Abstract
The gaseous hormone ethylene is perceived in Arabidopsis by a five member receptor family that consists of the subfamily 1 receptors ETR1 and ERS1 and the subfamily 2 receptors ETR2, ERS2, and EIN4. Previous work has demonstrated that the basic functional unit for the ethylene receptor, ETR1, is a disulfide-linked homodimer. We demonstrate here that ethylene receptors isolated from Arabidopsis also interact with each other through noncovalent interactions. Evidence that ETR1 associates with other ethylene receptors was obtained by co-purification of ETR1 with tagged versions of ERS1, ETR2, ERS2, and EIN4 from Arabidopsis membrane extracts. ETR1 preferentially associated with the subfamily 2 receptors compared with the subfamily 1 receptor ERS1, but ethylene treatment affected the interactions and relative composition of the receptor complexes. When transgenically expressed in yeast, ETR1 and ERS2 can form disulfide-linked heterodimers. In plant extracts, however, the association of ETR1 and ERS2 can be largely disrupted by treatment with SDS, supporting a higher order noncovalent interaction between the receptors. Yeast two-hybrid analysis demonstrated that the receptor GAF domains are capable of mediating heteromeric receptor interactions. Kinetic analysis of ethylene-insensitive mutants of ETR1 is consistent with their dominance being due in part to an ability to associate with other ethylene receptors. These data suggest that the ethylene receptors exist in plants as clusters in a manner potentially analogous to that found with the histidine kinase-linked chemoreceptors of bacteria and that interactions among receptors contribute to ethylene signal output.
The plant hormone ethylene plays an important role in plant growth and development (1). Ethylene regulates seed germination, seedling growth, leaf and petal abscission, fruit ripening, organ senescence, and pathogen responses. Arabidopsis can respond to ethylene as low as 0.2 nl/liter based on kinetic analysis of the growth inhibition of etiolated seedlings by ethylene (2), and up to 1000 μl/liter based on analysis of the induction of an ethylene-responsive reporter gene in stem tissues (3).
There are five ethylene receptors (ETR1, ERS1, ETR2, ERS2, and EIN4) in Arabidopsis (4, 5). These receptors have N-terminal transmembrane domains that contain an ethylene binding site (6–8), and also serve in localization of the receptor to the endoplasmic reticulum and possibly to the Golgi apparatus (9, 10). The C-terminal histidine kinase-like domain and receiver domain (ERS1 and ERS2 lack receiver domains) function in signal output (11, 12). The five-member ethylene receptor family is divided into two subfamilies. ETR1 and ERS1 belong to subfamily 1, and they have functional histidine kinase domains (13, 14). ETR2, ERS2, and EIN4 belong to subfamily 2, and lacking the necessary residues for histidine kinase activity are now thought to function as Ser/Thr kinases (14).
The basic functional unit for the ethylene receptors is a dimer. Ethylene receptors need copper ions to bind ethylene, and there is one copper ion, and thus the ability to bind one molecule of ethylene, per receptor dimer (7). Consistent with a dimer being the functional unit is the finding that two receptor monomers are maintained as a disulfide-linked dimer, two conserved Cys residues near the N terminus being implicated in forming the covalent linkage (15, 16).
Both gain-of-function and loss-of-function mutants have been isolated for the ethylene receptors. An ethylene receptor mutant etr1-1 that has lost its ethylene binding ability because of a single amino acid change exhibits a dominant ethylene-insensitive phenotype (6, 17, 18). Single loss-of-function mutants of ethylene receptors do not show substantially different phenotypes from wild type. However, combinations of ethylene receptor loss-of-function mutants show a constitutive ethylene response phenotype, suggesting that ethylene receptors are functionally redundant and negatively regulate ethylene responses (19).
Protein-protein interactions are important for ethylene signaling. Ethylene receptors interact with CTR1 to mediate ethylene signal transduction. CTR1 is a Raf-like kinase, and functions downstream of the ethylene receptors (20, 21). The association of CTR1 with ethylene receptors in Arabidopsis was demonstrated by the use of genetic and biochemical approaches (21–24). In this study we demonstrate that ETR1 can also physically interact with other ethylene receptors in Arabidopsis. ETR1 was found preferentially associated with the subfamily 2 ethylene receptors. The interaction between ethylene receptors is a higher order interaction, suggesting that the ethylene receptors exist in plants as clusters in a manner potentially analogous to that found with the histidine kinase-linked chemoreceptors of bacteria (25). The higher order interactions between ethylene receptors may provide an explanation for the broad range of ethylene responsiveness found in plants as well as the dominant nature of ethylene-insensitive mutations found in the receptors.
EXPERIMENTAL PROCEDURES
Constructs and Plant Transformation—For preparation of ETR2, ERS2, and EIN4 with C-terminal tandem affinity purification (TAP)3 tags, the vector pCAMBIA1380-SBP-TAP was used. For this vector, DNA encoding the streptavidin-binding peptide (SBP) along with an additional 267 bp containing BamHI and a HindIII restriction sites was amplified from the vector pTAG2K (26) and cloned into the PCR 2.1-TOPO vector (Invitrogen). The TAP tag was amplified and cloned into the BamHI and HindIII restriction sites of the TOPO vector. The SBP-TAP tag was then cloned into the SalI and HindIII restriction sites of the vector pCAMBIA1380 to make pCAMBIA-SBP-TAP. The region encoding ETR2 along with upstream promoter sequence was amplified from the Arabidopsis BAC clone K14B15 (GenBank™ accession no. AB025608) using primers 5′-GTCGACGCGATTCTGACATTCTGT-3′ and 5′-GTCGACGAAGTTGGTCAGCTTGCA-3′. The region encoding ERS2 along with upstream promoter sequence was amplified from the Arabidopsis BAC clone F19P19 (GenBank™ accession no. AC000104) using primers 5′-GTCGACGGTAAGAGTCCACGTAGG-3′ and 5′-GTCGACAGTGGCTAGTAGACGGAG-3′. The region encoding EIN4 along with upstream promoter sequence was amplified from the Arabidopsis BAC clone F7O18 (GenBank™ accession no. AC011437) using primers 5′-GTCGACGCTCTTCTCCGTTGTGGC-3′ and 5′-GTCGACACTCGCTCGCGGTCTGCA-3′. The PCR products were cloned into the SalI site of pCAMBIA-SBP-TAP. For preparation of ERS1 with a C-terminal TAP tag, the region encoding ERS1 along with upstream promoter sequence was amplified from the Arabidopsis BAC clone T20B5 (GenBank™ accession no. AC002409) using primers 5′-GGATCCCAGGGATGTGCACTGAAG-3′ and 5′-GGATCCACCAGTTCCACGGTCTGG-3′. The PCR product was cloned into the BamHI site of pCAMBIA2380-Myc-TAP vector (23) to yield a construct with the c-Myc epitope in tandem with the original TAP tag.
For transformation into Arabidopsis, constructs were introduced into Agrobacterium tumefacians strain GV3101 and used to transform Arabidopsis by the floral-dip method (27). The different ethylene receptor constructs were transformed into their respective receptor loss-of-function mutant backgrounds (19, 28, 29). In addition, to test for functionality of the tagged receptors, the ETR2-TAP construct was transformed into the etr1/etr2/ein4 triple mutant (19), and the ERS2-TAP and EIN4-TAP constructs were each transformed into the etr2/ers2/ein4 triple mutant (19).
Seedling Ethylene Response—Treatment and analysis of the triple response of Arabidopsis seedlings to ethylene was performed as described (29). The rapid short-term kinetic response of hypocotyl growth was analyzed as described (11).
