Abstract
Pseudomonas aeruginosa undergoes spontaneous mutation that impairs secretion of several extracellular enzymes during extended cultivation in vitro in rich media, as well as during long-term colonization of the cystic fibrosis lung. A frequent type of strong secretion deficiency is caused by inactivation of the quorum-sensing regulatory gene lasR. Here we analyzed a spontaneously emerging subline of strain PAO1 that exhibited moderate secretion deficiency and partial loss of quorum-sensing control. Using generalized transduction, we mapped the secretion defect to the vfr gene, which is known to control positively the expression of the lasR gene and type II secretion of several proteases. We confirmed this secretion defect by sequencing and complementation of the vfr mutation. In a reconstruction experiment conducted with a 1:1 mixture of wild-type strain PAO1 and a vfr mutant of PAO1, we observed that the vfr mutant had a selective advantage over the wild type after growth in static culture for 4 days. Under these conditions, spontaneous vfr emerged in a strain PAO1 population after four growth cycles, and these mutants accounted for more than 40% of the population after seven cycles. These results suggest that partial or complete loss of quorum sensing and secretion can be beneficial to P. aeruginosa under certain environmental conditions.
The gram-negative bacterium Pseudomonas aeruginosa lives as a saprophyte in a variety of humid habitats, and, as an opportunistic pathogen, it can infect many different host organisms. P. aeruginosa owes this versatility, in part, to powerful type I, II, and III secretion mechanisms. Together, these mechanisms help the cells secrete more than a dozen lytic enzymes and toxins (5, 33, 38). For instance, the extracellular protease elastase (LasB), which is secreted via a type II mechansim, enables P. aeruginosa to degrade skim milk. Thus, on nutrient agar amended with skim milk wild-type bacteria are surrounded by large clear halos, whereas secretion-deficient mutants form smaller halos or no halos (2). Clinical and environmental isolates of P. aeruginosa are normally secretion competent. Interestingly, however, a small percentage of all isolates turn out to be secretion defective on milk plates, and null mutations in the lasR gene are the most common cause of this phenotype (10). The lasR gene product is the master regulator of quorum-sensing control in P. aeruginosa. In lasR-negative mutants the quorum-sensing signals N-(3-oxododecanoyl)-homoserine lactone (oxo-C12-HSL) and N-butanoyl-homoserine lactone (C4-HSL) are produced at very low levels compared with the levels produced in the wild type. Oxo-C12-HSL binds to the LasR protein, which then activates transcription of numerous genes, notably the quorum-sensing genes lasI (oxo-C12-HSL synthase), rhlI (C4-HSL synthase), and rhlR (regulator activated by C4-HSL) (13, 18, 26, 34). As quorum sensing positively controls the expression of a large number of genes, including those for elastase (lasB) and the type II secretion apparatus, lasR mutants have a secretion-negative phenotype on milk plates.
In previous work, we found that spontaneous lasR mutants of P. aeruginosa strain PAO1 have a strong selective advantage over the wild type during stationary phase at alkaline pH, which results in enrichment of these mutants when cells are subcultured in a rich aerated medium with repeated cycles that are at least 2 days long (9). In a minimal medium containing caseinate as the sole carbon source, lasR mutants have been observed to emerge in growing cultures of strain PAO1 after several cycles of serial transfer (25). These mutants accumulate because they benefit from the proteolytic enzymes secreted by the lasR+ cells in the population (25). Interestingly, after extensive colonization of the cystic fibrosis lung by P. aeruginosa for several years, up to 60% of the bacterial isolates carry mutations in the lasR gene (3, 29). Thus, although quorum sensing is helpful to P. aeruginosa in most situations, under special conditions it can pay for the organism to get rid of it (3, 6, 10, 25). In the present work, we observed that secretion-defective mutants of P. aeruginosa PAO1 can also arise in a lasR+ background during serial transfer in the laboratory. We focused on an isolate that exhibits a residual secretion phenotype on milk agar, and we mapped the responsible mutation to the vfr gene. This gene, which is a homolog of crp in Escherichia coli, is known to regulate positively the expression of lasR, type II secretion, and pilus formation in P. aeruginosa (1, 36, 37). In a reconstitution experiment, we isolated spontaneous vfr mutants from a PAO1 population after several cycles of static growth. Our findings are intriguing because vfr mutants of P. aeruginosa also arise frequently in the cystic fibrosis lung during long-term colonization (29).
MATERIALS AND METHODS
Bacterial strains, bacteriophage, plasmids, and growth conditions.
The bacterial strains, bacteriophage, and plasmids used in this study are shown in Table 1. P. aeruginosa and E. coli strains were grown at 37°C in Luria-Bertani (LB) medium, tryptic soy broth (TSB), or nutrient yeast broth (NYB) (24, 32). Growth was measured by determining the optical density at 600 nm (OD600). Plasmids were transferred to P. aeruginosa by electroporation (28) or by using the conjugative properties of pRK2013 in triparental mating (4), with antibiotic selection on Pseudomonas isolation agar (Difco) containing 0.4% glycerol, on nutrient agar (NA), or on LB agar (32). The antibiotics used were carbenicillin (300 μg/ml), tetracycline (100 μg/ml), gentamicin (50 μg/ml), streptomycin (1,000 μg/ml), and chloramphenicol (10 μg/ml) for P. aeruginosa and tetracycline (15 μg/ml), gentamicin (10 μg/ml), and ampicillin (50 μg/ml) for E. coli. The soft agar used for phage F116L preparations consisted of NYB amended with 0.5% agar. Proteolytic activity was tested by growing colonies for 12 to 24 h on tryptic soy agar (TSA) plates containing 1.5% skim milk (Difco). Secretion-proficient revertants of strain PAO1P were isolated by the method described previously (35). Briefly, production of a toxic hybrid protein (LasB-pfColA) in the poorly secreting strain PAO1P allows positive selection of secretion-competent bacteria. In bacteria with a functional type II secretion system, LasB-pfColA is secreted into the medium, whereas accumulation of LasB-pfColA in the periplasm of a nonsecreting strain is lethal for the cells.
