Abstract
In bacteria, septum formation frequently initiates before the last steps of chromosome segregation. This is notably the case when chromosome dimers are formed by homologous recombination. Chromosome segregation then requires the activity of a double-stranded DNA transporter anchored at the septum by an integral membrane domain, FtsK. It was proposed that the transmembrane segments of proteins of the FtsK family form pores across lipid bilayers for the transport of DNA. Here, we show that truncated Escherichia coli FtsK proteins lacking all of the FtsK transmembrane segments allow for the efficient resolution of chromosome dimers if they are connected to a septal targeting peptide through a sufficiently long linker. These results indicate that FtsK does not need to transport DNA through a pore formed by its integral membrane domain. We propose therefore that FtsK transports DNA before membrane fusion, at a time when there is still an opening in the constricted septum.
Keywords: cell division, chromosome segregation, DNA transport, site-specific recombination
Introduction
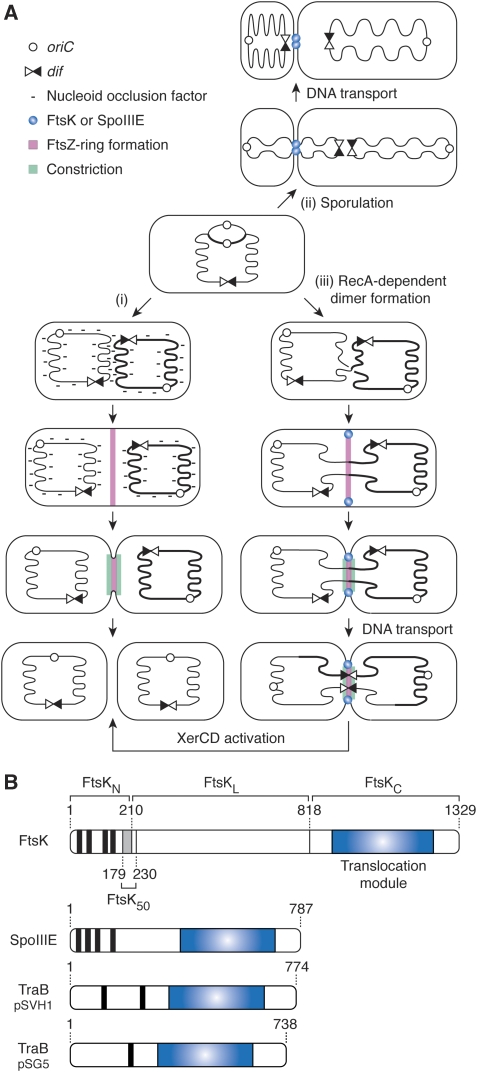
During cell proliferation, the timing of DNA synthesis, chromosome segregation and cell division must be coordinated to ensure the stable inheritance of the genetic material. In eukaryotes, this is achieved by the temporal separation of these processes and the existence of checkpoint mechanisms that delay certain steps until others are completed. Likewise, nucleoid occlusion normally prevents the initiation of septum formation in the presence of un-segregated DNA at the site of division in bacteria (Figure 1A (i); Wu and Errington, 2004; Bernhardt and de Boer, 2005). Nevertheless, septum formation frequently initiates before the last stages of chromosome segregation. This is notably the case during sporulation in Bacillus subtilis and during chromosome dimer resolution in Escherichia coli (Figure 1A (ii) and (iii); see Errington et al, 2001; Barre, 2007 for a review). The correct distribution of the genetic material is then achieved by the respective activities of two related double-stranded DNA transporters, SpoIIIE and FtsK (Bath et al, 2000; Aussel et al, 2002).
Figure 1.
