Abstract
The BRCA2 tumor suppressor functions in repair of DNA by homologous recombination through regulating the action of Rad51. In turn, BRCA2 appears to be regulated by other interacting proteins. Dss1, a small interacting protein that binds to the C-terminal domain, has a profound effect on activity as deduced from studies on the BRCA2-related protein Brh2 in Ustilago maydis. Evidence accumulating in mammalian systems suggests that BCCIP, another small interacting protein that binds to the C-terminal domain of BRCA2, also serves to regulate homologous recombination activity. Here we were interested in testing the role of the putative U. maydis BCCIP ortholog Bcp1 in DNA repair and recombination. In keeping with the mammalian paradigm, Bcp1 bound to the C-terminal region of Brh2. Mutants deleted of the gene were extremely slow growing, showed a delay passing through S phase and exhibited sensitivity to hydroxyurea, but were otherwise normal in DNA repair and homologous recombination. In the absence of Bcp1 cells were unable to maintain the wild type morphology when challenged by a DNA replication stress. These results suggest that Bcp1 could be involved in coordinating morphogenetic events with DNA processing during replication.
Keywords: BRCA2, Rad51, Dss1, BCCIP, recombination, repair
1. Introduction
BRCA2 is a 3418 residue protein that functions in maintenance of genome integrity and in cell proliferation. Individuals with mutation in the structural gene are predisposed to breast, ovarian, and other cancers [1]. Cells with compromised BRCA2 activity exhibit hypersensitivity to genotoxins, impaired cytokinesis, frequent chromosome aberrations, breakdown of replication forks, and deficiency in DNA repair directed by homologous recombination [1–5]. In the course of discovery an important clue to BRCA2’s function was the finding that it associated with Rad51, a central component directing the cellular recombinational repair system required for proficiency in homologous pairing and DNA strand exchange [6,7]. BRCA2 exerts its role in repair by governing the action of Rad51 through interactions with the BRC-repeat domain located medially plus a second unrelated domain at the extreme C-terminus [8]. BRCA2 enables Rad51 to gain access to DNA by mediating loading through a DNA binding domain comprised of a helix-rich region and tandem OB folds (oligonucleotide/oligosaccharide-binding) closely related to those present in RPA for association with DNA [9].
Much of the molecular intricacies of BRCA2’s function that have been illuminated concern its interaction with Rad51. But given BRCA2’s large size and apparent involvement in numerous genetic processes, it might be expected that BRCA2 would have additional interacting partners. Indeed, evidence has accumulated tying BCCIP [10], BRCA1 [11], BRAF35 [12], CDKs [8], DSS1 [13], EMSY [14], FANCD2 [15], FANCG [16], PALB2 [17], P/CAF [18], Plk1 [19], and USP11 [20] among others to BRCA2. These interacting proteins were discovered by and large through pulldown procedures, yeast two-hybrid methodology, or in some instances on the basis of identifying a modifying enzyme function. The occurrence of germline mutations in breast cancer patients has providing compelling evidence linking certain of these proteins with BRCA2 function, but with respect to the majority, the details of the underlying molecular mechanisms and the significance of their interaction with BRCA2 are not well delineated.
As the BRCA2/Rad51 system is conserved in the genetically amenable fungus Ustilago maydis, there is opportunity to probe for mechanistic information by means not always so expeditious in mammalian systems [21]. The BRCA2 related protein Brh2 is a streamlined version of the mammalian protein. It is composed of 1075 amino acid residues and contains a single N-terminal BRC element and a C-terminal region with extended similarity to the BRCA2 DBD (DNA/DSS1-binding domain), the latter corresponding to part of the helical domain (HD) and extending through the OB1 and OB2 folds. In previous work we showed that an ortholog of DSS1 is present in U. maydis and that it plays a role as an essential activator of Brh2 function [22]. Mutants deleted of the structural gene have a phenotype virtually identical to brh2 mutants, i.e., they are extremely sensitive to DNA damage, defective in recombination, disturbed in meiosis, and exhibit extreme genomic instability.
Here, we have focused on BCCIP, which was identified in a two hybrid screen using a region from the very conserved C-terminal domain of BRCA2 as bait [10]. Independently, BCCIP was also found by two hybrid screening as a p21Cip1 binding protein [23]. BLAST analysis revealed that the protein is highly conserved in eukaryotes and bears similarity to the calcium binding domains of calmodulin and M-calpain, suggesting it plays a role in calcium binding [10]. BCCIP was found to colocalize with BRCA2 in the nucleus and to associate with Rad51 complexes during co-immunopreciptation. RNAi silencing inhibited formation of BRCA2 and Rad51 nuclear foci formation after DNA damage and markedly reduced recombinational repair of DNA double-strand breaks [24]. In Saccharomyces cerevisiae, the putative BCCIP ortholog Bcp1, is essential for growth, but does not appear to play a direct role in recombinational repair. It was identified as a dosage suppressor of a mutant defective in MSS4, a gene required for synthesis of phosphoinositide PI4, 5P2, which is important for organization of the actin cytoskeleton and for normal endocytosis [25]. With the use of a conditional allele it was shown that Bcp1 serves as a nuclear export factor for Mss4, ensuring transport to its site of activity at the plasma membrane.
S. cerevisiae does not appear to have a protein homologous to BRCA2. Therefore it might be imagined that Bcp1 could be dedicated to a functional system other than recombinational repar. But if BCCIP and Bcp1 function similarly, then appropriate nuclear exit of BRCA2 might be an important means of regulating its activity. We were interested in exploring the notion that a BCCIP ortholog in U. maydis, if indeed present, could serve as an important auxiliary factor to Brh2. Here we report identifying a BCCIP ortholog in U. maydis. We studied its interaction with Brh2, generated mutants deleted of the structural gene, and characterized them for DNA repair, recombination, and morphology phenotypes.
