Abstract
During lagging-strand DNA replication in eukaryotic cells primers are removed from Okazaki fragments by the flap endonuclease and DNA ligase I joins nascent fragments. Both enzymes are brought to the replication fork by the sliding clamp proliferating cell nuclear antigen (PCNA). To understand the relationship among these three components, we have carried out a synthetic lethal screen with cdc9-p, a DNA ligase mutation with two substitutions (F43A/F44A) in its PCNA interaction domain. We recovered the flap endonuclease mutation rad27-K325* with a stop codon at residue 325. We created two additional rad27 alleles, rad27-A358* with a stop codon at residue 358 and rad27-pX8 with substitutions of all eight residues of the PCNA interaction domain. rad27-pX8 is temperature lethal and rad27-A358* grows slowly in combination with cdc9-p. Tests of mutation avoidance, DNA repair, and compatibility with DNA repair mutations showed that rad27-K325* confers severe phenotypes similar to rad27Δ, rad27-A358* confers mild phenotypes, and rad27-pX8 confers phenotypes intermediate between the other two alleles. High-copy expression of POL30 (PCNA) suppresses the canavanine mutation rate of all the rad27 alleles, including rad27Δ. These studies show the importance of the C terminus of the flap endonuclease in DNA replication and repair and, by virtue of the initial screen, show that this portion of the enzyme helps coordinate the entry of DNA ligase during Okazaki fragment maturation.
CELLULAR maintenance of genomic integrity is essential for the continued viability of all organisms. The fidelity of DNA replication has to be maintained and DNA insults have to be repaired to ensure that deleterious mutations are not passed on to progeny or cause cancerous growth. A number of cellular proteins have multiple roles in DNA replication, mutation avoidance, and repair. In Saccharomyces cerevisiae, the flap endonuclease, proliferating cell nuclear antigen (PCNA), and DNA ligase I encoded by RAD27, POL30, and CDC9, respectively, are all required for proper replication and also function to avoid mutation and to facilitate repair.
The flap endonuclease, FEN-1 in humans, is a highly conserved structure-specific nuclease that has both endonuclease and 5′–3′ exonuclease activity. During lagging-strand replication these activities function to remove primers from Okazaki fragments, either by endonucleolytic cleavage of a flap made by strand displacement (Liu et al. 2004) or by sequential exonucleolytic removal of single nucleotides at the 5′ end of the primer (Murante et al. 1994).
While deletion of RAD27 is not lethal to yeast cells, the rad27Δ mutant exhibits temperature-sensitive growth, is a mutator, and undergoes genomic instability (Johnson et al. 1995; Reagan et al. 1995; Tishkoff et al. 1997b; Chen and Kolodner 1999). In addition, its sensitivity to low doses of the methylating agent methylmethane sulfonate (MMS) implicates the participation of the enzyme in base excision repair (BER) (Reagan et al. 1995; Wu and Wang 1999). rad27Δ mutants have been reported to be either mildly sensitive to UV light or not sensitive to UV light (Reagan et al. 1995; Sommers et al. 1995). In the strain background that the mutant is mildly sensitive, its combination with rad2Δ yields a double mutant more sensitive than each single mutant, implying that the enzyme does not participate in RAD2-mediated nucleotide excision repair (NER) (Reagan et al. 1995). The flap endonuclease has also been implicated in double-strand break (DSB) repair by virtue of the incompatibility of rad27Δ with mutations of the DSB repair pathways (Tishkoff et al. 1997b; Symington 1998). In addition, either the yeast enzyme or its human ortholog has been shown to participate in reactions of homologous recombination, nonhomologous end joining, and telomere maintenance (Parenteau and Wellinger 1999, 2002; Wu et al. 1999; Wang et al. 2004; Kikuchi et al. 2005). Curiously, the rad27Δ mutant is not sensitive to gamma radiation but is sensitive to high doses of MMS that are thought to act as a radiomimetic agent (Reagan et al. 1995; Sommers et al. 1995).
PCNA is the replicative clamp that acts as a scaffold to facilitate the loading of DNA replication and repair proteins, including DNA ligase I and the flap endonuclease to DNA (Warbrick 2000, 2006; Maga and Hubscher 2003). PCNA (POL30) is essential for cell viability, which is indicative of its central role in DNA metabolism. Biochemical characterization of its effect on the flap endonuclease shows that it stimulates its activity ∼50-fold, evidencing the productive nature of the interaction (Gomes and Burgers 2000; Tom et al. 2000; Frank et al. 2001; Stucki et al. 2001). The ability of DNA ligase to efficiently catalyze the formation of phosphodiester bonds in the DNA backbone may also be facilitated by its binding to PCNA. Tom et al. (2001) showed that, in vitro, PCNA enhances the ligation reaction 5-fold and that the stable association of DNA ligase with nicked duplex DNA requires PCNA.
Both DNA ligase and the flap endonuclease bind to PCNA via their respective PCNA interactive peptide domains (PIP box). The PIP box is a conserved sequence motif of the amino acids QXXLXXFF. The PIP box fits into the interdomain connector loop (IDCL) of PCNA to provide a protein–protein interaction surface (Gomes and Burgers 2000; Chapados et al. 2004; Sakurai et al. 2005; Pascal et al. 2006). Mutations in the PIP box or the IDCL that impair the interaction of DNA ligase and the flap endonuclease to PCNA lead to genomic instability (Amin and Holm 1996; Eissenberg et al. 1997; Gary et al. 1999; Refsland and Livingston 2005; Subramanian et al. 2005). We have reported that the double mutants made by combinations of cdc9-p, rad27-p, and pol30-90—mutations with alterations of the PIP box or the IDCL in the respective proteins—have synergistic phenotypes with respect to MMS sensitivity and to trinucleotide repeat instability (Refsland and Livingston 2005). These results suggest that the two enzymes function in a concerted manner that is facilitated by PCNA.
The precise nature of how PCNA coordinates the entry of the flap endonuclease and DNA ligase into the replication fork is not well understood. Biochemical and structural studies have begun to elucidate a possible ordering of these PCNA-mediated interactions. The possibility of such an ordering is underscored by the observation that DNA ligase adopts a toroidal conformation by completely encircling duplex DNA while interacting with PCNA (Pascal et al. 2004). Moreover, both PCNA and DNA ligase may be loaded onto the DNA in a mechanism utilizing the replication clamp loader replication factor C (RFC) (Levin et al. 2004; Vijayakumar et al. 2009), again suggesting a complete encirclement of the DNA by DNA ligase as well as by PCNA. PCNA and DNA ligase are similar in size and their interaction is likely to extend along the face of PCNA in a manner that would prevent other proteins such as the flap endonuclease from binding to the IDCL (Pascal et al. 2004, 2006). A biochemical study with purified yeast proteins showed that the two enzymes cannot bind simultaneously to PCNA (Subramanian et al. 2005). These studies suggest that a coordinated sequential interaction among PCNA, DNA ligase, and the flap endonuclease is important for replication and repair.
Alternatively, both the flap endonuclease and DNA ligase may bind to the same molecule of PCNA. Since PCNA is a homotrimer, DNA ligase can potentially bind to one monomer while the flap endonuclease binds to another, using its extended C-terminal tail in a conformation allowing it to be tethered to PCNA concurrently with DNA ligase (Gomes and Burgers 2000; Sakurai et al. 2005). DNA ligase could also bind to PCNA in an extended conformation while the flap endonuclease cleaves the DNA. Sulfolobus solfataricus DNA ligase has been shown to have an open, extended conformation while binding to PCNA (Pascal et al. 2006). Presumably, once the flap endonuclease has removed the 5′ flap, DNA ligase acquires a closed, ring-shaped conformation to catalyze the joining of Okazaki fragments (Pascal et al. 2006).
Exactly how the interaction of these enzymes with PCNA is coordinated in vivo, whether singly or concurrently, is not well understood. To further elucidate how the interaction of DNA ligase with PCNA is ordered, we performed a genetic screen to identify mutations that are synthetically lethal with cdc9-p (F44A/F35A), an allele of DNA ligase that has impaired binding to PCNA (Refsland and Livingston 2005; Subramanian et al. 2005). We postulated that genes recovered from this screen would function in DNA repair, replication, and recombination or would be involved in ordering the DNA ligase–PCNA interaction. From the screen we recovered a truncated allele of RAD27, rad27-K325*. This allele encodes a protein that lacks the PIP box and the entire C-terminal domain of the enzyme but retains the N terminus containing the nuclease activities. We have characterized this allele and compared it to two other rad27 alleles in which we have created different alterations of the C-terminal end of the flap endonuclease.
MATERIALS AND METHODS
Yeast strains:
All S. cerevisiae strains used in this study, with the exception of SSL938, are isogenic to SSL204 (ura3 ade2 his3 trp1 leu2) and are listed in Table 1 (Dornfeld and Livingston 1991). Only differences between this strain and the derived strains are noted. Deletion strains were created in this background by single-step disruption (Rothstein 1983) and allelic substitutions were made using two-step transplacement (Scherer and Davis 1979). In some cases mutations vital to the synthetic-lethal screen, e.g., ura3 and ade2, are shown for clarity. Strain SSL938 was derived from ura3, his3, trp1, and CAN1 strains in our collection that did not contribute to the SSL204 lineage and was made rad27Δ by single-step disruption.
TABLE 1.
