcatggatc |
| X54942 | Cyclin-dependent kinase regulatory subunit 2 (CKS2) | agtctocggcgagttgttgcctg/gactctgttggacaccaagtctcc |
| U51004 | Hint protein; protein kinase C inhibitor 1 (Hint/PKCI-1) | gaagatcatocgcaaggaaataccag/cttattcaggcccagatcagcagc |
| M80563 | S100 calcium-binding protein A4(S100-A4) | gtttgatcctgactgctgtcatgg/gcatcaagcacgtgtctgaaggagc |
| U46461 | Dishevelled 1 (DVL1) | tcaccatcgccaatgccgtcatcg/tggagccactgttgaggttcaggg |
| U33821 | TAXI-binding protein 1 51 (TXBP151) | tcctgatcctccaagtcaacatttacg/caaacacctgctggtcatagtcagg |
| U56390 | Caspase-9 (CASP9) | agctggacgccatatctagtttgccc/ggtgcaagataaggcagggtgaggg |
| M15796 | Proliferating cyclic nuclear antigen (PCNA) | gaaggtgttggaggcactcaaggac/ggtgcttcaaatactagcgccaagg |
| L07515 | Heterochromatin protein homolog 1 (HP1) | gggcagacgttagcgtgagtgatc/atattccacttgtoccttaaocacgc |
| X74262 | Chromatin assembly factor 1 p48 subunit (CAF1) | agagtgcaaoccagacttgcgtctoc/ocaggaaacatcttctactactgccg |
| X03124 | Tissue metalloproteinase inhibitor 1 (TIMP1) | tgcaattccgacctcgtcatcagggc/agaaactcctcgctgcggttgtggg |
| U36223 | Fibroblast growth factor 8 | ccaacaagcgcatcaacgccatgg/cagccctcgtacttggcattctgc |
| X51420 | Tyrosinase-related protein 1 (TRP-1) | ctgcacggatgacttgatgggatcc/cttocaagcactgagcgacatcctg |
Figure 3.
RT-PCR confirmation of the alterations in gene expression identified by cDNA array analysis in NCI-H522 cell line. ERK6, ABL2, Myt1, RGC1, RAD21H, KRS2, hEGR1, ITGB8, and ARAF1 genes are downregulated, whereas CKS1, CKS2, S100-A4, TXBP151, CAF1, HP1, and TIMP-1 genes are upregulated by 1 µM 5-aza-dC treatment for 4 days.
Genes Showing Alteration in mRNA Expression in Multiple 5-Aza-dC-Treated NSCLC Cell Lines
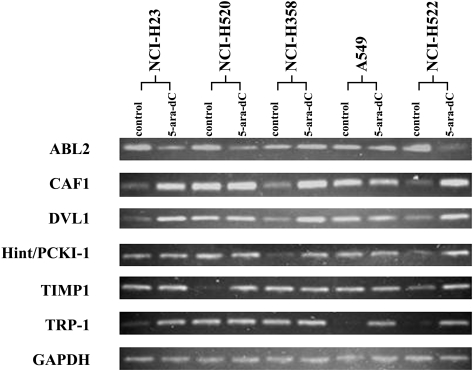
To determine which genes showed recurrent changes in gene expression in multiple cell lines, the detection of mRNA expression of all 30 candidate genes was expanded to NCI-H23, NCI-H358, HCI-H520, and A549 cell lines treated similarly with 5-aza-dC. All RNA samples used in these detections were equally quantified relative to the expression of the GAPDH gene. The downregulation of ABL2 gene and the upregulation of Hint/PKCI-1, CAF1, DVL1, TIMP-1, and TRP-1 genes were observed in three, two, three, three, two, and three of five cell lines treated with 5-aza-dC, respectively (Figure 4).
Figure 4.
RT-PCR to detect the expression of candidate genes in five human NSCLC cells lines treated by 1 µM 5-aza-dC. Downregulation of the ABL2 and upregulation of the CAF1, DVL1, Hint/PKCI-1, TIMP-1, and TRP-1 genes were found in three, three, three, two, two, and three of five cell lines, respectively. The expression of GAPDH was used as quantitative standard.