Isolation of Arabidopsis Membranes—Microsomal fractions were isolated from either dark-grown Arabidopsis seedlings (29) or Arabidopsis plants grown in liquid culture under continuous light (9). Aminovinylglycine (AVG), an inhibitor of ethylene biosynthesis, was included in growth medium for dark-grown seedlings as indicated. Plant material was homogenized in a buffer containing 30 mm Tris (pH 8.3 at 4 °C), 150 mm NaCl, 1 mm EDTA, and 20% (v/v) glycerol with protease inhibitors and then centrifuged at 8,000 × g for 15 min as described (9). The supernatant was then centrifuged at 100,000 × g for 30 min, and the resulting membrane pellet resuspended in 10 mm Tris (pH 7.6 at 22 °C), 150 mm NaCl, 0.1 mm EDTA, and 10% (v/v) glycerol with protease inhibitors (resuspension buffer).
Antibodies and Immunoblot Analysis—ETR1 was identified by the use of a polyclonal anti-ETR1 antibody generated against amino acids 401–738 of ETR1 (9). TAP-tagged proteins were detected based on the ability of the protein-A motif to bind rabbit anti-goat IgG antibody coupled to horseradish peroxidase (Santa Cruz Biotechnology) (TAP antibody). The anti-BiP antibody was a gift from R. Boston (North Carolina State University). GST fusion proteins were identified by the use of a monoclonal anti-GST antibody (Santa Cruz Biotechnology). Immunoblot analysis was performed as described (30). Protein concentration was determined by the use of the BCA reagent (Pierce) according to the manufacturer after first adding 0.2 ml 0.4% (w/v) deoxycholate to solubilize membrane proteins. Bovine serum albumin was used as a standard for protein assays. Prior to SDS-PAGE, protein samples were mixed with SDS-PAGE loading buffer (300 mm Tris (pH6.8), 15% glycerol, 6% SDS, 0.01% bromphenol blue), and incubated at 37 °C for 1 h or ramped from 37 to 73 °C over 40 min using a thermocyler, so as to prevent the aggregation of integral membrane proteins that can occur with boiling. For reductive SDS-PAGE, 300 mm DTT was added to the loading buffer. Following SDS-PAGE, proteins were electrotransferred to Immobilon nylon membrane (Millipore) for immunoblotting. Immunodecorated proteins were visualized by enhanced chemiluminescence detection according to the manufacturer (Pierce) and quantified using a Bio-Rad GS-800 densitometer and Quantity One software, quantification based on a set of exposure standards for the immunodecorated proteins of interest.
Purification of TAP-tagged Ethylene Receptors—Microsomes were brought to 1 mg/ml protein and incubated with 0.5% (w/v) lysophosphatidylcholine (LPC; 1-palmitoyl-2-hydroxy-sn-glycero-3-phosphocholine, from Avanti Polar-Lipids, Inc.) for 1 h at 4 °C, then centrifuged at 100,000 × g for 30 min. The supernatant was diluted to 0.25% (w/v) LPC and incubated with human IgG-agarose (Sigma) for 3 h at 4°C on a rocking platform. The beads were washed with resuspension buffer supplemented with 0.5% Nonidet P-40 (Sigma) to remove unbound proteins, and the bound proteins were eluted with 1% SDS and analyzed by immunoblot.
Disruption of Interactions between ETR1 and ERS2-TAP—Microsomes from 5 g of ERS2-TAP transgenic plants grown in liquid culture were solubilized with LPC. ERS2-TAP was pulled-down with human IgG-agarose and eluted with 1% SDS to disrupt noncovalent interactions. To disrupt ETR1-ERS2 heterodimers as well as noncovalent interactions, the IgG bead-bound proteins were eluted with 1% SDS and 10 mm DTT. The eluted proteins were diluted 10-fold with resuspension buffer supplemented with 1 mg/ml bovine serum albumin and 0.25% LPC. The diluted solution was incubated for 1 h at 4 °C and then centrifuged at 100,000 × g for 30 min to remove any precipitate. The supernatant was incubated with human IgG-agarose for 2 h at 4 °C. The beads were washed with resuspension buffer supplemented with 0.5% Nonidet P-40 and eluted with 1% SDS. The eluted proteins were analyzed by immunoblot.
Analysis of the Interaction between ETR1 and ERS2 in Yeast—For expression of ETR1-TAP in yeast, the vector pYES2 (Invitrogen) was used. The TAP tag was amplified from the vector pBS1479 (31) using primers 5′-GCTGGACTCGAGATGGAAAAGAGAAGATGG-3′ and 5′-CGATATTCTAGATCAGGTTGACTTCCCCGC-3′, and cloned into the XhoI and XbaI restriction sites of the vector pYES2 to make the pYES2-TAP vector. Full-length ETR1 was amplified from an ETR1 cDNA clone (15) using primers 5′-AAGCTTATGGAAGTCTGCAATTGT-3′ and 5′-GCGGCCGCATGCCCTCGTACAGTAC-3′, and the PCR product cloned into the HindIII and NotI sites of pYES2-TAP to make pYES2-ETR1-TAP. For expression of ERS2-(1–356)-GST in yeast, the HIS3 gene was amplified from a vector pBJ243 (gift from Dr. Charles K. Barlowe, Dartmouth) using primers 5′-GGGCCCTTCCCGTTTTAAGAGCTTGGT-3′ and 5′-GGGCCCAAAGGAAAGCGCGCCTCGTTC-3′. The PCR product was cloned into the ApaI site of pYES2 to make the pYES2(HIS3) vector. The GST tag was amplified from the vector pEG(KT) (32) using primers 5′-ATCACTCGAGTCGACTGTCCCCTATACTAGGTTATTGG-3′ and 5′-ATCATCTAGATTACAGATCCGATTTTGGAGGATG-3′. The PCR product was cloned into the XhoI and XbaI sites of the vector pYES2(HIS3) to make the pYES2(HIS3)-GST vector. The DNA sequence encoding the first 356 amino acids of ERS2 was amplified from an ERS2 cDNA clone (gift from Dr. Priti Krishna, University of Western Ontario, Canada) using primers 5′-GAGCTCATGTTAAAGACATTGTTAGTCC-3′ and 5′-GTCGACGAGATTCTTCAAGAATCACGGC-3′. The PCR product was cloned into the SacI and SalI sites of pYES2-GST-HIS3 to make pYES2(HIS3)-ERS2(1–356)-GST.
The pYES2-ETR1-TAP construct and the pYES2(HIS3)-ERS2(1–356)-GST construct were transformed into yeast (Saccharomyces cerevisiae) strain EGY188 (MATa ura3 his3 trp1 LexA-LEU2) (33). Standard media and procedures were used for growth (34), and 0.2% or 0.05% (w/v) galactose was used to induce protein expression for 6 h. An anti-GST antibody was using for detecting both ETR1-TAP and ERS2-(1–356)-GST by immunoblot.
Yeast Two-hybrid Analysis—The DNA binding domain fusion (bait) plasmids and transcriptional activation domain fusion (prey) plasmids were constructed as follows. To create the ETR1 prey, an EcoICRI-EcoRV blunt-ended restriction fragment of the ETR1 cDNA (in pBluescript SK) encoding the intracellular portion of ETR1 (129–738 end) was ligated into the EcoRI site of the prey vector pACTII (22), after filling in the EcoRI overhangs using T4 DNA polymerase. The EcoRV site of the ETR1 cDNA fragment was provided by the vector. To create the ETR1 GAF prey, a fragment encoding the ETR1 GAF domain (129–330) was PCR-amplified using primers 5′-GGCCATGGTCGATAGAGAAATGGGATTG-3′ and 5′-AAGGATCCCCCGGTTAAAGAGCAACATTCTG-3′, digested with NcoI and BamHI, and then cloned into the NcoI and BamHI sites of pACTII. For the ETR2 bait encoding the GAF domain to the end of the protein (143–773), a XhoI ETR2 cDNA fragment was cloned into the XhoI site of an existing ETR2 GAF domain clone in plasmid pBluescript SK, and confirmed by restriction digestion for the correct orientation. The entire ETR2 fragment was then released using SmaI and a XhoI partial digestion, and the XhoI end was filled in using T4 DNA polymerase to create a blunt end. The resulting fragment was cloned into the SmaI site of the bait vector plexA-NLS (pBTM116) (22). For the ETR2 GAF domain bait, a DNA fragment encoding the ETR2 GAF domain (143–381) was PCR-amplified using primers 5′-TCCCGGGGAAAGTTAAAGTTAGAGAG-3′ and 5′-CTTAACGCCTCATCCCTTC-3′, digested with SmaI and then cloned into the SmaI site of plexA-NLS.