Yeast strains
| Strain | Genotype | Source |
|---|---|---|
| SSL204 | his3Δ200 trp1 leu2 ade2-101 ura3-52 | Dornfeld and Livingston (1991) |
| SSL254 | MATalys5Δ∷HIS3 | This study |
| SSL533 | MATarad27Δ∷HIS3 | Schweitzer and Livingston (1998) |
| SSL534 | MATα rad27Δ∷HIS3 | Schweitzer and Livingston (1998) |
| SSL535 | MATarad2Δ∷URA3 | This study |
| SSL539 | MATaexo1Δ∷URA3 | This study |
| SSL543 | MATapol30-79 | Schweitzer and Livingston (1999) |
| SSL544 | MATapol30-90 | Schweitzer and Livingston (1999) |
| SSL545 | MATα pol30-90 | Schweitzer and Livingston (1999) |
| SSL612 | MATacdc9-1 | Ireland et al. (2000) |
| SSL615 | MATadna2-1 | Ireland et al. (2000) |
| SSL630 | MATacdc9Δ∷KanMX {pRS316.CDC9} | This study |
| SSL631 | MATα cdc9Δ∷KanMX {pRS316.CDC9} | This study |
| SSL632 | MATacdc9-p | Refsland and Livingston (2005) |
| SSL633 | MATα ade3Δ∷hisG | This study |
| SSL634 | MATaade3Δ∷hisG cdc9-p | This study |
| SSL635 | MATα ade3Δ∷hisG cdc9-p | This study |
| SSL636 | MATaade3Δ∷hisG cdc9-p lys5Δ∷HIS3 | This study |
| SSL637 | MATα ade3Δ∷hisG cdc9-p lys5Δ∷HIS3 | This study |
| SSL638 | MATaade3Δ∷hisG cdc9Δ∷KanMX {pRS316.CDC9} lys5Δ∷HIS3 | This study |
| SSL639 | MATα ade3Δ∷hisG cdc9Δ∷KanMX {pRS316.CDC9} lys5Δ∷HIS3 | This study |
| SSL642 | MATaFUR1∷HIS3∷FUR1 | This study |
| SSL643 | MATα FUR1∷HIS3∷FUR1 | This study |
| SSL644 | MATaade3Δ∷hisG cdc9-p FUR1∷HIS3∷FUR1 | This study |
| SSL651 | MATα ade3Δ∷hisG cdc9-p FUR1∷HIS3∷FUR1 | This study |
| SSL649 | MATarad27-p | Refsland and Livingston (2005) |
| SSL650 | MATα rad27-p | Refsland and Livingston (2005) |
| SSL667 | MATα cdc9-p rad27-p | Refsland and Livingston (2005) |
| SSL826 | MATarad51Δ∷ADE2 | Asleson and Livingston (2003) |
| SSL927 | MATarad27-A358*-isolate1 | This study |
| SSL928 | MATarad27-A358*-isolate2 | This study |
| SSL929 | MATarad27-pX8-isolate1 | This study |
| SSL930 | MATarad27-pX8-isolate2 | This study |
| SSL931 | MATacdc9-p rad27-pX8 | This study |
| SSL933 | MATacdc9-p rad27-A358* | This study |
| SSL935 | MATarad27-K325*-isolate1 | This study |
| SSL936 | MATα rad27-K325*-isolate2 | This study |
| SSL938 | MATarad27Δ∷HIS3 | This study |
| SSL944 | MATaexo1Δ∷URA3 rad27-pX8 | This study |
| SSL955 | MATaexo1Δ∷URA3 rad27p | This study |
| SSL957 | MATaexo1Δ∷URA3 rad27-358* | This study |
Media:
Media were prepared according to standard recipes (Burke et al. 2000) except that synthetic media were buffered with succinic acid–NaOH as previously described (Hartwell 1970). In particular, the omission media in these studies were made from a single batch of nutrients that were mixed from solid ingredients in the proportion previously described (Burke et al. 2000) but lacked uracil, leucine, and tryptophan. When necessary, these were added in the amount previously specified (Burke et al. 2000).
Synthetic lethal screen:
To screen for mutations that are synthetically lethal in combination with cdc9-p, a cdc9-p strain was modified to make it both ade3 and FUR1∷HIS3∷FUR1. Tester strains were also constructed to test for dominance (cdc9-p ade3Δ∷hisG lys5∷HIS3) and to test for inactivating mutations in CDC9 (cdc9Δ∷KanMX {pRS316.CDC9.ADE3} ade3Δ∷hisG lys5∷HIS3).
The synthetic lethal screen was carried out using a red/white colony-sectoring assay in an ade2 ade3 background (Kranz and Holm 1990). The strain was modified by duplicating FUR1 to eliminate false positive mutations (fur1) in uracil utilization (Koren et al. 2003). The modified cdc9-p strain was transformed with the plasmid pRS316.ADE3.CDC9. Approximately 300 transformed cells were spread on YPD media and mutagenized by exposure to 3.4 mW/cm2 of UV light (253.7 nm UV-C) to 60% survival. Cells that grew at 30° and did not appear to contain white sectors were streaked onto agar containing 5-FOA. Those that proved to require pRS316.ADE3.CDC9 for survival were tested for dominance and for the absence of inactivating mutations of CDC9. The tester strain for dominance could also be used to test the segregation pattern of the synthetic lethal mutation. At this point candidates that were recessive and segregated as a single mutation were outcrossed to the wild-type parent to recover the synthetic lethal mutation in a CDC9 background. Of the candidates that were recessive and segregated as a single mutation, we found two that also conferred temperature-sensitive growth at 35°. Because mutations in POL30 (PCNA) might have caused a synthetic lethal phenotype and temperature-sensitive growth, we transformed the two candidates with pRS316.POL30. One candidate grew better when transformed, and we chose this candidate for further study.
Identification of the synthetic lethal mutation:
To identify the mutation causing synthetic lethality with cdc9-p, we transformed the outcrossed mutant with a yeast genomic library (Jauert et al. 2005). We recovered plasmids with the genomic fragment containing APN1, RAD27, and ABF1. We took a candidate gene approach and transformed the isolate with pRS316.RAD27, which rescued the temperature lethality. The rad27 allele from the candidate was recovered by PCR and subjected to DNA sequencing. The copy had a stop codon at amino acid residue 325 in RAD27.
rad27 alleles:
We recreated the rad27 mutation, rad27-K325*, that we had recovered, in our wild-type strain along with a flanking restriction enzyme recognition sequence that did not further alter the gene. We also made two additional alleles, rad27-A358* and rad27-pX8 (339QGRLDGFF346–339AAAAKAAA346) by the same methods. Because of the propensity of rad27 mutations to undergo genetic changes, each of our mutations as well as rad27Δ was backcrossed to our wild-type parental strain. The resulting diploids were sporulated, and only strains that yielded a majority of complete tetrads were used. Mutant isolates were confirmed by PCR and restriction digestion. Two isolates (spore colonies) of each mutation were used in our studies, although only one is shown.
Mutation avoidance assays:
The rate of spontaneous forward mutation to canavanine resistance was measured in the rad27 mutants with or without the PCNA overexpression plasmids, pRS424.POL30 or pRS425.POL30. The canavanine mutagenesis assay was modified from methods described previously (Marsischky et al. 1996; Sia et al. 1997). At least three independent experiments were performed for each strain. Mutation rates were estimated using the method of the median (Lea and Coulson 1949). Statistical differences were determined by averaging three or more rate estimations and comparing values using Student's t-test.
To measure the rate of dinucleotide repeat instability, plasmid pSH44 was introduced into our strains, grown in media selective for tryptophan, and plated to medium containing 5-FOA as previously described (Henderson and Petes 1992; Kokoska et al. 1998). Mutation rates were estimated using the method of the median (Lea and Coulson 1949). Statistical differences were determined by averaging three or more rate estimations and comparing values using Student's t-test.
MMS sensitivity:
All rad27 strains with or without pRS424.POL30 or pRS426.POL30, and cdc9-p rad27-p with or without pRS426.rad27-K325*, were tested for MMS resistance at 25° on the basis of the method previously described (Boundy-Mills and Livingston 1993). The means of at least three independent experiments were used to generate survival curves.
Growth capability:
The temperature sensitivity of the rad27, cdc9-1, and dna2-1 strains with or without pRS424.POL30 or pRS426.rad27-K325* was examined by performing spotting assays. Log phase cultures (OD600 ∼ 0.6–1) were grown in appropriate media and serially diluted in a 96-well microtiter dish. Spots were made on media such that the highest-density spot contained 15,000 cells with 10-fold decreases in the following three consecutive spots. The plates were incubated at 25° and 35° for 4 days unless otherwise specified.
RESULTS
Identification of rad27-K325* in a synthetic lethal screen with cdc9-p:
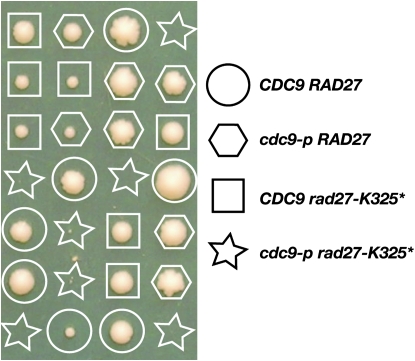
We screened for mutations that confer synthetic lethality with the DNA ligase I mutation cdc9-p that has substitutions of the two aromatic residues of the enzyme's PIP box (F43A/F44A). The cdc9-p mutant does not exhibit growth defects, but the mutated copy of DNA ligase I binds weakly to PCNA and causes instability of CAG repeat tracts (Refsland and Livingston 2005; Subramanian et al. 2005). To recover mutations that are synthetically lethal with cdc9-p, we introduced a wild-type copy of CDC9 on a URA3 ADE3-containing plasmid into the cdc9-p strain, mutagenized it, and screened for colonies that could not sector or grow on agar containing 5-FOA. We next screened the isolates for additional phenotypes that might suggest an involvement in DNA replication or repair. One isolate was temperature sensitive for growth and partially suppressible by high-level expression of PCNA (POL30) on YPD. Rescue of the temperature sensitivity with a yeast genomic library identified clones containing RAD27 encoding the flap endonuclease. DNA sequencing of the RAD27 and POL30 alleles in the isolate revealed a stop codon at position 325 in RAD27 and no mutations in POL30. The rad27 mutation rad27-K325* was recreated and used to replace the wild-type copy in our parental wild-type strain. To authenticate that rad27-K325* is sufficient to yield the synthetic lethal phenotype with cdc9-p, the rad27-K325* allele was mated to the parental cdc9-p strain and the diploid was sporulated. The spore colonies (Figure 1) show that each single mutant is able to germinate and to form colonies, but the rad27-K325* cdc9-p double mutant cannot.
Figure 1.—
Synthetic lethality of rad27-K325* combined with cdc9-p (F43A/F44A). A diploid heterozygous for both mutations was sporulated, and the spore colonies were germinated on YPD agar at 25°. The genotypes of viable spore colonies were determined by restriction enzyme digestion of PCR products of the RAD27 and CDC9 alleles.
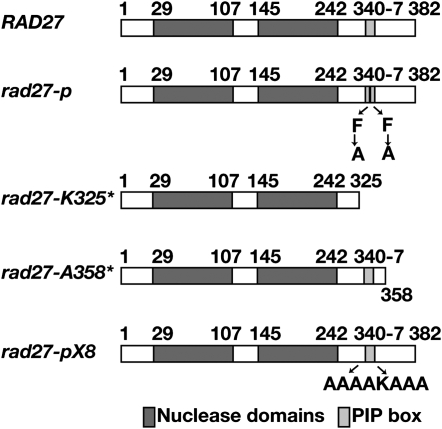
Because we already found a genetic interaction between cdc9-p and rad27-p (Refsland and Livingston 2005), the corresponding PIP box mutations of the aromatic residues of the flap endonuclease (F346A/F347A) (Gary et al. 1999) and the identification of an additional rad27 allele that disturbs the axis among the flap endonuclease, DNA ligase, and PCNA warranted further examination. The position of the stop codon in rad27-K325* leaves the nuclease domains of the flap endonuclease intact but removes other domains. The deleted domains include (1) a hinge region that permits rotation of PCNA-bound flap endonuclease to reach the DNA, (2) the PIP box, (3) residues C-terminal to the PIP box that bind to PCNA, and (4) additional C-terminal domains that may be necessary for nuclear import, protein–protein interactions, and post-translational modifications (Liu et al. 2004) (Figure 2). Co-crystallization of the human flap endonuclease with PCNA showed that the enzyme makes contacts with PCNA through a number of its domains. The flap endonuclease makes contacts with PCNA through its core domain, through residues N-terminal to the PIP box, through the PIP box, and through residues C-terminal of the PIP box (Sakurai et al. 2005). The most extensive interactions are between the PIP box of the flap endonuclease and the hydrophobic pocket of the IDCL of PCNA and between C-terminal residues of the flap endonuclease and additional residues of the IDCL. To better understand how these regions of the flap endonuclease contribute to its interaction with PCNA, we made two additional rad27 alleles. One allele substitutes α-helical forming residues for the eight amino acids of the PIP box (rad27-339AAAAKAAA346, named rad27-pX8) and the other truncates the enzyme beyond the C-terminal residues that interact with the IDCL of PCNA (rad27-A358*) (Figure 2). These alleles, as well as the complete deletion allele rad27Δ and the original flap endonuclease PIP box allele rad27-p, were used for subsequent characterizations and comparisons.