Upregulation of Hint/PKCI-1 Gene Was Directly Associated with Demethylation-Induced Cell Growth Inhibition
To determine the association of the alterations in gene expression of the candidate genes with DNA demethylation-induced cell growth inhibition, ABL2, Hint/PKCI-1, and TRP-1 were selected for functional analysis. The antisense oligo transfection for the ABL2 gene and full-length cDNA transfection for the Hint/PKCI-1 and TRP-1 genes were performed in cell lines that showed alterations in mRNA expression of these genes after DNA demethylation treatment. A significant cell growth inhibition was observed in ABL2 antisense oligo-transfected NCI-H522, NCI-H23, and NCI-H358 cell lines compared with each sense oligo-transfected cell line (Figure 5). No difference in ABL2 mRNA expression was observed between antisense oligo and sense oligo-transfected cells (data not shown). The attempt to detect the change in protein level after antisense oligo transfection failed due to the lack of antibody against ABL2 protein. In Hint/PKCI-1 full-length cDNA transfection study, a marked cell growth inhibition in cell proliferation assays using 2% serum was observed in both NCI-H522 and NCI-H358 cells lines (Figure 6). However, no obvious cell growth inhibition was observed in TRP-1 cDNA-transfected A549, NCI-H23, and NCI-H522 cell lines. The expression of the transfected gene was detected by RT-PCR only in cells transfected with forward-oriented cDNA in expression vector (data not shown).
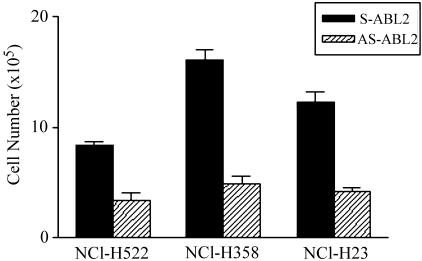
Figure 5.
Cell number counting after antisense oligo transfection for ABL2 gene in human NSLCLC cell lines showing downregulation of ABL2 gene after 5-aza-dC treatment. Significant differences in cell numbers between sense oligo-transfected (S-ABL2) and antisense oligo-transfected (AS-ABL2) cells were observed in NCI-H522, NCI-H358, and NCI-H23 cells.
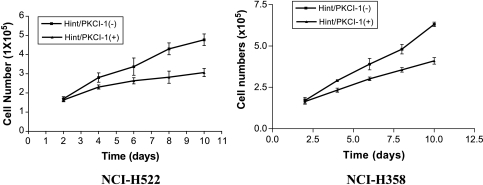
Figure 6.
Cell proliferation assays for NCI-H522 and NCI-H358 cell lines transfected with Hint/PKCI-1 cDNA. Slower growth was observed in both cell lines transfected with forward-oriented cDNA, indicated as Hint/PKCI-1(+), in pLXSN expression vector as compared with that transfected with reverse-oriented cDNA, labeled as Hint/PKCI-1(-).
Hint/PKCI-1 Gene Reduced In Vivo Tumorigenicity of NCI-H358 Cells
To assess the suppressive effect of the Hint/PKCI-1 gene on tumorigenic potential of both NCI-H522 and NCI-H358 cells, we tested the tumorigenicity of two stably transfected cell lines in nude mice. Animals that received NCI-H358 cells transfected with either pLXSN-Hint/PKCI-1(+) or pLXSN alone developed tumors during the first 4 weeks of the experiment. However, tumors from pLXSN alone continued to grow progressively during the observation period, whereas tumors from Hint/PKCI-1(+) transfection exhibited a significantly slower growth over the entire period of the examination. The difference in tumor size between two injection groups was very significant (P < .01). Histopathologic analysis of the tumors was consistent with lung adenocarcinoma. The NCI-H522 cell line, although derived from lung adenocarcinoma, did not produce tumors in nude mice. The nude mice in vivo tumorigenicity assays were repeated twice and the results are summarized in Table 2.
Table 2.
Suppression of In Vivo Tumorigenicity by Hint/PKCI-1 in NCI-H358 Cell Line.
| Cell Lines/Transfection | Latency (days) | Tumor Weight (g) | Number of Tumors/Mice |
| NCI-H358/vector | 18–24 | 0.310 ± 0.080 | 3/3 |
| NCI-H358/Hint/PKCI-1 | 18–24 | 0.120 ± 0.025 (P < .01) | 3/3 |
| NCI-H358/vector* | 17–22 | 0.350 ± 0.080 | 3/3 |
| NCI-H358/Hint/PKCI-1* | 19–24 | 0.130 ± 0.020 (P < .01) | 3/3 |
Numbers were obtained from the repeated experiments.
