Abstract
Macrophage migration inhibitory factor (MIF) is a pleiotropic cytokine that has been implicated in the pathogenesis of inflammatory disorders such as infection, sepsis, and autoimmune disease. MIF exists preformed in cytoplasmic pools and exhibits an intrinsic tautomerase and oxidoreductase activity. MIF levels are elevated in the serum of animals and patients with infection or different inflammatory disorders. To elucidate how MIF actions are controlled, we searched for endogenous MIF-interacting proteins with the potential to interfere with key MIF functions. Using in vivo biotin-tagging and endogenous co-immunoprecipitation, the ribosomal protein S19 (RPS19) was identified as a novel MIF binding partner. Surface plasmon resonance and pulldown experiments with wild type and mutant MIF revealed a direct physical interaction of the two proteins (KD = 1.3 × 10-6 m). As RPS19 is released in inflammatory lesions by apoptotic cells, we explored whether it affects MIF function and inhibits its binding to receptors CD74 and CXCR2. Low doses of RPS19 were found to strongly inhibit MIF-CD74 interaction. Furthermore, RPS19 significantly compromised CXCR2-dependent MIF-triggered adhesion of monocytes to endothelial cells under flow conditions. We, therefore, propose that RPS19 acts as an extracellular negative regulator of MIF.
A large body of evidence now shows that macrophage migration inhibitory factor (MIF)2 activates a range of intracellular pathways and plays a key role in host immune and inflammatory responses (1, 2). Certain of the MIF inflammatory functions also have been proposed to be the result of the unusual enzymatic properties of the protein, namely tautomerase and oxidoreductase activities (3-6). Inhibition or deletion of MIF attenuates disease progression in experimental models such as atherosclerosis, arthritis, glomerulonephritis, sepsis, autoimmune encephalitis, and autoimmune diabetes (7-13). A pivotal step in the inflammatory response is the chemokine-governed adherence of monocytes to the endothelial lining which is then followed by their egress from the vasculature at the affected site. Earlier data from MIF-/- mice illustrate a role of MIF in leukocyte recruitment that was recently substantiated by the finding that MIF serves as a chemoattractant for monocytes and T cells by directly binding to the chemokine receptors CXCR2 and CXCR4 (14, 15). On the cell surface MIF also associates with CD74 (invariant chain of major histocompatibility complex class II) which colocalizes with CXCR2 (14, 16). Interaction with different surface molecules is thought to partly explain the wide impact of MIF on cellular pathways.
Despite its role as a key mediator in immune and inflammatory diseases, very little is known of how MIF action is regulated and terminated. Accordingly, we searched for endogenous molecules with the ability to control key steps of MIF signaling (i.e. receptor binding and/or receptor-associated functions). In this study, we identified ribosomal protein S19 (RPS19), a component of the small ribosomal subunit that is also released by apoptotic cells (17), as such a candidate.
EXPERIMENTAL PROCEDURES
Identification of MIF-interacting Partners by Coimmunoprecipitation—Mouse NIH 3T3 fibroblasts were grown in Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated fetal calf serum (PAA Laboratories, Cölbe, Germany) and antibiotics. Cells were lysed in Nonidet P-40 (1% IGEPAL CA-630) buffer containing protease inhibitors. Lysed cells were disrupted by passage through a 21-gauge needle and subjected to sonication by two 10-s bursts at 200-300 watts separated by a 10-s cooling period. After centrifugation at 12,000 × g at 4 °C for 10 min, the supernatant was precleared with 30 μl of protein G-Sepharose 4B Fast Flow beads (GE Healthcare) on a rotating wheel at 4 °C for 1 h before incubation with either rabbit anti-rat MIF antibody or preimmune serum immobilized on 30 μl of protein G-Sepharose commenced at 4 °C for 2 h. After 5 washes with lysis buffer for 10 min each, beads were resuspended in Laemmli sample buffer and boiled for 10 min. Immunoprecipitates were separated on a NuPAGE 4-12% Novex Bis-Tris gel (Invitrogen) and stained with colloidal Coomassie staining solution (Sigma).
Identification of MIF-interacting Partners by in Vivo Biotinylation of Tagged MIF—The method of de Boer et al. (18) was employed using the modified tagging construct pN3-CTB developed by Rischitor (19). The rat MIF cDNA insert was produced by standard PCR using the upstream primer CGAATTCCGCCACCATGCCTATGTTCATCGTG (EcoRI site in bold) and the downstream primer GATGTCGACAGCGAAGGTGGAACCGTTCCA (SalI site in bold) and a pGEX-4T-2-MIF full-length expression construct (48) as template. pN3-CTB-MIF was constructed by cloning the EcoRI/SalI-restricted PCR fragment into pN3-CTB. For stable transfection, pBudCE4.1-birA (18) and pN3-CTB-MIF were linearized with XhoI and Eco 01091I, respectively, and transfected together or individually (pBudCE4.1-birA) into NIH 3T3 cells using Lipofectamine (Invitrogen). Antibiotic selection started with 800 μg/ml Geneticin (for pN3-CTB-MIF) and 600 μg/ml zeocin (for pBudCE4.1-birA) and was successively reduced in three steps down to 100 μg/ml Geneticin and 150 μg/ml zeocin after 4 weeks. After 5-8 weeks clones were isolated using small sterile filter discs soaked in trypsin solution. Clones were examined for expression of a biotinylated 22-kDa MIF fusion protein using immunoblots with rabbit anti-rat-MIF antiserum and streptavidin horseradish peroxidase conjugate (Dako, Hamburg, Germany).
One of the clones that strongly expressed the MIF fusion protein and one control clone expressing birA only were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, biotin (0.1 mg/liter), Geneticin (100 μg/ml), and zeocin (150 μg/ml) at 37 °C. Cells were lysed in 1% IGEPAL CA-630 (v/v), 150 mm NaCl, 50 mm Tris-HCl, pH 8, and proteinase inhibitors and applied to streptavidin-agarose beads (Novagen; 200 μl of bead slurry per mg of cell extract). After 1 h on a rocking platform, beads were washed 3 times with lysis buffer. Bound proteins were eluted by boiling for 10 min in SDS sample buffer, separated by SDS-PAGE on a 4-12% NuPAGE Bis-Tris gel (Invitrogen), and stained with colloidal Coomassie Blue (Sigma). Both lanes were cut into 12 gel slices, and proteins in all slices were digested with trypsin (20). Extracted peptides were separated and sequenced by liquid chromatography-coupled electrospray ionization-tandem mass spectroscopy on a quadrupole-time of flight (TOF) instrument (Q-TOF Ultima, Waters) under standard conditions. Proteins were identified by comparing peptide fragment spectra against all entries in the NCBI nr data base using MASCOT as search engine.