Figure 2.—
rad27 alleles.
Phenotypic characterizations of rad27alleles:
To learn whether these rad27 mutations possess unique phenotypes and to distinguish one from another and from the null mutation, we carried out assays for growth, mutagenesis, and DNA repair. Because rad27 mutants are genetically unstable, our characterizations were carried out with two isolates of each mutant, although only one is shown. In all tests the two isolates had the same phenotype, including statistical differences when calculated.
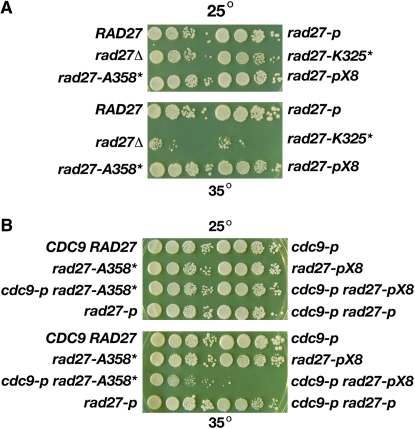
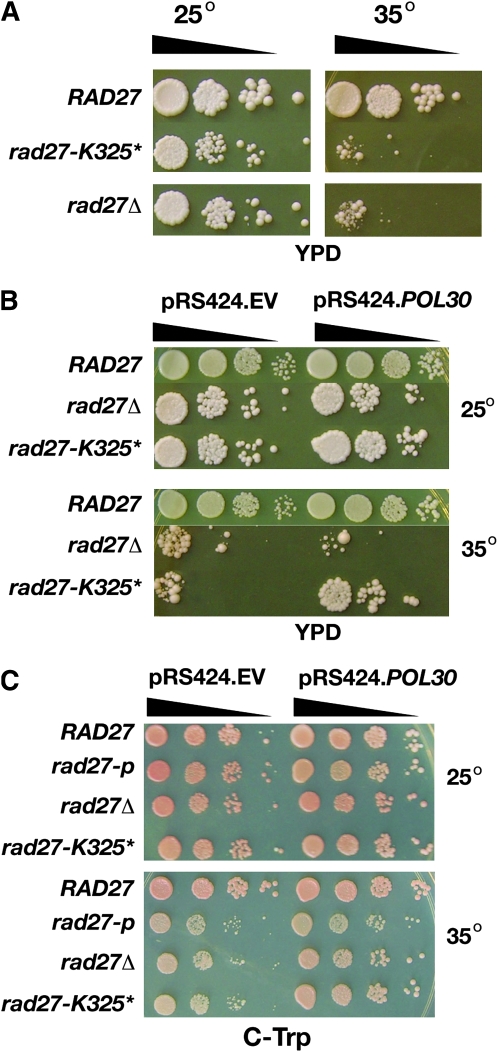
To begin, we determined whether the strains are temperature sensitive for growth at 35°. Both rad27Δ and rad27-K325* exhibit temperature-sensitive growth whereas rad27-A358* and rad27-pX8 grow robustly at 35° on YPD (Figure 3A). Thus, the loss of the C terminus that eliminates all PCNA interacting residues (save for the few interactions between the core of the enzyme and PCNA) is more deleterious than the other mutations that retain either the PIP box (rad27-A358*) or the residues that interact with the hydrophobic pocket of the IDCL (rad27-pX8).
Figure 3.—
Temperature-sensitive growth of rad27 mutants. (A) Growth of mutants at 25° and 35° on YPD agar. (B) Growth of cdc9-p rad27 double mutants at 25° and 35° on YPD agar.
While neither rad27-pX8 nor rad27-A358* confer temperature-sensitive growth as single mutations, rad27-pX8 exhibits temperature-sensitive growth as a double mutant with cdc9-p, and rad27-A358* confers slow growth at the restrictive temperature when combined with cdc9-p (Figure 3B). This also contrasts them with the rad27Δ and rad27-K325* mutants that have an absolute lethality with cdc9-p. The temperature-sensitive lethality of rad27-pX8 and the slow growth phenotype of rad27-A358* with cdc9-p are another indication of the balance that must be achieved among the flap endonuclease and DNA ligase I in binding to PCNA.
Mutation to canavanine resistance and repeat tract instability:
As part of our initial characterization of the rad27 alleles, we determined the rate of spontaneous forward mutation to canavanine resistance. The rate of mutagenesis is significantly higher in all the rad27 alleles tested, apart from rad27-p, when compared to the wild type (P < 0.05) (Table 2). The rad27Δ and rad27-K325* alleles confer mutation rates >100-fold that of the wild type. The other two rad27 alleles, rad27-A358* and rad27-pX8, have 3-fold and 17-fold higher mutation rates than that of the wild type, respectively. These results show the relative importance of the C-terminal domains of the flap endonuclease in mutation avoidance. In particular, the mutation rate of the rad27-pX8 mutant shows the necessity of a complete PIP box in mutation avoidance. The mutation rate of rad27-K325* is greater than the product of the mutation rates of rad27-A358* and rad27-pX8. The rad27-K325* mutation eliminates the PIP box and all residues C-terminal to it. In particular, it eliminates a set of residues C-terminal to the PIP box that are retained by the rad27-A358* mutation. These residues contact the IDCL of PCNA and their elimination in rad27-K325* may account for the mutant's synergistic mutation rate.
TABLE 2.
Mutation rates in rad27 mutants
| Strain | Mutation rate (×10−7 events/cell/generation) ± SDab |
|---|---|
| RAD27 | 1.1 ± 0.7 (1) |
| rad27-p | 1.2 ± 0.6 (1) |
| rad27Δ | 129 ± 77 (117) |
| rad27-K325* | 165 ± 50 (150) |
| rad27-A358* | 3.1 ± 1 (3) |
| rad27-pX8 | 18 ± 7 (17) |
Mutation to canavanine resistance for cells grown in YPD media is shown.
SD, standard deviation.
Values normalized to the RAD27 rate are shown in parentheses. All the rad27 mutants, apart from rad27-p, have mutation rates significantly higher (P < 0.05) than the wild-type rate.
We also made another test of mutation avoidance, using a standard assay for dinucleotide repeat instability. We introduced a plasmid containing a run of GT repeats embedded in frame in URA3 into the rad27 mutants (Henderson and Petes 1992). As in the canavanine resistance assay, the rad27-K325* mutation confers a mutation rate to Ura− that is as great, if not greater, than rad27Δ (Table 3). The rad27-pX8 and rad27-A358* mutations confer rates proportional to their mutation rate to canavanine resistance. While the product of the mutation rates to Ura− conferred by these two mutations is greater than the rate found for the rad27-K325* mutation, the sum of the two is less than the rad27-K325* rate, again suggesting that the extra portion of the flap endonuclease between residues 347 and 358 is important for mutation avoidance.
TABLE 3.
Dinucleotide repeat instability in rad27 alleles
| Strain | Mutation rate (×10−6 events/cell/generation) ± SDab |
|---|---|
| RAD27 | 1 ± 0.3 |
| rad27-p | 2 ± 1 |
| rad27Δ | 604 ± 309 |
| rad27-K325* | 810 ± 164 |
| rad27-A358* | 22 ± 1 |
| rad27-pX8 | 272 ± 21 |
Mutation to 5-FOA resistance for cells harboring pSH44 grown in omission media is shown.
SD, standard deviation.
All the rad27 mutants, apart from rad27-p, have mutation rates significantly higher (P < 0.05) than the wild-type rate.
Characterization of rad27 alleles in combination with exo1Δ, rad2Δ, and rad51Δ:
The involvement of the flap endonuclease in multiple DNA transactions is evidenced by the genetic interaction of rad27Δ with mutations of the repair and recombination genes EXO1, RAD2, and RAD51. To characterize and to distinguish among our rad27 alleles, we mated them to exo1Δ, rad2Δ, and rad51Δ. We sporulated the resulting diploids and germinated the spores at 25° on YPD. If the spores germinated, we then tested their growth at 35° (supporting information, Table S1).
EXO1 encodes exonuclease I, a 5′–3′ exonuclease (Tishkoff et al. 1997a). rad27Δ is synthetically lethal in combination with exo1Δ (Tishkoff et al. 1997a). This lethality is thought to occur because Exo1 has 5′-flap cleavage capability that is redundant with the flap endonuclease activity of the enzyme (Tran et al. 2002). In addition, high-level expression of the nucleases encoded by EXO1 suppresses the hypermutation phenotype of rad27Δ (Sun et al. 2003). Like the rad27Δ exo1Δ double mutant, the rad27-K325* exo1Δ double mutant is inviable (Table S1). rad27-pX8 exo1Δ is viable at 25° but exhibits a temperature-sensitive phenotype at 35°. rad27-A358*, the mutant with the least severe phenotype, is viable in combination with exo1Δ at both temperatures. These data show that the rad27Δ and rad27-K325* mutants, which are temperature sensitive and have high mutation rates, have an absolute requirement for EXO1. The EXO1 requirement for growth of rad27-pX8 at 35° shows the need for this nuclease to support viability of the flap endonuclease mutant lacking all the residues of its PIP box.
RAD2 encodes an endonuclease homologous to the flap endonuclease and the EXO1 encoded nuclease that is required for nucleotide excision repair. In contrast to the inviability of the exo1Δ rad27Δ double mutant, the rad2Δ rad27Δ double mutant is viable, indicating that other nucleases can support DNA replication and repair (Reagan et al. 1995; Sommers et al. 1995). Like the suppression of rad27Δ by high-level expression of EXO1, so too can high-level expression of RAD2 suppress the mutation avoidance phenotype of rad27Δ (Sun et al. 2003). When we combined rad2Δ with our rad27 alleles, all the double mutants are viable at 25°. However, at 35°, the rad2Δ double mutants with rad27Δ and rad27-K325* are temperature sensitive. The double mutants of the less severe mutations, rad27-pX8 and rad27-A358*, are viable at 35° (Table S1). Because both rad27Δ and rad27-325* are temperature sensitive as single mutants, the inviability of the rad2Δ double mutants does not yield information on the contribution of RAD2 to their inviability.