TABLE 1.
Bacterial strains, plasmids, and bacteriophage used in this study
| Strain, bacteriophage, or plasmid | Relevant characteristicsa | Reference(s) or origin |
|---|---|---|
| P. aeruginosa strains | ||
| PAO1 | Prototroph, chl-2, otherwise wild type | 11 |
| PAO1P | Spontaneous vfr-193 mutant of PAO1 | This study |
| PAO1P-rif | Spontaneous rifampin-resistant mutant of PAO1P | This study |
| PAO1PR | PAO1P vfr+ revertant | This study |
| PAO6382 | ΔpvdF mutant of PAO1 | C. Reimmann, unpublished |
| PAO6301 | PAO1 vfr::ΩSp/Sm; Spr Smr | This study |
| PAO6661 | ΔpvdFtrpG::Tn5Gm; Gmr | This study |
| E. coli strains | ||
| BW20767 | leu-63::IS10 recA1 zbf-5 creB510 hsdR17 endA1 thi uidA(ΔMluI::pir+) chromosome::RP4-2 Tc::Mu Km::Tn7; Tpr Smr | 16 |
| S17-1/λpir | prothihsdRrecA chromosome::RP4-2 Tc::Mu Km::Tn7/λpir; Tpr Smr | 22, 27 |
| TG1 | supEhsdΔ5 thi Δ(lac-proAB)/F′ traD36 proAB+lacIqlacZΔM15 | 24 |
| Bacteriophage | ||
| F116L | Temperate, transducing | 15 |
| Plasmids | ||
| pLM1 | Tn5Gm delivery vector, Gmr derivative of pRL27; Gmr Apr | S. Heeb |
| pME6032 | lacIq ptac expression vector; Tcr | 7 |
| pME9603 | pME6032 containing vfr+ | This study |
| pMAL.R | lasR′-lacZ transcriptional fusion in pMP220; Tcr | 17 |
| pMAL.V | rhlR′-lacZ transcriptional fusion in pMP220; Tcr | 17 |
| pMP220 | IncP replicon for lacZ transcriptional fusions; Tcr | 31 |
| pRK2013 | ColE1 with tra-1 mob-1 of RK2; Kmr | 4 |
| pRL27 | Tn5 delivery vector; Kmr Apr | 16 |
| pRV700 | lasB-pfcolA cloned under ptac in pMMB67EH; Apr | 35 |
Apr, ampicillin resistant; Gmr, gentamicin resistant; Kmr, kanamycin resistant; Smr, streptomycin resistant; Spr, spectinomycin resistant; Tcr, tetracycline resistant.
Protein analysis.
P. aeruginosa cells grown in TSB were collected at the transition between the late exponential and early stationary phases. Cells at a concentration corresponding to 4 OD600 units were harvested by centrifugation, and the supernatant was precipitated at 4°C with 10% (wt/vol) (final concentration) trichloroacetic acid for 2 h. Precipitated proteins were washed twice with 90% (vol/vol) acetone in water, solubilized in 100 μl of sodium dodecyl sulfate-polyacrylamide gel electrophoresis sample buffer (19), and heated at 95°C for 10 min. Samples (10 μl) were separated by electrophoresis on an 11% sodium dodecyl sulfate-polyacrylamide gel (Bio-Rad III). Gels were stained with Coomassie brilliant blue R-250, and protein bands were identified by matrix-assisted laser desorption ionization-time of flight mass spectrometry.
β-Galactosidase assays.
Expression of the quorum-sensing genes lasR and rhlR during growth was studied by measuring the β-galactosidase activities of lacZ transcriptional fusions carried on pMAL.R or pMAL.V. Overnight cultures of P. aeruginosa harboring either of these lacZ fusion constructs were diluted to obtain an OD600 of 0.01 in LB medium containing tetracycline. Samples were harvested at intervals for determination of the OD600 and β-galactosidase activities, which were assayed as previously described (17). Briefly, 1 OD600 unit of a bacterial culture was removed at each time point, and the cells were harvested and permeabilized in 200 μl (final volume) of Z buffer in a microtiter well. A414 values were determined with a microtiter plate reader (Labsystems Multiskan MCC/340). One unit of β-galactosidase activity corresponded to the enzyme activity liberating 10−9 mol of o-nitrophenol from o-nitrophenylgalactoside per min at 28°C. All experiments were performed three times, and averages and standard deviations are presented below.
Transposon Tn5Gm mutagenesis.
About 3,000 random Tn5Gm insertions in strain PAO1PR were generated by five separate biparental matings between E. coli BW20767/pLM1 and P. aeruginosa PAO1PR, as described previously (16). Transposon insertion mutants were selected on NA containing 50 μg/ml gentamicin and 10 μg/ml chloramphenicol and were stored in NYB containing 15% (vol/vol) glycerol and 50 μg/ml gentamicin at −80°C. Genomic DNA was extracted from a protease secretion-positive PAO1P-rif transductant following F116L-mediated transduction with a PAO1PR:Tn5Gm donor (see below). This DNA was subjected to restriction digestion with NcoI, self-ligated, and introduced into E. coli S17-1/λpir by electroporation, with selection for gentamicin resistance. After isolation of the plasmid containing Tn5Gm, the transposon insertion site was determined by nucleotide sequencing with the transposon-specific primer tnpRL17-1 (5′-AACAAGCCAGGGATGTAACG-3′) and was localized on the PAO1 chromosome (http://www.pseudomonas.com/) using BLASTN analysis. The trpG::Tn5Gm mutant PAO6661 (Table 1) was obtained and mapped similarly.