Identification of RPS19 by Matrix-assisted Laser Desorption/Ionization (MALDI)-Mass Spectroscopy Fingerprint Analysis and Peptide Matching—A 16-kDa band identified by co-immunoprecipitation was excised from the gel. The gel piece was washed once with water and twice with 50 mm ammonium hydrogen carbonate:acetonitrile (1:1) and acetonitrile, alternately. Gel pieces were re-swollen in a minimal volume of a 10 ng/μl trypsin solution (sequencing grade, Roche Diagnostics) in 25 mm ammonium hydrogen carbonate and incubated for 16 h at 37 °C. Peptides were extracted with 5 μl of 1% (v/v) trifluoroacetic acid containing 5 mm octylglycoside. 2 μl of the solution were applied to a thin layer of α-cyano-4-hydroxycinnamic acid on an AnchorChip target (Bruker Daltonik, Bremen, Germany). After 10 min the supernatant was removed, and the spot was washed twice with 2 μl of 0.1% (v/v) trifluoroacetic acid. Mass fingerprints of tryptic digests were obtained by MALDI-TOF mass spectrometry using an Ultraflex™ TOF/TOF mass spectrometer (Bruker Daltonik). The identified protein was verified by analyzing selected peptides in LIFT mode (tandem mass spectroscopy), and fragment masses were also submitted to MASCOT.
Expression and Purification of Wild Type and Mutant MIF Proteins—Recombinant rat MIF that differs from mouse MIF by only one amino acid (rat MIF, Ser-54; mouse MIF, Asn-54) was expressed and purified as previously described (21). Possible endotoxin contamination was removed using Detoxi-Gel (Pierce) which was also applied to the wild type human MIF preparation (see below). Human MIF and MIF mutant proteins were obtained from 1 liter of LB cultures of Escherichia coli BL21(DE3) transformed with constructs for wild type (wt) MIF and mutants P2A MIF (N-terminal amino acid alanine 2 exchanged for proline), Δ4 MIF (N-terminal four amino acids deleted), and C60S MIF (amino acid cysteine 60 replaced by serine) (22). Expression was induced with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside for 3 h. Cells were harvested, and a lysate was prepared exactly as described (23). Cell lysates were sonicated on ice by five 10-s bursts using a microtip (Sonoplus, Bandelin, Berlin, Germany) and centrifuged at 17,000 × g at 4 °C for 30 min. wtMIF and P2A MIF and Δ4 MIF mutants were purified from the soluble fraction of the lysate, whereas the C60S MIF mutant was purified via an inclusion body preparation. The Δ4 MIF mutant was present in comparable portions in soluble and inclusion body fraction alike. Soluble proteins were precipitated with 70% ammonium sulfate (saturation), taken up in PBS, and chromatographed on a Sephacryl S100 HiPrep 16/60 gel filtration column (GE Healthcare) using an ÄKTAbasic UPC10 HPLC system (GE Healthcare). Positive fractions were pooled and passed over directly coupled 1-ml Resource S/Mono Q 5/50 columns (GE Healthcare). In PBS wtMIF, P2A MIF and Δ4 MIF proteins are in the flow-through with the majority of impurities binding to the column.
For the C60S MIF mutant the pellet of the initial lysate was resuspended in 25 ml of 50 mm Tris-HCl, pH 8.0, 10 mm EDTA, 100 mm NaCl, 0.5% (v/v) Triton X-100, and washed 4 times in the same buffer. The washed inclusion bodies were denatured in 6 m guanidine hydrochloride, 100 mm dithiothreitol in PBS. The solubilized protein was dialyzed against 5 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride in PBS, cleared by centrifugation, and passed over directly coupled Resource S/Mono Q columns as described above. All proteins had a purity of more than 98% as assessed by SDS-PAGE.
The identity of all purified proteins was confirmed by MALDI-TOF mass spectrometry as described above. Human wtMIF, P2A MIF, and C60S MIF had the N-terminal methionine removed due to a second cleavable residue as predicted (24), whereas the Δ4 MIF mutant had retained this methionine. Because of the internal mutation, the C60S mutant could not be confirmed.
Expression and Purification of GST-RPS19 and RPS19-His—Mouse RPS19 cDNA clone IRAKp961E1430Q was obtained from ImaGenes (Berlin, Germany). The RPS19 cDNA was amplified by PCR with Pfu polymerase (Promega) using forward primer 5′-CGAGGAATTCCCATGCCCGGAGTTACTG-3′ and reverse primer 5′-CGCCTCGAGTAATGCTTCTTGTTGGC-3′ for the glutathione S-transferase (GST) tag vector and forward primer 5′-CGCCATATGCCCGGAGTTACTGTAAAA-3′ and reverse primer 5′-GCGAAGCTTATGCTTCTTGTTGGCAGC-3′ for the His tag vector (introduced restriction sites are underlined). EcoRI- and XhoI-restricted PCR fragments were ligated into pGEX-4T-2 (GE Healthcare) yielding pGST-RPS19. Because of an internal NdeI site within the RPS19 cDNA, pET21a(+) (Merck) was restricted with NdeI, blunted, restricted with HindIII, and ligated to the HindIII-restricted PCR fragment. Both inserts plus flanking regions were validated by DNA sequencing (Seqlab, Göttingen, Germany). GST-RPS19 and RPS19-His were expressed in E. coli BL21(DE3) by induction with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside at 37 °C for 3 h. For the GST-tagged RPS19 protein, cells were lysed in PBS by sonication and treated with 1% Triton X-100 for 30 min. After centrifugation at 12,000 × g for 15 min, the supernatant containing GST-RPS19 was subjected to glutathione-Sepharose 4B (GE Healthcare) chromatography. The purity of the eluted protein was higher than 95% as estimated by SDS-PAGE.
For the His-tagged protein, bacterial cells were lysed with lysozyme followed by sonication, and the native protein was purified from the lysate by standard Ni-NTA chromatography. Bound protein was eluted with 50 mm NaH2PO4, 300 mm NaCl, 250 mm imidazole, pH 8.0, and dialyzed against PBS, pH 7.8, containing 0.5 mm phenylmethylsulfonyl fluoride and 1 mm dithiothreitol.
Surface Plasmon Resonance—Biosensor analyses were performed on a BIACORE X system (GE Healthcare). Recombinant MIF dissolved in 0.01 m sodium acetate, pH 4.5, was covalently attached to a CM5 sensor chip (GE Healthcare) by the amine coupling method according to the manufacturer's instructions. Final levels of immobilization were ∼5000 response units. 1000 response units correspond to 10 μg/μl on a CM5 chip. Analyses were performed at 25 °C using 0.01 m HEPES, pH 7.4, 0.15 m NaCl, and 0.005% surfactant P20 as a driving buffer at a flow rate of 20 μl/min. All experiments were carried out at 25 °C at a constant flow rate of 20 μl/min HBS-EP buffer. 40 μl of the analyte (RPS19) diluted in HBS-EP buffer (GE Healthcare) were injected over the immobilized MIF followed by a 2-min period when buffer was passed over the surface. Five concentrations of RPS19 were passed over the chip (62.5, 125, 250, 500, and 1000 nm).
Preparation of Antibodies against RPS19 and MIF and Double Immunofluorescence—Antibodies against His-tagged RPS19 or wild type rat MIF were raised in New Zealand White rabbits. In the case of anti-RPS19, serum samples were affinity-purified using His-tagged RPS19 immobilized on Ni-NTA-agarose (25). The purified RPS19 immunoglobulins were dialyzed against water for 1 h and against PBS overnight at 4 °C. A second chicken anti-RPS19 antibody was obtained from Lydie Da Costa (26). The MIF antibody is available through Invitrogen (#36-7401).