RAD51 encodes the strand exchange protein that facilitates double-strand break repair. Previous studies have shown that the lesions caused by the absence of the flap endonuclease are repaired via the recombinational repair pathway (Symington 1998). When we attempted to make double mutants of each of our rad27 alleles with rad51Δ, we found that, with the exception of the two alleles with the least severe phenotypes, rad27-p and rad27-A358*, all are inviable at 25° (Table S1 and Refsland and Livingston 2005). The rad27-A358* rad51Δ double mutant is temperature sensitive at 35° (Table S1). This temperature-induced lethality is interesting because neither the rad51Δ nor the rad27-A358* single mutant is temperature sensitive. Furthermore, the rad27-A358* mutant exhibits slight sensitivity to MMS and has a slightly elevated mutation rate. The synthetic lethality of the rad27-A358* rad51Δ double mutant at 35° suggests that the DSB repair pathway is essential to repair the few lesions generated in the rad27-A358* mutant.
MMS sensitivity of rad27 mutants:
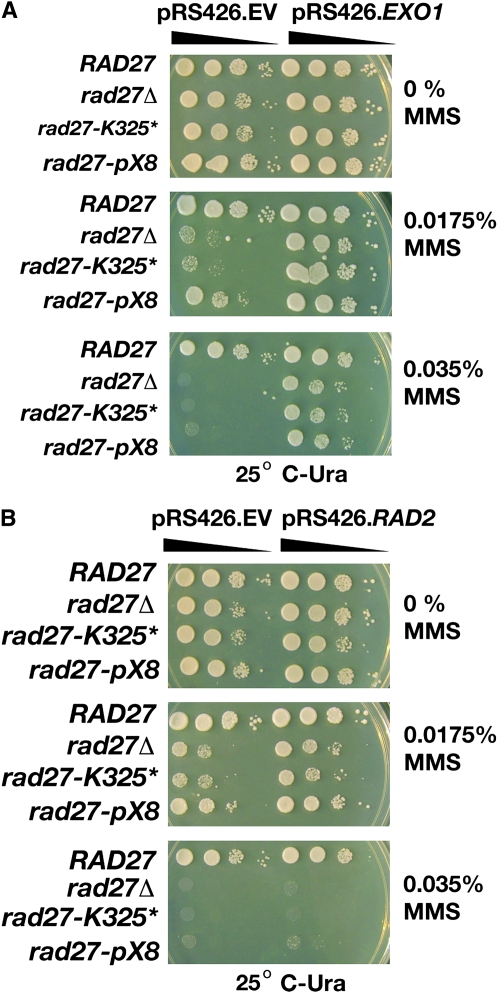
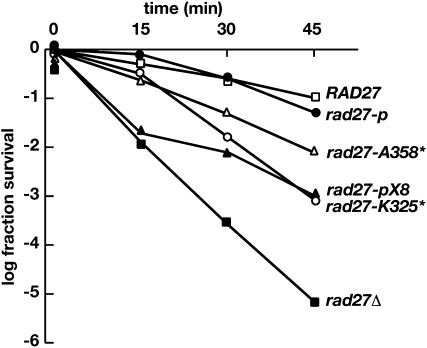
We also examined the sensitivity of the rad27-K325* and rad27-pX8 mutants to two tests of MMS sensitivity, one at relatively low concentrations in agar that reports base excision repair and one at a high concentration where the chemical acts as a radiomimetic agent. rad27-K325* has a sensitivity much like rad27Δ at concentrations of MMS of 0.0175 and 0.035% (Figure 4). The rad27-pX8 mutant is partially resistant at the lower of these two concentrations (Figure 4). The sensitivity can be suppressed by high-copy expression of EXO1 (Figure 4A) and to a lesser extent by high-copy expression of RAD2 (Figure 4B).
Figure 4.—
Growth of rad27 mutants on MMS agar. The mutants were spotted on selective agar containing MMS at 25°. (A) Strains were transformed with either pRS426 (EV empty vector) or pRS426.EXO1. (B) Strains were transformed with either pRS246 (EV empty vector) or pRS426.RAD2.
We also incubated mutants in a higher concentration of MMS that produces double-strand breaks in the DNA. All of the rad27 mutants are susceptible to DNA damage induced by this concentration of MMS (Figure 5). The mutants exhibit a gradient in their sensitivity. rad27-p is nearly as resistant to MMS as the wild-type parent as previously observed (Gary et al. 1999; Refsland and Livingston 2005). rad27-A358* is sensitive but more resistant than either rad27-pX8 or rad27-K325*. Both the rad27-pX8 and rad27-K325* mutants retain partial resistance. In the case of the rad27-pX8 mutant, the decrease in viability describes a biphasic curve that was observed for both isolates that we examined (data not shown). Of greater interest to us is the partial resistance of the rad27-K325* mutant in comparison to the null mutant rad27Δ. This is an indication that the truncated protein product of the rad27-K325* allele retains partial biological function.
Figure 5.—
Survival of rad27 mutants to exposure to 0.5% MMS. Multiple survival studies were carried out for the strains contemporaneously, and the results were averaged. For clarity, the error for each time point is not shown.
High-level expression of rad27-K325* suppresses the temperature and MMS sensitivity phenotypes of the cdc9-p rad27-p mutant:
While the temperature sensitivity of its growth on YPD, its synthetic lethality in combination with cdc9-p, exo1Δ, rad2Δ, and rad51Δ, and its mutation rate do not distinguish the rad27-K325* mutant from the null, its partial resistance to MMS suggests that the mutant protein retains partial biological function. The truncated enzyme retains its nuclease domain, and other studies have shown that mutation of its PCNA binding motifs does not destroy its nuclease activity (Frank et al. 2001).
Another indication of the biological activity remaining in the rad27-K325* mutant is our observation that this mutant is capable of growth at 35° when plated on selective media (C−Ura) (Figure S1). Its ability to grow on selective media contrasts it to the null, which is unable to grow at on these media at 35°. In addition, when rad27-K325* is expressed in the rad27Δ strain, it confers growth (Figure S1). Thus, the rad27-K325* mutant enzyme retains partial function.
We have also measured whether the protein product of rad27-K325* is as abundant as the wild-type protein. Using immunoblotting to detect epitope-tagged proteins, we observed that both the wild-type and the mutant proteins are expressed at similar levels (Figure S2). Thus, partial activities that are retained by the rad27-K325* mutant are not likely the result of an unstable protein.
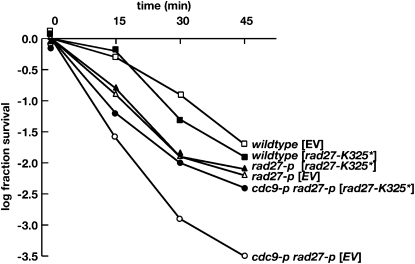
Finally, we transformed the rad27-K325* allele expressed from a high-copy plasmid into a strain with partial resistance to MMS to ask whether it could enhance or reduce the partial resistance. The strain we chose was the cdc9-p rad27-p double mutant that we had already characterized to have partial MMS resistance (Refsland and Livingston 2005). We tested the transformed strain for temperature and MMS sensitivity. The cdc9-p rad27-p double mutant grows weakly at 35°, and high-level expression of rad27-K325* suppresses the temperature-sensitive growth defect although the growth is not as robust as that of the wild type (Figure S3). In addition, high-level expression of rad27-K325* increases the MMS resistance of the double mutant nearly to wild-type resistance (Figure 6). These results suggest that the rad27-K325* enzyme, despite having its entire C terminus truncated, retains partial biological function. Furthermore, while the product of rad27-K325* might have interfered with the weakened binding of the rad27-p product to reduce the MMS resistance of the double mutant, it did not do so. While we cannot rule out that a physical interaction between the products of rad27-K325* and rad27-p accounts for the increase in function, the most straightforward conclusion is that the rad27-K325* enzyme retains partial activity.
Figure 6.—
Survival of the cdc9-p rad27-p double mutant to exposure to 0.5% MMS. Multiple survival studies were carried out for the strains contemporaneously, and the results were averaged. For clarity, the error for each time point is not shown. For these studies, the strains were grown in selective media, exposed to MMS, and plated on selective agar at 25°.
High-level expression of PCNA suppresses the temperature and hypermutagenic phenotypes of rad27-K325* and rad27Δ mutants:
Next, we revisited our initial observation that high-copy expression of POL30 (PCNA) partially suppresses the temperature-sensitive growth of the rad27-K325* mutant on YPD. As our initial observation was done using the mutagenized strain from which we identified the rad27-K325* mutation, we transformed the reconstituted rad27-K325* mutant with a high-copy plasmid expressing POL30 and assayed for growth at 25° and 35°. On YPD agar, overexpression of POL30 partially suppresses the temperature-sensitive growth defect of the rad27-K325* mutant (Figure 7, A and B). When the analyses were conducted on selective media, we observed that neither the null mutant nor the rad27-K325* mutant that had been transformed with the empty vector exhibited as profound a temperature growth deficit as they did on YPD agar at 35° (Figure 7C). (We note the variation in growth of rad27 mutants on rich and synthetic media in Figure 7 and previous figures. This variation among different media is reproducible.) When transformed with the plasmid that expresses PCNA, each mutant grows better as seen by the larger colonies that form (Figure 7C). Thus, a high level of PCNA has a suppressive effect on both mutations.
Figure 7.—
PCNA suppression of rad27 mutant growth defects on YPD and selective agar. (A) Temperature sensitive growth on YPD agar. (B) YPD agar with pRS424 or pRS424.POL30. (C) Selective agar with pRS424 or pRS424.POL30.
In an attempt to provide a quantitative demonstration of the suppressive effects of POL30 on the two alleles, we measured doubling times. High-level expression of POL30 reduced significantly (P < 0.05) the doubling time of two rad27-K325* isolates by 51 and 22 min when grown at 36° in liquid culture. In the case of the null allele, we did not measure a reduction in doubling time when PCNA was overexpressed. The results show that suppression of rad27-K325* by high-level expression of PCNA is more pronounced than suppression of rad27Δ. Thus, while high-level expression of PCNA ameliorates the growth phenotypes of both rad27-K325* and rad27Δ, the differences in the degree of suppression suggest that more than one mechanism of suppression may be operating in the POL30 suppression of rad27-K235*.