Transduction with phage F116L.
The phage F116L preparations used for transduction were prepared as follows. One hundred microliters of an overnight culture of a P. aeruginosa strain and 100 μl of phage stock (approximately 105 PFU/ml) were added to 3 ml of soft agar. The mixture was overlaid on NA plates and incubated at 37°C for 12 h. Semiconfluent plaques were then scraped from the plates and transferred into 2 ml of TNM buffer (10 mM Tris-HCl [pH 7.4], 150 mM NaCl, 10 mM MgSO4). The lysate was vortexed, centrifuged at 20,000 × g for 10 min, and filtered through a 0.45-μm filter. Phage titers (PFU/ml) were determined by spotting 5-μl portions of appropriate dilutions onto a lawn of PAO1 and counting the resulting plaques after 12 h of incubation at 37°C. Phage stocks were routinely stored at 4°C. Generalized transduction using F116L was performed as described by Krishnapillai (15). Pools of Tn5Gm transposon mutants (48 transposon mutants per pool) were grown overnight in NYB containing 50 μg/ml gentamicin and 10 μg/ml chloramphenicol. F116L transducing lysates were prepared for 21 separate pools. A spontaneous rifampin-resistant PAO1P derivative (designated PAO1P-rif) was generated by plating PAO1P on NA containing 200 μg/ml rifampin. An overnight culture of PAO1P-rif (500 μl) was resuspended in 500 μl TNM buffer and incubated with F116L transducing lysate (5 × 108 PFU/ml) at 37°C for 15 min. Nonadsorbed phage was removed by two washes in TNM buffer. Transductants were selected on NA containing 100 μg/ml rifampin and 50 μg/ml gentamicin, purified by plating on selective medium, and screened for protease production on TSA-milk plates.
Nucleotide sequencing of the trpG-argC region and construction of a vfr insertion mutant.
Chromosomal DNA from strains PAO1P and PAOPR was prepared by using a Nucleospin C+T kit (Macherey-Nagel) and was sequenced commercially (Fasteris Life Sciences, Plan-les-Ouates, Switzerland). The sole difference from the PAO1 sequence was a T-to-C transition at position 193 in the vfr gene of PAO1P. The vfr:ΩSp/Sm mutant PAO6301 was constructed by insertion of an ΩSp/Sm cassette as previously described (23).
Complementation of the vfr-193 mutation in strain PAO1P.
The vfr+ gene with flanking regions was isolated on a 1.05-kb fragment by PCR using PAO1PR chromosomal DNA as a template and primers vfrF (5′-CGCGAATTCGGTCACCGAGAGCGGTATTC-3′) and vfrR (5′-CCCAGATCTCGACCTTCATGGTCCGTCTG-3′). The vfr+ fragment was cut with EcoRI and BglII and ligated into the expression vector pME6032, resulting in plasmid pME9603. This construct was introduced into PAO1P via electroporation. Transformants were selected on NA containing 150 μg/ml tetracycline, and protease production was tested on TSA-milk plates containing 1 mM isopropyl-β-d-thiogalactoside (IPTG).
Competition experiments.
The recovery of vfr mutants and the wild type was assessed in shaking and static broth cultures at 37°C. NYB was inoculated with a 50:50 mixture of strains PAO1P and PAO1PR or strains PAO1 and PAO6301 obtained from overnight NYB cultures of the individual strains; the inoculum concentrations were estimated from OD600 values. Immediately following inoculation, 100-μl samples were removed, and appropriate serial dilutions were plated on NA to verify that equivalent proportions of the bacterial populations were present. At intervals, 100-μl samples were taken, and appropriate serial dilutions were spread on NA plates and incubated overnight at 37°C. Discrimination of the strains was based on the difference in colony morphology between the wild type (fuzzy edges) and the vfr mutants (smaller colonies with smooth edges). The individual population sizes were expressed as percentages of the total population.
For competition experiments on solid media, NYB was inoculated with a 50:50 mixture of strains PAO1P and PAO1PR or strains PAO1 and PAO6301 as described above, and the presence of equivalent proportions of the two populations was verified by immediately plating an appropriate dilution on NA plates. Concurrently, 100-μl samples of the same liquid culture were spread on NA plates, which were then incubated at room temperature. At indicated time points, bacterial samples were obtained by removing agar plugs, which were placed in 1 ml of 0.9% NaCl, vortexed vigorously for 1 min, mechanically broken with the aid of a pipette tip, and vortexed again for 1 min. Serial dilutions were plated on NA and incubated overnight at 37°C. As described above, strain discrimination was based on differences in colony morphology, and the individual population sizes were expressed as percentages of the total population.
Selection and isolation of vfr mutants under static culture conditions.