Cells were cultured on coverslips and fixed with ice-cold methanol for 10 min. Blocking for 1 h in 5% bovine serum albumin (w/v) and 5% (v/v) normal horse serum was followed by incubation with rabbit anti-mouse RPS19 (1:200) decorated with donkey anti-rabbit IgG conjugated with Cy3 (1:1000) and mouse anti-MIF (1:200, clone 3D9, available by NIH resource sharing) detected by donkey anti-mouse IgG conjugated with fluorescein isothiocyanate (1:1000). Primary antibodies were applied overnight at 4 °C, and both secondary antibodies were incubated for 1 h at room temperature. 4,6-Diamidino-2-phenylindole was used for nuclear staining. Images were acquired with a confocal laser scanning microscope (Leica TCS SP2).
Pulldown Assays—Biotinylation of rat MIF was performed using the ECL protein biotinylation module (GE Healthcare) according to the recommendations of the manufacturer. Non-reacted succinimide ester was separated from biotinylated MIF using Sephadex G-25 columns. Biotinylated MIF was eluted with PBS, pH 7.5, and 2.5 μg were immobilized on 30 μl (50% slurry) of monomeric avidin beads (Pierce) by incubation in 500 μl of PBS at room temperature on a rotating wheel for 30 min. To completely remove free biotinylated MIF, beads were washed with PBS and then incubated in 500 μl of lysis buffer (50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1% IGEPAL CA-630) with increasing amounts of GST-RPS19 (50, 100, and 200 ng) on a rotating wheel at 4 °C for 1 h. As a control, uncoated avidin beads were incubated with the same amounts of GST-RPS19 alone. Beads were then washed 5 times with lysis buffer and finally boiled in Laemmli sample buffer for 5 min. Protein complexes were separated by SDS-PAGE, transferred onto nitrocellulose membrane, and detected with anti-GST antibody conjugated with peroxidase.
Similarly, RPS19-His (2 μg) was immobilized on Ni-NTA-agarose beads, washed with PBS, and incubated for 1 h with different amounts of recombinant rat MIF or 2 μg of human wild type MIF or P2A MIF, C60S MIF, and Δ4 MIF proteins. As a control, MIF proteins alone were incubated with Ni-NTA-agarose beads. After extensive washing with lysis buffer as described previously or radioimmune precipitation assay buffer (150 mm NaCl, 1% IGEPAL CA-630, 0.5% sodium deoxycholate, 0.1% SDS, 50 mm Tris-HCl, pH 8.0, 1 mm phenylmethylsulfonyl fluoride) in the case of human MIF and MIF mutants, proteins bound to the beads were boiled in Laemmli sample buffer, resolved by SDS-PAGE, and stained with colloidal Coomassie (Sigma) or blotted and probed for RPS19, stripped, and reprobed for MIF.
l-Dopachrome Methyl Ester Tautomerase Assay—The substrate l-dopachrome methyl ester was freshly prepared before each measurement as described but without HPLC purification (27). Enzymatic activity was determined in an 800-μl assay reaction obtained by mixing 400 μl of PBS containing recombinant rat wtMIF at a concentration of 1 μm with 400 μl of crude l-dopachrome methyl ester substrate. In reactions that contained MIF and RPS19, both proteins were preincubated in 400 μl of PBS for 1 h before measurement. As control, SCGB 2A1-His (28), a protein of similar size and with the same tag, was used. Enzyme activity was measured by monitoring the reaction kinetics at 475 nm in an Ultrospec 2100 pro spectrophotometer (GE Healthcare). These conditions resulted in fast and non-linear kinetics leading to a quantitative turnover of the substrate over a time period of 1 min after reaction start. Therefore, the decrease in absorbance from 0 to 4 s, i.e. the initial reaction rate, was calculated and defined as tautomerase activity. Unpaired t tests were performed to compare the reaction rates +/- RPS19. Differences with a value of p < 0.05 were considered statistically different.
Flow Chamber Adhesion Assay—Human aortic endothelial cells (HAoEC; PromoCell, Heidelberg, Germany) were maintained in PromoCell medium and used at passages 3-5. Mono-Mac6 cells (a gift of Prof. H. W. L. Ziegler-Heitbrock, University of Leicester) were cultured in RPMI 1640 medium supplemented with 10% fetal calf serum, 2 mm l-glutamine, 1% non-essential amino acids, 1 mm sodium pyruvate, and 10 μg/ml human insulin as described (29). The laminar flow assays were performed as described previously (14). Briefly, HAoEC were grown to confluence in 35-mm dishes and preincubated with MIF (50 ng/ml) and RPS19-His (6 μg/ml) or control buffer for 2 h at 37 °C, 5% CO2. The dishes were assembled at the bottom of a parallel wall flow chamber and mounted on the stage of an Olympus IX71 inverted microscope with 20× and 40× phase contrast objectives. MonoMac6 cells (1 × 106/ml) labeled with calcein-AM were pretreated with a blocking antibody against CXCR2 (R&D Systems, Minneapolis, MN) or matching isotype control IgG (3 μg/ml) and resuspended in assay buffer (1 × Hanks' balanced salt solution, 10 mm Hepes, pH 7.4, 0.5% bovine serum albumin). The cell suspension was supplemented at 37 °C with 1 mm Ca2+/Mg2+ shortly before perfusing 5 × 105 cells/ml into the flow chamber at a shear rate of 1.5 dyn/cm2 for 2 min. The number of adherent monocytes was analyzed in multiple high-power fields using the cell M software (Olympus, Tokyo, Japan). Data are expressed as the means ± S.E. Student's t tests (two-sided, unpaired) were performed to compare experimental groups. Differences with a value of p < 0.05 were considered statistically different.
MIF-RPS19 Capture Assay—The assay was performed as published (30). Briefly, 96-well plates were coated with 60 μl/well of 26 ng/μl purified soluble CD74 (amino acids 73-232) at 4 °C overnight. After washing 4 times with 250 μl/well Tris-buffered saline, the plates were incubated with 100 μl/well Superblock (Pierce) at 4 °C overnight. Various concentrations of RPS19 were preincubated with 2 ng/μl biotin-MIF (biotin labeling kit from Roche Applied Science) for 1 h at room temperature in the dark. The Superblock was removed and replaced with 120 μl/well of the RPS19 protein/biotin-MIF pre-incubated mixture, and incubation was continued in the dark at 4 °C for overnight. After washing the plate 4 times, 60 μl/well of streptavidin-alkaline phosphatase (R&D) was added for 1 h at room temperature in the dark followed by washing of the plate before adding 60 μl/well of PNPP (Sigma) and allowing color to develop in the dark at room temperature and reading at 405 nm. For control, human MIF was denatured by incubation at 100 °C for 5 min.