We next tested whether high-copy expression of POL30 can suppress the canavanine mutation rate of the rad27 alleles. We found that overexpressing PCNA significantly (P < 0.05) decreases the rates of spontaneous mutation to canavanine resistance in all the rad27 mutants tested, including the null (Table 4). (rad27-A358* and rad27-p were not tested because they have a low rate of mutagenesis.) Overexpression of PCNA reduces the spontaneous mutation rate of canavanine resistance fourfold in the rad27-K325* mutant. Similarly, overexpression of PCNA reduces the spontaneous mutation rate fivefold in the rad27-pX8 mutant.
TABLE 4.
Mutation rates in rad27 mutants
| Mutation rate (×10−7 events/cell/generation) ± SDa
|
||
|---|---|---|
| Strain | Empty vectorb | POL30b |
| RAD27 | 1.8 ± .3 (1) | 2.3 ± 1 (1) |
| rad27Δ | 257 ± 65 (143) | 31 ± 4 (13) |
| rad27-K325* | 134 ± 22 (74) | 36 ± 9 (16) |
| rad27-pX8 | 62 ± 14 (35) | 12 ± 3 (5) |
| rad27Δc | 200 ± 38 | 49 ± 20 |
Mutation to canavanine resistance for cells grown in omission media.
SD, standard deviation.
Values normalized to the RAD27 rates for empty vector (pRS424) or for POL30, as appropriate, are shown in parentheses. For all strains except RAD27, the pRS424.POL30 rate is significantly lower (P < 0.05) than the empty vector rate.
Strain SSL938.
In accord with the suppressive effects of PCNA on colony growth, high levels of PCNA also significantly reduce the mutation rate in the null mutant (Table 4). To exclude the possibility that this PCNA suppression results from complementation of a hypomorphic pol30 mutation in our parental strain, we recovered and sequenced the endogenous POL30 gene, including its upstream region. Sequencing showed that our parental strain does not harbor any pol30 mutations. As further assurance, we repeated this study in a rad27Δ strain derived from a different strain background. The suppressive effect of high-level expression of PCNA on canavanine resistance was also seen in strain SSL938, a rad27Δ strain that is not isogenic to our rad27 alleles (Table 4). The ability of PCNA to suppress both hypomorphic and null rad27 alleles suggests that additional PCNA helps mutant cells by increasing flap endonuclease function in the hypomorphic mutants, rad27-K325* and rad27-pX8, or by providing an opportunity for another nuclease to take the place of the flap endonuclease or both.
We have also have tested whether high-copy expression of PCNA could suppress in other assays of mutation avoidance and DNA repair. High-copy expression of PCNA suppresses the incidence of GT repeat tract instability (Table S2). (Note that the plasmid expressing POL30 in the repeat tract assay carried a different nutritional selection marker from that in the other assay, LEU2 instead of TRP1. Thus, the suppressive response is not likely influenced by nutritional selection.) We did not find suppression of MMS sensitivity either on agar or in liquid culture (data not shown). The suppression recorded in some assays and not others may indicate differences in the protein and enzyme requirements in each assay and in their respective interactions with PCNA.
The high-copy suppression of rad27-pX8 afforded us the opportunity to examine the behavior of the rad27-pX8 exo1Δ double mutant. Both mutations have a mild mutator phenotype in comparison to rad27Δ. When combined, they produce a synergistic mutation rate that is noteworthy because of the low mutation rate of the exo1Δ mutant (Table 5). This genetic interaction is consistent with the temperature lethality of the double mutant although the mutation rates were determined at the permissive temperature (25°). Next, we introduced POL30 into these strains to determine the extent of suppression. High-copy expression of POL30 has previously been reported to suppress exo1Δ although the results show very slight suppression (Amin et al. 2001). Because the mutation rate to canavanine resistance is only slightly elevated in our exo1Δ mutant over the wild-type rate, we were unable to detect POL30 suppression. POL30 is able to suppress the rad27-pX8 exo1Δ double mutant, but it is unable to bring it down to the level of the most severe single mutant rad27-pX8 (Table 5). The results suggest that the synergism underlies a necessity for each enzyme to act on the other's deficiency and that the suppression by POL30 cannot make up for this necessity.
TABLE 5.
Mutation rates in exo1 and rad27-pX8 mutants
| Mutation rate (×10−7 events/cell/generation) ± SDa
|
||
|---|---|---|
| Strain | Empty vectorb | POL30b |
| RAD27 | 2.8 ± 0.7 (1) | 5.8 ± 3 (1) |
| exo1Δ | 12 ± 3 (4) | 11 ± 1 (2) |
| rad27-pX8 | 53 ± 7 (19) | 12 ± 3 (2) |
| exo1Δ rad27-pX8 | 289 ± 59 (103) | 99 ± 26 (17) |
Mutation to canavanine resistance for cells grown in omission media is shown.
SD, standard deviation.
Values normalized to the RAD27 rates for empty vector (pRS424) or for POL30, as appropriate, are shown in parentheses. The pRS424.POL30 rate is significantly lower (P < 0.05) than the empty vector rate for the rad27-pX8 and the exo1Δ rad27-pX8 mutants.
High-level expression of PCNA does not suppress the temperature-sensitive phenotypes of the DNA ligase I mutant cdc9-1 or the helicase–endonuclease mutant dna2-1:
To examine whether suppression by high-level expression of PCNA is specific to rad27 mutations, we tested two additional replication mutations. We chose mutations of the enzymes most closely associated with the activity of the flap endonuclease in DNA replication. During DNA replication, the flap endonuclease is thought to bring the DNA2 helicase–nuclease to the replication fork because the helicase–nuclease binds to the flap endonuclease (Budd and Campbell 1997; Stewart et al. 2006). However, PCNA is not known to bind to the DNA2-encoded helicase–nuclease. The DNA2 nuclease may participate in primer removal along with the flap endonuclease (Bae et al. 1998, 2002; Budd et al. 2000; Stewart et al. 2006). The role of the DNA2 helicase activity remains enigmatic, although it, along with the nuclease activity, is essential to the yeast cell (Budd et al. 1995, 2000). High-level expression of DNA2 suppresses rad27Δ (Budd and Campbell 1997). For our studies on PCNA suppression we chose to test the dna2-1 mutant that has an alteration in the enzyme's helicase domain and is temperature sensitive for growth (Budd and Campbell 1995). This dna2 mutation is synthetically lethal with rad27Δ (Budd and Campbell 1997). We also tested the DNA ligase I mutation cdc9-1 that is temperature sensitive for growth.
Each mutant was transformed with a high-copy plasmid harboring POL30 and grown at 25° and 35°. High-copy expression of POL30 does not suppress temperature-sensitive lethality of either mutant (data not shown). Comparisons among temperature-sensitive mutations of multiple genes are difficult because of potential differences in phenotypic characteristics. Nevertheless, these results show that the suppressive effect of PCNA is specific to the flap endonuclease mutants rather than a generic, ameliorating effect that high levels of PCNA might have on other DNA replication proteins.
rad27-K325* is synthetically lethal with pol30-79 and pol30-90:
Finally, because our mutations have altered the PCNA binding capacity of the flap endonuclease, we were interested in whether they could be combined with pol30 mutations that are known to affect the flap endonuclease. Mutational analysis of POL30 has identified two mutations of PCNA that have profound effects on flap endonuclease binding and activation (Eissenberg et al. 1997). pol30-79 (L126A/I128A) and pol30-90 (P252A/K253A) are mutations in the hydrophobic pocket of the IDCL and the C-terminal end of PCNA, respectively. In vitro, the pol30-79 mutant protein fails to interact with the flap endonuclease but manages to stimulate the nuclease activity. In contrast, the pol30-90 mutant protein binds to the flap endonuclease but fails to stimulate its nuclease activity (Gomes and Burgers 2000). In genetic studies, Eissenberg et al. (1997) reported that the pol30-79 rad27Δ and pol30-90 rad27Δ double mutants are viable but exhibit poor growth at 23°.
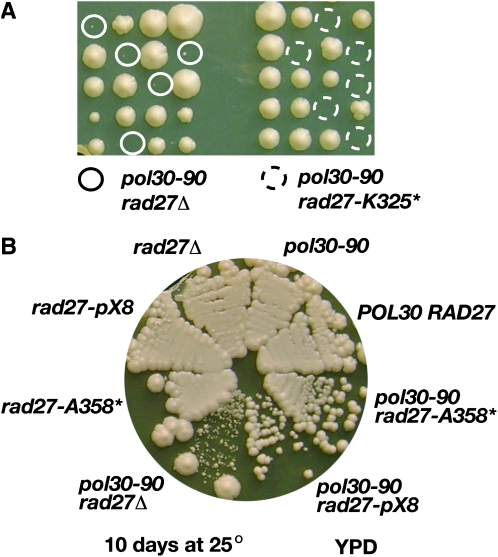
To determine whether our rad27 alleles are viable in combination with these PCNA mutations, we mated pol30-79 and pol30-90 with the rad27 alleles. The diploids were sporulated and dissected onto YPD at 25°. In our strain background both rad27Δ and rad27-K325* proved inviable with pol30-79, while the rad27-A358* pol30-79 and rad27-pX8 pol30-79 double mutants were viable but grew slower than either single mutant (data not shown). More interesting to us was the observation that the rad27Δ pol30-90 double mutant is viable and grows slowly at 25° [as previously observed (Eissenberg et al. 1997)], whereas the combination of rad27-K325* with pol30-90 is inviable (Figure 8A). We were able to streak out the rad27Δ pol30-90 spore colonies onto rich media where they formed microcolonies after 10 days at 25° (Figure 8B). As expected, the combinations of rad27-A358* and rad27-pX8 with pol30-90 yielded viable spores. When grown on rich media, these double mutants exhibit growth defects that are commensurate with the severity of their mutator and repair phenotypes (Figure 8B). For example, pol30-90 rad27-pX8 grows slower than does pol30-90 rad27-A358*. Thus, rad27-K325* is anomalous in that it exhibits synthetic lethality with pol30-90 that is not shown by the null mutation.
Figure 8.—
Interaction between rad27 mutations and the pol30-90 mutation. (A) Germination of spore colonies that have the genotype rad27Δ pol30-90 or rad27-K325* pol30-90. (B) The difference in colony size among the rad27 pol30-90 double mutants after 10 days of incubation at 25°.
DISCUSSION
PCNA is responsible for bringing the flap endonuclease and DNA ligase I into lagging-strand synthesis during DNA replication. How PCNA orders these final steps in lagging-strand synthesis is not well understood. The recovery of a C-terminal truncated mutation of the flap endonuclease missing its PCNA-interacting domains that is synthetically lethal with a mutation in DNA ligase I that weakens its interaction with PCNA contributes to our understanding of how the interactions among PCNA, the flap endonuclease, and DNA ligase I occur. The recovery of the rad27-K325* allele in a synthetic lethal screen with an allele of DNA ligase I, cdc9-p, and the mutant's genetic interactions with pol30-90 might not have been unexpected. Prior studies have shown a genetic interaction between mutations of the flap endonuclease and DNA ligase that weaken each enzyme's binding to PCNA, rad27-p and cdc9-p (Refsland and Livingston 2005), and between rad27Δ and various pol30 alleles (Eissenberg et al. 1997).