Yeast two-hybrid analysis was performed using yeast strain L40 as described (22), except that yeast transformants were grown overnight in minimal liquid medium lacking tryptophan and leucine (to select for the bait and prey plasmids, respectively) with shaking at 30 °C, and then serial 10-fold dilutions were spotted in 6-μl drops onto agar medium.
Real-time PCR—Real-time PCR was performed as described (35), using primer sets specific for ERS1 (At2g40940) (5′-ACCTATGTGTGCAGGTGAAGGACA-3′ and 5′-AGCCCGACAAACCGTTTACAGAGA-3′), ETR2 (At3g23150) (5′-AGAGAAACTCGGGTGCGATGT-3′ and 5′-TCACTGTCGTCGCCACAATC-3′), ERS2 (At1g04310) (5′-TCAAGAAGCGGTTTGGCGACATTG-3′ and 5′-TAGACCGTCCTCAACAACCCGAAT-3′, and β-tubulin (At5g62700) (5′-CGTAAGCTTGCTGTGAATCTCATC-3′ and 5′-CTGCTCGTCAACTTCCTTTGTG-3′).
RESULTS
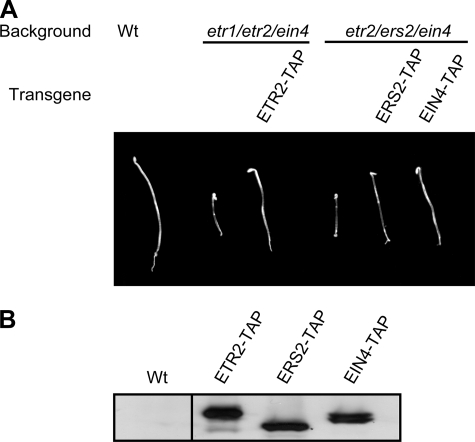
TAP-tagged Ethylene Receptors Are Functional—To characterize ethylene receptors from Arabidopsis at the protein level, a TAP tag (31) was fused to the C terminus of the ethylene receptors ETR2, ERS2, EIN4, and ERS1. The native promoter and genomic sequence of each ethylene receptor gene were used for the constructs to allow for normal levels and patterns of expression. The ethylene receptor constructs were transformed into corresponding single loss-of-function mutant backgrounds to use for biochemical analysis. In addition, the ETR2-TAP construct was transformed into an etr1/etr2/ein4 triple mutant (19), and the ERS2-TAP and EIN4-TAP constructs were each transformed into an etr2/ers2/ein4 triple mutant (29), to determine whether TAP-tagged ethylene receptors were functional. The TAP-tagged ethylene receptors rescued the constitutive ethylene-response phenotype of the triple mutants (Fig. 1A), indicating that the TAP tag does not disrupt receptor function. Consistent with rescue, the TAP-tagged ethylene receptors were detectable by immunoblot in membranes isolated from the transgenic plants (Fig. 1B).
FIGURE 1.
Functionality of TAP-tagged ethylene receptors. A, rescue of the constitutive ethylene-response phenotype of triple knock-out mutants by TAP-tagged receptors. ETR2-TAP was transformed into an etr1/etr2/ein4 triple mutant. ERS2-TAP and EIN4-TAP were transformed into an etr2/ers2/ein4 triple mutant. B, protein expression of the TAP-tagged ethylene receptors from the transgenic plants shown in A. Immunoblot analysis was performed with 30 μg of membrane protein. TAP-tagged ethylene receptors were detected by use of a goat anti-rabbit IgG coupled with horseradish peroxidase. Lanes are from the same immunoblot exposure.
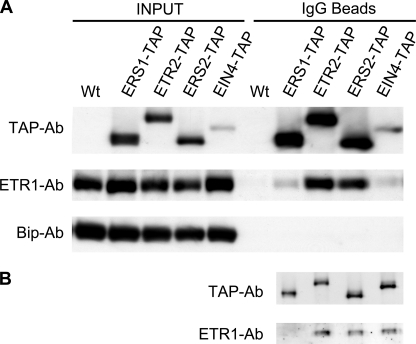
ETR1 Co-purifies with Other TAP-tagged Ethylene Receptors from Arabidopsis—The transgenic plants that expressed TAP-tagged ethylene receptors in their corresponding single loss-of-function mutant backgrounds were used to determine if ETR1 interacted with the other ethylene receptors in Arabidopsis. The TAP-tagged ethylene receptors were purified by incubating lysophosphatidylcholine-solubilized membrane proteins with human IgG beads. The IgG beads bind the protein-A portion of the TAP tag, resulting in affinity purification of the TAP-tagged ethylene receptors. An anti-ETR1 antibody was then used to determine if ETR1 co-purified with the TAP-tagged ethylene receptors.
The pull-down experiments demonstrated that ETR1 co-purified with TAP-tagged ERS1, ETR2, ERS2, and EIN4 (Fig. 2). Controls confirmed that IgG beads did not pull-down ETR1 from the wild-type control, nor did IgG beads pull-down BiP, a resident protein of the plant endoplasmic reticulum (36), indicating that the co-purification of ETR1 was mediated by the TAP-tagged ethylene receptors. By comparing the enrichment factor of the TAP-tagged receptor to the enrichment factor of ETR1 by IgG beads, we estimated that ∼25% of the total amount of ETR1 co-purified with ETR2 or ERS2, and ∼3% of the total amount of ETR1 co-purified with ERS1 or EIN4 (Fig. 2A).
FIGURE 2.
Interaction of ETR1 with other ethylene receptors of Arabidopsis. A, co-purification of ETR1 with TAP-tagged ethylene receptors. Transgenic plants that express TAP-tagged ethylene receptors in corresponding single loss-of-function mutants were used. Membrane proteins (1 mg/ml) from wild type and transgenic plants grown in liquid culture were solubilized with 0.5% (w/v) lysophosphatidylcholine. The soluble supernatants were incubated with IgG beads to pull-down TAP-tagged ethylene receptors. The amounts of TAP-tagged ethylene receptors and ETR1 before IgG binding (INPUT) and those bound on IgG beads (IgG Beads) were detected with an anti-TAP antibody and an anti-ETR1 antibody. BiP was detected with an anti-BiP antibody, and served as an internal control that should not bind to the IgG beads. B, efficiencies of each ethylene receptor for co-purification of ETR1 based on equivalent loading of each TAP-tagged receptor from the IgG-bead fractions shown in A.
Expression of the TAP-tagged receptors varied, resulting in differing yields following pull-down with IgG beads (Fig. 2A). Therefore, we also ran gels of the same purified fractions, loading these so as to have equivalent intensity of the TAP-tagged receptors (Fig. 2B). The amount of ETR1 co-purified with the TAP-tagged ethylene receptors was then compared to determine the efficiency of co-purification. Based on this analysis, the subfamily 2 ethylene receptors ETR2, ERS2, and EIN4 have similar efficiency for co-purification of ETR1, whereas the subfamily 1 member ERS1 has a lower efficiency for co-purification of ETR1.