DNA traffic in bacteria. (A) Coordination between chromosome segregation and cell division in B. subtilis and E. coli. (i) Normal vegetative cell division in E. coli or B. subtilis. When the bulk of the sister chromosomes remains at mid-cell, nucleoid occlusion factors prevent FtsZ-ring formation. Segregation of the nucleoids away from mid-cell allows the recruitment of the division machinery. (ii) Sporulation in B. subtilis. Only a third of the chromosome is trapped in the prespore compartment during the asymmetric cell division of sporulation. SpoIIIE transports the remaining two-third of the chromosome. (iii) Chromosome dimer resolution in E. coli. RecA-mediated homologous recombination during replication can lead to the formation of dimers of the circular chromosome. Replication and segregation proceed until the bulk of the sister chromosomes are distributed in each daughter cell, but the two nucleoids remain linked by DNA passing through the division septum. FtsK-dependent DNA transport brings dif sites together. FtsK subsequently activates Xer recombination to resolve the dimer into monomers. (B) Schematic representation of FtsK, SpoIIIE and the Tra proteins of two Streptomycetes conjugative plasmids. Transmembrane domains are depicted as vertical black bars and translocation modules as blue rectangles. Numbers indicate the position of amino acid residues. A grey rectangle indicates the part of FtsKN that belongs to a ∼50 amino acid region (FtsK50; aa 179–230) that increases the efficiency with which FtsK peptides carrying an intact C-terminal domain activate Xer recombination under low expression levels.
The assembly of SpoIIIE at the sporulation septum of B. subtilis (Figure 1A (ii)) correlates with the establishment of a barrier to protein and membrane dye diffusion between the mother cell and the prespore compartments (Wu and Errington, 1994; Wu et al, 1995; Liu et al, 2006; Burton et al, 2007), which suggested that SpoIIIE transports DNA after septal membrane fusion (Liu et al, 2006; Becker and Pogliano, 2007; Burton et al, 2007). In E. coli and in most proteobacteria, chromosome dimers are resolved into monomers by two tyrosine recombinases, XerC and XerD, which add a crossover at a specific chromosomal site, dif (Bigot et al, 2007; Val et al, 2008). FtsK has two successive roles in this process: it mobilizes DNA to bring together the two dif sites carried by a dimer and it activates Xer recombination at dif by a direct contact with XerD (Figure 1A (iii)). Although FtsK is recruited at an early stage of septum formation, Xer recombination at dif occurs at a later stage, concomitantly with cell wall constriction (Figure 1A (iii); Kennedy et al, 2008). This observation suggested that FtsK might initiate DNA transport after membrane fusion like SpoIIIE.
The carboxy-terminal domains of SpoIIIE and FtsK include a RecA-type ATPase fold (Figure 1B, translocation module), which forms hexamers around double-stranded DNA (Massey et al, 2006) and uses the energy of ATP to translocate (Saleh et al, 2004, 2005). The same module is found in the transfer (Tra) proteins of various conjugative plasmids of Streptomycetes (Figure 1B; (Grohmann et al, 2003)). SpoIIIE, FtsK and the Tra proteins also share a common structural organization, which includes an integral membrane domain (Figure 1B). In view of this organization, it was proposed that their integral membrane domains form a pore in lipid bilayers across which DNA is transported in a process akin to conjugation (Liu et al, 2006; Becker and Pogliano, 2007; Burton et al, 2007).
Previous structural and functional analysis indicated that E. coli FtsK is composed of an amino-terminal domain with four transmembrane segments that is essential for cell division (FtsKN, aa 1–210; Figure 1B; Begg et al, 1995; Diez et al, 1997; Draper et al, 1998; Liu et al, 1998; Wang and Lutkenhaus, 1998; Yu et al, 1998; Dorazi and Dewar, 2000), a long linker region of low complexity (FtsKL; aa 211–817; Figure 1B; Bigot et al, 2004) and a carboxy-terminal domain that contains all the necessary determinants for DNA translocation and Xer recombination activation (FtsKC; aa 818–1329; Figure 1B; Barre et al, 2000; Aussel et al, 2002; Yates et al, 2003; Bigot et al, 2004). It was observed that a truncation of FtsK lacking most of FsKN and FtsKL did not support chromosome dimer resolution even if it could efficiently process plasmid dimers in vivo (Aussel et al, 2002), which fitted with the idea that FtsKN and FtsKL might be implicated in the formation of a pore for the transport of chromosomal DNA at cell division.