Discussion
Our experiments show that 5-aza-dC can significantly induce the inhibition of in vitro growth of human NSCLC cell lines through DNA demethylation-induced changes in gene expression. The observations of cell apoptosis and cell cycle arrest in S-phase in 5-aza-dC-treated NCI-H522 cells, two contributing factors to drug-induced cell growth inhibition, are consistent with the general understanding of the consequence of 5-aza-dC-induced DNA demethylation. These changes in tumor cell phenotypes are believed to be based on the induction of alterations in gene expression. During the course of growth inhibition of NCI-H522 cells, the mRNA expression of 16 genes was upregulated, whereas that of 14 genes was downregulated in over 1000 genes tested in cDNA array analysis. These genes belong to different categories in terms of regulation of cell functions. The CKS1 gene, for example, is involved in the regulation of cell cycle [26]; the CASP9 gene is implicated in the regulation of cell apoptosis [27]; and the ERK6 gene may contribute to cell transformation [28]. However, the genes with recurrent alteration in other 5-aza-dC-treated cell lines are also more likely involved in the regulation of cell growth. The expression of the ABL2, Hint/PKCI-1, DVL1, CAF1, TIMP-1, and TRP-1 genes was found to be altered in more than two NSCLC cell lines used in this study, implying a contribution of these genes to cell growth inhibition in multiple NSCLCs.
Our data strongly suggest that the Hint/PKCI-1 gene functions as a negative regulator of tumor cell growth because 5-aza-dC-induced upregulation of the Hint/PKCI-1 was found in two of five NSCLC cell lines. Overexpression of the Hint/PKCI-1 by cDNA transfection resulted in in vitro cell growth inhibition of these two NSCLC cell lines. In addition, stable transfection of Hint/PKCI-1 cDNA significantly reduced in vivo tumorigenicity of NCI-H358 cells. Hint/PKCI-1 is the only other characterized human histidine triad (HIT) nucleotide-binding protein in addition to tumor-suppressor gene FHIT [29], which is a frequent target of genomic deletion and aberrant DNA methylation in human lung cancer as well as other kinds of cancer [30–32]. Like FHIT, the Hint/PKCI-1 gene is also involved in nucleotide metabolism by binding nucleotide molecules and hydrolyzing ADP in vitro [33]. However, the involvement of the Hint/PKCI-1 gene in carcinogenesis has not been investigated. The Hint/PKCI-1 gene is localized at chromosome 7q21–22, a region of genomic deletion reported in radon-induced rat lung cancer [34] and in more invasive human NSCLCs [35]. In addition, microcell transfer studies suggested the existence of a metastasis-suppressor gene in this region [36]. Thus, Hint/PKCI-1 might represent a negative regulator of tumor cell properties in this region. The examination of Hint/PKCI-1 expression in primary tumors would provide more convincing evidence on the role of this gene in NSCLCs. In addition, it would be interesting to see whether gene expression of both Hint/PKCI-1 and FHIT is altered in NSCLCs.
Downregulation of ABL2 was detected in three of five NSCLC cell lines treated with 5-aza-dC. In addition, antisense oligo transfection against ABL2 caused a significant in vitro cell growth inhibition in these three cell lines. However, in the absence of evidence showing that antisense strategy resulted in the decrease of protein expression of ABL2, one cannot speculate that this protooncogene is involved in the pathogenesis of NSCLC.
Functional exploration of the ABL2, Hint/PKCI-1, and TRP-1 genes in this study is part of identifying genes in DNA demethylation-treated NSCLC cells that may be relevant in the pathogenesis of lung cancer. The other candidate gene selected for future functional characterizations is CAF1, which was first found to be a retinoblastoma protein (Rb)-binding protein and a negative regulator of Ras in yeast [37]. It was later found to be a component of the histone deacetylase complex recruited by Rb and to mediate transcriptional repression of E2F1 [38].
In summary, our observations demonstrate that 5-aza-dC can significantly induce the growth inhibition of human lung cancer cells by DNA demethylation and the consequent changes in mRNA expression of multiple genes. Also, our approach of characterizing the variation of gene expression during the transition of DNA methylation thus is apparently efficient in identifying new cancer-related genes. Promoter methylation status will be characterized to determine whether the changes in gene expression of these genes are caused by their own aberrant promoter methylation, or are derived from the regulation of other genes, which are primarily regulated by DNA demethylation. These studies should shed more insight into the functional understanding of the candidate genes identified in this study.