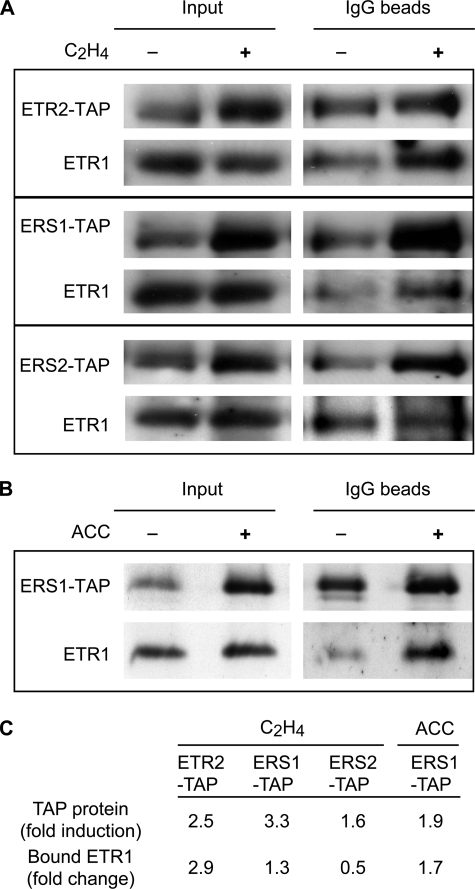
Effect of Ethylene upon ETR1 Interactions with TAP-tagged Ethylene Receptors—To determine whether ethylene affects the interactions between ETR1 and the other ethylene receptors, we used etiolated seedlings, because they show a pronounced response to ethylene treatment. Interactions were examined with ERS1-TAP, ETR2-TAP, and ERS2-TAP, each of which is induced at the transcriptional level by ethylene (37). Aminoethoxyvinylglycine, an inhibitor of ACC synthase, was included in the growth medium to block endogenous production of ethylene (38).
Protein levels of ERS1-TAP, ETR2-TAP, and ERS2-TAP all increased following 4-h treatment with 10 μl/liter ethylene (Fig. 3), consistent with their transcriptional regulation by ethylene. The pull-downs revealed differing levels of ETR1 interactions with the TAP-tagged receptors. In the absence of ethylene, we estimated that ∼16% of the total amount of ETR1 co-purified with ETR2, 43% with ERS2, and 11% with ERS1. Following ethylene treatment, the total amount of ETR1 associated with ETR2-TAP increased at a level coincident with the increase in ETR2-TAP levels (Fig. 3, A and C). The total amount of ETR1 associated with ERS1-TAP also showed a small increase following the short-term ethylene treatment (Fig. 3, A and C). Longer-term ethylene treatment of the ERS1-TAP line resulted in a total level of ETR1 association with ERS1-TAP coincident with the increase in ERS1-TAP levels (Fig. 3, B and C). In contrast to the effect of ethylene upon ETR1 association with ETR2 and ERS1, decreased levels of ETR1 were found associated with ERS2 following ethylene treatment (Fig. 3, A and C). These results indicate that receptor complexes exist in both the absence and presence of ethylene, but the kinetics of formation and relative composition for the receptor complexes may vary following ethylene treatment.
FIGURE 3.
Interaction of ETR1 with other ethylene receptors in the absence and presence of ethylene. Etiolated seedlings that express ERS1-TAP, ETR2-TAP, or ERS2-TAP were grown in the absence or presence of 10 μl/liter ethylene for 4 h (A), or continuously in the absence or presence of 50 μm ACC, the biosynthetic precursor of ethylene (B). Protein levels of ETR1, ERS1-TAP, ETR2-TAP, and ERS2-TAP in the transgenic plants are shown (Input) as well as after co-purification of ETR1 with ERS1-TAP, ETR2-TAP, or ERS2-TAP (IgG beads). C, quantification of immunoblot data. The fold induction for each TAP-tagged receptor (TAP protein) following ethylene treatment is based upon the induction found in the input lanes and is normalized against the ETR1 protein level. The fold change for the total amount of ETR1 bound by the TAP-tagged receptors is determined based upon the amount found after pull-down on the IgG beads.
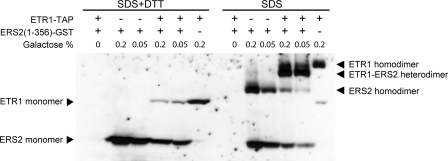
Formation of Heterodimers by ETR1 and ERS2 When Transgenically Expressed in Yeast—Previous data indicate that ETR1 exists as a disulfide-linked dimer, and that Cys-4 and Cys-6 are the sites that participate in formation of disulfide bonds in the homodimer (15). The Cys-4 and Cys-6 residues are conserved in all ethylene receptors, and the N termini of the five receptors have high amino acid identity. Evidence indicates that ethylene receptors can form homodimers in plants (15, 39), but the presence of heterodimers between ethylene receptors has not been demonstrated.
To determine whether ETR1 can interact with other ethylene receptors as a disulfide-linked heterodimer, we used a transgenic yeast system for receptor expression. We co-transformed yeast with a full-length tagged version of ETR1 (ETR1-TAP) and a truncated tagged version of ERS2 (ERS2-(1–356)-GST). ERS2-(1–356) contains just the transmembrane domains and the GAF domain of ERS2, and thus has a significantly different molecular mass from full-length ETR1. Thus, it should be possible to resolve the disulfide-linked heterodimer of ETR1-ERS2 from the ETR1 homodimer and the ERS2 homodimer by nonreducing SDS-PAGE electrophoresis.
Expression of ETR1-TAP and ERS2-(1–356)-GST was induced with galactose. Both ETR1-TAP and ERS2-(1–356)-GST can be detected by immunoblot when using the anti-GST monoclonal antibody, because of a weak interaction between the TAP tag and the mouse and goat IgGs (Fig. 4). In the presence of reducing reagent, ERS2-(1–356)-GST migrated at 65 kDa, consistent with its predicted molecular mass of 65 kDa, and ETR1-TAP migrated at 110 kDa, consistent with its predicted molecular mass of 102 kDa. In the absence of reducing reagent, both ERS2-(1–356)-GST and ETR1-TAP were found to form homodimers when expressed individually. The homodimer of ERS2-(1–356)-GST migrated at 140 kDa, consistent with its predicted molecular mass of 130 KDa, and the homodimer of ETR1-TAP migrated at 210 kDa, consistent with its predicted molecular mass of 204 kDa. When the two proteins were co-expressed in yeast, a new band at a size of 180 kDa was detected between the homodimer bands of ETR1-TAP and ERS2-(1–356)-GST, consistent with a predicted heterodimer molecular mass of 167 kDa (Fig. 4). Interestingly, the truncated ERS2 protein preferentially formed the ERS2-ETR1 heterodimer rather than the ERS2 homodimer. This may relate to a natural propensity for ERS2 to interact with subfamily 1 receptors. Alternatively it may be an indirect effect of the truncated ERS2 lacking its histidine kinase domain, as this domain may facilitate dimerization.
FIGURE 4.
Formation of disulfide-linked heterodimers between ETR1 and ERS2 following transgenic expression in yeast. ETR1-TAP and ERS2-(1–356)-GST were co-transformed or independently transformed into yeast, and induced with 0.2% (w/v) or 0.05% (w/v) galactose for 6 h. Total protein (40 μg) was subjected to reducing (+DTT) and nonreducing (–DTT) SDS-PAGE electrophoresis on an 8% (w/v) gel. An anti-GST antibody was used to detect both ETR1-TAP and ERS2-(1–356)-GST.