To gain further insights into the function of FtsKN and FtsKL in DNA transport, we decided to investigate the efficiency of chromosome dimer resolution of E. coli cells carrying new mutant ftsK alleles in which the coding regions of FtsKN and FtsKL were modified, but in which the coding region of FtsKC was left intact. We report that FtsK peptides lacking all of the transmembrane segments of FtsKN allow for the efficient resolution of chromosome dimers if they are connected to a septal targeting peptide through a sufficiently long linker. These results indicate that the transport of chromosomal DNA across lipid bilayers during the last stage of chromosome segregation in E. coli does not require the formation of a pore by FtsKN and FtsKL. We propose therefore that E. coli FtsK most likely transports DNA before membrane fusion, at a time when there is still an opening in the constricted septum.
Results
Experimental strategy
The role of FtsKN and FtsKL in DNA transport could not be directly addressed because both domains are implicated in cell division and because FtsKN is essential (Bigot et al, 2004). To circumvent this difficulty, we made use of a hyperactive mutant of FtsA, FtsA*, which can partially bypass the need for FtsKN in cell division, so that the entire ftsK gene can be deleted (Geissler and Margolin, 2005; Geissler et al, 2007). FtsK mutants were introduced in ftsK− ftsA* cells on a low copy pSC101 vector and under the natural ftsK promoter to prevent overproduction or deregulation of their expression (Bigot et al, 2004; Kennedy et al, 2008). We could thus monitor, in the absence of any additional functional copy of FtsKN, if FtsK peptides mutated in FtsKN and FtsKL supported DNA transport by measuring chromosome dimer resolution.
A good estimation of the efficaciousness of chromosome dimer resolution in a given strain is provided by the excess number of viable cells produced at each generation when compared with an isogenic strain totally deficient in chromosome dimer resolution. We therefore monitored the growth advantage that mutant ftsK alleles provide to ftsA* ftsK− cells compared with ftsA* ftsK−dif− cells, in which the deletion of dif abolishes dimer resolution. Note that the difference in fitness of ftsA* ftsK− and ftsA* ftsK−dif− cells producing a given FtsK peptide only reflects the ability of this peptide to sustain chromosome dimer resolution as any other effect that its production might have on growth, such as by interfering with cell division, will be identical in the two cell lines.
The expression and the localization of the different ftsK alleles were checked using fusions with green and/or yellow fluorescent proteins (GFP and YFP) expressed from a low copy pSC101 vector under the natural ftsK promoter. Their ability to activate Xer recombination, which reflects the formation of stable and active FtsKC hexamers on DNA, was checked using a pseudo-dimer plasmid recombination assay (Barre et al, 2000). To this aim, the ftsK alleles were expressed from an arabinose promoter on a high copy vector and we measured the frequency of excision of a DNA segment placed between two direct tandem repeats of dif on a pSC101 plasmid.