Abbreviations
- NSCLC
non small cell lung carcinoma
- 5-aza-dC
5-aza-2′-deoxycytidine
- ABL2
tyrosine protein kinase ABL2
- Hint/PKCI-1
hint protein/protein kinase C inhibitor 1
References
- 1.Jemal A, Chu KC, Tarone RE. Recent trends in lung cancer mortality in the United States. J Natl Cancer Inst. 2001;93:277–283. doi: 10.1093/jnci/93.4.277. [DOI] [PubMed] [Google Scholar]
- 2.Steenland K, Loomis D, Shy C, Simonsen N. Review of occupational lung carcinogens. Am J Ind Med. 1996;29:474–490. doi: 10.1002/(SICI)1097-0274(199605)29:5<474::AID-AJIM6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 3.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 4.Belinsky SA, Palmisano WA, Gilliland FD, Crooks LA, Divine KK, Winters SA, Grimes MJ, Harms HJ, Tellez CS, Smith TM, Moots PP, Lechner JF, Stidley CA, Crowell RE. Aberrant promoter methylation in bronchial epithelium and sputum from current and former smokers. Cancer Res. 2002;62:2370–2377. [PubMed] [Google Scholar]
- 5.Soria JC, Rodriguez M, Liu DD, Lee JJ, Hong WK, Mao L. Aberrant promoter methylation of multiple genes in bronchial brush samples from former cigarette smokers. Cancer Res. 2002;62:351–355. [PubMed] [Google Scholar]
- 6.Zochbauer-Muller S, Fong KM, Maitra A, Lam S, Geradts J, Ashfaq R, Virmani AK, Milchgrub S, Gazdar AF, Minna JD. 5′ CpG island methylation of the FHIT gene is correlated with loss of gene expression in lung and breast cancer. Cancer Res. 2001;61:3581–3585. [PubMed] [Google Scholar]
- 7.Paz MF, Fraga MF, Avila S, Guo M, Pollan M, Herman JG, Esteller M. A systematic profile of DNA methylation in human cancer cell lines. Cancer Res. 2003;63:1114–1121. [PubMed] [Google Scholar]
- 8.Shi H, Marier S, Nimmerich I, Yan PS, Caldwell CW, Olek A, Huang TH. Oligonucleotide-based microarray for DNA methylation analysis: principles and applications. J Cell Biochem. 2003;88:138–143. doi: 10.1002/jcb.10313. [DOI] [PubMed] [Google Scholar]
- 9.Pinto A, Attadia V, Fusco A, Ferrara F, Spada OA, Di Fiore PP. 5-Aza-2′-deoxycytidine induces terminal differentiation of leukemic blasts from patients with acute myeloid leukemias. Blood. 1984;64:922–929. [PubMed] [Google Scholar]
- 10.Fulda S, Kufer MU, Meyer E, van Valen F, Dockhorn-Dworniczak B, Debatin KM. Sensitization for death receptor- or drug-induced apoptosis by re-expression of caspase-8 through demethylation or gene transfer. Oncogene. 2001;20:5865–5877. doi: 10.1038/sj.onc.1204750. [DOI] [PubMed] [Google Scholar]
- 11.Robertson KD, Jones PA. DNA methylation: past, present and future directions. Carcinogenesis. 2000;21:461–467. doi: 10.1093/carcin/21.3.461. [DOI] [PubMed] [Google Scholar]
- 12.Weiser TS, Guo ZS, Ohnmacht GA, Parkhurst ML, Tong-On P, Marincola FM, Fischette MR, Yu X, Chen GA, Hong JA, Stewart JH, Nguyen DM, Rosenberg SA, Schrump DS. Sequential 5-aza-2 deoxycytidine-depsipeptide FR901228 treatment induces apoptosis preferentially in cancer cells and facilitates their recognition by cytolytic T lymphocytes specific for NY-ESO-1. J Immunother. 2001;24:151–161. doi: 10.1097/00002371-200103000-00010. [DOI] [PubMed] [Google Scholar]
- 13.Zhu WG, Lakshmanan RR, Beal MD, Otterson GA. DNA methyltransferase inhibition enhances apoptosis induced by histone deacetylase inhibitors. Cancer Res. 2001;61:1327–1333. [PubMed] [Google Scholar]
