Abstract
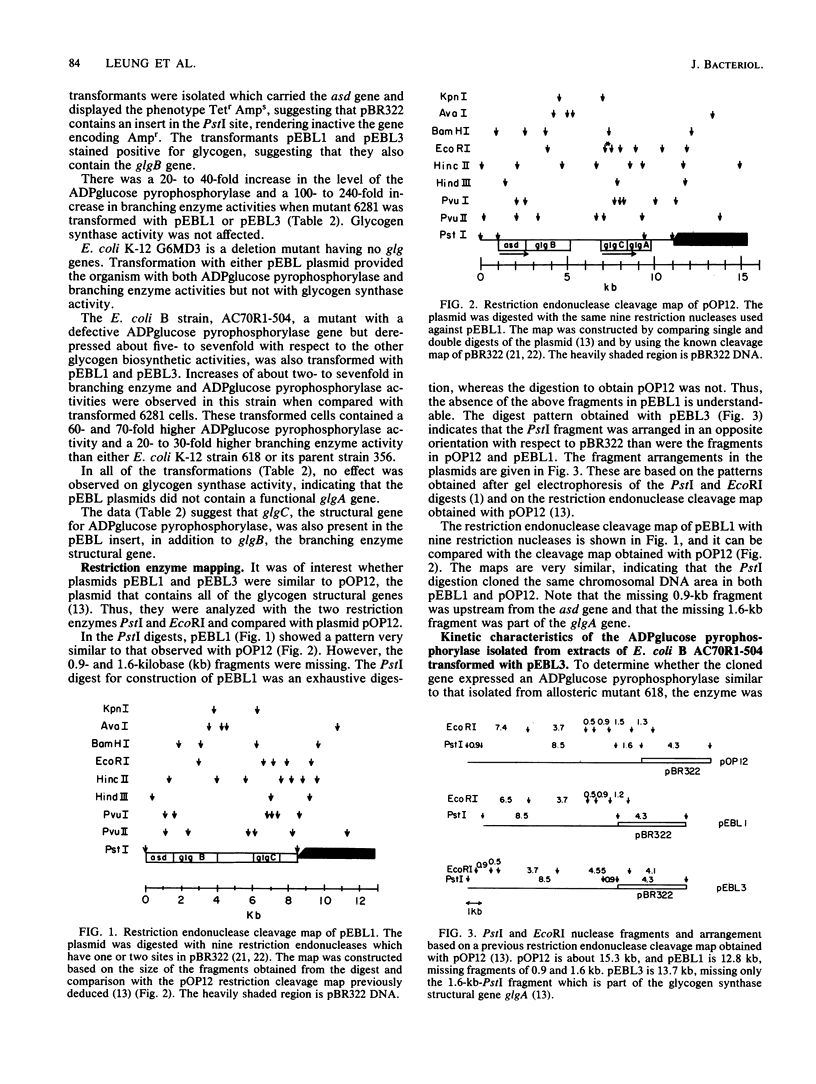
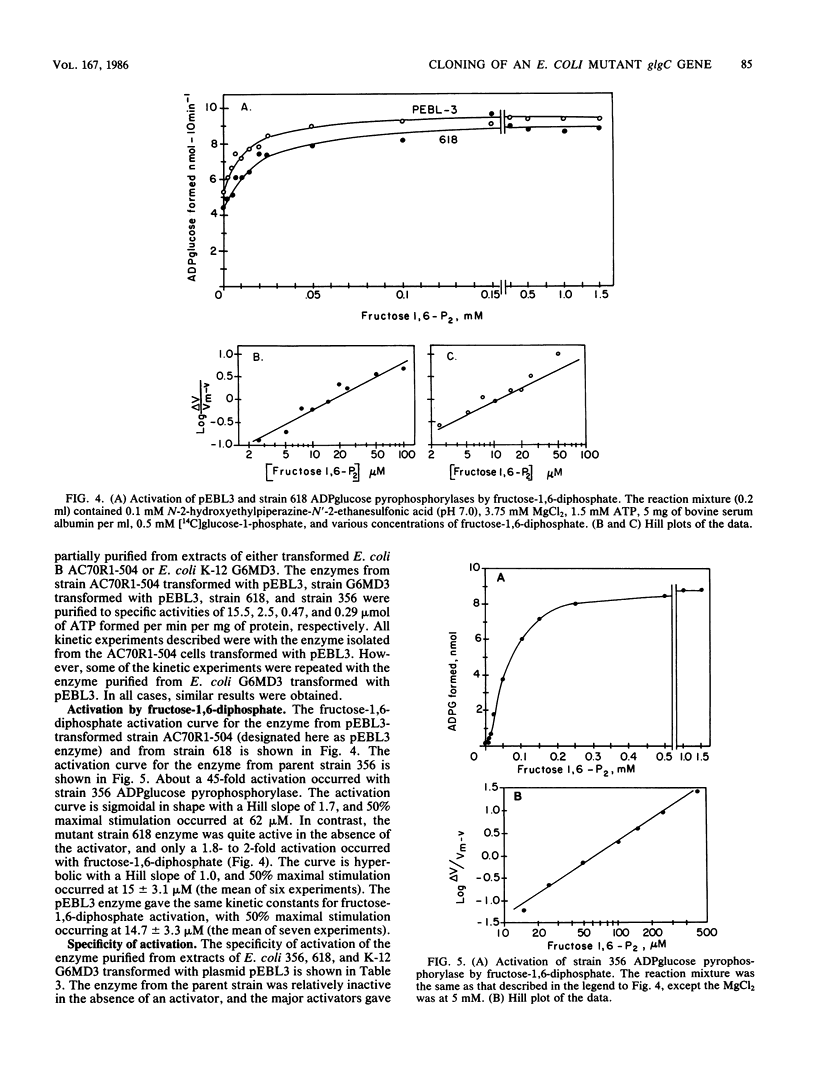
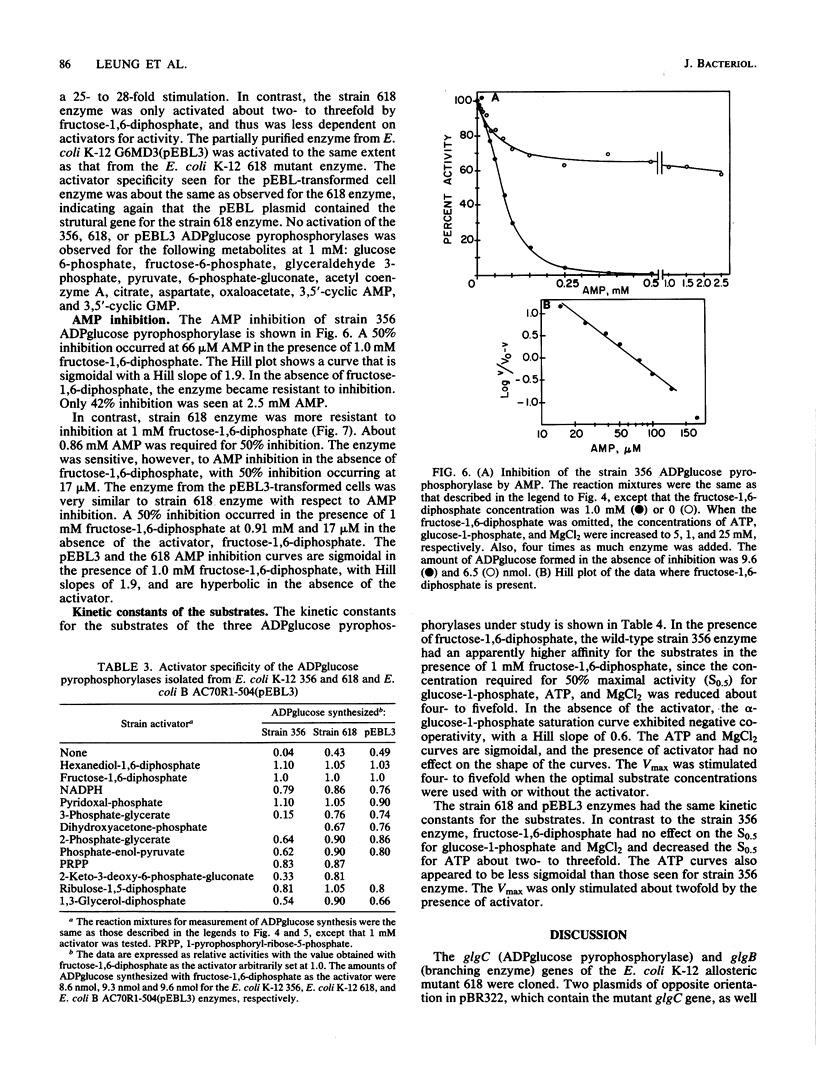
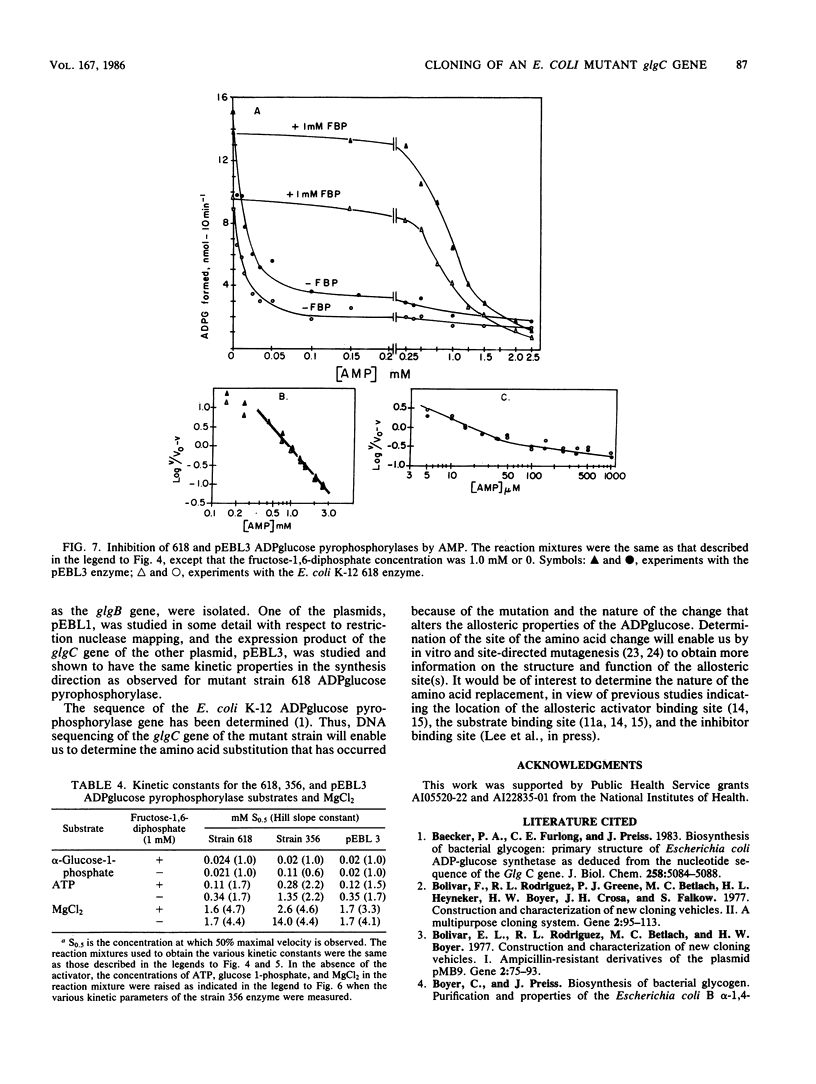
A mutant strain of Escherichia coli K-12, designated 618, accumulates glycogen at a faster rate than wild-type strain 356. The mutation affects the ADPglucose pyrophosphorylase regulatory properties (N. Creuzat-Sigal, M. Latil-Damotte, J. Cattaneo, and J. Puig, p. 647-680, in R. Piras and H. G. Pontis, ed., Biochemistry of the Glycocide Linkage, 1972). The enzyme is less dependent on the activator, fructose 1,6 bis-phosphate for activity and is less sensitive to inhibition by the inhibitor, 5'-AMP. The structural gene, glgC, for this allosteric mutant enzyme was cloned into the bacterial plasmid pBR322 by inserting the chromosomal DNA at the PstI site. The glycogen biosynthetic genes were selected by cotransformation of the neighboring asd gene into an E. coli mutant also defective in branching enzyme (glgB) activity. Two recombinant plasmids, pEBL1 and pEBL3, that had PstI chromosomal DNA inserts containing glgC and glgB were isolated. Branching enzyme and ADPglucose pyrophosphorylase activities were increased 240- and 40-fold, respectively, in the asd glgB mutant, E. coli K-12 6281. The E. coli K-12 618 mutant glgC gene product was characterized after transformation of an E. coli B ADPglucose pyrophosphorylase mutant with the recombinant plasmid pEBL3. The kinetic properties of the cloned ADPglucose pyrophosphorylase were similar to those of the E. coli K-12 618 enzyme. The inserted DNA in pEBL1 was arranged in opposite orientation to that in pEBL3.