Spontaneous vfr mutants were selected as follows. A wild-type PAO1 culture was used to inoculate 10 ml of TSB with 1 × 108 bacteria per ml. After incubation at 37°C for 24 h, a culture sample was taken and used to reinoculate 10 ml of fresh TSB with 1 × 108 bacteria per ml, and the resulting culture was incubated for 24 h. A total of seven incubation cycles were carried out. For each cycle, 100-μl samples were taken and appropriate serial dilutions were spread on LB plates and incubated overnight at 37°C. Colonies were then screened for the specific vfr morphotype (smaller colonies with smooth edges). The first colonies having a vfr morphotype were observed at the fourth cycle, and the proportion of such colonies increased during the following cycles.
RESULTS
Isolation of a secretion-deficient PAO1 mutant and a revertant of this mutant.
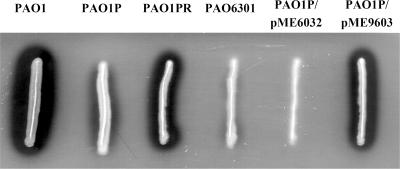
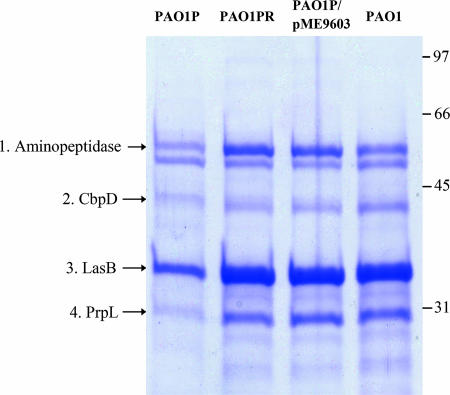
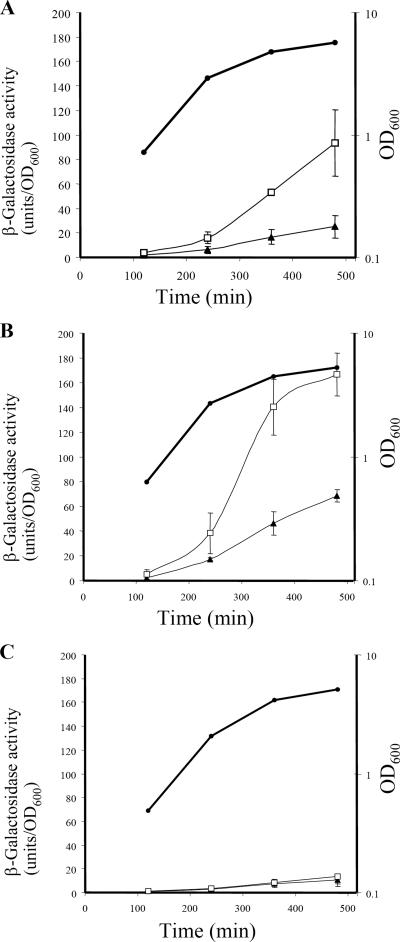
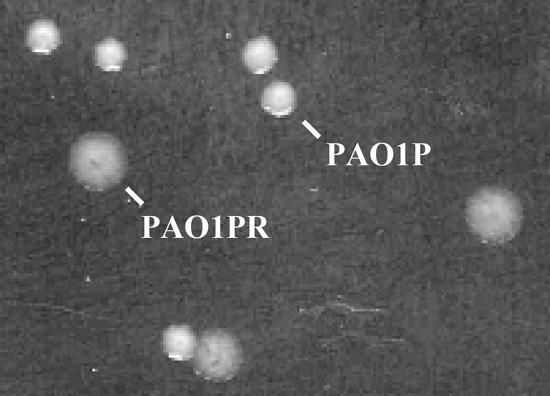
Secretion-deficient derivatives of P. aeruginosa wild-type strain PAO1 were observed to emerge after serial transfer of colonies on rich solid media and streaking on milk-containing plates. One isolate, designated PAO1P, which was partially secretion deficient on TSA amended with milk (Fig. 1), was chosen for this investigation. During growth in LB medium, PAO1P produced both oxo-C12-HSL and C4-HSL (data not shown), suggesting that the lasR gene was not mutated, as a lasR mutation results in severely diminished production of both of these quorum-sensing signals (9). Secretion-proficient revertants of strain PAO1P were selected by introducing plasmid pRV700 expressing the toxic hybrid protein LasB-pfColA under control of the tac promoter. We have shown in previous work that unless LasB-pfColA is effectively secreted, it is toxic to the cell (35). Of the PAO1P derivatives which survived this selection procedure, one isolate having a wild-type secretion phenotype on TSA-milk was kept for further analysis, cured of pRV700 (35), and designated PAO1PR (Fig. 1). A sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of the secreted proteins showed that strain PAO1PR exhibited enhanced secretion of several proteins, including elastase (LasB, PA3724), aminopeptidase (PA2939), chitin binding protein (CbpD, PA0825), and protease IV (PrpL, PA4175), and the pattern closely resembled that of wild-type strain PAO1 (Fig. 2). Both lasR′-lacZ and rhlR′-lacZ transcriptional fusions were upregulated in strain PAO1PR compared with the low levels of expression seen in strain PAO1P (Fig. 3), suggesting that the reduced secretion ability of strain PAO1P might be related, directly or indirectly, to downregulation of the quorum-sensing system. This experiment also indicated that strain PAO1P is not defective in lasR. Such a defect would strongly affect the expression of rhlR′-lacZ but not that of lasR′-lacZ (17).
FIG. 1.
Lytic activity on a TSA-milk plate containing 1 mM IPTG after 14 h of incubation at room temperature of strains PAO1 (wild type), PAO1P (secretion-deficient subline), PAO1PR (secretion-proficient revertant of PAO1P isolated via LasB-pfColA genetic selection), PAO6301 (vfr mutant), PAO1P/pME6032 (secretion-deficient subline with an empty vector), and PAO1P/pME9603 (secretion-deficient subline complemented by a plasmid expressing the vfr gene).