Chimaeras between FtsKLC and ZipA or FtsW support chromosome dimer resolution
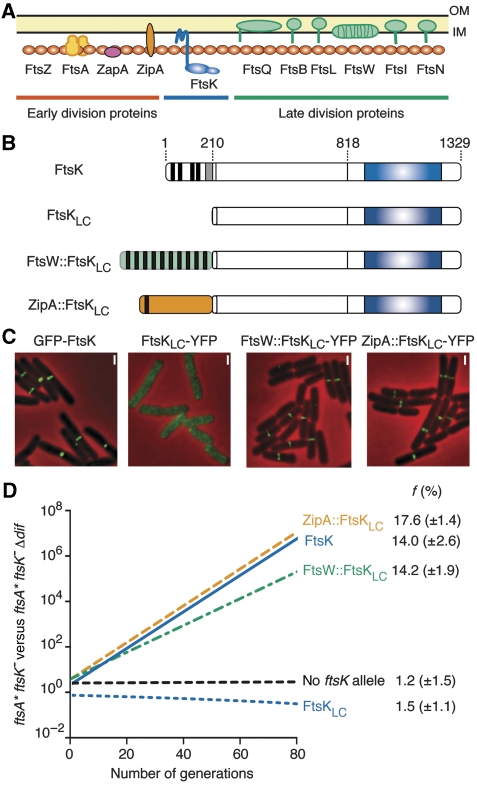
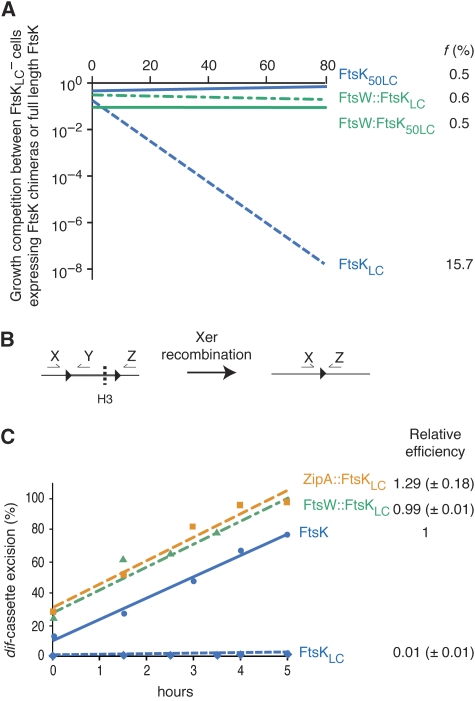
We reported earlier that a truncated protein lacking the whole of the N-terminal domain of FtsK, FtsKLC (Figure 2B), supports dif-recombination on plasmids (Barre et al, 2000). FtsKLC is efficiently produced from a pSC101 vector under the ftsK promoter and is stable (Supplementary Figure 1). However, it provided no growth advantage to ftsA* ftsK− cells compared with ftsA* ftsK−dif− cells (Figure 2D), indicating that it is completely inactive in chromosome dimer resolution. Actually, FtsKLC∷YFP yielded a diffuse cytoplasmic signal (Figure 2C), indicating that FtsKL and FtsKC do not carry sufficient determinants for septum localization. We decided therefore to test the activity of FtsK chimaeras that are anchored at the site of division by a fusion to another integral membrane cell division protein. To this aim, we fused FtsKLC to FtsW and ZipA, the only two integral membrane cell division proteins that have a C-terminal cytoplasmic tail like FtsK (Figure 2A). FtsW is anchored to the membrane by 10 transmembrane helices and arrives at the septum after FtsK. ZipA is anchored to the membrane by a single transmembrane helix and arrives at the septum before FtsK (Harry et al, 2006). ZipA∷FtsKLC and FtsW∷FtsKLC (Figure 2B) displayed the characteristic localization pattern of cell division proteins, that is one band at the middle of the cell in pre-dividing cells and one focus at the middle of the cell in dividing cells (Figure 2C). In growth competition, they supported chromosome dimer resolution to a similar level than full-length FtsK, indicating that they allowed for the eventual resolution of all occuring dimers (Figure 2D). Similar results were obtained in FtsKLC− cells, indicating that the activity of ZipA∷FtsKLC and FtsW∷FtsKLC was not limited to the particular ftsA* genetic context (data not shown). We conclude that anchoring FtsKLC in the cytoplasmic membrane and targeting it to the septum is sufficient for the resolution of chromosome dimers in E. coli, which argues against the idea that formation of a pore by FtsKN is essential for the transport of DNA across lipid bilayers.
Figure 2.
Localization and chromosome dimer resolution activity of FtsKLC chimaeras. (A) Schematic view of septum assembly. The protein icons are ordered from left to right according to the commonly accepted assembly sequence. OM, outer membrane; IM, inner membrane. (B) Schematic representation of the FtsKLC chimaeras as in Figure 1B. (C) Localization of FtsKLC chimaeras fused at their C-terminal ends to YFP and of full-length FtsK fused at its N-terminal end to GFP in ftsK+ cells. A similar localization pattern was observed in ftsA* ftsK− and in ftsKLC− cells (data not shown). (C) Pictures of ftsK+ cells are shown because of their simpler cell division phenotype. Scale bars (white bars), 1 μm. (D) Growth competition between ftsA* ftsK− and ftsA* ftsK− Δdif cells. The graph of a typical result is shown. f, mean and standard deviation of the growth advantage given by the ectopic production of each FtsKLC chimaera (result from three independent experiments).