2. Methods and Materials
2.1. U. maydis strains and methods
Manipulations with U. maydis, culture methods, diploid construction, gene transfer procedures, survival after irradiation, and recombination assays have been described previously (see [26] and references therein). The gene encoding Bcp1 was identified as um03018 in the annotated Ustilago maydis genome database (http://mips.gsf.de/genre/proj/ustilago/) (GenBank accession number EF382649). Gene disruption was performed as described by standard methodology [27,28] using the hygromycin resistance cassette on plasmid pBS-hhn obtained from Dr. Jörg Kämper, Marburg University as a source for selectable marker. Primers for constructing the knockout vector and for confirming the knockout as shown in Fig. 3A are listed as follows. A arm forward--CGTCTCGCATTGCATCGTCTTGTG; A arm reverse--GAAGGCCTGAGTGGCCGTCTGTATTGTGCGACTTGGACC; B arm forward--GAAGGCCATCTAGGCCGCACCTCAGTAGCAACATTGC; B arm reverse--CGTGATCGCTGATGGAGTAGCAC; p-wt--GCTTCGAGGCTCCCCTCTTTG; p-rev--CGTCACGAACGGAACTCTCGG; p-mut--TCAGGCTCTCGCTGAATTCCC. U. maydis strains included haploids UCM350 (pan1-1 nar1-6 a1b1), UCM565 (Δbrh2 pan1-1 nar1-6 a1b1), UCM567 (ino1-3 nar1-1 a2b2), UCM639 (Δbcp1 pan1-1 nar1-6 a1b1), UCM640 (Δbcp1 ino1-3 nar1-1 a2b2), UCM642 (Δbcp1 Δbrh2 pan1-1 nar1-6 a1b1) and diploids UCM556 (nar1-1/nar1-6 pan1-1/+ met1-2/+), UCM577 (Δbrh2/Δbrh2 nar1-1/nar1-6 pan1-1/+ met1-2/+), UCM641 (Δbcp1/Δbcp1 nar1-1/nar1-6 pan1-1/+ ino1-3/+). pan, ino, met, nar, and ab indicate requirements for pantothenate, inositol, methionine, inability to reduce nitrate, and mating loci, respectively.
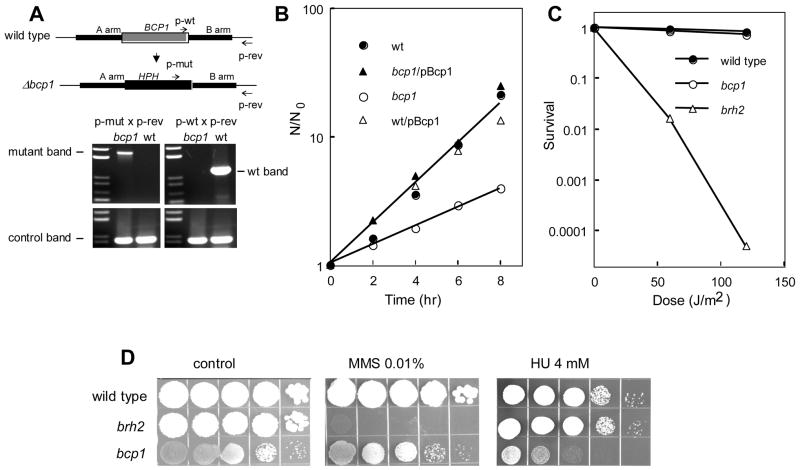
Fig. 3.
Phenotype analysis of bcp1 mutant. A. BCP1 gene disruption. PCR fragments of approximately 1 kbp each representing the BCP1 flanking sequences (A arm and B arm, respectively) were amplified and linked to the hygromycin resistance cassette HPH to generate a knockout vector. HygR transformants were screened for precise deletion by a PCR method that used primers arranged in the configuration shown to confirm the loss of the wild type gene (in gray) and the replacement by the HPH cassette (in black). The control band was an unrelated fragment flanking the BRH2 gene. B. Growth rates were determined in cultures seeded at a starting density of about 2 × 106 cells per ml. Samples were removed at 2 hr intervals and cell densities determined by counting under a microscope using a hemocytometer. Values shown are averages of 2–4 independent cultures. C. Survival of cells after irradiation with the indicated doses of UV. D. Spot tests showing survival on medium containing the indicated concentrations of methylmethane sulfonate (MMS) or hydroyxurea (HU). From left to right are serial 10-fold dilutions from a starting cell density of about 2 × 107 per ml.
FACS analysis was performed as described previously [29]. To analyze cultures carrying a mixture of aberrant cell clusters and individual cells, we initially centrifuged the culture at low speed (2000 rpm) and for a short time (5 min) in a high viscosity medium (i. e., containing 1M sorbitol). Under these conditions single cells sedimented rapidly to the bottom of the tube, while cell aggregates remained in suspension. Microscopy was carried out using a Nikon CF600 microscope. Standard FITC and DAPI filter sets were used for epifluorescence analysis of nuclear staining with DAPI, and calcofluor staining was performed as described [30]. Photomicrographs were obtained with a ORCA G digital camera (Hamamatsu) and the images were processed with Metamorph software.
2.2. Co-precipitation methodology
Expression of MBP-Brh2 and His-Bcp1 fusion proteins in E. coli strain BL21(DE3), and co-precipitation or pull-down analyses have been described previously [26,31]. As deletions removing up to 213 amino acid residues from the N-terminus of Brh2 make no difference in ability to complement the radiation sensitivity of the brh2 null mutant [32], we cut into the coding sequence for this nonessential region to expedite cloning of Brh2 truncation alleles and to enhance solubility of their protein products. For expression in E. coli, the allele representing completely functional Brh2 started at residue 106 (see Figure 2). Western blotting was performed using anti-MBP antiserum (New England BioLabs), or His-Tag monoclonal antibody (Novagen).