To exclude the possibility that GST mediates the heterodimer formation between ETR1-TAP and ERS2-(1–356)-GST, we co-expressed GST and ETR1-TAP in yeast. Results indicate that GST does not form a disulfide-linked heterodimer with ETR1-TAP (data not shown). Because the TAP tag does not contain cysteine, the possibility of it forming a disulfide-linked heterodimer with ERS2-(1–356) is excluded. The formation of a heterodimer between ETR1 and ERS2 in yeast supports the possibility that such heterodimer interactions could occur between ETR1 and the other ethylene receptors of Arabidopsis.
Higher Order Interaction of ETR1 with ERS2 in Arabidopsis—Because ETR1 co-purifies with ERS2-TAP from Arabidopsis, we considered two possibilities by which ETR1 could interact with ERS2-TAP. First, ETR1 and ERS2-TAP could form disulfide-linked heterodimers. This possibility is supported by our finding that ETR1 and ERS2 can form heterodimers in yeast (Fig. 4). According to this hypothesis, the interaction of ETR1 with ERS2-TAP would not be disrupted by treatment with SDS, which would be unable to break the covalent disulfide bond. Second, an ETR1 homodimer could interact with an ERS2-TAP homodimer through noncovalent interactions. Such an interaction would represent a higher order interaction and, being noncovalent, should be capable of being destroyed by denaturing treatment with SDS.
To determine whether ETR1 interacts with ERS2 by forming heterodimers or through higher order interactions in Arabidopsis, IgG-bound ERS2-TAP along with its associated ETR1 were eluted with 1% (w/v) SDS. SDS treatment can disrupt most protein-protein interactions maintained by noncovalent interactions, and thus the ETR1-ERS2 interaction will be disrupted if it is maintained by higher order interactions. The eluted solution was then diluted 10-fold to reduce the SDS concentration to 0.1% and brought to 0.25% (w/v) LPC, thus allowing IgG beads to pull-down ERS2-TAP again. ETR1 was then examined on the IgG beads to determine whether ETR1 still co-purified with ERS2-TAP after the SDS treatment.
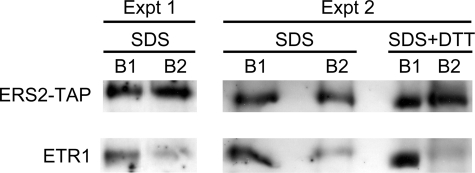
Two independent replicates of the experiment indicate that the 1% SDS treatment reduced the co-purification of ETR1 with ERS2-TAP by ∼80% (Fig. 5), demonstrating that the interaction of ETR1 and ERS2-TAP in Arabidopsis is maintained by higher order interactions. The fact that a small amount of ETR1 still co-purified with ERS2-TAP after the SDS treatment indicates that ETR1-ERS2 heterodimers may also exist in Arabidopsis (Fig. 5). To test this hypothesis, ERS2-TAP and ETR1 bound by IgG beads were eluted with 1% SDS and 10 mm DTT, thereby allowing for the disruption of disulfide-linked heterodimers and higher order interactions. This treatment almost completely abolished the co-purification of ETR1 with ERS2-TAP (Fig. 5).
FIGURE 5.
The interaction of ETR1 and ERS2-TAP is disrupted by SDS. Membrane proteins from 5 g of ERS2-TAP transgenic Arabidopsis plants were solubilized with lysophosphatidylcholine, and pulled-down by IgG beads. The IgG bead-bound proteins were eluted with either 1% SDS, or 1% SDS and 10 mm DTT. The eluted protein (marked as B1) was diluted 10-fold with resuspension buffer supplemented with 0.25% lysophosphatidylcholine and 1 mg/ml bovine serum albumin. The eluted protein was then incubated with IgG beads to pull-down ERS2-TAP. ERS2-TAP and ETR1 bound by IgG beads (marked as B2) were detected by an anti-TAP antibody and an anti-ETR1 antibody. The analysis with the 1% SDS elution was repeated twice (Expt 1 and Expt 2).
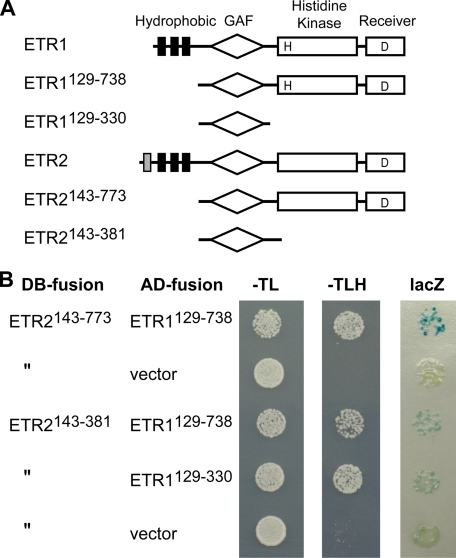
The GAF Domain Facilitates Physical Interactions between Receptors—We performed yeast two-hybrid analysis to determine which regions of the receptors were capable of mediating heteromeric interactions, examining interactions between the subfamily 1 receptor ETR1 and the subfamily 2 receptor ETR2 (Fig. 6). Constructs containing the entire soluble domains but lacking the N-terminal transmembrane domains were tested and determined to interact based on two different reporters (HIS3+ and LacZ). Similar results were also obtained when we examined interaction between the soluble domains of ETR1 and ERS2 (data not shown). The soluble region of greatest sequence similarity between the subfamily 1 and 2 receptors is the GAF domain. GAF domains have been shown to mediate cGMP binding and light regulation in some proteins, but their function in the ethylene receptors is unknown (40). We found through the examination of additional truncated versions of the ETR1 and ETR2 receptors that the GAF domain was sufficient to mediate their interaction, although based on the LacZ reporter analysis the strength of this interaction was reduced compared with that observed with the entire soluble domains (Fig. 6).
FIGURE 6.
Yeast two-hybrid analysis of receptor interactions. A, structure of ETR1, ETR2, and constructs used for analysis. The hydrophobic ethylene-sensing domain (hydrophobic segments indicated by black and gray bars), the GAF domain, the His kinase domain, and the receiver domain are indicated. H and D indicate putative phosphorylation sites. The ETR1 and ETR2 full-length receptors consist of 738 and 773 residues, respectively. B, results of yeast two-hybrid analysis. The portions of the ethylene receptors fused with the DNA binding domain (DB-fusion) and transcriptional activation domain (AD-fusion) are indicated. Cells were spotted onto agar medium from 1/100 dilutions of liquid overnight cultures and grown for 3 days. Medium lacking tryptophan and leucine (–TL) selects for the DB-fusion and AD-fusion plasmids, respectively. Protein interactions are shown by growth on medium lacking histidine (–TLH) and by the X-gal filter assay (lacZ). The X-gal filter assay was performed on the same overnight cultures grown on –TL medium, but spotted at a dilution of 1/1000, with staining shown after a 4-h, 22 °C incubation. As a negative control, no activation of reporter genes was detected when the ETR1 AD-fusions were tested with a lamin DB-fusion (22) (data not shown).
Kinetic Analysis of the Truncated etr1-1-(1–349) Receptor—etr1-1 is a dominant gain-of-function mutation that results in a C65Y amino acid change in the receptor such that the mutant receptor no longer binds ethylene (6, 7). This results in a gain-of-function because the etr1-1 receptor appears locked into a functional conformation that constitutively activates CTR1 to suppress ethylene responses. Surprisingly, a truncated version of the mutant receptor, etr1-1-(1–349), which lacks its signal output domain still confers on plants a partial ethylene-insensitive phenotype when introduced into the etr1-7 genetic background that lacks ETR1 (30, 41). This dominance could arise because of an ability of etr1-1-(1–349) to interact with and convert other receptors to an ethylene-insensitive conformation. Consistent with this possibility, etr1-1-(1–349) contains the GAF domain, which we found sufficient for facilitating physical interaction with other members of the ethylene receptor family (Fig. 6).