- 14.Juttermann R, Li E, Jaenisch R. Toxicity of 5-aza-2′-deoxycytidine to mammalian cells is mediated primarily by covalent trapping of DNA methyltransferase rather than DNA demethylation. Proc Natl Acad Sci USA. 1994;91:11797–11801. doi: 10.1073/pnas.91.25.11797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Macleod AR, Szyf M. Expression of antisense to DNA methyltransferase mRNA induces DNA demethylation and inhibits tumorigenesis. J Biol Chem. 1995;270:8037–8043. doi: 10.1074/jbc.270.14.8037. [DOI] [PubMed] [Google Scholar]
- 16.Santi DV, Norment A, Garrett CE. Covalent bond formation between a DNA-cytosine methyltransferase and DNA containing 5-azacytosine. Proc Natl Acad Sci USA. 1984;81:6993–6997. doi: 10.1073/pnas.81.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Merlo A, Herman JG, Mao L, Lee DJ, Gabrielson E, Burger PC, Baylin B, Sidransky D. 5′ CpG island methylation is associated with transcriptional silencing of the tumour suppressor p16/CDKN2/MTS1 in human cancers. Nat Med. 1995;1:686–692. doi: 10.1038/nm0795-686. [DOI] [PubMed] [Google Scholar]
- 18.Yoshiura K, Kanai Y, Ochiai A, Shimoyama Y, Sugimura T, Hirohashi S. Silencing of the E-cadherin invasion-suppressor gene by CpG methylation in human carcinomas. Proc Natl Acad Sci USA. 1995;92:7416–7419. doi: 10.1073/pnas.92.16.7416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hiltunen MO, Alhonen L, Koistinaho J, Myohanen S, Paakkonen M, Marin S, Kosma VM, Janne J. Hypermethylation of the APC (adenomatous polyposis coli) gene promoter region in human colorectal carcinoma. Int J Cancer. 1997;70:644–648. doi: 10.1002/(sici)1097-0215(19970317)70:6<644::aid-ijc3>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 20.Herman JG, Latif F, Weng Y, Lerman MI, Zbar B, Liu S, Samid D, Duan DS, Gnarra JR, Linehan WM, Baylin SB. Silencing of the VHL tumor-suppressor gene by DNA methylation in renal carcinoma. Proc Natl Acad Sci USA. 1994;91:9700–9704. doi: 10.1073/pnas.91.21.9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cote S, Momparler RL. Activation of the retinoic acid receptor beta gene by 5-aza-2′-deoxycytidine in human DLD-1 colon carcinoma cells. Anticancer Drugs. 1997;8:56–61. doi: 10.1097/00001813-199701000-00007. [DOI] [PubMed] [Google Scholar]
- 22.Dobrovic A, Simpfendorfer D. Methylation of the BRCA1 gene in sporadic breast cancer. Cancer Res. 1997;57:3347–3350. [PubMed] [Google Scholar]
- 23.Herman JG, Umar A, Polyak K, Graff JR, Ahuja N, Issa JP, Markowitz S, Willson JK, Hamilton SR, Kinzler KW, Kane MF, Kolodner RD, Vogelstein B, Kunkel TA, Baylin SB. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci USA. 1998;95:6870–6875. doi: 10.1073/pnas.95.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yuan BZ, Durkin ME, Popescu NC. Promoter hypermethylation of DLC-1, a candidate tumor suppressor gene, in several common human cancers. Cancer Genet Cytogenet. 2003;140:113–117. doi: 10.1016/s0165-4608(02)00674-x. [DOI] [PubMed] [Google Scholar]
- 25.Yuan BZ, Jefferson AM, Baldwin KT, Thorgeirsson SS, Popescu NC, Reynolds SH. DLC-1 operates as a tumor suppressor gene in human non-small cell lung carcinomas. Oncogene. 1999;23:1405–1411. doi: 10.1038/sj.onc.1207291. [DOI] [PubMed] [Google Scholar]