FIG. 2.
Extracellular proteins from strains PAO1P, PAO1PR, PAO1P/pME9603, and PAO1. The arrows indicate the four secreted proteins whose amounts are significantly reduced in PAO1P compared to the other three strains. The molecular masses (in kDa) are indicated on the right. The following proteins were identified by mass spectrometry: 1, PA2939 (aminopeptidase); 2, PA0825 (chitin binding protein CbpD); 3, PA3724 (elastase LasB); and 4, PA4175 (protease IV PrpL).
FIG. 3.
Downregulation of quorum-sensing gene expression in strain PAO1P. (A) Transcription of lasR as monitored with plasmid pMAL.R (lasR′-lacZ) in PAO1P and PAO1PR. (B) Transcription of rhlR as monitored with pMAL.V (rhlR′-lacZ) in the same strains. (C) Vector pMP220, which served as a negative control. The β-galactosidase activities of PAO1P (▴) and PAO1PR (□) were assayed in duplicate. Experiments were performed three times with cultures growing in LB medium supplemented with tetracycline at 37°C. Cell growth was monitored by measuring the OD600 (•).
Mapping of the locus affecting secretion in strains PAO1P and PAO1PR.
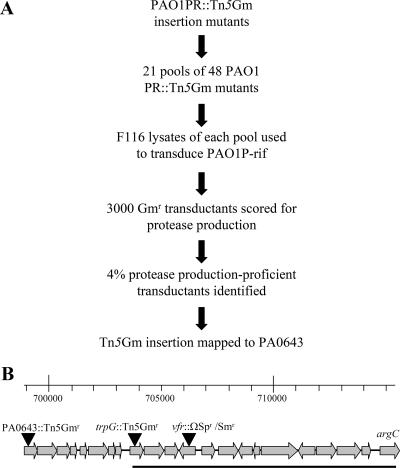
As there are many genes that influence the expression of the quorum-sensing system in P. aeruginosa (8, 13, 18, 23, 26), we decided to map the locus determining the secretion phenotypes in strains PAO1P and PAO1PR by generalized transduction using phage F116L (12). This was done in several steps, as summarized in Fig. 4A. First, random insertions of Tn5Gm (carrying a gentamicin resistance determinant) were generated in strain PAO1PR. F116L transducing lysates were prepared for 21 pools, each containing 48 Tn5Gm insertion mutants. In transductions with strain PAO1P-rif as the recipient, one pool contained 4% gentamicin-resistant transductants that showed restored secretion ability on milk-containing agar. The sequence flanking the transposon insertion in the transductants was determined and found to be part of the PA0643 gene, which is located in a pyocin R gene cluster (21) (Fig. 4B). This result indicated that the locus affecting secretion was located within cotransduction distance (<50 kb) of PA0643. Inspection of this region revealed that the vfr gene was a candidate locus (Fig. 4B). Mutations in the vfr gene result in pleiotropic effects, including reduced expression of the lasR gene and type II secretion (1, 37). To verify linkage of the PAO1P secretion locus with vfr, we transduced strain PAO1P with an F116L lysate prepared with PAO6661 (vfr+ trpG:Tn5Gm) and selected gentamicin-resistant transductants. In 97% (155 of 160) of the transductants, secretion was restored. This showed that the secretion locus was close to trpG and possibly within vfr.
FIG. 4.
(A) Outline of transductional mapping of the locus affecting secretion in PAO1P. The arrows indicate the chronological order. (B) Diagram of the vfr gene region in P. aeruginosa PAO1. Map coordinates (in bp) are indicated on the scale. The solid line indicates the 11.5-kb segment sequenced in this work. Transposon or resistance cassette insertions are represented by filled triangles.
Secretion defect in PAO1P is linked to a vfr mutation.
The entire 11.5-kb trpG-vfr-argC region was sequenced in strain PAO1PR, whose sequence was indistinguishable from that of wild-type strain PAO1, and in strain PAO1P, which had a single point mutation in vfr but otherwise was identical to the parental strain. The vfr-193 point mutation in PAO1P is a T-to-C transition at position 193 in the vfr gene (corresponding to position 706480 on the chromosome map), resulting in substitution of Tyr-65 for a histidine residue in the Vfr protein. This mutation affected the function but not the production of Vfr, as the same amount of Vfr protein was found in both strains by Western blotting (data not shown). In conclusion, these data document that PAO1P is a PAO1 mutant producing an inactive Vfr protein and that PAO1PR is a true vfr+ revertant of PAO1P. Further support for this conclusion was obtained from inspection of colony morphology. As mutation in vfr abrogates pilus synthesis and therefore twitching motility, strain PAO1P formed round, even colonies, whereas strains PAO1 and PAO1PR produced larger colonies with fuzzy edges (Fig. 5), in agreement with a previous study (36).
FIG. 5.
Colony morphology of strains PAO1P and PAO1PR.
Complementation of the vfr-193 mutation.
To demonstrate that the Y65H mutation in Vfr is involved in the secretion deficit in PAO1P, we introduced a recombinant plasmid containing the wild-type vfr allele, pME9603, into this strain. The complemented mutant had a restored secretion phenotype on milk-containing agar, and a control showed that the empty vector pME6032 had no influence on secretion in a vfr mutant (Fig. 1). The complemented vfr mutant PAO1P/pME9603 also showed restoration of secreted proteins (Fig. 2). In conclusion, it appears that the Y65H point mutation is involved in the secretion phenotype of strain PAO1P.