If etr1-1-(1–349) requires additional ethylene receptors to confer ethylene insensitivity, then its effectiveness may vary in a manner dependent upon the expression levels of the other receptors. We therefore examined the effectiveness of etr1-1-(1–349) compared with etr1-1 by kinetic analysis following treatment of seedlings with ethylene, making use of transgenic lines in which the truncated and full-length versions of etr1-1 are expressed at similar levels (30). Unlike ETR1, which is constitutively expressed, three members of the ethylene receptor family (ERS1, ETR2, and ERS2) are induced by ethylene treatment in wild-type seedlings (11, 37). The total level of ETR1 associated with ETR2-TAP and ERS1-TAP increases following ethylene treatment (Fig. 3) and thus, if etr1-1-(1–349) requires interaction with ETR2 and ERS1 to confer ethylene insensitivity, it should be more effective following ethylene treatment of seedlings.
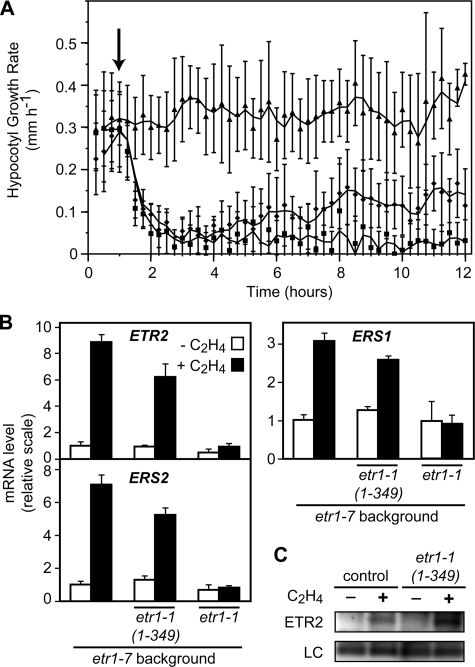
The genetic background used (etr1-7) shows a kinetic response to 10 μl/liter exogenous ethylene similar to that found in wild-type seedlings, displaying a rapid decrease in the hypocotyl growth rate that stabilizes at a new steady-state growth rate by 2 h following the initial ethylene application (Fig. 7A) (11). A transgenic etr1-7 line containing a full-length version of the etr1-1 receptor displays no change in its growth rate upon ethylene treatment, thereby demonstrating the ethylene insensitivity conferred on the seedlings by introduction of this mutant receptor (Fig. 7A). Because the mutant etr1-1 receptor is full-length, it contains its own signal output domain to suppress ethylene responses and so should not be dependent upon other ethylene receptors to exert its effect. The truncated etr1-1-(1–349) transgenic line initially behaves similarly to the background etr1-7 line in response to ethylene, showing a rapid decrease in hypocotyl growth rate that closely matches that of etr1-7 (Fig. 7A). But rather than plateauing at a new steady state growth rate, the growth rate starts to increase again at ∼2 h after ethylene treatment, reaching a new growth rate intermediate between that of the etr1-7 background and the transgenic line containing full-length etr1-1. An etr1-7 line containing the truncated wild-type receptor construct ETR1-(1–349) showed a kinetic response similar to that of etr1-7 (data not shown), demonstrating that the altered kinetic response of etr1-1-(1–349) is not due simply to the introduction of a truncated receptor but that it also requires the C65Y (etr1-1) mutation. Analysis by real-time PCR confirmed that the transcriptionally regulated ethylene receptor genes ERS1 (subfamily 1), as well as ETR2 and ERS2 (subfamily 2) were induced by 4-h ethylene treatment in etr1-7 and the etr1-1-(1–349) transgenic line, but were not induced in the transgenic line containing full-length etr1-1 (Fig. 7B). The change in receptor transcript levels following ethylene treatment is reflected at the protein level based on immunoblot analysis using an anti-ETR2 antibody (Fig. 7C).
FIGURE 7.
Effect of etr1-1 and etr1-1-(1–349) upon the short-term ethylene response in etiolated seedlings. A, kinetic analysis of hypocotyl growth for etr1-7 (squares), etr1-1 in the etr1-7 background (triangles), and etr1-1-(1–349) in the etr1-7 background (diamonds). The arrow indicates the time at which the 10 μl/liter ethylene treatment was initiated. The mean at each time point with S.E. are shown. B, expression of ETR2, ERS2, and ERS1 in the different lines based on real-time PCR analysis following 4 h in the absence or presence of 10 μl/liter ethylene. C, protein levels of ETR2 in the etr1-7 background (control) and the etr1-1-(1–349) transgenic line in the etr1-7 background, following 4 h in the absence or presence of 10 μl/liter ethylene. ETR2 was detected with an anti-ETR2 antibody. The H+-ATPase was detected with an anti-ATPase antibody and served as a loading control (LC).
DISCUSSION
Protein-protein interactions are important for signaling by ethylene receptors. Both genetic and biochemical evidence indicate that ethylene receptors interact with CTR1 to mediate ethylene signal transduction (21–23). According to the current model, when plants grow in air (absence of ethylene), the kinase domain of CTR1 actively represses ethylene responses. In the presence of ethylene, binding of ethylene to the receptors leads to a conformational change in CTR1 that inactivates its kinase domain, thereby relieving the suppression of ethylene responses. The physical association of the ethylene receptors with CTR1 is essential for its regulation as a mutation that abolishes this interaction results in a constitutive ethylene response (21, 23).
Co-purification of ETR1 with other TAP-tagged ethylene receptors demonstrates that receptor-receptor interactions also occur in Arabidopsis. Our results suggest that 60–70% of ETR1 is physically associated with other receptors. This number cannot be considered an exact representation of the levels of receptor-receptor interactions for several reasons. First, the amount of ETR1 found associated with the TAP-tagged receptors is likely to be dependent upon the transgenic expression level of these receptors. Although we expressed the transgenes from their native promoters, we previously observed variability in levels of transgenically expressed ETR1 protein when compared with the native protein expression level (9). Second, our analysis of receptor interactions required solubilization of the receptors from membranes, a process that may disrupt protein-protein interactions and thus result in an underestimation of the actual association levels. Third, our analysis of ethylene effects on receptor associations suggests dynamic changes in the composition of receptor-receptor complexes. Nevertheless our data on ETR1 associations with other receptors, along with the finding that ERS2-Myc can co-purify with ERS1-TAP and EIN4-TAP,4 suggest a substantial level of interaction between the ethylene receptors.
The interaction between ETR1 and ERS2-TAP isolated from plants was disrupted by SDS treatment, indicating that the interaction between ethylene receptors is a noncovalent higher order interaction, and that the ethylene receptors may thus exist as multimeric clusters. The capacity for receptors to form covalent disulfide-linked heterodimers was supported by use of a transgenic yeast system, but such heterodimers do not appear common in plants based on our analysis here, as well as prior analysis of ETR1 truncations in plants in which we only found clear evidence for disulfide-linked homodimers (30). The formation of the disufide-linked heterodimers in yeast could represent an artifact arising from the high levels of receptor expression and points to the necessity of analyzing such receptor interactions under native expression conditions. Our data are consistent with the recent report that Arabidopsis ethylene receptors can form homomeric and heteromeric associations with each other at the ER in transiently transformed tobacco leaf cells (42), and extend this analysis to indicate that in planta such interactions are predominantly higher order.