Chromosome dimer resolution by an FtsK truncation lacking transmembrane segments
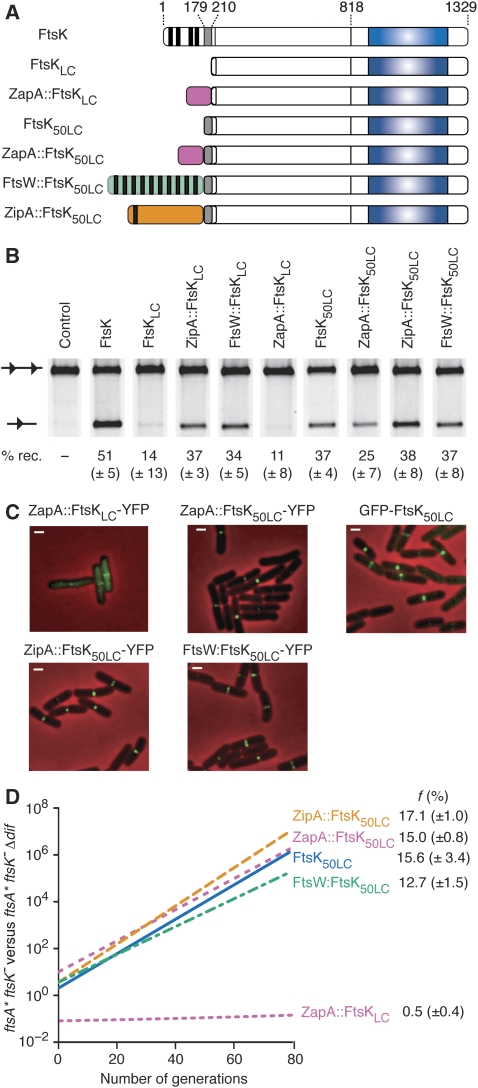
To rule out the unlikely possibility that the transmembrane segments of ZipA and FtsW could substitute for those of FtsKN in the formation of a DNA channel across lipid bilayers, we searched for a way to target FtsKLC to the septum in the absence of any transmembrane segments (Figure 3). To this aim, we fused it to ZapA, a cell division protein that is fully cytoplasmic (Figure 2A; Harry et al, 2006). The zapA∷ftsKLC allele (Figure 3A) is totally inactive in chromosome dimer resolution (Figure 3D). However, zapA∷ftsKLC activates dif-recombination on plasmids less efficiently than full-length ftsK, zipA∷ftsKLC or ftsW∷ftsKLC under low expression levels (Figure 3B). In addition, it is imperfectly recruited to the septum, much of the fluorescent signal observed with its YFP derivative remaining diffuse and cytoplasmic (Figure 3C). Taken together, these results suggested that the fusion of FtsKLC to ZapA interfered either with the stability of the protein, its septum localization and/or its capacity to activate Xer recombination.
Figure 3.
Localization and chromosome dimer resolution activity of FtsK50LC chimaeras. (A) Schematic representation of the FtsK50LC chimaeras as in Figure 1B. (B) In vivo pseudo-dimer resolution assay on plasmids. A scheme of the plasmid-borne substrate and product is shown on the left of the gel. dif sites are represented as black triangles. The efficiency of recombination was determined after 4 h of growth in 0.2% arabinose. (C) Localization of FtsK50LC chimaeras fused at their C-terminal ends to YFP as in Figure 2B. (D) Growth competition between ftsA* ftsK− and ftsA* ftsK− Δdif cells ectopically producing the FtsK50LC chimaeras as in Figure 2C.