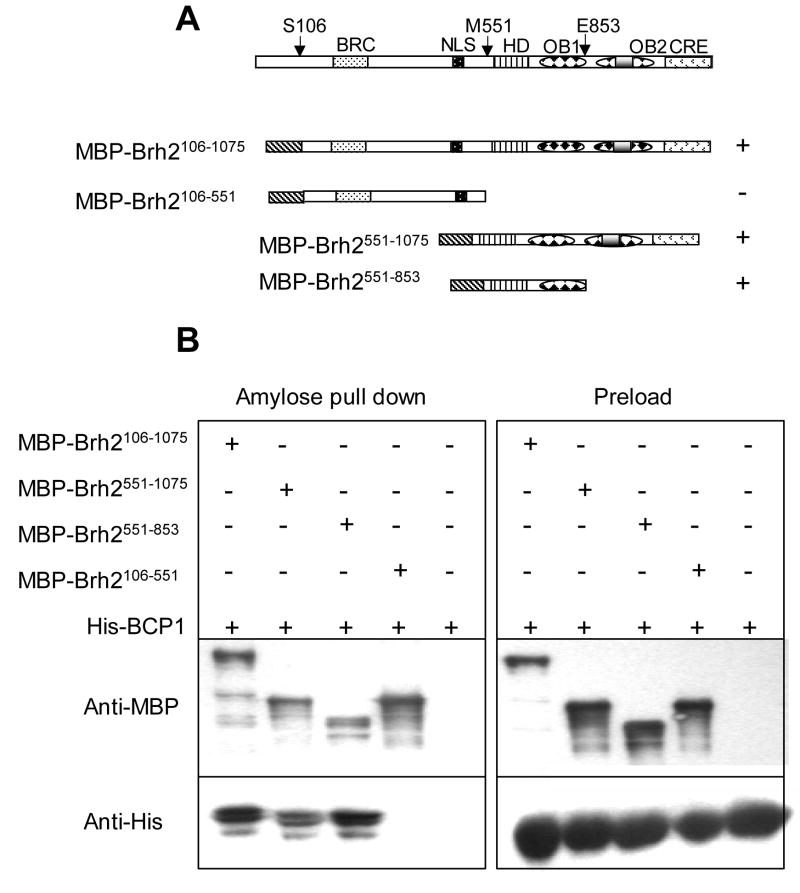
Fig. 2.
Bcp1 interaction with Brh2. A. Brh2 regions are illustrated showing the Rad51-interacting regions BRC and CRE [31}, the nuclear localization signal NLS, the helix rich domain and two OB folds in the DNA binding domain, and the amino acid residue at the fusion or truncation sites. The pluses and minus on the right summarize the Bcp1/Brh2 interactions. B. E. coli strains co-expressing the indicated affinity-tagged proteins shown schematically were processed as described in Methods. MBP-tagged Brh2 derivatives were pulled down from extracts using amylose beads. After washing, beads were eluted with maltose-containing buffer and samples were electrophoresed in 10% polyacrylamide gels containing sodium dodecyl sulfate. Gels were analyzed by western blotting using anti-MBP or anti-His antibodies to identify MBP-Brh2 or His-Bcp1 tagged derivatives.
2.3. U. maydis plasmids
The plasmid for protein expression in U. maydis is a pUC19 derivative [33] containing an U. maydis ARS, a fragment of the glyceraldehyde-3-phosphate dehydrogenase promoter (gap) for driving expression of the Bcp1 coding sequence, and a gene expressing resistance to carboxin (CbxR) for use as a selectable marker. Gap repair was performed using ARS plasmid pCM1057 containing the CbxR resistance gene deleted of a 0.25 kbp BglII fragment to score as a marker. The plasmid was cut with BglII prior to transformation to reveal the gap. Transformation control was performed using pCM1052 containing the intact CbxR resistance gene.
3. Results
3.1. Bcp1 interacts with Brh2
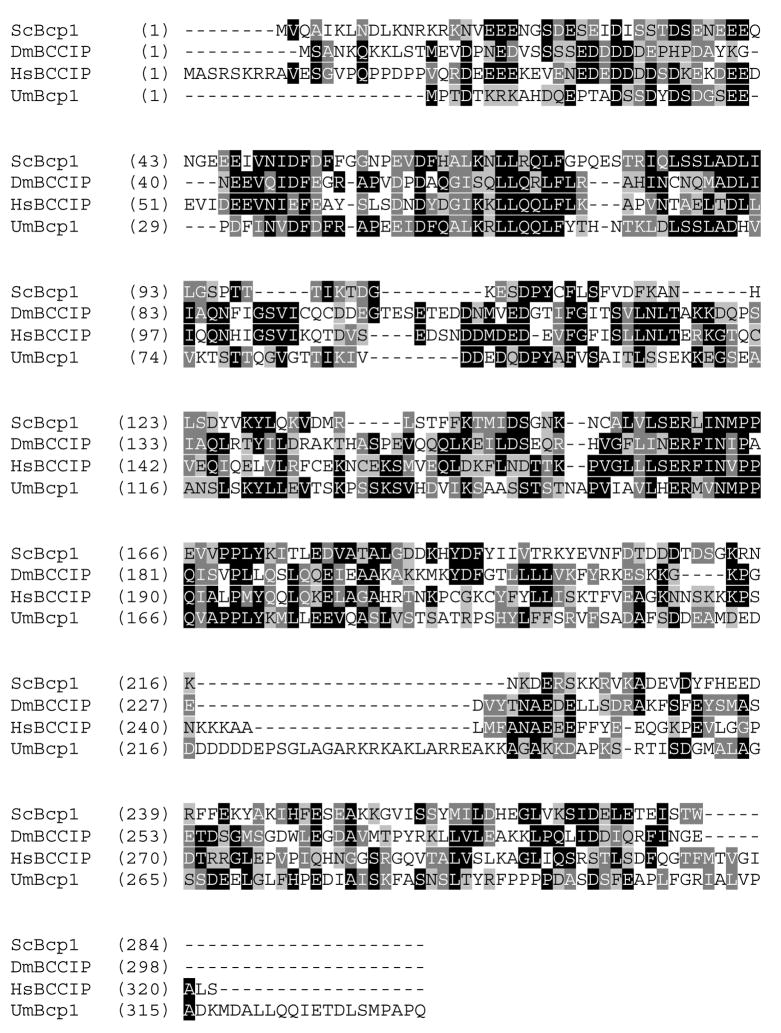
Using human BCCIP sequence as a search query we identified a convincing ortholog (hereafter, Bcp1) in the U. maydis genome database. Bcp1 is 335 amino acids residues in length compared with 322 residues for BCCIP and features an acidic N-terminal region and a highly conserved internal region similar to the other BCCIP orthologs (Fig. 1). The residues corresponding to BCCIP 61-173, which were mapped to contain the minimum BRCA2 interacting region, are conserved in Bcp1. To confirm that the BCCIP/BRCA2 interaction is recapitulated, Bcp1 was tested directly for interaction by coprecipitating MBP-tagged Brh2 with hexahistidine-tagged Bcp1 after expression in E. coli (Fig. 2) Bcp1 was pulled down by a fusion representing the fully functional Brh2 (Brh2106–1075), by the C-terminal DBD Brh2551–1075and by a smaller region of the C-terminal-DBD (Brh2551–853), but not the N-terminal region (Brh2106–551). This localizes the interacting domain in Brh2 to a region that roughly corresponds to the homologous region in BRCA2 that was mapped to interact with BCCIP.