Ethylene receptors contain several domains that may play roles in maintaining receptor-receptor interactions. We demonstrate here that the GAF domains of the receptors can physically interact. GAF domains are widespread in nature and, although best known for their capacity to bind a diversity of small molecules, have also been found to mediate the dimerization of phosphodiesterase 2A (40, 43). All the ethylene receptors also contain histidine-kinase-like domains, and bacterial histidine-kinase domains have been found to dimerize through coiled-coil interactions (44). In addition, three of the receptors (ETR1, ETR2, EIN4) contain receiver domains and the crystal structure of the ETR1 receiver domain reveals it to form a dimer (45). Whether these interactions are responsible for the formation of homodimers or higher order interactions is currently unknown and it is possible that the same domain could function for both purposes. In addition, the analysis of domain-domain interactions of the ethylene receptors has to date relied upon bacterial and yeast overexpression systems, and it remains to be determined how well these represent the associations found in the plant.
Higher order interactions between ethylene receptors suggest a mechanism as to how Arabidopsis can display such a high sensitivity for ethylene. When transgenically expressed in yeast, ETR1 binds ethylene with a Kd of 2.4 nm (6). The other ethylene receptors appear to bind ethylene with similar affinity to ETR1 (46). Interestingly, Arabidopsis responds to ethylene levels as low as 0.2 nl/liter (0.09 nm), which is 300-fold lower than the Kd value (2). The higher order interaction between ethylene receptors would allow for the ethylene signal to be amplified by lateral receptor interactions. By transmitting the ligand-bound conformation to proximate ligand-free receptors, one ethylene molecule can modulate signaling by several ethylene receptors. This signal amplification mechanism has been demonstrated in bacterial chemotaxis signaling. The bacterial chemoreceptors form higher order clusters through direct physical interactions, and respond to ligand binding in a coordinated way (25). Such cooperative behavior can be considered in terms of a model in which the receptor cluster exists in equilibrium between T (tense) and R (relaxed) states, with ligand binding to a subset of receptors shifting the equilibrium of the entire cluster toward the R state.
The interactions between ethylene receptors may fine tune the signal output from the receptors. Although all receptors appear to have similar ethylene binding abilities, several lines of evidence indicate that different receptors may have different signal output abilities. First, their C-terminal signal output domains are different. Second, subfamily 1 ethylene receptors of Arabidopsis have a greater level of signal output ability than subfamily 2 receptors (28, 29, 47), potentially due to differing affinities for the Raf-like kinase CTR1. Thus, interactions between different receptors would provide multiple signal output combinations that allow plants to alter the degree to which they respond to ethylene binding.
Interactions between receptors can also explain the dominance exhibited by ethylene-insensitive mutations in the receptors such as the etr1-1 mutation (17). The etr1-1 mutant receptor appears locked into a functional conformation that constitutively activates CTR1 to suppress ethylene responses, conferring an ethylene-insensitive phenotype. etr1-1 is a dominant mutation, suggesting the etr1-1 receptor produces sufficient signal output to suppress ethylene responses. However, the native ETR1 does not appear able to produce enough signal output to suppress ethylene responses by itself, based on the finding that a triple receptor knock-out mutant (etr2/ers2/ein4) that only has ETR1 and ERS1 shows a constitutive ethylene response phenotype (19). The higher order interaction model can explain the effectiveness of the etr1-1 receptor in suppressing the ethylene responses. According to this model, etr1-1 is locked in its signal output conformation and is unresponsive to ethylene binding by neighboring receptors. The dominant etr1-1 receptor may instead amplify its signal output by conferring upon other receptors a similar conformation (i.e. shift equilibrium of the receptor cluster toward the T-state). Our kinetic analysis of etr1-1-(1–349) seedlings is consistent with this model, as the mutant line became progressively less sensitive to ethylene following an ethylene treatment that induced transcription of additional receptors. Also consistent with this model is the previous finding that etr1-1-(1–349) is less effective at conferring ethylene insensitivity in genetic backgrounds null for other members of the receptor family, in particular subfamily 1 receptors such as ERS1 (41). The etr1-1-(1–349) receptor, although lacking histidine kinase and receiver domains, could be capable of interacting with other receptors by its GAF domain, which we show here is sufficient for mediating receptor-receptor interactions. This result suggests that direct GAF-GAF interactions may be important for mediating signaling between receptors.
Previous genetic analyses indicate that synergistic interactions exist between subfamily 1 receptors and subfamily 2 receptors. A double loss-of-function mutant (etr1/ers1) which lacks subfamily 1 ethylene receptors shows a severe constitutive ethylene response phenotype, and transforming genes of subfamily 2 receptors cannot rescue the phenotype (28, 29, 47). These data suggest that subfamily 1 receptors are absolutely needed to suppress ethylene responses in Arabidopsis, and their functions cannot be replaced by subfamily 2 receptors. However, a triple knock-out mutant etr2/ers2/ein4 that lacks the subfamily 2 receptors also shows a constitutive ethylene response phenotype (19, 29), indicating that subfamily 1 receptors are unable to efficiently suppress ethylene signal transduction by themselves. These two genetic observations suggest that subfamily 1 members and subfamily 2 members may function synergistically to suppress ethylene responses. In wild-type plants, subfamily 2 receptors may recruit CTR1 (23), but subfamily 1 receptors may help subfamily 2 receptors activate their associated CTR1 by receptor-receptor interactions, thus allowing enough signal output to suppress ethylene responses.
The finding that the subfamily 1 receptor ETR1 interacts with subfamily 2 receptors suggests that signal transduction between ethylene receptors of subfamilies 1 and 2 may occur in part through a two-component phosphorelay. Several reports indicate that such a phosphorelay, although not required for signaling, may modulate the ethylene response (11, 12, 28, 48). A significant difference between subfamily 1 and subfamily 2 members is that subfamily 1 receptors have a functional histidine kinase domain, whereas subfamily 2 receptors have Ser/Thr kinase activity (4, 14). The histidine kinase domain is a classic component in a two-component signal transduction system (49). In this system, signals are mediated through a His-Asp phosphorelay between proteins containing a histidine kinase domain and proteins containing a receiver domain in which a conserved Asp amino acid can be phosphorylated by the His-Asp phosphorelay. In Arabidopsis, the subfamily 1 receptors ETR1 and ERS1 each have functional histidine kinase domains, and ETR1, ETR2, and EIN4 each have receiver domains. Upon autophosphorylation on histidine, the subfamily 1 receptors may transfer the phosphate to the aspartate within the receiver domains of the subfamily 2 receptors ETR2 and EIN4, both of which we found capable of interacting with ETR1.
Acknowledgments
We thank B. Séraphin for the vector containing the TAP tag, P. Krishna for the ERS2 cDNA clone, and the ABRC for additional DNA clones. We thank R. Boston for the BiP antibody.
This work was supported, in whole or in part, by National Institutes of Health Grant 1R01GM071855 (to C. C.). This work was also supported by Department of Energy Grants DE-FG02-05ER15704 (to G. E. S.) and DE-FG02-99ER20329 (to C. C.), National Science Foundation Grant MCB-0430191 (to G. E. S.), and the University of Maryland Agricultural Experiment Station. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: TAP, tandem affinity purification; GST, glutathione S-transferase; DTT, dithiothreitol; SBP, streptavidin-binding peptide; X-gal, 5-bromo-4-chloro-3-indolyl-β-d -galactopyranoside; ACC, 1-aminocyclopropane-1-carboxylic acid; Wt, wild type.