We characterized earlier a region of ∼50 amino acids (FtsK50; aa 179–230; Figure 1B) that increase the efficiency with which FtsK peptides carrying an intact C-terminal domain activate Xer recombination under low expression levels (Aussel et al, 2002). Only half of this region was present in our initial chimaeras (aa 211–230; Figure 3A). We decided therefore to include the rest of this region (aa 179–210) in our ZapA fusion to increase its capacity to activate Xer recombination. The zapA∷ftsK50LC allele yielded 25% (±7) of cassette excision compared with 11% (±8) for zapA∷ftsKLC in the plasmid Xer recombination assay (Figure 3B). We checked by SDS–PAGE that this was not linked to a major change in the stability and/or production of the chimaera (Supplementary Figure 2), which prompted us to test its capacity to resolve chromosome dimers. Indeed, the zapA∷ftsK50LC allele fully supported chromosome dimer resolution (Figure 3D). In addition, we noticed that the presence of FtsK50 also corrected the imperfect localization of ZapA∷FtsKLC (Figure 3C), which suggested that FtsK50 could be involved in the septal recruitment of FtsK. We decided therefore to investigate the localization and activity of FtsK50LC. Surprisingly, FtsK50LC displayed a typical cell division localization pattern and supported chromosome dimer resolution (Figure 3C and D). It is very unlikely that FtsK50 contains a full transmembrane segment as previous characterization of the membrane topology of FtsK indicated that all the amino acid residues after 146 are in the cytoplasm (Dorazi and Dewar, 2000). Further evidence for this can be taken from our observation that neither the presence of FtsK50 alter the pattern of localization of chimaeras between FtsKLC and ZipA or FtsW nor their capacity to resolve chromosome dimers (Figure 3C and D), whereas the addition of an extra transmembrane segment would have either led to the re-location of FtsKLC in the periplasm or to the inversion of the membrane topology of ZipA or FtsW. Thus, we have identified an FtsK truncation, FtsK50LC, which allows for the eventual resolution of all occuring dimers even if it lacks transmembrane segments.
Effective dimer resolution depends on the length of the linker
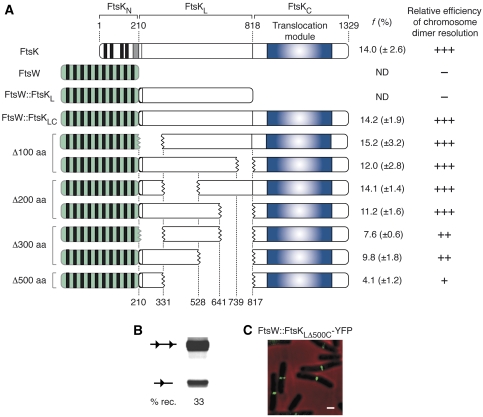
We next investigated the importance of FtsKL in chromosome dimer resolution. We reported earlier that the direct fusion of FtsKN to FtsKC (FtsKNC) led to a 70% diminution in chromosome dimer resolution (Bigot et al, 2004), even if the chimaera supported dif-recombination on plasmids (Barre, unpublished results). However, ftsKNC cells displayed a filamentous phenotype that was independent from their deficiency in chromosome dimer resolution (Bigot et al, 2004), suggesting that this fusion interfered with the function of FtsKN in septum and/or pore formation. We therefore decided to introduce linker deletions in the coding region of the ftsW∷ftsKLC allele and to test their activity in the ftsA* ftsK− background, that is in a genetic context in which the loss of fitness cannot be attributed to the role(s) played by FtsKN and FtsKL in cell division and/or pore formation (Figure 4A). A 500 aa deletion of the linker led to a 70% reduction of chromosome dimer resolution (Figure 4A), even if the truncated peptide supported dif-recombination on plasmids to a similar level than full-length ftsK (Figure 4B) and was efficiently recruited to the septum (Figure 4C). However, shorter deletions did not, or only slightly, affected chromosome dimer resolution, independently of their location in FtsKL (Figure 4; Δ100–200 aa and Δ300 aa deletions). Similar results were obtained with deletions in the linker domain of the FtsW∷FtsK50LC chimaera (data not shown). We conclude that no specific region of FtsKL is implicated in chromosome dimer resolution, but that efficacious chromosome dimer resolution depends on the total length of the linker domain.
Figure 4.
Role of linker in chromosome dimer resolution. (A) Scheme of the FtsK peptides tested for their chromosome dimer resolution ability. Numbers on the bottom refer to the amino acid position residues delimiting the different FtsKL deletions. The range of the deletion size is indicated on the left. f, mean and standard deviation of the growth advantage given by the ectopic production of each FtsKLC chimaera (result from three independent experiments). The relative efficiency of chromosome dimer resolution is indicated as follows: >70% (+++); 35–70% (++); <35% (+); 0% (–). ND, not determined. (B) In vivo pseudo-dimer resolution assay on plasmids for the FtsW∷FtsKLΔ500C chimaera as in Figure 3B. (C) Localization of the YFP derivative of FtsW∷FtsKLΔ500C as in Figure 2C.