Fig. 1.
Identification of a BCCIP ortholog in U. maydis. Human and yeast sequences were used as queries to search the U. maydis genome (http://www.broad.mit.edu/annotation/genome/ustilago_maydis/Home.html). Multiple alignment shows sequences from U. maydis Bcp1 (GenBank accession number EF382649), human BCCIP (CAI12093), Drosophila melanogaster (AAL39431), and S. cerevisiae (Q06338). Residues identical in two of four sequences have black backgrounds, while those conserved are in gray.
3.2. Bcp1 is important for growth
The entire coding sequence for Bcp1 was deleted in haploid tester strains by a standard disruption method [27,28]. The mutants obtained were confirmed to be deleted of the gene and replaced with a drug resistance marker by a PCR-based procedure (Fig. 3A). The bcp1 knockouts were viable but slow growing (Fig. 3B). This finding contrasts with the phenotype of the corresponding bcp1 mutant in Saccharomyces cerevisiae (http://www.yeastgenome.org/), which was determined to be inviable [25]. To confirm that the mutants were indeed deleted of the structural gene, the cloned BCP1 gene expressed under control of a heterologous promoter was reintroduced into the bcp1 mutant on a self-replicating plasmid. The growth defect was fully complemented by ectopic expression of the cloned gene. Introduction of the cloned gene into the wild type strain had no adverse effect on growth.
3.3. Bcp1 has no direct role in DNA repair or recombination
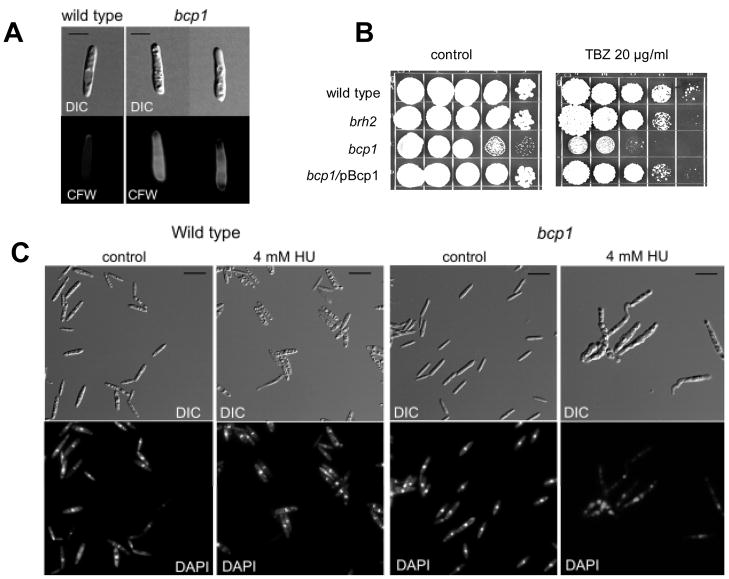
Loss of Rad51, Brh2, or the Brh2-interacting protein Dss1 was shown previously to have profound effects on the DNA repair system [21,22]. Mutants were found to exhibit extreme sensitivity to ultraviolet light (UV), ionizing radiation and DNA genotoxins such as chemical alkylating agents. To determine if bcp1 exhibited a similar sensitivity, we tested whether it was sensitive to UV or methylmethanesulfonate (MMS). At doses that promoted extensive killing of brh2, the bcp1 mutant was unaffected (Fig. 3C & D). On the other hand it was interesting to note that bcp1 was moderately sensitive to hydroxyurea, while brh2 was not sensitive (Fig. 3D). As hydroxyurea blocks DNA synthesis by inhibiting ribonucleotide reductase, bcp1’s hydroxyurea sensitivity suggests there could be some connection between the growth defect and replication, although the effect could be secondary, or simply an additive effect of further reduction in growth rate. Following the findings in S. cerevisiae that the yeast BCP1 gene might ultimately have an impact on cytoskeleton organization and maintenance [25], we also tested the effect of thiabendazole (TBZ), a microtubule poison, and found the bcp1 mutant to be mildly sensitive. In all of the untreated controls above, the slow growth phenotype of bcp1 was obvious by the small colony size. The wildtype gene complemented both the slow growth phenotype and the TBZ when reintroduced into the mutant.
Recombination proficiency was measured by two different methods (Table 1). In the first, we investigated DNA double-strand-break repair using a plasmid-based transformation system that relies on homologous recombination between an ectopic marker on the plasmid and an endogenous chromosomal allele. The assay measures carboxin resistance (CbxR) after introduction of a self-replicating plasmid containing the cloned variant of the succinic dehydrogenase iron-sulfur gene subunit conferring resistance to carboxin, which has been inactivated by deleting a 0.25 kbp fragment from the coding sequence. Repair of the deleted sequence and concomitant transformation to CbxR requires recombination with the endogenous chromosomal allele to utilize it as a template to direct repair. As previously established gap repair is highly dependent upon Rad51, Brh2, and Dss1[21,22]. However, in the case of the bcp1mutant there is no loss in gap repair proficiency.
Table 1.