The rad27 allele we recovered from our screen, rad27-K325*, and the ones we created, rad27-A358* and rad27-pX8, have a spectrum of phenotypes that are consistent among most of the tests of growth, mutation avoidance, and DNA repair. In general, they fall within a gradient that extends from the null mutant with the most severe display of a phenotype to the wild type. In between, the rad27-K325* mutant is closest to the null mutant and the rad27-A358* mutant and the rad27-p mutant are closest to the wild type in their phenotypes. The rad27-pX8 mutant occupies a middle ground. The combinations of these rad27 alleles with rad2Δ, exo1Δ, cdc9-p, and rad51Δ exemplify a novel example of this gradient. While all rad27 mutations are viable in combination with rad2Δ at 25°, the most severe ones, rad27Δ and rad27-K325*, are temperature sensitive for growth, while the two others, rad27-pX8 and rad27-A358*, are not. This again suggests that rad2Δ does not exacerbate the mutant phenotypes of rad27 mutations. Next, while the most severe mutations are inviable when combined with exo1Δ, the rad27-pX8 double mutant is temperature sensitive for growth and the rad27-A358* mutant grows at the restrictive temperature. When the alleles are combined with cdc9-p, not only does the rad27-pX8 double mutant become inviable at 35° but also the rad27-A358* double mutant exhibits a growth defect at that temperature. Finally, while all mutations except rad27-A358* are inviable in combination with rad51Δ, this weakest mutation becomes temperature sensitive. The consistency among tests suggests that progressive loss or mutational substitution in the C-terminal tail plays nearly evenly among the myriad of functions that the flap endonuclease fulfills in the cell.
While the rad27 mutations fall into a pattern, each has properties that are unique. First, the rad27-K325* mutant, while it shares a number of phenotypes with rad27Δ, is different from rad27Δ. Like rad27Δ, rad27-K325* is lethal in combination with cdc9-p, confers temperature-sensitive growth, and is as mutagenic as rad27Δ. Similarly, both are partially suppressible by high-level expression of PCNA. Furthermore, when we combined rad27Δ and rad27-K325* with mutations of genes required for DNA repair, exo1Δ and rad2Δ, and recombination, rad51Δ, they proved to behave similarly. Both are synthetically lethal when combined with exo1Δ and rad51Δ and are temperature sensitive with rad2Δ.
rad27-K325* can be distinguished from rad27Δ in tests that show that the truncated protein is likely to retain some biological activity and in other tests that show that the mutant protein may confer a degree of toxicity to the cell. First, the rad27-K325* mutant is more resistant to MMS exposure than is the null mutant. The partial resistance suggests that the mutant protein is partially capable in the repair of the damage caused by the radiomimetic agent. Retention of partial activity might be expected because the mutant retains all the nuclease domains of the enzyme and the mutant protein appears as stable as the wild-type protein. Retention of partial activity in one assay (MMS resistance) and not in another (mutation avoidance) is likely a reflection of the multiple activities that the enzyme possesses. Depending on the substrate presented to the purified enzyme, it acts as a flap endonuclease, as an exonuclease, or as a gap endonuclease (Zheng et al. 2005; Singh et al. 2007). Furthermore, flap endonuclease mutations differentially affect the multiple activities (Xie et al. 2001; Zheng et al. 2005; Singh et al. 2007). Thus, the difference in phenotypic penetrance conferred by rad27-K325* may reflect the possibility that the mutant protein can act on some chromosomal substrates but not on others.
We have found that while high-level expression of rad27-K325* did not increase the level of MMS resistance of the null mutant beyond the level seen in the rad27-K325* strain (data not shown), high-level expression of rad27-K325* increases the resistance of the cdc9-p rad27-p double mutant to MMS exposure. Why the product of the rad27-K325* allele increases the MMS resistance of the cdc9-p rad27-p double mutant, where the PIP box of both enzymes has been mutated, is not obvious. Such results might be accounted for by physical interactions between the two mutated types of the flap endonuclease or by a partition of function between the two mutant types. The first explanation does not appear to be plausible because the flap endonuclease is not known to form multimers, the product of the rad27-K325* allele appears to be stable, and the product of the rad27-p allele is presumed to be stable by virtue of its near wild-type MMS resistance. The second explanation of a partition of function cannot be ruled out, but what function one mutant form could confer to the other is not apparent. We suggest that complementation may result from the manner in which the two mutant forms of the flap endonuclease bind to PCNA. Binding studies show that removal of the PIP box, as created in the rad27-K325* allele, surprisingly increases the affinity of the flap endonuclease for PCNA (Guo et al. 2008). Possibly, in the suppressed double mutant, the product of rad27-K325* binds first to PCNA and alters the conformation of PCNA so that the product of rad27-p is able to bind in an active conformation by a mechanism first suggested by Gomes and Burgers (2000).
The other assay that shows the difference between the rad27-K325* allele and the null allele is the lethality that the former has with pol30-90. While rad27Δ pol30-90 grows slowly, it is not lethal as is rad27-K325* pol30-90. This not only shows a difference between the two alleles but also shows that rad27-K325* may confer a slight toxicity to the cell when PCNA is mutated. We also note that in measures of mutation avoidance, the rad27-K325* mutant exhibits slightly greater promiscuity than the null mutant (Tables 2 and 3). Toxicity is not unprecedented, as a rad27 allele, rad27-n, with a mutation of the nuclease catalytic site is toxic to the yeast cell. This deleterious allele is detoxified by mutating the PIP box (Gary et al. 1999). Thus, some mutant forms of the flap endonuclease appear to bind to PCNA in a manner that inhibits the binding of other enzymes, in turn inhibiting DNA replication.
The rad27-pX8 mutation that substitutes α-helical forming residues for the eight amino acids of the PIP box is interesting because of the severity of its phenotypes in comparison to rad27-p, a mutation of two of the eight PIP box residues. It is distinctly more sensitive to MMS than rad27-p and lethal in combination rad51Δ. These phenotypes suggest a strong need for interaction between the flap endonuclease and PCNA in the incidence of DSBs or in recovery from DSBs. Furthermore, it becomes temperature sensitive when combined with exo1Δ. In the case of the exo1Δ rad27-pX8 double mutant, we note that its synergistic mutation rate at 25° places this rad27 mutation in a class of mutations uncovered in a screen for enhancers of the weak exo1Δ mutator phenotype (Amin et al. 2001). The simplest explanation for the temperature sensitivity of the exo1Δ rad27-pX8 double mutant is that the mutant enzyme becomes inactive, and the synthetic lethality reflects the synthetic lethality of the exo1Δ rad27Δ double mutant. Considering that the rad27-pX8 mutant is viable at 35° and, consequently, likely to retain partial function, the synthetic lethality may reflect the sensitivity of the exo1Δ mutant to diminished RAD27 function.
The rad27-A358* mutant is interesting because of its seemingly mild phenotypes in combination with its absolute requirement for CDC9 and RAD51 at 35°. In particular, its requirement for RAD51 implies that the mutant protein introduces DNA lesions that have an absolute requirement for the DSB repair system.
One result that comments strongly on the role of PCNA in mediating the events of DNA replication and repair is that overexpression of PCNA reduces the rates of spontaneous mutations not only in the two partial hypomorphic mutants rad27-K325* and rad27-pX8 but also in the null mutant. A possible explanation for suppression of rad27Δ is that the large quantity of PCNA recruits a novel nuclease that suppresses the mutant phenotype. Potential candidates for suppression of the null are the nucleases encoded by EXO1 and RAD2 and the nuclease activity of the DNA2 helicase–nuclease. High levels of Exo1 compensate for the loss of the flap endonuclease in rad27Δ mutants by suppressing the temperature-induced lethality and mutation avoidance phenotypes (Tishkoff et al. 1997a; Sun et al. 2003). High-level expression of RAD2 also compensates (Sun et al. 2003). While both Rad2 family members suppress the temperature-sensitive growth and mutator activity of rad27Δ, only RAD2 high-level expression suppresses its MMS sensitivity (Sun et al. 2003). In addition, high-level expression of DNA2 partially compensates for the absence of the flap endonuclease (Budd and Campbell 1997). While all three activities might compensate, we note that none of the three have been shown to directly interact with PCNA.
Suppression of rad27-K325* and rad27-pX8 by overexpression of PCNA may occur by a second mechanism that does not solely involve compensation by another nuclease. Instead, suppression may occur, either in part or in whole, by increased binding of the mutant nucleases to PCNA by mass action. In discussing this possibility, we take into account the many contacts that the human flap endonuclease ortholog makes with PCNA and the mechanism by which the nuclease is thought to reach DNA substrates. Co-crystallization of human FEN-1 with PCNA revealed that the flap endonuclease makes contacts throughout its length, including the core nuclease domain that remains in our rad27-K325* mutant (Sakurai et al. 2005). The multiple contacts also include segments of FEN-1 that extend on either side its PIP box. In addition, a truncated mutant of the human flap endonuclease missing its PIP box is capable of binding to PCNA (Guo et al. 2008). Thus, all mutant forms that we have created should be capable of binding to PCNA with reduced affinity. Increasing the amount of PCNA should increase the amount of binding, and that may be partially responsible for suppression of the mutant phenotypes.
The activity of the flap endonuclease is greatly increased by its binding to PCNA (Li et al. 1995; Jonsson et al. 1998; Gomes and Burgers 2000; Tom et al. 2000; Frank et al. 2001). The mechanism that accounts for this activation suggests that the enzyme first binds to PCNA through its PIP box in a manner that precludes its binding to DNA substrates and then rotates through a hinge region between its core and the PIP box to allow the enzyme to meet its substrates (Gomes and Burgers 2000; Sakurai et al. 2005). This two-step mechanism was first proposed by the difference between the binding affinity and nuclease activation of the pol30-79 and pol30-90 mutant copies of PCNA (Gomes and Burgers 2000). The pol30-79 mutation lies in the hydrophobic pocket of the IDCL and interacts directly with the PIP box, while the pol30-90 mutation is adjacent to the hinge domain and may be important for rotation. The pol30-79 mutant protein does not bind strongly to the flap endonuclease but can still stimulate its activity, while the pol30-90 mutant protein binds more strongly but does not stimulate the nuclease activity. The two modes of flap endonuclease binding to PCNA are supported by the co-crystallization results that demonstrate a hinge permitting rotation of the nuclease across the front face into the PCNA-encircled DNA (Sakurai et al. 2005). The difference in behavior between the two pol30 mutations suggests that while initial binding is important, it is not necessary for stimulation, while rotation through the hinge is necessary to achieve stimulation.