RESULTS
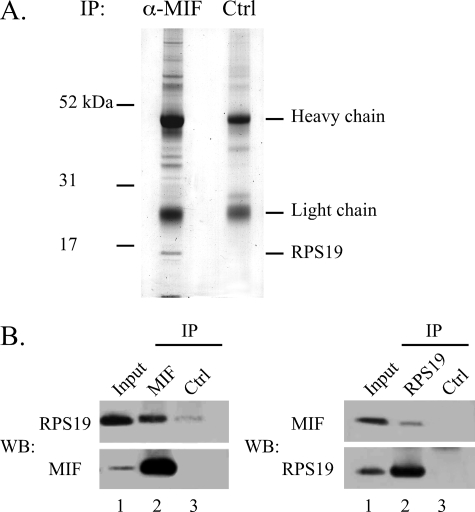
Identification of RPS19 as a MIF-interacting Protein—A far Western analysis was performed under reducing conditions to evaluate possible sources for the identification of MIF-interacting proteins. As each cell type (PC12, NIH 3T3) or tissue extract (rat and mouse testis) examined exhibited a similar pattern of reactive bands (data not shown), co-immunoprecipitation (co-IP) experiments were performed with endogenous lysates of NIH 3T3 fibroblasts. In co-IPs with a polyclonal MIF antibody a number of putative MIF-interacting proteins were co-precipitated. A prominent band at 16 kDa was readily identified in the anti-MIF precipitate but not in a reaction with an isotype control antibody (Fig. 1A). This 16-kDa protein was analyzed by tryptic in-gel digestion followed by MALDI-TOF mass spectrometry and identified as ribosomal protein S19 (RPS19). Confirmation of this finding was provided when the same IP samples were blotted and probed with an RPS19 antibody, stripped, and then re-probed with an antibody against MIF (Fig. 1B). Both proteins were detected in the anti-MIF co-IP sample, whereas the control-IP was negative.
FIGURE 1.
Co-immunoprecipitation of MIF and RPS19. A, extracts from mouse NIH 3T3 fibroblasts were incubated with a polyclonal rabbit anti-rat MIF antibody or with a rabbit IgG polyclonal isotype control antibody before immobilization on protein G-Sepharose beads. Co-immunoprecipitated proteins were separated on a NuPAGE 4-12% Novex Bis-Tris gel and stained with Coomassie. The MIF band is visible at the very bottom of the gel (left lane). The band at ∼16 kDa was excised from the gel and analyzed by tryptic digestion and MALDI-TOF mass spectrometry. B, lysates from NIH 3T3 cells were immunoprecipitated (IP) with anti-rat MIF antibody (MIF, left panel) and anti RPS19 antibody (RPS19, right panel) in comparison to an isotype control antibody (Ctrl). Separated immunoprecipitates were immunoblotted (WB) and probed for RPS19 and, after stripping the membrane, for MIF (left panel) or vice versa (right panel).
Further verification was obtained when RPS19 could also be identified as a MIF-interacting protein in a different screen utilizing a tagged MIF fusion protein that was expressed in NIH 3T3 cells. The C-terminal tag was a peptide that is recognized and biotinylated in vivo by the bacterial birA biotin ligase that was stably co-expressed in the same NIH 3T3 clone (18). Biotinylated MIF and associated MIF-interacting proteins were purified in a single step by binding to streptavidin-agarose beads and separated by SDS-PAGE. A stable NIH 3T3 clone that expresses birA ligase only was used as control. Both SDS-PAGE lanes were cut into 12 slices, and proteins within each slice were identified by mass spectrometry. RPS19 was identified among the proteins that were purified from cells expressing biotinylated tagged MIF but not from cells expressing the biotin ligase only. The finding that two independent assays identify RPS19 as an MIF-interacting protein provides robust evidence for a bona fide interaction of both proteins.
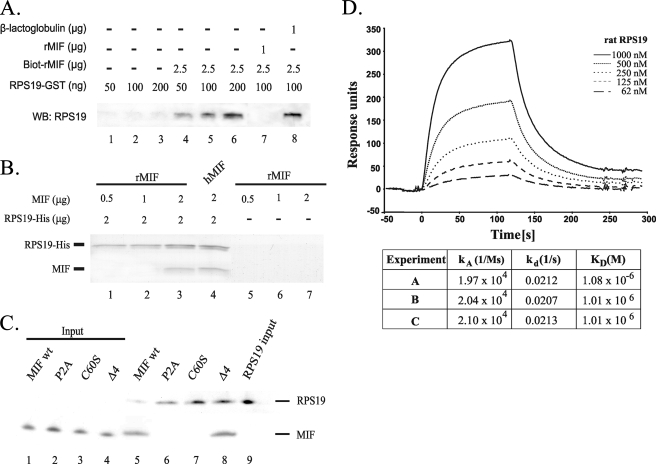
MIF Directly Interacts with RPS19 in Vitro—Pulldown assays were performed to determine whether the interaction between MIF and RPS19 was direct or indirect (Fig. 2A). Biotinylated-MIF immobilized on monomeric avidin beads was incubated with increasing amounts of RPS19-GST (lanes 4-6). As a control, unloaded avidin beads were incubated with the same amount of RPS19-GST alone (lanes 1-3). Detection of recovered RPS19-GST by GST-immunoblotting confirmed that MIF directly interacted with RPS19, and accordingly, the more RPS19 was added, the more was bound to and recovered from the coated beads. The specificity of this interaction was determined by a competition experiment using rat MIF and β-lactoglobulin. RPS19 and a 10-fold molar excess of unlabeled MIF (lane 7) or lactoglobulin (lane 8) were preincubated together before the mixtures were added to avidin beads coated with biotinylated MIF. When RPS19 was preincubated with unlabeled MIF, no RPS19 could be pulled down anymore (lane 7), whereas the unspecific competitor lactoglobulin did not affect the interaction of RPS19 with immobilized biotinylated MIF (lane 8).
FIGURE 2.
Direct interaction between MIF and RPS19 in vitro using pulldown and surface plasmon resonance assays. A, biotinylated rat MIF (Biot-rMIF) immobilized on avidin beads was incubated with increasing amounts of RPS19-GST (lanes 4-6). As control, unloaded avidin beads were incubated with the same amounts of RPS19-GST (lanes 1-3). Preincubation of RPS19 with a 10-fold molar excess of unlabeled soluble MIF prevented a pulldown (lane 7), which was not observed when β-lactoglobulin was used in the preincubation (lane 8). Detection of recovered RPS19-GST was performed using Western blot (WB) analysis with an anti-GST antibody. B, His-tagged RPS19 was immobilized on Ni-NTA-agarose beads and incubated with different amounts of recombinant rat MIF (lane 1-3) or with human MIF (lane 4). As control, recombinant rat MIF was incubated with naked Ni-NTA beads (lane 5-7). The immobilized proteins were resolved by SDS-PAGE and stained with Coomassie. C, equal amounts of His-tagged RPS19 were immobilized on Ni-NTA-agarose beads and incubated with 2 μg of recombinant human MIF or with human MIF mutants P2A MIF, C60S MIF, and Δ4 MIF (lanes 5-8), respectively. As control, all proteins were incubated with naked Ni-NTA matrix (data not shown). Immobilized proteins were resolved by SDS-PAGE, and MIF and RPS19 were detected by immunoblotting. D, interaction between RPS19 and recombinant MIF monitored in real time by biosensor analysis. Increasing concentrations of RPS19 (62-1000 nm) were passed over a sensor chip with immobilized MIF for 120 s (association phase) before the flow was switched to buffer alone for another 120 s (dissociation phase). Association and dissociation rate constants were derived using BIAevaluation 4.1 software and a 1:1 curve fitting model. The table provides association and dissociation rate constants (ka and kd) and the dissociation equilibrium constant (KD) derived from three biosensor experiments.