Selective advantage of vfr mutants in vitro.
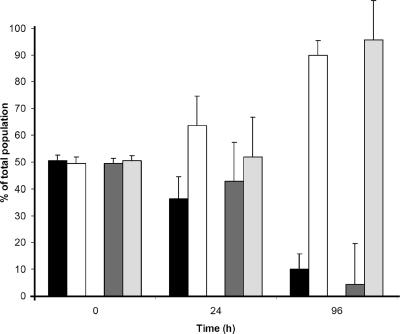
We wondered whether the emergence of vfr mutants would be favored by some selective pressure under laboratory conditions. We first tested growth and survival during stationary phase in well-aerated NYB. Under these conditions, lasR mutants have a competitive advantage over the wild type after >24 h of incubation at 37°C (9). Unexpectedly, under the same experimental conditions, PAO1P and PAO1PR behaved differently; in a 50:50 mixture, viable cells of the vfr mutant PAO1P were recovered from the culture less frequently than revertant PAO1PR cells (data not shown). In a static broth culture, by contrast, the vfr mutant PAO1P was the predominant strain recovered. A similar result was obtained with a 50:50 mixture of wild-type strain PAO1 and the vfr insertion mutant PAO6301 (Fig. 6). Although the selective advantage of the vfr mutants was small at the end of growth after 24 h, the vfr mutants were isolated preferentially over the vfr+ strains after 96 h of incubation at 37°C (Fig. 6). At that time the culture medium had a pH of about 9. Moreover, when the 50:50 mixtures of PAO1P/PAO1PR and PAO6301/PAO1 were cultivated on NA and viable cells were removed from the agar surface by vigorous shaking in saline medium, the vfr mutants were isolated preferentially (ratio, 60:40) over the wild type after 1 h of incubation, and the ratios remained similar to this ratio after 96 h for both strain pairs (data not shown). We concluded that vfr mutants are enriched relative to the wild type when P. aeruginosa strains are serially transferred between static cultures or on solid growth media. In the case of transfer between solid media, the differential recovery may be explained by the fact that the wild type adheres more strongly to the agar surface than a vfr mutant. This is probably a consequence of reduced expression of type IV pili in the mutant (37).
FIG. 6.
Growth of vfr-positive and -negative strains in mixed culture under static culture conditions. Cells were incubated in NYB at 37°C and plated following serial dilution on NA. PAO1P and PAO6301 were distinguished from PAO1PR and PAO1 on the basis of colony morphology. The bars indicate the means of three independent measurements, and the error bars indicate the standard deviations. Black bars, PAO1PR; open bars, PAO1P; dark gray bars, PAO1; light gray bars, PAO6301 (vfr).
Generation of spontaneous vfr mutants in vitro.
We reasoned that the selective advantage of Vfr-negative cells should allow us to isolate spontaneous vfr mutants from a PAO1 population grown in static culture. This was done by serially transferring strain PAO1 as described in Materials and Methods. After each cycle, we screened for colonies having the typical vfr morphotype, as shown in Fig. 5. Such colonies first appeared after four cycles; their proportion increased to 15% after six cycles and to more than 40% after seven cycles. Fourteen colonies having a vfr morphotype were collected at cycles 4, 6, and 7 and screened further for twitching motility and protease secretion on TSA-milk plates; 9 of these 14 isolates were similar to the vfr mutant PAO6301. None of them produced the Vfr protein according to a Western blot analysis using a Vfr antibody (data not shown). Sequencing of the vfr allele in the nine isolates revealed a unique deletion of cytosine at position 377 in the vfr gene, indicating that all of the isolates are siblings. This experiment demonstrated the positive selection that acts on vfr mutants under certain growth conditions.
DISCUSSION
This study reports the third case of a PAO1 subline that shows deficiencies in quorum-sensing-controlled gene expression after subculturing in the laboratory. An examination of the first two cases by Heurlier et al. (9) revealed spontaneous mutations in the lasR gene, resulting in a complete loss of quorum-sensing control and type II secretion. The results obtained here for PAO1P can be attributed to a mutation in the vfr global regulatory gene. The consequences of this defect are reduced quorum-sensing control (Fig. 3), reduced type II secretion (Fig. 1 and 2), and reduced twitching motility (Fig. 5). The secretion-proficient strain PAO1PR was isolated by our positive selection method, which is based on a LasB-pfColA hybrid protein (35). It is remarkable that with this powerful method a rare true revertant could be selected in a secretion-defective background.
An intriguing question is which selective forces favor the emergence of such sublines. In rich media, lasR mutants were found to have a survival advantage over the wild type in agitated 2-day-old cultures at alkaline pH (9), whereas the vfr mutants had an advantage in unshaken 4-day-old cultures at alkaline pH (Fig. 6). During exponential growth, there seemed to be no major fitness difference between the wild type and the lasR and vfr mutants, and the selective advantage of both types of mutants appeared in late stationary phase (9) (Fig. 6). Thus, the cost of keeping the quorum-sensing machinery operational in the wild type is probably not the most decisive selection factor, contrary to what might be assumed (14) and what is indeed observed in caseinate medium (25). Rather, it appears that cells with impaired quorum sensing and secretion have a better chance to survive in stationary phase because they are more resistant to cell lysis (9, 10). The survival strategies involved are not clear, and it is puzzling that differential survival was observed in shake cultures in one case (lasR) and in static cultures in the other case (vfr). In shake cultures propagated by serial transfer, spontaneous PAO1 lasR mutants appeared after 6 to 10 days at frequencies of several percent (9). We conducted a similar experiment with static liquid cultures of strain PAO1 by screening for colonies having a vfr morphotype (Fig. 5). After 4 days of subculturing, we observed the first vfr mutant, and static growth during additional cycles enriched for this mutant, confirming the selective advantage of the mutant over the wild type. This experiment also highlights the danger of using serial transfers for the maintenance of P. aeruginosa stocks.