The behavior of our mutations can be interpreted on the basis of the proposed mechanism. We suggest that the product of rad27-K325* retains partial biological activity and binds weakly to PCNA. Because it is missing the hinge, it cannot reach most DNA substrates. The extra toxicity of rad27-K325* when combined with pol30-90 in comparison to the viability of the rad27Δ pol30-90 double mutant supports our view that the mutant protein binds to PCNA through it core domain and excludes binding by other PCNA binding proteins. In essence, the product of rad27-K325* may act as a competitive inhibitor, and this effect is exacerbated in the pol30-90 mutant that weakens the interaction between PCNA and its other partners. This competitive effect may also operate to prevent the product of cdc9-p from successfully binding to PCNA, and this exclusion may lead to the synthetic lethality between cdc9-p and rad27-K325*. On the basis of the rotation model, we interpret our results to suggest that the product of rad27-pX8 has difficulty with the initial steps of binding to PCNA but possesses the hinge region and C-terminal residues so it is able to rotate and complete catalysis if binding occurs. Combining rad27-pX8 with pol30-90 diminishes both the initial binding and possibly the rotation event as the substitutions in rad27-pX8 change the glutamine residue that interacts with the amino acid residues of PCNA that are mutated in pol30-90. These additive changes make the double mutant grow slowly. Finally, the product of the rad27-A358* mutant possesses all the binding regions and is nearly completely functional. Thus, the behavior of our flap endonuclease alleles is consistent with the binding studies and the proposed rotation model.
Finally, we return to the hypothesis that propelled these studies. On the basis of our studies of CAG repeat tract expansions, we suggested that PCNA mediates the ordered entry of the flap endonuclease and DNA ligase I into replication (Refsland and Livingston 2005). We began this study with a synthetic lethal screen. As befitting a synthetic lethal screen, we used a cdc9 allele that can best be characterized as having a mild phenotype, particularly as it grows throughout a range of temperatures and does not exhibit a severe sensitivity to MMS exposure that is seen for other cdc9 mutations (Hartwell et al. 1973; Montelone et al. 1981; Refsland and Livingston 2005). Our synthetic lethal screen with cdc9-p yielded a rad27 mutation. Considering that rad27Δ is synthetically lethal with many yeast genes, especially those involved in DNA transactions, the appearance of a flap endonuclease mutation with seemingly little activity might not have been unexpected (Tong et al. 2004; Loeillet et al. 2005; Pan et al. 2006). While we have not systematically screened the collection of yeast gene deletions or combined cdc9-p with a panel of other replication and repair mutations, the finding of a new rad27 allele might not have been by chance alone. Previously, we found that we could combine rad27Δ with cdc9-1 at a permissive temperature (Ireland et al. 2000). Thus, while direct comparisons between cdc9-1 and cdc9-p have not been made, the absolute lethality of rad27-K325* with cdc9-p is novel. The novelty is that the combination of the cdc9 and rad27 alleles with the less severe phenotypes (cdc9-p and rad27-K325*) is lethal while the combination of the alleles with more severe phenotypes (cdc9-1 and rad27Δ) is not.
More important, the absolute lethality of rad27Δ and rad27-K325*, the temperature-sensitive lethality of rad27-pX8, and the temperature-impaired growth of rad27-A358* and rad27-p when combined with cdc9-p suggest a strong cellular requirement that both enzymes need to be able to interact with PCNA to complete DNA replication and repair. Thus, the inability of the double mutants to grow is best described as a synergistic effect. The relationship is clearly not one of epistasis. We argue that the effect is greater than additive, making it synergistic. We might have expected the double mutants to grow weakly if their combination were additive. In support of our contention, we note that the triple mutant of cdc9-p rad27-p pol30-90 is viable at 30° (Refsland and Livingston 2005), whereas the cdc9-p rad27-K325* double mutant is not. By this view, the triple mutant shows an additive effect and the cdc9-p rad27-K325* double mutant is synergistic. While our arguments may be circumstantial, our results warrant further examination of the role PCNA plays in ordering the entry of the flap endonuclease and DNA ligase I into replication and repair.
Acknowledgments
We thank Medora Huseby for discussion of helical forming amino acid sequences, Jill Schweitzer for construction of some of the strains, and David Kirkpatrick for his yeast gene library. This work was supported by a National Institutes of Health grant and by the Department of Biochemistry, Molecular Biology, and Biophysics at the University of Minnesota.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.103937/DC1.
References
- Amin, N. S., and C. H. Holm, 1996. In vivo analysis reveals that the interdomain region of the yeast proliferating cell nuclear antigen is important for DNA replication and repair. Genetics 144: 479–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin, N. S., M. N. Nguyen, S. Oh and R. D. Kolodner, 2001. Exo1-dependent mutator mutations: model system for studying functional interactions in mismatch repair. Mol. Cell. Biol. 21: 5142–5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asleson, E. N., and D. M. Livingston, 2003. Investigation of the stability of rad52 mutant proteins uncovers post-translational and transcriptional regulation of Rad52p. Genetics 163: 91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae, S.-H., E. Choi, K.-H. Lee, J. S. Park, S.-H. Lee et al., 1998. Dna2 of Saccharomyces cerevisiae possess a single-stranded DNA-specific endonuclease activity that is able to act on double-stranded DNA in the presence of ATP. J. Biol. Chem. 273: 26880–26890. [DOI] [PubMed] [Google Scholar]
- Bae, S. H., D. W. Kim, J. Kim, J. H. Kim, D. H. Kim et al., 2002. Coupling of DNA helicase and endonuclease activities of yeast Dna2 facilitates Okazaki fragment processing. J. Biol. Chem. 277: 26632–26641. [DOI] [PubMed] [Google Scholar]
- Boundy-Mills, K. L., and D. M. Livingston, 1993. A Saccharomyces cerevisiae RAD52 allele expressing a C-terminal truncation protein: activities and intragenic complementation of missense mutations. Genetics 133: 39–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budd, M. E., and J. L. Campbell, 1995. A yeast gene required for DNA replication encodes a protein with homology to DNA helicases. Proc. Natl. Acad. Sci. USA 92: 7642–7646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budd, M. E., and J. L. Campbell, 1997. A yeast replicative helicase, DNA2 helicase, interacts with yeast FEN-1 nuclease in carrying out its essential function. Mol. Cell. Biol. 17: 2136–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budd, M. E., W. C. Choe and J. L. Campbell, 1995. DNA2 encodes a DNA helicase essential for replication of eukaryotic chromosomes. J. Biol. Chem. 270: 26766–26769. [DOI] [PubMed] [Google Scholar]
- Budd, M. E., W. C. Choe and J. L. Campbell, 2000. The nuclease activity of the yeast Dna2 protein, which is related to the RecB-like nucleases, is essential in vivo. J. Biol. Chem. 275: 16518–16529. [DOI] [PubMed] [Google Scholar]
- Burke, D., D. Dawson and T. Stearns, 2000. Methods in Yeast Genetics, pp. 171–181. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Chapados, B. R., D. J. Hosfield, S. Han, J. Qiu, B. Yelent et al., 2004. Structural basis for FEN-1 substrate specificity and PCNA-mediated activation in DNA replication and repair. Cell 116: 39–50. [DOI] [PubMed] [Google Scholar]
- Chen, C., and R. D. Kolodner, 1999. Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants. Nat. Genet. 23: 81–85. [DOI] [PubMed] [Google Scholar]
- Dornfeld, K. J., and D. M. Livingston, 1991. Effects of controlled RAD52 expression on repair and recombination in Saccharomyces cerevisiae. Mol. Cell. Biol. 11: 2013–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg, J. C., R. Ayyagari, X. V. Gomes and P. M. J. Burgers, 1997. Mutations in yeast proliferating cell nuclear antigen define distinct sites for interaction with DNA polymerase δ and DNA polymerase ɛ. Mol. Cell. Biol. 17: 6367–6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank, G., J. Qiu, L. Zheng and B. Shen, 2001. Stimulation of eukaryotic flap endonuclease-1 activities by proliferating cell nuclear antigen (PCNA) is independent of its in vitro interaction via a consensus PCNA binding region. J. Biol. Chem. 276: 36295–36302. [DOI] [PubMed] [Google Scholar]
- Gary, R., M. S. Park, J. P. Nolan, H. L. Cornelius, O. G. Kozyreva et al., 1999. A novel role in DNA metabolism for the binding of Fen1/Rad27 to PCNA and implications for genetic risk. Mol. Cell. Biol. 19: 5373–5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes, X. V., and P. M. Burgers, 2000. Two modes of FEN1 binding to PCNA regulated by DNA. EMBO J. 19: 3811–3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, Z., V. Chavez, P. Singh, L. D. Finger, H. Hang et al., 2008. Comprehensive mapping of the C-terminus of flap endonuclease-1 reveals distinct interaction sites for five proteins that represent different DNA replication and repair pathways. J. Mol. Biol. 377: 679–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell, L. H., 1970. Periodic density fluctuation during the yeast cell cycle and the selection of synchronous cultures. J. Bacteriol. 104: 1280–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell, L. H., R. K. Mortimer, J. Culotti and M. Culotti, 1973. Genetic control of the cell division cycle in yeast. V. Genetic analysis of cdc mutants. Genetics 74: 267–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson, S. T., and T. D. Petes, 1992. Instability of simple sequence DNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 12: 2749–2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ireland, M. J., S. S. Reinke and D. M. Livingston, 2000. The impact of lagging strand replication mutations on the stability of CAG repeat tracts in yeast. Genetics 155: 1657–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jauert, P. A., L. E. Jensen and D. T. Kirkpatrick, 2005. A novel yeast genomic DNA library on a geneticin-resistance vector. Yeast 22: 653–657. [DOI] [PubMed] [Google Scholar]
- Johnson, R. E., G. K. Kovvali, L. Prakash and S. Prakash, 1995. Requirement of the yeast RTH1 5′ to 3′ exonuclease for the stability of simple repetitive DNA. Science 269: 238–240. [DOI] [PubMed] [Google Scholar]