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baecker P. A., Furlong C. E., Preiss J. Biosynthesis of bacterial glycogen. Primary structure of Escherichia coli ADP-glucose synthetase as deduced from the nucleotide sequence of the glg C gene. J Biol Chem. 1983 Apr 25;258(8):5084–5088. [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Betlach M. C., Boyer H. W. Construction and characterization of new cloning vehicles. I. Ampicillin-resistant derivatives of the plasmid pMB9. Gene. 1977;2(2):75–93. doi: 10.1016/0378-1119(77)90074-9. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Carlson C. A., Parsons T. F., Preiss J. Biosynthesis of bacterial glycogen. Activator-induced oligomerization of a mutant Escherichia coli ADP-glucose synthase. J Biol Chem. 1976 Dec 25;251(24):7886–7892. [PubMed] [Google Scholar]
- Govons S., Gentner N., Greenberg E., Preiss J. Biosynthesis of bacterial glycogen. XI. Kinetic characterization of an altered adenosine diphosphate-glucose synthase from a "glycogen-excess" mutant of Escherichia coli B. J Biol Chem. 1973 Mar 10;248(5):1731–1740. [PubMed] [Google Scholar]
- Govons S., Vinopal R., Ingraham J., Preiss J. Isolation of mutants of Escherichia coli B altered in their ability to synthesize glycogen. J Bacteriol. 1969 Feb;97(2):970–972. doi: 10.1128/jb.97.2.970-972.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappel W. K., Preiss J. Biosynthesis of bacterial glycogen: purification and characterization of ADPglucose pyrophosphorylase with modified regulatory properties from Escherichia coli B mutant CL1136-504. Arch Biochem Biophys. 1981 Jun;209(1):15–28. doi: 10.1016/0003-9861(81)90252-6. [DOI] [PubMed] [Google Scholar]