Recombination phenotype comparison of bcp1
| Gap repaira | Allelic recombinationb (× 10−5) | ||
|---|---|---|---|
| CbxR (× 10−4) | Spontaneous | UV induced | |
| Wild type | 1.4 | 0.4 ± 0.05 | 21 ± 4 |
| bcp1 | 1.8 | 0.2 ± 0.04 | 18 ± 10 |
| brh2 | <0.02 | <0.01 | <0.01 |
Gap repair frequency is the ratio of the CbxR transformants arising per microgram DNA using the gapped plasmid substrate pCM1057 compared to CbxR transformants per microgram of intact pCM1052 DNA, a self-replicating plasmid expressing CbxR. Strains employed were UCM359 (wildtype). UCM565 (brh2), and UCM639 (bcp1).
Nar+ allelic recombination frequency per viable cell. The spontaneous level was determined with UCM556 (wild type), UCM577(brh2), and UCM641 (bcp1) by fluctuation analysis performed in triplicate with 9 independent cultures. The UV dose was 120 J/m2. Values were determined from triplicate cultures.
By the second procedure, we used the more traditional method of investigating heteroallelic recombination in diploids. Here a homozygous diploid bcp1/bcp1 strain heteroallelic at the nitrate reductase locus (nar1) was assayed for the spontaneous levels and in response to DNA damage. In mutants lacking Rad51, Brh2, or Dss1, the spontaneous levels and the damage-induced levels of recombination are greatly reduced [21,22]. In contrast, loss of Bcp1 function had little to no effect on allelic recombination. Thus, by two different methodologies, it appears that Bcp1 function is not required for recombinational repair.
3.4. Genetic interaction between bcp1 and brh2
To define relationships between bcp1 and brh2, we constructed a bcp1 brh2 double mutant and characterized it with respect to growth. We found that growth was slightly improved by comparison with the bcp1 single mutant (Fig. 4A). However, neither the UV sensitivity nor MMS sensitivity was suppressed by comparison with the brh2 single mutant (not shown).
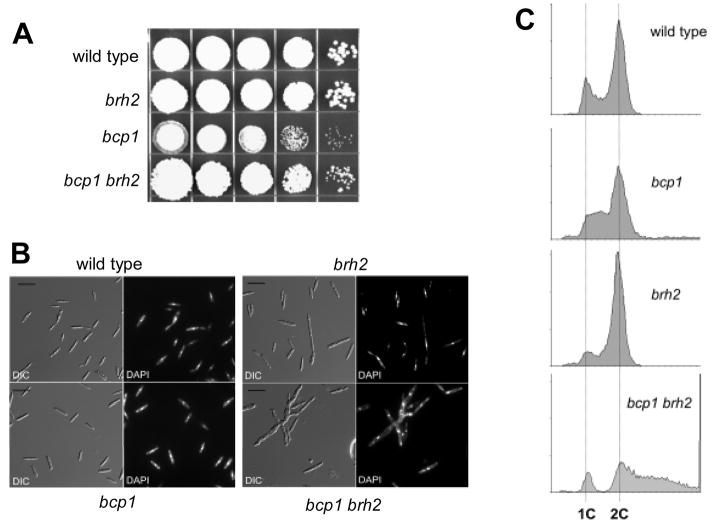
Fig. 4.
Genetic interactions between bcp1 and brh2. A. Spot tests showing growth ability of strains indicated on the left. From left to right are serial 10-fold dilutions from a starting cell density of about 2 × 107 per ml. B. Morphology of U. maydis strains. Cell suspensions were removed during log phase growth of cell cultures, applied to glass slides and examined microscopically under a 60X objective lens. The bar indicates approximately 10 microns. C. DNA content of the different mutant cells. Histograms indicate DNA content (x-axis) as 1C or 2C corresponding to G1 or G2 content, and cell number (y-axis). Asynchronous cultures were removed during log phase growth for FACS analysis. In the case of bcp1 brh2 double mutant strain, to enrich for the population of single cells, cultures were centrifuged at low speed in 1M sorbitol-containing medium and cells at the bottom of the tube were used for FACS analysis. DNA content higher than 2C in this strain corresponds to complex cell aggregates.
We analyzed the appearance of the cells in liquid culture (Fig. 4B) and found that bcp1 cells showed a low frequency of budding cells in comparison to wild type cells (9% of cell population in mutant cells versus 42% in wild type cells). In contrast, brh2 cells showed a higher frequency of budding cells (64%) and frequently showed elongated buds. Interestingly, bcp1 brh2 double mutant cells are commonly misshapen, have somewhat elongated buds, and fail to complete cytokinesis, resulting in highly branched cell clusters (Fig. 4B, Table 2). In the population of bcp1 brh2 cells that did not show aberrant morphology, the frequency of budding cells appears more similar to that showed by wild-type cells (around 36% of the cells showed buds).
Table 2.
Aberrant cell separation
| genotype | Wild type | brh2 | bcp1 | bcp1 brh2 |
|---|---|---|---|---|
| complex cells (%)a | <2 | <2 | <2 | 33 |
Cell suspensions were applied to a hemocytometer and viewed under a microscope. Cells were counted as complex if there were more than 4–5 branch structures evident. Approximately 100 cells from each strain were scored.
Studies correlating nuclear density and cell morphology in U. maydis showed that cells complete DNA synthesis before beginning to form buds. That is, bud formation takes place at the onset of G2 phase [34]. The accumulation of unbudded cells in bcp1 mutants therefore suggests either a delay in G1 or S phase. To add further support to this idea, we analyzed the DNA content of the different strains by fluorescence-activated cell sorter (FACS) analysis (Figure 4C). We found that bcp1 cells showed a substantial accumulation of cells with a DNA content between 1C and 2C (i. e., cells traversing S-phase). In contrast brh2 cells were accumulated in 2C content, which together with the presence of cells with elongated buds suggests a delay in exiting G2 phase, as would be expected for cells deficient in repair of DNA double-strand breaks but proficient in activation of the DNA damage checkpoint. The presence of cell aggregates formed in bcp1 brh2 mutants made analysis of the DNA content of difficult. Therefore, we enriched the population in non-aberrant forms by differential centrifugation (see Material and Methods) and found that bcp1 brh2 single cells did not show the apparent delay in S-phase (Fig. 4C). Together these results suggest that Bcp1 and Brh2 act somehow in opposing directions, with respect to their effects in cell cycle and morphology.