Z. Gao and G. E. Schaller, unpublished data.
References
- 1.Abeles, F. B., Morgan, P. W., and Saltveit, M. E., Jr. (1992) Ethylene in Plant Biology, 2nd Ed., Academic Press, San Diego
- 2.Binder, B. M., Mortimore, L. A., Stepanova, A. N., Ecker, J. R., and Bleecker, A. B. (2004) Plant Physiol. 136 2921–2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, Q. G., and Bleecker, A. B. (1995) Plant Physiol. 108 597–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schaller, G. E., and Kieber, J. J. (2002) The Arabidopsis Book, 1–18 [DOI] [PMC free article] [PubMed]
- 5.Chen, Y. F., Etheridge, N., and Schaller, G. E. (2005) Ann. Bot. (Lond) 95 901–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schaller, G. E., and Bleecker, A. B. (1995) Science 270 1809–1811 [DOI] [PubMed] [Google Scholar]
- 7.Rodriguez, F. I., Esch, J. J., Hall, A. E., Binder, B. M., Schaller, G. E., and Bleecker, A. B. (1999) Science 283 996–998 [DOI] [PubMed] [Google Scholar]
- 8.Hall, A. E., Chen, Q. G., Findell, J. L., Schaller, G. E., and Bleecker, A. B. (1999) Plant Physiol. 121 291–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, Y. F., Randlett, M. D., Findell, J. L., and Schaller, G. E. (2002) J. Biol. Chem. 277 19861–19866 [DOI] [PubMed] [Google Scholar]
- 10.Dong, C.-H., Rivarola, M., Resnick, J. S., Maggin, B. D., and Chang, C. (2008) Plant J. 53 275–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Binder, B. M., O'Malley, R. C., Wang, W., Moore, J. M., Parks, B. M., Spalding, E. P., and Bleecker, A. B. (2004) Plant Physiol. 136 2913–2920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qu, X., and Schaller, G. E. (2004) Plant Physiol. 136 2961–2970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gamble, R. L., Coonfield, M. L., and Schaller, G. E. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 7825–7829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moussatche, P., and Klee, H. J. (2004) J. Biol. Chem. 279 48734–48741 [DOI] [PubMed] [Google Scholar]
- 15.Schaller, G. E., Ladd, A. N., Lanahan, M. B., Spanbauer, J. M., and Bleecker, A. B. (1995) J. Biol. Chem. 270 12526–12530 [DOI] [PubMed] [Google Scholar]
- 16.Hall, A. E., Findell, J. L., Schaller, G. E., Sisler, E. C., and Bleecker, A. B. (2000) Plant Physiol. 123 1449–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bleecker, A. B., Estelle, M. A., Somerville, C., and Kende, H. (1988) Science 241 1086–1089 [DOI] [PubMed] [Google Scholar]
- 18.Chang, C., Kwok, S. F., Bleecker, A. B., and Meyerowitz, E. M. (1993) Science 262 539–544 [DOI] [PubMed] [Google Scholar]
- 19.Hua, J., and Meyerowitz, E. M. (1998) Cell 94 261–271 [DOI] [PubMed] [Google Scholar]
- 20.Kieber, J. J., Rothenberg, M., Roman, G., Feldman, K. A., and Ecker, J. R. (1993) Cell 72 427–441 [DOI] [PubMed] [Google Scholar]
- 21.Huang, Y., Li, H., Hutchison, C. E., Laskey, J., and Kieber, J. J. (2003) Plant J. 33 221–233 [DOI] [PubMed] [Google Scholar]
- 22.Clark, K. L., Larsen, P. B., Wang, X., and Chang, C. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 5401–5406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao, Z., Chen, Y. F., Randlett, M. D., Zhao, X. C., Findell, J. L., Kieber, J. J., and Schaller, G. E. (2003) J. Biol. Chem. 278 34725–34732 [DOI] [PubMed] [Google Scholar]
- 24.Cancel, J. D., and Larsen, P. B. (2002) Plant Physiol. 129 1557–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baker, M. D., Wolanin, P. M., and Stock, J. B. (2006) Bioessays 28 9–22 [DOI] [PubMed] [Google Scholar]
- 26.Keefe, A. D., Wilson, D. S., Seelig, B., and Szostak, J. W. (2001) Protein Expr. Purif. 23 440–446 [DOI] [PubMed] [Google Scholar]
- 27.Clough, S. J., and Bent, A. F. (1998) Plant J. 16 735–743 [DOI] [PubMed] [Google Scholar]
- 28.Wang, W., Hall, A. E., O'Malley, R., and Bleecker, A. B. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 352–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao, X. C., Qu, X., Mathews, D. E., and Schaller, G. E. (2002) Plant Physiol. 130 1983–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gamble, R. L., Qu, X., and Schaller, G. E. (2002) Plant Physiol. 128 1428–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rigaut, G., Shevchenko, A., Rutz, B., Wilm, M., Mann, M., and Séraphin, B. (1999) Nat. Biotech. 17 1030–1032 [DOI] [PubMed] [Google Scholar]
- 32.Mitchell, D. A., Marshall, T. K., and Deschenes, R. J. (1993) Yeast 9 715–723 [DOI] [PubMed] [Google Scholar]
- 33.Schiestl, R. H., and Gietz, R. D. (1989) Curr. Genet. 16 339–346 [DOI] [PubMed] [Google Scholar]
- 34.Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A., and Strohl, K. (eds) (1994) Current Protocols in Molecular Biology, John Wiley & Sons, Inc., New York
- 35.Chen, Y.-F., Shakeel, S. N., Bowers, J., Zhao, X.-C., Etheridge, N., and Schaller, G. E. (2007) J. Biol. Chem. 282 24752–24758 [DOI] [PubMed] [Google Scholar]
- 36.Höfte, H., and Chrispeels, M. J. (1992) Plant Cell 4 995–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hua, J., Sakai, H., Nourizadeh, S., Chen, Q. G., Bleecker, A. B., Ecker, J. R., and Meyerowitz, E. M. (1998) Plant Cell 10 1321–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang, S. F., and Hoffman, N. E. (1984) Annu. Rev. Plant Physiol. 35 155–189 [Google Scholar]
- 39.Takahashi, H., Kobayashi, T., Sato-Nara, K., Tomita, K. O., and Ezura, H. (2002) J. Exp. Bot. 53 415–422 [DOI] [PubMed] [Google Scholar]
- 40.Aravind, L., and Ponting, C. P. (1997) Trends Biochem. Sci. 22 458–459 [DOI] [PubMed] [Google Scholar]
- 41.Xie, F., Liu, Q., and Wen, C.-K. (2006) Plant Physiol. 142 492–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grefen, C., Städele, K., Ružička, K., Obrdlik, P., Harter, K., and Horák, J. (2008) Mol. Plant 1 308–320 [DOI] [PubMed] [Google Scholar]
- 43.Martinez, S. E., Wu, A. Y., Glavas, N. A., Tang, X.-B., Turley, S., Hol, W. G. J., and Beavo, J. A. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 13260–13265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wolanin, P. M., Thomason, P. A., and Stock, J. B. (2002) Genome Biol. 3 Reviews 3013.1–3013.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muller-Dieckmann, H. J., Grantz, A. A., and Kim, S. H. (1999) Structure Fold. Des. 7 1547–1556 [DOI] [PubMed] [Google Scholar]
- 46.O'Malley, R. C., Rodriguez, F. I., Esch, J. J., Binder, B. M., O'Donnell, P., Klee, H. J., and Bleecker, A. B. (2005) Plant J. 41 651–659 [DOI] [PubMed] [Google Scholar]
- 47.Qu, X., Hall, B. P., Gao, Z., and Schaller, G. E. (2007) BMC Plant Biol. 7 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cho, Y. H., and Yoo, S. D. (2007) Plant Physiol. 143 612–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schaller, G. E. (2000) Adv. Bot. Res. 32 109–148 [Google Scholar]