- Kawaguchi K., Fox J., Holmes E., Boyer C., Preiss J. De novo synthesis of Escherichia coli glycogen is due to primer associated with glycogen synthase and activation by branching enzyme. Arch Biochem Biophys. 1978 Oct;190(2):385–397. doi: 10.1016/0003-9861(78)90291-6. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Latil-Damotte M., Lares C. Relative order of glg mutations affecting glycogen biosynthesis in Escherichia coli K12. Mol Gen Genet. 1977 Feb 15;150(3):325–328. doi: 10.1007/BF00268132. [DOI] [PubMed] [Google Scholar]
- Lee Y. M., Preiss J. Covalent modification of substrate-binding sites of Escherichia coli ADP-glucose synthetase. Isolation and structural characterization of 8-azido-ADP-glucose-incorporated peptides. J Biol Chem. 1986 Jan 25;261(3):1058–1064. [PubMed] [Google Scholar]
- Okita T. W., Rodriguez R. L., Preiss J. Biosynthesis of bacterial glycogen. Cloning of the glycogen biosynthetic enzyme structural genes of Escherichia coli. J Biol Chem. 1981 Jul 10;256(13):6944–6952. [PubMed] [Google Scholar]
- Parsons T. F., Preiss J. Biosynthesis of bacterial glycogen. Incorporation of pyridoxal phosphate into the allosteric activator site and an ADP-glucose-protected pyridoxal phosphate binding site of Escherichia coli B ADP-glucose synthase. J Biol Chem. 1978 Sep 10;253(17):6197–6202. [PubMed] [Google Scholar]