- 26.Kaiser P, Moncollin V, Clarke DJ, Watson MH, Bertolaet BL, Reed SI, Bailly E. Cyclin-dependent kinase and Cks/Suc1 interact with the proteasome in yeast to control proteolysis of M-phase targets. Genes Dev. 1999;13:1190–1202. doi: 10.1101/gad.13.9.1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li P, Nijhawan D, Budihardjo I, Srinivasula SM, Ahmad M, Alnemri ES, Wang X. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- 28.Marinissen MJ, Chiariello M, Gutkind JS. Regulation of gene expression by the small GTPase Rho through the ERK6 (p38 gamma) MAP kinase pathway. Genes Dev. 2001;15:535–553. doi: 10.1101/gad.855801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klein MG, Yao Y, Slosberg ED, Lima CD, Doki Y, Weinstein IB. Characterization of PKCI and comparative studies with FHIT, related members of the HIT protein family. Exp Cell Res. 1998;244:26–32. doi: 10.1006/excr.1998.4153. [DOI] [PubMed] [Google Scholar]
- 30.Ohta M, Inoue H, Cotticelli MG, Kastury K, Baffa R, Palazzo J, Siprashvili Z, Mori M, McCue P, Druck T, Croce CM, Huebner K. The FHIT gene, spanning thechromosome3p142fragile site and renal carcinoma-associated t(3;8) breakpoint, is abnormal in digestive tract cancers. Cell. 1996;84:587–597. doi: 10.1016/s0092-8674(00)81034-x. [DOI] [PubMed] [Google Scholar]
- 31.Sozzi G, Veronese ML, Negrini M, Baffa R, Cotticelli MG, Inoue H, Tornielli S, Pilotti S, De Gregorio L, Pastorino U, Pierotti MA, Ohta M, Huebner K, Croce CM. The FHIT gene 3p142 is abnormal in lung cancer. Cell. 1996;85:17–26. doi: 10.1016/s0092-8674(00)81078-8. [DOI] [PubMed] [Google Scholar]
- 32.Yuan BZ, Keck-Waggoner C, Zimonjic DB, Thorgeirsson SS, Popescu NC. Alterations of the FHIT gene in human hepatocellular carcinoma. Cancer Res. 2000;60:1049–1053. [PubMed] [Google Scholar]
- 33.Lima CD, Klein MG, Hendrickson WA. Structure-based analysis of catalysis and substrate definition in the HIT protein family. Science. 1997;278:286–290. doi: 10.1126/science.278.5336.286. [DOI] [PubMed] [Google Scholar]
- 34.Dano L, Guilly MN, Muleris M, Morlier JP, Altmeyer S, Vielh P, El-Naggar AK, Monchaux G, Dutrillaux B, Chevillard S. CGH analysis of radon-induced rat lung tumors indicates similarities with human lung cancers. Genes Chromosomes Cancer. 2000;29:1–8. doi: 10.1002/1098-2264(2000)9999:9999<000::aid-gcc1000>3.3.co;2-s. [DOI] [PubMed] [Google Scholar]
- 35.Lukeis R, Ball D, Irving L, Garson OM, Hasthorpe S. Chromosome abnormalities in non-small cell lung cancer pleural effusions: cytogenetic indicators of disease subgroups. Genes Chromosomes Cancer. 1993;8:262–269. doi: 10.1002/gcc.2870080409. [DOI] [PubMed] [Google Scholar]
- 36.Ichikawa T, Hosoki S, Suzuki H, Akakura K, Igarashi T, Furuya Y, Oshimura M, Rinker-Schaeffer CW, Nihei N, Barrett JC, Isaacs JT, Ito H. Mapping of metastasis suppressor genes for prostate cancer by microcell-mediated chromosome transfer. Asian J Androl. 2000;2:167–171. [PubMed] [Google Scholar]
- 37.Qian YW, Wang YC, Hollingsworth RE, Jr, Jones D, Ling N, Lee EY. A retinoblastoma-binding protein related to a negative regulator of Ras in yeast. Nature. 1993;364:648–652. doi: 10.1038/364648a0. [DOI] [PubMed] [Google Scholar]
- 38.Nicolas E, Morales V, Magnaghi-Jaulin L, Harel-Bellan A, Richard-Foy H, Trouche D. RbAp48 belongs to the histone deacetylase complex that associates with the retinoblastoma protein. J Biol Chem. 2000;275:9797–9804. doi: 10.1074/jbc.275.13.9797. [DOI] [PubMed] [Google Scholar]