In an independent pulldown assay (Fig. 2B), His-tagged RPS19 was immobilized on Ni-NTA-agarose beads and incubated with either increasing amounts of rat MIF (0.5-2 μg) or 2 μg of human MIF. Protein complexes bound to the beads were separated by SDS-PAGE and stained with Coomassie. Rat MIF did not bind to the Ni-NTA matrix (lanes 5-7) but was retained on RPS19-coated beads (lanes 1-3). RPS19 also directly interacted with human MIF (lane 4), which is 95.7% homologous to the rat protein.
To further characterize the nature of the interaction, we also used three MIF mutants in the latter pulldown assay (P2A MIF, Δ4 MIF, and C60S MIF) (22). Whereas the Δ4 MIF mutant was pulled down like the wild type MIF, the P2A MIF and C60S MIF mutants were not bound to RPS19-coated beads (Fig. 2C).
Thermodynamic parameters of the formation of the MIF-RPS19 complex were quantified employing surface plasmon resonance. The kinetic data (Fig. 2D) were used to determine the association (ka = 2.04 × 104 ± 6.5 × 102 (Ms)-1) and dissociation rate constants (kd = 0.021 ± 3.2 × 10-4 s-1) and the dissociation constant KD = 1.3 × 10-6 ± 4 × 10-8 m. In summary, interaction of MIF with RPS19 has been shown by four independent methods, namely endogenous co-IP, in vivo biotin tagging, pulldown experiments, and surface plasmon resonance.
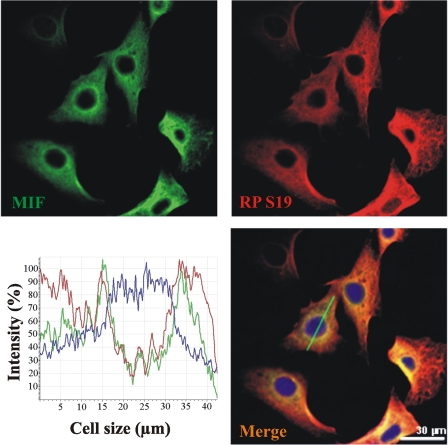
MIF and RPS19 Co-localize in NIH 3T3 Cells—Two-color immunofluorescence confocal microscopy of NIH 3T3 cells showed that MIF co-localized with RPS19 (Fig. 3). Colocalization of both factors is most prominent in the perinuclear region with varying overlap in the more peripheral cytoplasm. Little (RPS19) or no staining (MIF) was evident in the nucleus. MIF and RPS19 fluorescence intensities monitored along virtual cellular cross-sections indicate overlapping signals (see a typical example in Fig. 3, merged image).
FIGURE 3.
MIF and RPS19 co-localize in NIH 3T3 cells. Double-labeling immunofluorescence of MIF and RPS19 reveals co-localization in the cytoplasmic compartment of NIH 3T3 cells. Distribution of MIF is shown in green (upper left picture) and of RPS19 in red (upper right picture). The yellow areas in the merged image indicate colocalization of MIF and RPS19. Cell nuclei are stained blue with 4,6-diamidino-2-phenylindole. The graph illustrates the distribution of fluorescence intensities over a virtual cellular cross section (green line in the merged image). Colocalization is mostly prominent in the perinuclear region.
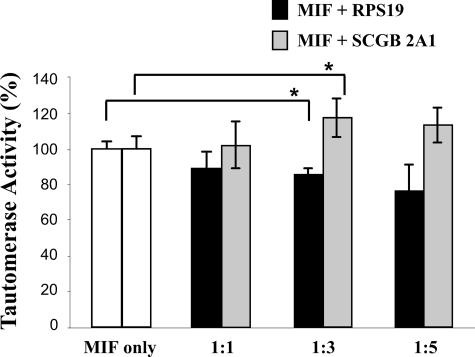
MIF Tautomerase Activity Is Only Moderately Affected by RPS19—Because the tautomerase activity of MIF may play a role in several of its functions as a cytokine (31), we investigated if RPS19 binding to MIF modulates its enzymatic activity. Tautomerization of l-dopachrome methyl ester to 5,6-dihydroxyindole-2-carboxylic acid by MIF was measured in the absence or presence of His-tagged RPS19 (Fig. 4). Increasing concentrations of RPS19 resulted in a dose-dependent but moderate decrease of MIF tautomerase activity, which was statistically significant only at a 3-fold molar excess of RPS19 over MIF but not at any other ratio used. When an excess of His-tagged secretoglobin 2A1 (32), a protein of similar size and without enzymatic activities, was used as control, MIF tautomerase activity was moderately increased, which was statistically significant also at a 3-fold molar excess over MIF but not at a 5-fold molar excess. Therefore, we conclude that overall both proteins have only a marginal effect on MIF tautomerase activity.
FIGURE 4.
PRS19 has only a moderate effect on the tautomerase activity of MIF. MIF catalyzes tautomerization of l-dopachrome methyl ester to 5,6-dihydroxyindole-2-carboxylic acid. Reaction kinetics were spectrophotometrically recorded at 475 nm over 1 min. The reaction rate within the initial 4 s was calculated and defined as tautomerase activity (see “Experimental Procedures”). The activity of MIF in the absence of RPS19 was set to 100%. The assay was performed with 1 μm MIF and after preincubation of MIF with a 1-, 3- or 5-fold molar excess (1:1, 1:3, 1:5) of His-tagged RPS19 or His-tagged SCGB 2A1 for 1 h. The control protein SCGB 2A1 is of a similar size like MIF and has no enzymatic activity. Data are expressed as the mean ± S.E. Differences with a value of p < 0.05 were considered statistically different and are marked with an asterisk.
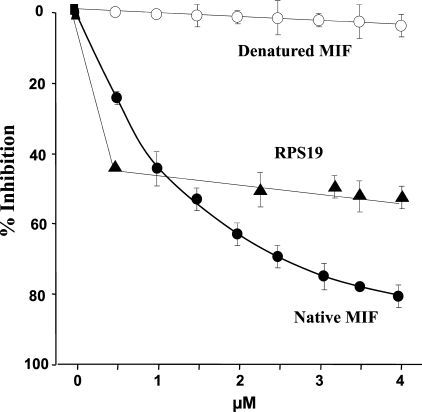
RPS19 Inhibits MIF Binding to Its Receptor CD74—To investigate if RPS19 can interfere with the binding of MIF to its receptor CD74, a sensitive capture competition assay system was used. Preincubation of RPS19 to biotin-MIF inhibited the interaction of biotin-MIF with CD74 by 55% at the highest dose. Interestingly, the lowest concentration of RPS19 applied reached already close to maximal inhibition rates (43%), which improved only marginally at higher doses (Fig. 5). As a control, increasing concentrations of unlabeled human MIF (0.5-4 μm) suppressed the binding of biotin-MIF to immobilized CD74 by 80%, whereas denatured human MIF showed no inhibitory effect (Fig. 5). Of note, the inhibitory effect of RPS19 at the lowest concentration tested (0.5 μm; 43% inhibition) was substantially more effective than the same concentration of human MIF (25% inhibition).