- Parsons T. F., Preiss J. Biosynthesis of bacterial glycogen. Isolation and characterization of the pyridoxal-P allosteric activator site and the ADP-glucose-protected pyridoxal-P binding site of Escherichia coli B ADP-glucose synthase. J Biol Chem. 1978 Nov 10;253(21):7638–7645. [PubMed] [Google Scholar]
- Preiss J., Greenberg E., Sabraw A. Biosynthesis of bacterial glycogen. Kinetic studies of a glucose-1-phosphate adenylyltransferase (EC 2.7.7.27) from a glycogen-deficient mutant of Escherichia coli B. J Biol Chem. 1975 Oct 10;250(19):7631–7638. [PubMed] [Google Scholar]
- Preiss J. Regulation of adenosine diphosphate glucose pyrophosphorylase. Adv Enzymol Relat Areas Mol Biol. 1978;46:317–381. doi: 10.1002/9780470122914.ch5. [DOI] [PubMed] [Google Scholar]
- Preiss J., Shen L., Greenberg E., Gentner N. Biosynthesis of bacterial glycogen. IV. Activation and inhibition of the adenosine diphosphate glucose pyrophosphorylase of Escherichia coli B. Biochemistry. 1966 Jun;5(6):1833–1845. doi: 10.1021/bi00870a008. [DOI] [PubMed] [Google Scholar]
- Schwartz M. Location of the maltose A and B loci on the genetic map of Escherichia coli. J Bacteriol. 1966 Oct;92(4):1083–1089. doi: 10.1128/jb.92.4.1083-1089.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. pBR322 restriction map derived from the DNA sequence: accurate DNA size markers up to 4361 nucleotide pairs long. Nucleic Acids Res. 1978 Aug;5(8):2721–2728. doi: 10.1093/nar/5.8.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoller M. J., Smith M. Oligonucleotide-directed mutagenesis of DNA fragments cloned into M13 vectors. Methods Enzymol. 1983;100:468–500. doi: 10.1016/0076-6879(83)00074-9. [DOI] [PubMed] [Google Scholar]