3.5. Requirement of bcp1 for proper morphology after DNA replication stress
In addition to the low frequency of bud formation in bcp1 mutant cells, we observed that they exhibited a slightly blunt-ended appearance that was distinguishable from wild type cells (Fig. 5A). This observation together with the strong morphological defect of the bcp1 brh2 mutant implies that Bcp1 may have some connection with proper morphogenesis. Two observations support this idea. First, bcp1 cells show an abnormal distribution of cell wall components. In wild type cells, the fluorochrome calcofluor stains chitin at the tip of the cell where the bud will be produced. In contrast, in bcp1 cells calcofluor staining disappears from the tip, but is abundantly associated with the lateral cell wall (Fig. 5A). This latter observation suggests bcp1 is defective in cytoskeleton organization and/or maintenance. In support of this notion, we found bcp1 to be mildly sensitive to thiabendazole (TBZ), a microtubule poison (Fig. 5B).
Fig. 5.
Bcp1 is required for proper morphogenesis. A. Chitin distribution in wild-type and bcp1 mutant cells (two different examples are shown). Cells were stained with calcofluor and observed with epifluorescence microscopy. The bar indicates approximately 3 microns. B. Spot tests showing survival on medium containing the microtubule poison thiabendazole (TBZ). From left to right are serial 10-fold dilutions from a starting cell density of about 2 × 107 per ml. C. Morphology of the cells after 12 h of incubation in the presence of HU. Wild-type cells arrest mainly as unbudded cells while bcp1 mutants display aberrant morphologies.
Recently, connections between morphogenetic events and the response to DNA damage and DNA replication stress have been uncovered in S. cerevisiae [35]. Accordingly, we wondered whether Bcp1 might be dedicated to a role in maintaining proper morphogenesis in response to conditions of DNA replication stress. Therefore, we investigated the morphological response of the bcp1 mutant to hydroxyurea. After treatment with hydroxyurea wild type cells become arrested in an unbudded form with a single nucleus (Fig. 5C). As bud formation in U. maydis does not start until DNA replication is complete [29], this hydroxyurea-induced arrest is consistent with the notion that proper morphogenesis and cell cycle progression are coordinated. However, in bcp1 cells this coordination seems to be disturbed as cells treated with hydroxyurea formed aberrant structures, many with more than one nucleus (Fig. 5C). These results support a role for Bcp1 in the cellular pathways connecting the DNA replication checkpoint with morphogenesis.
4. Discussion
BCCIP, like DSS1, was originally identified as a BRCA2-interacting protein in a two-hybrid screen. Both are small proteins containing acidic regions, but they appear to bind to different sites on BRCA2. Mapping studies have placed the BCCIP binding site somewhat distal to that of DSS1[10]. Because BRCA2 plays a major role governing homologous recombination through its interaction with Rad51, it can easily be imagined that other proteins could exert some additional means of control on the process through interaction with BRCA2. Indeed evidence has accumulated supporting a role for both BCCIP and DSS1 in recombinational repair [2,24], although in neither case has a hypomorphic mutant been identified nor has a gene knockout been reported to provide direct proof for a role. Both proteins are highly conserved and their orthologs are present throughout the eukaryotic kingdom including genetically amenable yeasts such as Sacchaomyces cerevisiae and Schizosaccharomyces pombe. But since the homologous recombination systems of these latter organisms are not mediated through a BRCA2-related protein, it was of interest to evaluate the contribution of DSS1 and BCCIP in a microbial system that does function in a BRCA2-dependent manner.
Previously we established that the DSS1 ortholog Dss1 plays a pivotal role in DNA repair, recombination, and genome stability in U. maydis and that the dss1 null mutant appears as a brh2 phenocopy [22]. The BCCIP-related protein Bcp1 appears to have a different role. Deletion of the gene causes a profound effect on growth rate and disturbs cellular morphology, but does not diminish the resistance to radiation nor reduce the capacity for recombination. Thus, physical association with Brh2 per se is not necessarily a hallmark of recombinational repair function. However, dss1 and bcp1 mutants are both defective in cell growth ability, although dss1 is not as strongly disturbed as bcp1 (compare this work with [22]). Similar to the previously reported observation that the dss1 brh2 double mutant grows better than the dss1 single mutant [31], here we also noted that the bcp1 brh2 double mutant grew more robustly than the bcp1 single mutant. In the case of dss1 we interpreted the observations to mean that Brh2 confers toxicity in the absence of Dss1, perhaps by interfering with the normal DNA processing systems [31]. Since Dss1 has been reported to play a role as a nuclear transporter [36], the notion occurred that one means of regulating Brh2 and mitigating this toxicity might be for Dss1 to shuttle Brh2 out of the nucleus. As it has been established in S. cerevisiae that Bcp1 functions in nuclear export of a protein dedicated to phosphoinositide synthesis [25], it could well be that Bcp1 serves in an export capacity in U. maydis promoting nuclear-cytoplasm trafficking of certain proteins including Brh2 as a possible means of regulation.
Alternatively, Bcp1 might well serve in conjunction with Brh2 in some process other than DNA repair and recombination. For instance BRCA2 has been reported to localize to the chromosome passenger complex in the late stages of cytokinesis to regulate nuclear division [37]. Perhaps Bcp1 links Brh2 in some way that is not yet clear with dynamic processes involving cytoskeleton. In light of Bcp1’s role in cell proliferation and homeostasis, its association with Brh2 might represent a nexus for communication or signaling between pathways for maintaining genome integrity and cell growth. Recently, a connection between morphogenesis and DNA replication stress has been uncovered in S. cerevisiae [35]. Transient relocalization of the replication-checkpoint protein Rad53 from the nucleus to the bud neck seems to be a determinant for proper morphogenetic response to DNA stress. By analogy Bcp1 could be envisioned as mediating communication between genome maintenance and morphogenic programs by regulating the subcellular localization of such proteins as Brh2.