FIGURE 5.
RPS19 inhibits MIF binding to its receptor CD74. Purified soluble CD74 was immobilized on a plastic matrix. Binding of biotinylated MIF to immobilized CD74 was measured by enzyme-linked immunosorbent assay after preincubation of biotin MIF with increasing amounts of native MIF, denatured human MIF, or RPS19. Data are expressed as the mean ± S.D. of three different experiments. For data points without the error bar the error is less than the width of the symbol.
MIF-induced Monocyte Arrest through CXCR2 Is Compromised by RPS19—Recently, it was shown that MIF controls inflammatory and atherogenic leukocyte recruitment through Gαi-coupled activities of chemokine receptors CXCR2 and CXCR4 (14).
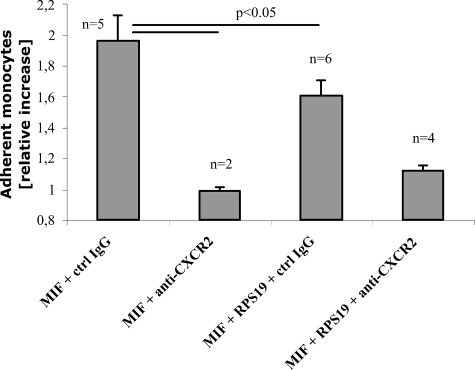
We used this assay to explore if the number of arrested mononuclear cells can be modified by RPS19. Calcein AM-labeled mononuclear MonoMac6 cells were passed over HAoECs with a defined shear rate. When HAoECs were pretreated with wild type human MIF in the presence of anti-CXCR2 antibodies, the basal level of arrested mononuclear cells was defined (Fig. 6, second column from left). Replacement of the specific anti-CXCR2 antibody by mouse IgG control antibodies resulted in a 2-fold MIF-induced increase in the number of arrested cells (left-most column). After pretreatment of HAoECs with MIF and RPS19, the number of arrested cells decreased by 39% (third column from left). This effect was dose-dependent and statistically significant at a 100-fold molar excess of RPS19 over MIF (data not shown). The addition of anti-CXCR2 antibodies to MIF and RPS19 during pretreatment reduced the number of arrested cells to almost basal levels. This demonstrates that induction of arrest of mononuclear cells by MIF as well as MIF blockage by RPS19 is CXCR2-mediated (Fig. 6, right-most column).
FIGURE 6.
RPS19 inhibits MIF-triggered mononuclear cell arrest. HAoECs were preincubated with MIF and control IgG, RPS19, and/or anti-CXCR2 antibodies as indicated. After preincubation HAoECs were perfused with calcein-AM labeled mononuclear MonoMac6 cells, and the number of adherent cells was determined. Data are presented as relative increase compared with pretreatment with MIF and anti-CXCR2 antibodies, which was set to 1. Data are expressed as the mean ± S.E. of the indicated number of experiments. Differences with a value of p < 0.05 were considered statistically different.
DISCUSSION
In the past 20 years significant progress has been made in our understanding of the role MIF plays in normal cellular physiology and in a variety of pathological conditions ranging from infection to autoimmunity and cancer (33, 34). In preclinical studies neutralization of MIF whether by antibodies, gene deletion, or small molecule inhibitors has shown promise as a potential treatment of these diseases (35, 36). As MIF functions within the cytokine cascade to control the initiation and progression of an inflammatory response, any shift toward MIF up-regulation increases the likelihood of systemic inflammation (37). Consequently, the identification of endogenous proteins that inhibit excess MIF activity is of great interest. Using an established in vivo biotinylation tagging approach (18), the RPS19 was identified as a new MIF-interacting partner in NIH 3T3 cells. By using endogenous co-immunoprecipitation as an independent method for the detection of protein-protein interactions, we could confirm RPS19 as an MIF binding partner. The validity of our co-IP approach was corroborated by the co-precipitation of JAB1, a well established MIF-interacting partner (38) (data not shown).
Direct physical interaction was shown by pulldown experiments of purified recombinant proteins and real-time binding analysis by surface plasmon resonance. The dissociation constant of formation of the MIF-RPS19 complex (KD = 1.3 × 10-6m) is well within the range of commonly observed and biologically relevant interactions, for instance in intracellular signal transduction cascades or in the binding of peptides to the T cell receptor or major histocompatibility complex. The dissociation rate indicates that 2% of the MIF-RPS19 complex decays per second. This together with a KD in the micromolar range is strongly indicative of not only a fast but also reversible adjustment of the equilibrium, thereby allowing rapid changes of bioavailable MIF rather than an irreversible blockage of MIF function.
Certain pro-inflammatory activities of MIF are known to be impaired by mutations affecting its enzymatic activity (3, 22, 27). This prompted us to investigate whether critical MIF mutations that abolish these activities impact on RPS19 binding. MIF tautomerase activity is dependent on Pro2, whereas its thiol-protein oxidoreductase activity is based on the Cys57-Ala-Leu-Cys60 (CALC) motif (22).
Pulldown assays showed that the Δ4 MIF mutant had a similar binding affinity as the wild type protein, whereas the P2A and C60S mutants did not bind to RPS19. This suggests that Cys60 is essential for a direct interaction. Although the Δ4 and P2A mutants are both missing Pro2 and, hence, have lost their tautomerase activity (22), they differ in binding to RPS19. Whereas the loss of the first four amino acids does not affect RPS19 binding, an exchange of Pro2 for alanine prevents binding, indicating that this is caused by the introduced alanine instead by the missing proline. These results suggest that Pro2 is not required for interaction, which is supported by our results showing that RPS19 only moderately interferes with MIF tautomerase activity. This is in contrast to the synthetic MIF inhibitor ISO-1 which specifically addresses the N terminus of the cytokine (4).
Although considered primarily as a component of the 40 S small subunit of the ribosome and, hence, an integral part of the protein translation machinery, RPS19 also exists in a free form in the cytosol (39). As such, RPS19 has important extraribosomal functions exemplified by its ability to interfere with growth factor signaling via its association with internalized fibroblast growth factor 2 (39, 40) and the PIM-1 oncoprotein (41). A comprehensive analysis of RPS19-binding proteins that were affinity-purified on immobilized GST-RPS19 identified a total of 159 proteins with many non-nucleolar and non-ribosomal factors (42). The list, which comprised many previously identified RPS19 interactors (e.g. RPS8), did not include PIM1, fibroblast growth factor 2, or MIF, however. This discrepancy can be attributed to the different methods employed to detect the interacting partners (i.e. yeast two-hybrid, co-IP, in vivo biotin-tagging).
Interestingly, a transglutaminase cross-linked RPS19 dimer has been described as a selective monocyte chemotactic factor in human rheumatoid arthritis when released by apoptotic cells into the extracellular fluid (43). Like MIF itself, the RPS19 dimer exerts a strong chemotactic stimulus on monocytes by mimicking the complement factor C5a and binding as a ligand to the C5a receptor (CD88) (44). This observation led to the inclusion of RPS19 in the family of chemokine-like function chemokines which also includes C5a and MIF (14, 45). Chemokine-like functions are characteristic for a group of proteins which, although they do not show typical structural chemokine features such as the chemokine-fold or the eponymous cysteine residues, have chemokine-like functions (14, 45). Therefore, we also investigated if MIF can bind to an RPS19 homodimer prepared in a type II transglutaminase reaction with monomeric protein (42). As we could produce only analytical amounts of the dimer (data not shown), we were unable to further investigate interaction of the PRS19 dimer with MIF.