Fully efficient resolution of chromosome dimers by chimaeras lacking the FtsK transmembrane segments
The competition assay between ftsA* ftsK− cells and ftsA* ftsK−dif− cells provides a good estimate of the ability of a protein to support dimeric chromosome resolution. However, it does not provide a direct estimate of the efficiency of the process as the latter requires an assessment of the time and energy that are spent to eventually achieve it. Nevertheless, we noticed in the course of our growth competition experiments that dif+ ftsA* ftsK− cells expressing FstK chimaeras that were fully active in dimer resolution yielded a similar number of colony forming units after 4 days of growth than dif+ ftsA* ftsK− cells expressing full-length FtsK, which suggested that these chimaeras efficiently resolved chromosome dimers (data not shown).
To further investigate the efficiency with which FtsK chimaeras resolved chromosome dimers, we decided to directly compare the growth of cells expressing them with the growth of cells expressing full-length FtsK. However, this must not be done in the ftsA* ftsK− background because these chimaeras do not necessarily compensate for the division defect of ftsA* ftsK− cells in contrast to full-length FtsK. Consequently, we compared the growth of FtsKLC− cells ectopically expressing FtsK chimaeras from a low copy pSC101 plasmid under the ftsK promoter to the growth of FtsKLC− cells ectopically expressing full-length FtsK (Figure 5A). FtsKLC− cells expressing FtsKLC had a growth disadvantage of 15.7% per cell per generation over cells expressing full-length FtsK (Figure 5A), which fits with the estimated rate of dimer formation (Steiner and Kuempel, 1998; Perals et al, 2001). In contrast, FtsKLC− cells expressing FtsW∷FtsKLC, FtsW∷FtsK50LC and FtsK50LC had the same fitness than cells expressing full-length FtsK, suggesting that these three chimaeras are fully efficient in chromosome dimer resolution (Figure 5B). Note that this result also indicates that the ectopic expression of these chimaeras did not interfere with other cellular processes than chromosome dimer resolution such as cell division.
Figure 5.
Fully efficient chromosome dimer resolution by FtsK chimaeras. (A) Growth competition between ftsKLC− cells ectopically expressing wild-type FtsK or FtsK chimaeras from a low copy vector under the ftsK promoter. f represents the growth disadvantage of the strain producing the FtsK chimaera compared to the strain producing wild-type FtsK. (B) Scheme of the chromosomal dif-cassette (left drawing) and of the recombinant product (right drawing) that is used to monitor the efficiency with which FtsK chimaeras induce Xer recombination at the normal dif locus on the chromosome. dif sites are represented by black triangles. Arrows indicate the positions of the primers used in the qPCR assay. Primers X/Z serve to quantify the relative number of chromosomes in which the DNA cassette between the two repeated dif sites is excised (right drawing). To this aim, genomic DNA needs to be digested with HindIII (dashed line, H3) to prevent contamination by larger products encompassing the cassette. Primers X/Y serve to quantify the relative number of chromosomes that still harbour the dif-cassette. (C) Chromosomal dif-cassette excision on ectopic production of full-length FtsK or FtsK chimaeras. The % of cassette excision was monitored by qPCR at different times after the induction of the production of the ftsK alleles. The actual data points and best-fit lines from a typical experiment are shown on the left. The slopes of the best-fit lines provide a good estimate of the rate of cassette excision as a function of time, which allowed us to calculate the relative efficiency of cassette excision of each chimaera when compared with full-length FtsK. The mean relative efficiencies are indicated on the right (results from three independent experiments for ZipA∷FtsKLC and FtsW∷FtsKLC and from two independent experiments for FtsKLC).