Acknowledgments
We thank Dr. Lorraine Symington (Columbia University) for comments on the manuscript. This work received support from the National Institutes of Health grant GM42482 and the Milo Gladstein Foundation to W.K.H., and by grant BIO2005-02998 from the Spanish government to J. P.-M.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nathanson KL, Wooster R, Weber BL. Breast cancer genetics: what we know and what we need. Nature Med. 2001;7:552–556. doi: 10.1038/87876. [DOI] [PubMed] [Google Scholar]
- 2.Gretarsdottir S, Thorlacius S, Valgardsdottir R, Gudlaugsdottir S, Sigurdsson S, Steinarsdottir M, Jonasson JG, Anamthawat-Jonsson K, Eyfjord JE. BRCA2 and p53 mutations in primary breast cancer in relation to genetic instability. Cancer Res. 1998;58:859–862. [PubMed] [Google Scholar]
- 3.Moynahan ME, Pierce AJ, Jasin M. BRCA2 is required for homology-directed repair of chromosomal breaks. Molecular Cell. 2001;7:263–272. doi: 10.1016/s1097-2765(01)00174-5. [DOI] [PubMed] [Google Scholar]
- 4.Tutt A, Gabriel A, Bertwistle D, Connor F, Paterson H, Peacock J, Ross G, Ashworth A. Absence of Brca2 causes genome instability by chromosome breakage and loss associated with centrosome amplification. Curr Biol. 1999;9:1107–1110. doi: 10.1016/s0960-9822(99)80479-5. [DOI] [PubMed] [Google Scholar]
- 5.Patel KJ, Yu VP, Lee H, Corcoran A, Thistlethwaite FC, Evans MJ, Colledge WH, Friedman LS, Ponder BA, Venkitaraman AR. Involvement of Brca2 in DNA repair. Mol Cell. 1998;1:347–357. doi: 10.1016/s1097-2765(00)80035-0. [DOI] [PubMed] [Google Scholar]
- 6.Pellegrini L, Venkitaraman A. Emerging functions of BRCA2 in DNA recombination. Trends Biochem Sci. 2004;29:310–316. doi: 10.1016/j.tibs.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 7.West SC. Molecular view of recombination proteins and their control. Nat Rev Mol Cell Biol. 2003;4:1–11. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- 8.Esashi F, Christ N, Gannon J, Liu Y, Hunt T, Jasin M, West SC. CDK-dependent phosphorylation of BRCA2 as a regulatory mechanism for recombinational repair. Nature. 2005;434:598–604. doi: 10.1038/nature03404. [DOI] [PubMed] [Google Scholar]
- 9.Yang H, Jeffrey PD, Miller J, Kinnucan E, Sun Y, Thoma NH, Zheng N, Chen PL, Lee WH, Pavletich NP. BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science. 2002;297:1837–1848. doi: 10.1126/science.297.5588.1837. [DOI] [PubMed] [Google Scholar]
- 10.Liu J, Yuan Y, Huan J, Shen Z. Inhibition of breast and brain cancer cell growth by BCCIPalpha, an evolutionarily conserved nuclear protein that interacts with BRCA2. Oncogene. 2001;20:336–345. doi: 10.1038/sj.onc.1204098. [DOI] [PubMed] [Google Scholar]
- 11.Chen J, Silver DP, Walpita D, Cantor SB, Gazdar AF, Tomlinson G, Couch FJ, Weber BL, Ashley T, Livingston DM, Scully R. Stable interaction between the products of the BRCA1 and BRCA2 tumor suppressor genes in mitotic and meiotic cells. Mol Cell. 1998;2:317–328. doi: 10.1016/s1097-2765(00)80276-2. [DOI] [PubMed] [Google Scholar]
- 12.Marmorstein LY, Kinev AV, Chan GK, Bochar DA, Beniya H, Epstein JA, Yen TJ, Shiekhattar R. A human BRCA2 complex containing a structural DNA binding component influences cell cycle progression. Cell. 2001;104:247–257. doi: 10.1016/s0092-8674(01)00209-4. [DOI] [PubMed] [Google Scholar]
- 13.Marston NJ, Richards WJ, Hughes D, Bertwistle D, Marshall CJ, Ashworth A. Interaction between the product of the breast cancer susceptibility gene BRCA2 and DSS1, a protein functionally conserved from yeast to mammals. Mol Cell Biol. 1999;19:4633–4642. doi: 10.1128/mcb.19.7.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hughes-Davies L, Huntsman D, Ruas M, Fuks F, Bye J, Chin SF, Milner J, Brown LA, Hsu F, Gilks B, Nielsen T, Schulzer M, Chia S, Ragaz J, Cahn A, Linger L, Ozdag H, Cattaneo E, Jordanova ES, Schuuring E, Yu DS, Venkitaraman A, Ponder B, Doherty A, Aparicio S, Bentley D, Theillet C, Ponting CP, Caldas C, Kouzarides T. EMSY links the BRCA2 pathway to sporadic breast and ovarian cancer. Cell. 2003;115:523–535. doi: 10.1016/s0092-8674(03)00930-9. [DOI] [PubMed] [Google Scholar]
- 15.Hussain S, Wilson JB, Medhurst AL, Hejna J, Witt E, Ananth S, Davies A, Masson JY, Moses R, West SC, de Winter JP, Ashworth A, Jones NJ, Mathew CG. Direct interaction of FANCD2 with BRCA2 in DNA damage response pathways. Hum Mol Gen. 2004;13:1241–1248. doi: 10.1093/hmg/ddh135. [DOI] [PubMed] [Google Scholar]
- 16.Hussain S, Witt E, Huber PA, Medhurst AL, Ashworth A, Mathew CG. Direct interaction of the Fanconi anaemia protein FANCG with BRCA2/FANCD1. Hum Mol Gen. 2003;12:2503–2510. doi: 10.1093/hmg/ddg266. [DOI] [PubMed] [Google Scholar]
- 17.Xia B, Sheng Q, Nakanishi K, Ohashi A, Wu J, Chirst N, Liu X, Jasin M, Couch FJ, Linvingston DM. Control of BRCA2 cellular and clinical funcitons by a nuclear partner, PALB2. Mol Cell. 2006;22:719–729. doi: 10.1016/j.molcel.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 18.Fuks F, Milner J, Kouzarides T. BRCA2 associates with acetyltransferase activity when bound to P/CAF. Oncogene. 1998;17:2531–2534. doi: 10.1038/sj.onc.1202475. [DOI] [PubMed] [Google Scholar]