Evidence that RPS19 can indeed be released from cells into extracellular fluids is supported by studies showing that another ribosomal protein, L4, is found in serum of ovarian cancer patients (46). It is tempting to speculate that by inhibiting MIF cytokine activity, monomeric RPS19 may, thus, limit an excessive inflammatory response. This view is supported by the finding that already low concentrations of RPS19 (0.5 μm) resulted in a significant blockage of MIF binding to its receptor CD74 comparable with the concentration of unlabeled MIF required to obtain the same level of inhibition (1 μm). We obtained further support for our hypothesis by investigating the effect of RPS19 on the recently discovered function of MIF as a non-cognate ligand of chemokine receptors CXCR2 and CXCR4 (14). MIF was found to promote the recruitment of monocytes and T lymphocytes by interacting with CXCR2 and CXCR4, a process that also involves CD74. In our flow chamber assays RPS19 significantly inhibited the MIF-dependent adhesion of monocytes to aortic endothelial cells, suggesting that RPS19 limits extracellular bioavailability of MIF for receptor binding. In analogy, Nm23H1, which inhibits MIF-mediated suppression of p53 activity, was recently identified as an intracellular inhibitor of MIF function (47).
In conclusion, our data suggest that RPS19 functions as an extracellular inhibitor of MIF. A shift in equilibrium from free MIF to a MIF·RPS19 complex may counteract excessive MIF function at sites of inflammation and, thus, decrease the likelihood of tissue damage, septic shock, and autoimmune reaction.
Acknowledgments
We thank Dr. Con Mallidis for carefully reading the manuscript, Prof. Guntram Suske for providing pN3-CTB, Dr. Harald Braun for organizing pBudCE4.1-birA, Prof. Gregor Bein for providing buffy coats, Dr. Lydie Da Costa for sending a sample of chicken anti-RPS19 antibody, Dr. Gabriella Krasteva for help at the confocal microscope, Dr. Henning Urlaub for protein identification by mass spectrometry, and Eva Schneider and Dr. Hongqi Lue for expert technical assistance.
This work was supported by Deutsche Forschungsgemeinschaft Grants Me 1323/2-4 (to A. M.) and SFB 542/TP A7 (J. B.) and a research grant of the University Medical Centre Giessen and Marburg (to A. M.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: MIF, migration inhibitory factor; Bis-Tris, 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol; PBS, phosphate-buffered saline; HPLC, high performance liquid chromatography; RPS19, ribosomal protein S19; MALDI, matrix-assisted laser desorption/ionization; TOF, time of flight; wt, wild type; GST, glutathione S-transferase; HAoEC, human aortic endothelial cells; Ni-NTA, nickel-nitrilotriacetic acid; co-IP, co-immunoprecipitation.
References
- 1.Calandra, T., and Roger, T. (2003) Nat. Rev. Immunol. 3 791-800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoi, A. Y., Iskander, M. N., and Morand, E. F. (2007) Inflamm. Allergy Drug Targets 6 183-190 [DOI] [PubMed] [Google Scholar]
- 3.Swope, M., Sun, H. W., Blake, P. R., and Lolis, E. (1998) EMBO J. 17 3534-3541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Al-Abed, Y., Dabideen, D., Aljabari, B., Valster, A., Messmer, D., Ochani, M., Tanovic, M., Ochani, K., Bacher, M., Nicoletti, F., Metz, C. N., Pavlov, V. A., Miller, E. J., and Tracey, K. J. (2005) J. Biol. Chem. 280 36541-36544 [DOI] [PubMed] [Google Scholar]
- 5.Rosengren, E., Bucala, R., Aman, P., Jacobsson, L., Odh, G., Metz, C. N., and Rorsman, H. (1996) Mol. Med. 2 143-149 [PMC free article] [PubMed] [Google Scholar]
- 6.Thiele, M., and Bernhagen, J. (2005) Antioxid. Redox Signal. 7 1234-1248 [DOI] [PubMed] [Google Scholar]
- 7.Lan, H. Y., Bacher, M., Yang, N., Mu, W., Nikolic-Paterson, D. J., Metz, C., Meinhardt, A., Bucala, R., and Atkins, R. C. (1997) J. Exp. Med. 185 1455-1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bernhagen, J., Calandra, T., Mitchell, R. A., Martin, S. B., Tracey, K. J., Voelter, W., Manogue, K. R., Cerami, A., and Bucala, R. (1993) Nature 365 756-759 [DOI] [PubMed] [Google Scholar]
- 9.Pan, J. H., Sukhova, G. K., Yang, J. T., Wang, B., Xie, T., Fu, H., Zhang, Y., Satoskar, A. R., David, J. R., Metz, C. N., Bucala, R., Fang, K., Simon, D. I., Chapman, H. A., Libby, P., and Shi, G. P. (2004) Circulation 109 3149-3153 [DOI] [PubMed] [Google Scholar]
- 10.Leech, M., Metz, C., Santos, L., Peng, T., Holdsworth, S. R., Bucala, R., and Morand, E. F. (1998) Arthritis Rheum. 41 910-917 [DOI] [PubMed] [Google Scholar]
- 11.Denkinger, C. M., Denkinger, M., Kort, J. J., Metz, C., and Forsthuber, T. G. (2003) J. Immunol. 170 1274-1282 [DOI] [PubMed] [Google Scholar]
- 12.Stosic-Grujicic, S., Stojanovic, I., Maksimovic-Ivanic, D., Momcilovic, M., Popadic, D., Harhaji, L., Miljkovic, D., Metz, C., Mangano, K., Papaccio, G., Al-Abed, Y., and Nicoletti, F. (2008) J. Cell. Physiol. 215 665-675 [DOI] [PubMed] [Google Scholar]
- 13.Hoi, A. Y., Hickey, M. J., Hall, P., Yamana, J., O'Sullivan, K. M., Santos, L. L., James, W. G., Kitching, A. R., and Morand, E. F. (2006) J. Immunol. 177 5687-5696 [DOI] [PubMed] [Google Scholar]
- 14.Bernhagen, J., Krohn, R., Lue, H., Gregory, J. L., Zernecke, A., Koenen, R. R., Dewor, M., Georgiev, I., Schober, A., Leng, L., Kooistra, T., Fingerle-Rowson, G., Ghezzi, P., Kleemann, R., McColl, S. R., Bucala, R., Hickey, M. J., and Weber, C. (2007) Nat. Med. 13 587-596 [DOI] [PubMed] [Google Scholar]