These observations made in the laboratory show an interesting correlation with studies of long-term P. aeruginosa infections. P. aeruginosa strains isolated at early stages from the cystic fibrosis lung are generally secretion competent (29), suggesting that establishment of infection requires type III secretion and subsequently, when the bacterial population has reached a “quorum,” type I and II secretion (20, 26, 30). The picture changes dramatically at later stages of lung infection. In a genetic study (29), a majority of P. aeruginosa isolates was found to be lasR negative, and moreover, about 30% of all isolates were vfr negative after several months or years of infection. Thus, persistence of P. aeruginosa in the cystic fibrosis lung appears to be favored following mutational loss of secretion ability. In other types of infection, lasR and other quorum-sensing-defective mutants appear at frequencies of up to 20% (10). Although no systematic time course studies have been conducted in these cases, the impression is that the frequencies of such mutants increase with increasing duration of infection. Our observation that the proportion of vfr-negative mutants increased to almost one-half of a P. aeruginosa PAO1 population after seven cycles of static growth in the laboratory correlates positively with the long-term colonization effects observed in the studies mentioned above. However, the selective forces behind this evolution are not necessarily the same in vitro and in vivo.
Acknowledgments
We thank K. Forest for providing Vfr antibody, Emanuela Frangipani for providing advice on transduction experiments, and Laween Meran for providing plasmid pLM1.
This work was supported by European project NANOFOLDEX (grant QLK3-CT-2002-0286) and the Bettencourt-Schueller Foundation.
Footnotes
Published ahead of print on 18 January 2008.
REFERENCES
- 1.Albus, A. M., E. C. Pesci, L. J. Runyen-Janecky, S. E. West, and B. H. Iglewski. 1997. Vfr controls quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 179:3928-3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown, M. R. W., and J. J. S. Foster. 1970. A simple diagnostic milk medium for Pseudomonas aeruginosa. J. Clin. Pathol. 23:172-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.D'Argenio, D. A., M. Wu, L. R. Hoffman, H. D. Kulasekara, E. Déziel, E. E. Smith, H. Nguyen, R. K. Ernst, T. J. L. Freeman, D. H. Spencer, M. Brittnacher, H. S. Hayden, S. Selgrade, M. Klausen, D. R. Goodlett, J. L. Burns, B. W. Ramsay, and S. L. Miller. 2007. Growth phenotypes of Pseudomonas aeruginosa lasR mutants adapted to the airways of cystic fibrosis patients. Mol. Microbiol. 64:512-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Filloux, A. 2004. The underlying mechanisms of type II protein secretion. Biochim. Biophys. Acta 1694:163-179. [DOI] [PubMed] [Google Scholar]
- 6.Haas, D. 2006. Cost of cell-cell signalling in Pseudomonas aeruginosa: why it can pay to be signal-blind. Nat. Rev. Microbiol. 4:562. [DOI] [PubMed] [Google Scholar]
- 7.Heeb, S., C. Blumer, and D. Haas. 2002. A regulatory RNA as a mediator in GacA/RsmA-dependent global control of exoproduct formation in Pseudomonas fluorescens CHA0. J. Bacteriol. 184:1046-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heurlier, K., V. Dénervaud, G. Pessi, C. Reimmann, and D. Haas. 2003. Negative control of quorum sensing by RpoN (σ54) in Pseudomonas aeruginosa PAO1. J. Bacteriol. 185:2227-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heurlier, K., V. Dénervaud, M. Haenni, L. Guy, V. Krishnapillai, and D. Haas. 2005. Quorum sensing-negative (lasR) mutants of Pseudomonas aeruginosa avoid cell lysis and death. J. Bacteriol. 187:4875-4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heurlier, K., V. Dénervaud, and D. Haas. 2006. Impact of quorum sensing on fitness of Pseudomonas aeruginosa. Int. J. Med. Microbiol. 296:93-102. [DOI] [PubMed] [Google Scholar]
- 11.Holloway, B. W. 1955. Genetic recombination in Pseudomonas aeruginosa. J. Gen. Microbiol. 13:572-581. [DOI] [PubMed] [Google Scholar]
- 12.Holloway, B. W., and V. Krishnapillai. 1975. Bacteriophages and bacteriocins, p. 99-132. In P. H. Clarke and M. H. Richmond (ed.), Genetics and biochemistry of Pseudomonas. John Wiley and Sons, London, United Kingdom.