- 19.Lee M, Daniels MJ, Venkitaraman AR. Phosphorylation of BRCA2 by the Polo-like kinase Plk1 is regulated by DNA damage and mitotic progression. Oncogene. 2004;23:865–872. doi: 10.1038/sj.onc.1207223. [DOI] [PubMed] [Google Scholar]
- 20.Schoenfeld AR, Apgar S, Dolios G, Wang R, Aaronson SA. BRCA2 is ubiquitinated in vivo and interacts with USP11, a deubiquitinating enzyme that exhibits prosurvival function in the cellular response to DNA damage. Mol Cell Biol. 2004;24:7444–7455. doi: 10.1128/MCB.24.17.7444-7455.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kojic M, Kostrub CF, Buchman AR, Holloman WK. BRCA2 homolog required for proficiency in DNA repair, recombination, and genome stability in Ustilago maydis. Mol Cell. 2002;10:683–691. doi: 10.1016/s1097-2765(02)00632-9. [DOI] [PubMed] [Google Scholar]
- 22.Kojic M, Yang H, Kostrub CF, Pavletich NP, Holloman WK. The BRCA2-interacting protein DSS1 is vital for DNA repair, recombination, and genome stability in Ustilago maydis. Mol Cell. 2003;12:1043–1049. doi: 10.1016/s1097-2765(03)00367-8. [DOI] [PubMed] [Google Scholar]
- 23.Ono T, Kitaura H, Ugai H, Murata T, Yokoyama KK, Iguchi-Ariga SM, Ariga H. TOK-1, a novel p21Cip1-binding protein that cooperatively enhances p21-dependent inhibitory activity toward CDK2 kinase. J Biol Chem. 2000;275:31145–31154. doi: 10.1074/jbc.M003031200. [DOI] [PubMed] [Google Scholar]
- 24.Lu H, Guo X, Meng X, Liu J, Allen C, Wray J, Nickoloff JA, Shen Z. The BRCA2-interacting protein BCCIP functions in RAD51 and BRCA2 focus formation and homologous recombinational repair. Mol Cell Biol. 2005;25:1949–1957. doi: 10.1128/MCB.25.5.1949-1957.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Audhya A, Emr SD. Regulation of PI4, 5P2 synthesis by nuclear-cytoplasmic shuttling of the Mss4 lipid kinase. EMBO J. 2003;22:4223–4236. doi: 10.1093/emboj/cdg397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kojic M, Zhou Q, Lisby M, Holloman WK. Rec2 interplay with both Brh2 and Rad51 balances recombinational repair in Ustilago maydis. Mol Cell Biol. 2006;26:678–688. doi: 10.1128/MCB.26.2.678-688.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kamper J. A PCR-based system for highly efficient generation of gene replacement mutants in Ustilago maydis. Mol Gen Genom. 2004;271:103–110. doi: 10.1007/s00438-003-0962-8. [DOI] [PubMed] [Google Scholar]
- 28.Brachmann A, Konig J, Julius C, Feldbrugge M. A reverse genetic approach for generating gene replacement mutants in Ustilago maydis. Mol Gen Genom. 2004;272:216–226. doi: 10.1007/s00438-004-1047-z. [DOI] [PubMed] [Google Scholar]
- 29.Garcia-Muse T, Steinberg G, Perez-Martin J. Pheromone-induced G2 arrest in the phytopathogenic fungus Ustilago maydis. Eukaryotic Cell. 2003;2:494–500. doi: 10.1128/EC.2.3.494-500.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Castillo-Lluva S, Perez-Martin J. The induction of the mating program in the phytopathogen Ustilago maydis is controlled by a G1 cyclin. Plant Cell. 2005;17:3544–3560. doi: 10.1105/tpc.105.036319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou Q, Kojic M, Cao Z, Lisby M, Mazloum N, Holloman WK. Dss1 interaction with Brh2 as a regulatory mechanism for recombinational repair. Mol Biol Cell. 2007;27:2512–2526. doi: 10.1128/MCB.01907-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kojic M, Zhou Q, Lisby M, Holloman WK. Brh2-Dss1 interplay enables properly controlled recombination in Ustilago maydis. Mol Cell Biol. 2005;25:2547–2557. doi: 10.1128/MCB.25.7.2547-2557.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kojic M, Holloman WK. Shuttle vectors for genetic manipulations in Ustilago maydis. Can J Microbiol. 2000;46:333–338. doi: 10.1139/w00-002. [DOI] [PubMed] [Google Scholar]
- 34.Perez-Martin J, Castillo-Lluva S, Sgarlata C, Flor-Parra I, Mielnichuk N, Torreblanca J, Carbo N. Pathocycles: Ustilago maydis as a model to study the relationships between cell cycle and virulence in pathogenic fungi. Mol Gen Genom. 2006;276:211–229. doi: 10.1007/s00438-006-0152-6. [DOI] [PubMed] [Google Scholar]
- 35.Enserink JM, Smolka MB, Zhou H, Kolodner RD. Checkpoint proteins control morphogenetic events during DNA replication stress in Saccharomyces cerevisiae. J Cell Biol. 2006;175:729–741. doi: 10.1083/jcb.200605080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thakurta AG, Gopal G, Yoon JH, Kozar L, Dhar R. Homolog of BRCA2-interacting Dss1p and Uap56p link Mlo3p and Rae1p for mRNA export in fission yeast. EMBO J. 2005;24:2512–2523. doi: 10.1038/sj.emboj.7600713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Daniels MJ, Wang Y, Lee M, Venkitaraman AR. Abnormal cytokinesis in cells deficient in the breast cancer susceptibility protein BRCA2. Science. 2004;306:876–879. doi: 10.1126/science.1102574. [DOI] [PubMed] [Google Scholar]