- 15.Gregory, J. L., Morand, E. F., McKeown, S. J., Ralph, J. A., Hall, P., Yang, Y. H., McColl, S. R., and Hickey, M. J. (2006) J. Immunol. 177 8072-8079 [DOI] [PubMed] [Google Scholar]
- 16.Leng, L., Metz, C. N., Fang, Y., Xu, J., Donnelly, S., Baugh, J., Delohery, T., Chen, Y., Mitchell, R. A., and Bucala, R. (2003) J. Exp. Med. 197 1467-1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nishimura, T., Horino, K., Nishiura, H., Shibuya, Y., Hiraoka, T., Tanase, S., and Yamamoto, T. (2001) J. Biochem. (Tokyo) 129 445-454 [DOI] [PubMed] [Google Scholar]
- 18.de Boer, E., Rodriguez, P., Bonte, E., Krijgsveld, J., Katsantoni, E., Heck, A., Grosveld, F., and Strouboulis, J. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 7480-7485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rischitor, G. (2005) Transcription Factor Sp3 as Target for SUMOylation in Vivo. Ph.D. thesis, Philipps-Universität Marburg, Marburg, Germany
- 20.Shevchenko, A., Wilm, M., Vorm, O., and Mann, M. (1996) Anal. Chem. 68 850-858 [DOI] [PubMed] [Google Scholar]
- 21.Berndt, K., Kim, M., Meinhardt, A., and Klug, J. (2008) Mol. Cell. Biochem. 307 265-271 [DOI] [PubMed] [Google Scholar]
- 22.Kleemann, R., Rorsman, H., Rosengren, E., Mischke, R., Mai, N. T., and Bernhagen, J. (2000) Eur. J. Biochem. 267 7183-7193 [DOI] [PubMed] [Google Scholar]
- 23.Sambrook, J., and Russell, D. W. (2001) Molecular Cloning, 3rd Ed., pp. 15.51, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York
- 24.Sherman, F., Stewart, J. W., and Tsunasawa, S. (1985) BioEssays 3 27-31 [DOI] [PubMed] [Google Scholar]
- 25.Gu, J., Stephenson, C. G., and Iadarola, M. J. (1994) Biotechniques 17 257, 260, and 262 [PubMed] [Google Scholar]
- 26.Da Costa, L., Tchernia, G., Gascard, P., Lo, A., Meerpohl, J., Niemeyer, C., Chasis, J. A., Fixler, J., and Mohandas, N. (2003) Blood 101 5039-5045 [DOI] [PubMed] [Google Scholar]
- 27.Bendrat, K., Al-Abed, Y., Callaway, D. J., Peng, T., Calandra, T., Metz, C. N., and Bucala, R. (1997) Biochemistry 36 15356-15362 [DOI] [PubMed] [Google Scholar]
- 28.Xiao, F., Mirwald, A., Papaioannou, M., Baniahmad, A., and Klug, J. (2005) Mol. Endocrinol. 19 2964-2978 [DOI] [PubMed] [Google Scholar]
- 29.Weber, C., Aepfelbacher, M., Haag, H., Ziegler-Heitbrock, H. W., and Weber, P. C. (1993) Eur. J. Immunol. 23 852-859 [DOI] [PubMed] [Google Scholar]
- 30.Kamir, D., Zierow, S., Leng, L., Cho, Y., Diaz, Y., Griffith, J., McDonald, C., Merk, M., Mitchell, R. A., Trent, J., Chen, Y., Kwong, Y. K., Xiong, H., Vermeire, J., Cappello, M., McMahon-Pratt, D., Walker, J., Bernhagen, J., Lolis, E., and Bucala, R. (2008) J. Immunol. 180 8250-8261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Swope, M. D., and Lolis, E. (1999) Rev. Physiol. Biochem. Pharmacol. 139 1-32 [DOI] [PubMed] [Google Scholar]
- 32.Ni, J., Kalff-Suske, M., Gentz, R., Schageman, J., Beato, M., and Klug, J. (2000) Ann. N. Y. Acad. Sci. 923 25-42 [DOI] [PubMed] [Google Scholar]
- 33.Bucala, R., and Lolis, E. (2005) Drug News Perspect. 18 417-426 [DOI] [PubMed] [Google Scholar]
- 34.Morand, E. F., Leech, M., and Bernhagen, J. (2006) Nat. Rev. Drug Discov. 5 399-410 [DOI] [PubMed] [Google Scholar]
- 35.Leech, M., Metz, C., Bucala, R., and Morand, E. F. (2000) Arthritis Rheum. 43 827-833 [DOI] [PubMed] [Google Scholar]
- 36.Santos, L., Hall, P., Metz, C., Bucala, R., and Morand, E. F. (2001) Clin. Exp. Immunol. 123 309-314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baugh, J. A., and Bucala, R. (2002) Crit. Care Med. 30 27-35 [Google Scholar]
- 38.Kleemann, R., Hausser, A., Geiger, G., Mischke, R., Burger-Kentischer, A., Flieger, O., Johannes, F. J., Roger, T., Calandra, T., Kapurniotu, A., Grell, M., Finkelmeier, D., Brunner, H., and Bernhagen, J. (2000) Nature 408 211-216 [DOI] [PubMed] [Google Scholar]
- 39.Soulet, F., Al Saati, T., Roga, S., Amalric, F., and Bouche, G. (2001) Biochem. Biophys. Res. Commun. 289 591-596 [DOI] [PubMed] [Google Scholar]
- 40.Wool, I. G. (1996) Trends Biochem. Sci. 21 164-165 [PubMed] [Google Scholar]
- 41.Chiocchetti, A., Gibello, L., Carando, A., Aspesi, A., Secco, P., Garelli, E., Loreni, F., Angelini, M., Biava, A., Dahl, N., Dianzani, U., Ramenghi, U., Santoro, C., and Dianzani, I. (2005) Haematologica 90 1453-1462 [PubMed] [Google Scholar]
- 42.Orru, S., Aspesi, A., Armiraglio, M., Caterino, M., Loreni, F., Ruoppolo, M., Santoro, C., and Dianzani, I. (2007) Mol. Cell. Proteomics 6 382-393 [DOI] [PubMed] [Google Scholar]
- 43.Nishiura, H., Shibuya, Y., Matsubara, S., Tanase, S., Kambara, T., and Yamamoto, T. (1996) J. Biol. Chem. 271 878-882 [DOI] [PubMed] [Google Scholar]
- 44.Nishiura, H., Shibuya, Y., and Yamamoto, T. (1998) Lab. Investig. 78 1615-1623 [PubMed] [Google Scholar]
- 45.Degryse, B., and de Virgilio, M. (2003) FEBS Lett. 553 11-17 [DOI] [PubMed] [Google Scholar]
- 46.Chatterjee, M., Mohapatra, S., Ionan, A., Bawa, G., Ali-Fehmi, R., Wang, X., Nowak, J., Ye, B., Nahhas, F. A., Lu, K., Witkin, S. S., Fishman, D., Munkarah, A., Morris, R., Levin, N. K., Shirley, N. N., Tromp, G., Abrams, J., Draghici, S., and Tainsky, M. A. (2006) Cancer Res. 66 1181-1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jung, H., Seong, H. A., and Ha, H. (2008) J. Biol. Chem. 283 32669-32679 [DOI] [PubMed] [Google Scholar]
- 48.Kim, M. (2003) Expression Cloning and Purification of Macrophage Migration Inhibitory Factor. Diploma thesis (M.Sc) Philipps-University, Marburg, Germany