- 13.Juhas, M., L. Eberl, and B. Tümmler. 2005. Quorum sensing: the power of cooperation in the world of Pseudomonas. Environ. Microbiol. 7:459-471. [DOI] [PubMed] [Google Scholar]
- 14.Keller, L., and M. G. Surette. 2006. Communication in bacteria: an ecological and evolutionary perspective. Nat. Rev. Microbiol. 4:249-258. [DOI] [PubMed] [Google Scholar]
- 15.Krishnapillai, V. 1971. A novel transducing phage. Its role in recognition of a possible new host-controlled modification system in Pseudomonas aeruginosa. Mol. Gen. Genet. 114:134-143. [DOI] [PubMed] [Google Scholar]
- 16.Larsen, R. A., M. M. Wilson, A. M. Guss, and W. W. Metcalf. 2002. Genetic analysis of pigment biosynthesis in Xanthobacter autotrophicus Py2 using a new, highly efficient transposon mutagenesis system that is functional in a wide variety of bacteria. Arch. Microbiol. 178:193-201. [DOI] [PubMed] [Google Scholar]
- 17.Latifi, A., M. Foglino, K. Tanaka, P. Williams, and A. Lazdunski. 1996. A hierarchical quorum-sensing cascade in Pseudomonas aeruginosa links the transcriptional activators LasR and RhIR (VsmR) to expression of the stationary-phase sigma factor RpoS. Mol. Microbiol. 21:1137-1146. [DOI] [PubMed] [Google Scholar]
- 18.Lazdunski, A. M., I. Ventre, and J. N. Sturgis. 2004. Regulatory circuits and communication in Gram-negative bacteria. Nat. Rev. Microbiol. 2:581-592. [DOI] [PubMed] [Google Scholar]
- 19.Lugtenberg, B., J. Meijers, R. Peters, P. van der Hoek, and L. van Alphen. 1975. Electrophoretic resolution of the “major outer membrane protein” of Escherichia coli K12 into four bands. FEBS Lett. 58:254-258. [DOI] [PubMed] [Google Scholar]
- 20.Lyczak, J. B., C. L. Cannon, and G. B. Pier. 2000. Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect. 2:1051-1060. [DOI] [PubMed] [Google Scholar]
- 21.Michel-Briand, Y., and C. Baysse. 2002. The pyocins of Pseudomonas aeruginosa. Biochimie 84:499-510. [DOI] [PubMed] [Google Scholar]
- 22.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reimmann, C., M. Beyeler, A. Latifi, H. Winteler, M. Foglino, A. Lazdunski, and D. Haas. 1997. The global activator GacA of Pseudomonas aeruginosa positively controls the production of the autoinducer N-butyryl-homoserine lactone and the formation of the virulence factors pyocyanin, cyanide, and lipase. Mol. Microbiol. 24:309-319. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 25.Sandoz, K. M., S. M. Mitzimberg, and M. Schuster. 2007. Social cheating in Pseudomonas aeruginosa quorum sensing. Proc. Natl. Acad. Sci. USA 104:15876-15881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schuster, M., and E. P. Greenberg. 2006. A network of networks: quorum-sensing gene regulation in Pseudomonas aeruginosa. Int. J. Med. Microbiol. 296:73-81. [DOI] [PubMed] [Google Scholar]
- 27.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering transposon mutagenesis in gram-negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 28.Smith, A. W., and B. H. Iglewski. 1989. Transformation of Pseudomonas aeruginosa by electroporation. Nucleic Acids Res. 17:10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith, E. E., D. G. Buckley, Z. Wu, C. Saenphimmachak, L. R. Hoffman, D. A. D'Argenio, S. I. Miller, B. W. Ramsey, D. P. Speert, S. M. Moskowitz, J. L. Burns, R. Kaul, and M. V. Olson. 2006. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc. Natl. Acad. Sci. USA 103:8487-8492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith, R. S., and B. H. Iglewski. 2003. P. aeruginosa quorum-sensing systems and virulence. Curr. Opin. Microbiol. 6:56-60. [DOI] [PubMed] [Google Scholar]
- 31.Spaink, H. P., R. J. H. Okker, C. A. Wijffelman, E. Pees, and B. J. J. Lugtenberg. 1987. Promoters in the nodulation region of the Rhizobium leguminosarum Sym plasmid pRL1JI. Plant Mol. Biol. 9:27-39. [DOI] [PubMed] [Google Scholar]
- 32.Stanisich, V. A., and B. W. Holloway. 1972. A mutant sex factor of Pseudomonas aeruginosa. Genet. Res. 19:91-108. [DOI] [PubMed] [Google Scholar]
- 33.Tommassen, J., A. Filloux, M. Bally, M. Murgier, and A. Lazdunski. 1992. Protein secretion in Pseudomonas aeruginosa. FEMS Microbiol. Rev. 9:73-90. [DOI] [PubMed] [Google Scholar]
- 34.Venturi, V. 2006. Regulation of quorum sensing in Pseudomonas. FEMS Microbiol. Rev. 30:274-291. [DOI] [PubMed] [Google Scholar]
- 35.Voulhoux, R., A. Lazdunski, and A. Filloux. 2001. Colicin A hybrids: a genetic tool for selection of type II secretion-proficient Pseudomonas strains. EMBO Rep. 2:49-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Whitchurch, C. B., S. A. Beatson, J. C. Comolli, T. Jakobsen, J. L. Sargent, J. J. Bertrand, J. West, M. Klausen, L. L. Waite, P. J. Kang, T. Tolker-Nielsen, J. S. Mattick, and J. N. Engel. 2005. Pseudomonas aeruginosa fimL regulates multiple virulence functions by intersecting with Vfr-modulated pathways. Mol. Microbiol. 55:1357-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wolfgang, M. C., V. T. Lee, M. E. Gilmore, and S. Lory. 2003. Coordinate regulation of bacterial virulence genes by a novel adenylate cyclase-dependent signaling pathway. Dev. Cell 4:253-263. [DOI] [PubMed] [Google Scholar]
- 38.Yahr, T. L., and M. C. Wolfgang. 2006. Transcriptional regulation of the Pseudomonas aeruginosa type III secretion system. Mol. Microbiol. 62:631-640. [DOI] [PubMed] [Google Scholar]