- Jonsson, Z. O., R. Hindges and U. Hubscher, 1998. Regulation of DNA replication and repair proteins through interaction with the front side of proliferating cell nuclear antigen. EMBO J. 17: 2412–2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi, K., Y. Taniguchi, A. Hatanaka, E. Sonoda, H. Hochegger et al., 2005. Fen-1 facilitates homologous recombination by removing divergent sequences at DNA break ends. Mol. Cell. Biol. 25: 6948–6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokoska, R. J., L. Stefanovic, H. T. Tran, M. A. Resnick, D. A. Gordenin et al., 1998. Destabilization of yeast micro- and minisatellite DNA sequences by mutations affecting a nuclease involved in Okazaki fragment processing (rad27) and DNA polymerase δ (pol3-t). Mol. Cell. Biol. 18: 2779–2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren, A., S. Ben-Aroya, R. Steinlauf and M. Kupiec, 2003. Pitfalls of the synthetic lethality screen in Saccharomyces cerevisiae: an improved design. Curr. Genet. 43: 62–69. [DOI] [PubMed] [Google Scholar]
- Kranz, J. E., and C. Holm, 1990. Cloning by function: an alternative approach for identifying yeast homologs of genes from other organisms. Proc. Natl. Acad. Sci. USA 87: 6629–6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea, D. E., and C. A. Coulson, 1949. The distribution of the numbers of mutants in bacterial populations. J. Genet. 49: 264–285. [DOI] [PubMed] [Google Scholar]
- Levin, D. S., S. Vijayakumar, X. Liu, V. P. Bermudez, J. Hurwitz et al., 2004. A conserved interaction between the replicative clamp loader and DNA ligase in eukaryotes; implications for Okazaki fragment joining. J. Biol. Chem. 279: 55196–55201. [DOI] [PubMed] [Google Scholar]
- Li, X., J. Li, J. Harrington, M. R. Lieber and P. M. Burgers, 1995. Lagging strand DNA synthesis at the eukaryotic replication fork involves binding and stimulation of FEN-1 by PCNA. J. Biol. Chem. 270: 22109–22112. [DOI] [PubMed] [Google Scholar]
- Liu, Y., H. I. Kao and R. A. Bambara, 2004. Flap endonuclease 1: a central component of DNA metabolism. Annu. Rev. Biochem. 73: 589–615. [DOI] [PubMed] [Google Scholar]
- Loeillet, S., B. Palancade, M. Cartron, A. Thierry, G. F. Richard et al., 2005. Genetic network interactions among replication, repair and nuclear pore deficiencies in yeast. DNA Repair 4: 459–468. [DOI] [PubMed] [Google Scholar]
- Maga, G., and U. Hubscher, 2003. Proliferating cell nuclear antigen (PCNA): a dancer with many partners. J. Cell Sci. 116: 3051–3060. [DOI] [PubMed] [Google Scholar]
- Marsischky, G. T., N. Filosi, M. F. Kane and R. Kolodner, 1996. Redundancy of Saccharomyces cerevisiae MSH3 and MSH6 in MSH2-dependent mismatch repair. Genes Dev. 10: 407–420. [DOI] [PubMed] [Google Scholar]
- Montelone, B. A., S. Prakash and L. Prakash, 1981. Spontaneous mitotic recombination in mms8–1, an allele of the CDC9 gene of Saccharomyces cerevisiae. J. Bacteriol. 147: 517–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murante, R. S., L. Huang, J. J. Turchi and R. A. Bambara, 1994. The calf 5′- to 3′-exonuclease is also an endonuclease with both activities dependent on primers annealed upstream of the point of cleavage. J. Biol. Chem. 269: 1191–1196. [PubMed] [Google Scholar]
- Pan, X., P. Ye, D. S. Yuan, X. Wang, J. S. Bader et al., 2006. A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell 124: 1069–1081. [DOI] [PubMed] [Google Scholar]
- Parenteau, J., and R. J. Wellinger, 1999. Accumulation of single-stranded DNA and destabilization of telomeric repeats in yeast mutant strains carrying a deletion of RAD27. Mol. Cell. Biol. 19: 4143–4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parenteau, J., and R. J. Wellinger, 2002. Differential processing of leading- and lagging-strand ends at Saccharomyces cerevisiae telomeres revealed by the absence of Rad27p nuclease. Genetics 162: 1583–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascal, J. M., P. J. O'Brien, A. E. Tomkinson and T. Ellenberger, 2004. Human DNA ligase I completely encircles and partially unwinds nicked DNA. Nature 432: 473–478. [DOI] [PubMed] [Google Scholar]
- Pascal, J. M., O. V. Tsodikov, G. L. Hura, W. Song, E. A. Cotner et al., 2006. A flexible interface between DNA ligase and PCNA supports conformational switching and efficient ligation of DNA. Mol. Cell 24: 279–291. [DOI] [PubMed] [Google Scholar]
- Reagan, M. S., C. Pittenger, W. Siede and E. C. Friedberg, 1995. Characterization of a mutant strain of Saccharomyces cerevisiae with a deletion of the RAD27 gene, a structural homolog of the RAD2 nucleotide excision repair gene. J. Bacteriol. 177: 364–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Refsland, E. W., and D. M. Livingston, 2005. Interactions among DNA ligase I, the flap endonuclease and proliferating cell nuclear antigen in the expansion and contraction of CAG repeat tracts. Genetics 171: 923–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein, R. J., 1983. One-step gene disruption in yeast. Methods Enzymol. 101: 202–211. [DOI] [PubMed] [Google Scholar]
- Sakurai, S., K. Kitano, H. Yamaguchi, K. Hamada, K. Okada et al., 2005. Structural basis for recruitment of human flap endonuclease 1 to PCNA. EMBO J. 24: 683–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer, S., and R. W. Davis, 1979. Replacement of chromosome segments with altered DNA sequences constructed in vitro. Proc. Natl. Acad. Sci. USA 76: 4951–4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweitzer, J. K., and D. M. Livingston, 1998. Expansions of CAG repeat tracts are frequent in a yeast mutant defective in Okazaki fragment maturation. Hum. Mol. Genet. 7: 69–74. [DOI] [PubMed] [Google Scholar]
- Schweitzer, J. K., and D. M. Livingston, 1999. The effect of DNA replication mutations on CAG tract instability in yeast. Genetics 152: 953–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia, E. A., R. J. Kokoska, M. Dominska, P. Greenwell and T. D. Petes, 1997. Microsatellite instability in yeast: dependence on repeat unit size and DNA mismatch repair genes. Mol. Cell. Biol. 17: 2851–2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, P., L. Zheng, V. Chavez, J. Qiu and B. Shen, 2007. Concerted action of exonuclease and Gap-dependent endonuclease activities of FEN-1 contributes to the resolution of triplet repeat sequences (CTG)n- and (GAA)n-derived secondary structures formed during maturation of Okazaki fragments. J. Biol. Chem. 282: 3465–3477. [DOI] [PubMed] [Google Scholar]
- Sommers, C. H., E. J. Miller, B. Dujon, S. Prakash and L. Prakash, 1995. Conditional lethality of null mutations in RTH1 that encodes the yeast counterpart of a mammalian 5′- to 3′-exonuclease required for lagging strand DNA synthesis in reconstituted systems. J. Biol. Chem. 270: 4193–4196. [DOI] [PubMed] [Google Scholar]
- Stewart, J. A., J. L. Campbell and R. A. Bambara, 2006. Flap endonuclease disengages Dna2 helicase/nuclease from Okazaki fragment flaps. J. Biol. Chem. 281: 38565–38572. [DOI] [PubMed] [Google Scholar]
- Stucki, M., Z. O. Jonsson and U. Hubscher, 2001. In eukaryotic flap endonuclease 1, the C terminus is essential for substrate binding. J. Biol. Chem. 276: 7843–7849. [DOI] [PubMed] [Google Scholar]
- Subramanian, J., S. Vijayakumar, A. E. Tomkinson and N. Arnheim, 2005. Genetic instability induced by overexpression of DNA ligase I in budding yeast. Genetics 105: 427–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, X., D. Thrower, J. Qiu, P. Wu, L. Zheng et al., 2003. Complementary functions of the Saccharomyces cerevisiae Rad2 family nucleases in Okazaki fragment maturation, mutation avoidance, and chromosome stability. DNA Repair 2: 925–940. [DOI] [PubMed] [Google Scholar]
- Symington, L. S., 1998. Homologous recombination is required for the viability of rad27 mutants. Nucleic Acids Res. 26: 5589–5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff, D. X., A. L. Boerger, P. Bertrand, N. Filosi, G. M. Gaida et al., 1997. a Identification and characterization of Saccharomyces cerevisiae EXO1, a gene encoding an exonuclease that interacts with MSH2. Proc. Natl. Acad. Sci. USA 94: 7487–7492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff, D. X., N. Filosi, G. M. Gaida and R. D. Kolodner, 1997. b A novel mutation avoidance mechanism dependent on S. cerevisiae RAD27 is distinct from DNA mismatch repair. Cell 88: 253–263. [DOI] [PubMed] [Google Scholar]
- Tom, S., L. A. Henricksen and R. A. Bambara, 2000. Mechanism whereby proliferating cell nuclear antigen stimulates flap endonuclease I. J. Biol. Chem. 275: 10498–10505. [DOI] [PubMed] [Google Scholar]
- Tom, S., L. A. Henricksen, M. S. Park and R. A. Bambara, 2001. DNA ligase I and proliferating cell nuclear antigen form a functional complex. J. Biol. Chem. 276: 24817–24825. [DOI] [PubMed] [Google Scholar]
- Tong, A. H., G. Lesage, G. D. Bader, H. Ding, H. Xu et al., 2004. Global mapping of the yeast genetic interaction network. Science 303: 808–813. [DOI] [PubMed] [Google Scholar]
- Tran, P. T., N. Erdeniz, S. Dudley and R. M. Liskay, 2002. Characterization of nuclease-dependent functions of Exo1p in Saccharomyces cerevisiae. DNA Repair 1: 895–912. [DOI] [PubMed] [Google Scholar]
- Vijayakumar, S., B. Dziegielewska, D. S. Levin, W. Song, J. Yin et al., 2009. Phosphorylation of human DNA ligase I regulates its interaction with replication factor C and its participation in DNA replication and DNA repair. Mol. Cell. Biol. 29: 2042–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X., G. Ira, J. A. Tercero, A. M. Holmes, J. F. Diffley et al., 2004. Role of DNA replication proteins in double-strand break-induced recombination in Saccharomyces cerevisiae. Mol. Cell. Biol. 24: 6891–6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warbrick, E., 2000. The puzzle of PCNA's many partners. BioEssays 22: 997–1006. [DOI] [PubMed] [Google Scholar]
- Warbrick, E., 2006. A functional analysis of PCNA-binding peptides derived from protein sequence, interaction screening and rational design. Oncogene 25: 2850–2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, X., and Z. Wang, 1999. Relationships between yeast Rad27 and Apn1 in response to apurinic/apyrimidinic (AP) sites in DNA. Nucleic Acids Res. 27: 956–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, X., T. E. Wilson and M. R. Lieber, 1999. A role for FEN-1 in nonhomologous DNA end joining: the order of strand annealing and nucleolytic processing events. Proc. Natl. Acad. Sci. USA 96: 1303–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Y., Y. Liu, J. L. Argueso, L. A. Henricksen, H.-I. Kao et al., 2001. Identification of rad27 mutations that confer differential defects in mutation avoidance, repeat tract instability, and flap cleavage. Mol. Cell. Biol. 21: 4889–4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, L., M. Zhou, Q. Chai, J. Parrish, D. Xue et al., 2005. Novel function of the flap endonuclease 1 complex in processing stalled DNA replication forks. EMBO Rep. 6: 83–89. [DOI] [PMC free article] [PubMed] [Google Scholar]