The phenotype of FtsKLC− cells ectopically expressing ZipA∷FtsKLC or ZipA∷FtsK50LC from a low copy pSC101 vector under the ftsK promoter suggested that the resulting overexpression of ZipA altered the process of division (data not shown), which would confound the growth competition result. Consequently, we had to use a second approach to assess the efficiency with which ZipA∷FtsKLC processes chromosome dimers. We described earlier how Xer recombination can be monitored in real time using quantitative PCR in FtsKATP− cells habouring two dif sites in direct tandem repeat at the normal dif locus (Kennedy et al, 2008). Briefly, activation of Xer recombination leads to the excision of the dif-cassette, which can be detected by PCR using two pairs of primers (Figure 5B). Previous work indicated that in the majority of cases, this only occurs in the presence of a chromosome dimer (Barre et al, 2000; Perals et al, 2001; Kennedy et al, 2008) and at the time of cell division (Kennedy et al, 2008). Therefore, this assay allows us to directly address the time needed for the alignment of dif sites carried on a chromosome dimer and the subsequent activation of Xer recombination. The assay is only reliable when the production of FtsK is precisely controlled (Kennedy et al, 2008). To this aim, full-length FtsK, FtsW∷FtsKLC, ZipA∷FtsKLCand FtsKLC were produced from a low copy pSC101 plasmid under the arabinose promoter. Under these conditions, ZipA∷FtsKLC proved to be as efficient as full-length FtsK and FtsW∷FtsKLC (Figure 5C). In contrast, FtsKLC did not promote excision of the chromosomal dif-cassette (Figure 5C).
Taken together, these results suggest that anchoring FtsKLC in the cytoplasmic membrane and targeting it to the septum is sufficient for the fully efficient resolution of chromosome dimers in E. coli.
Discussion
E. coli FtsK transports DNA across a ‘pre-existing opening' in the septum
Our in vivo analysis of the role(s) played by FtsKN and FtsKL in DNA transport in E. coli is based on the fact that during chromosome dimer resolution, FtsK needs to bring together the two dif sites carried by a chromosome dimer through oriented DNA transport, in addition to activating Xer recombination. Consequently, our observation that FtsW∷FtsKLC, ZipA∷FtsKLC, ZapA∷FtsK50LC and FtsK50LC support chromosome dimer resolution indicates that these chimaeras are active in DNA transport, even if FtsW and ZipA share no homology with FtsKN and if ZapA∷FtsK50LC and FtsK50LC lack transmembrane segments (Figures 2 and 3). Likewise, no specific region of FtsKL was essential for chromosome dimer resolution (Figure 4). We conclude that chimaeric FtsK can transport DNA through a ‘pre-existing opening' between the two future daughter cells.
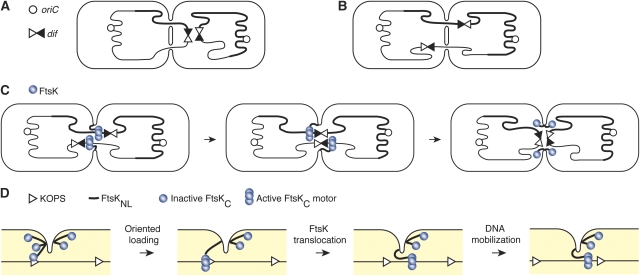
The nature of this ‘pre-existing opening' is subject to speculations. We cannot exclude that chimaeric FtsK proteins use a pore created by another septal membrane protein than FtsK. However, the most likely explanation for the fully efficient chromosome dimer resolution activity of ZipA∷FtsKLC, FtsW∷FtsKLC, FtsW∷FtsK50LC and FtsK50LC (Figure 5) is that FtsK transports DNA before the final closure of the septum, at a time when there is still a rather large opening between the two daughter cells. In this model, the length of FtsKL is important for DNA accessibility. Accordingly, we found that the efficiency of chromosome dimer resolution depended on the length of the linker, independently of its role in cell division (Figure 4). This model also explains our previous observation that Xer recombination activation depends on the initiation of constriction (Kennedy et al, 2008) as FtsKC, which is connected to the integral membrane of FtsK by a 600 aa linker that can only span 200 nm, should have a limited access to chromosomal DNA when the protein is distributed along the 800 nm-wide pre-division ring. In addition, this model answers two questions raised by the previous hypothesis of FtsK-dependent DNA transport across a closed septum: (i) how the last segment of a circular double-stranded DNA molecule is transferred (as depicted in Figure 6A and C) and (ii) how a recombination synapse between two dif sites is formed when they are segregated on either side of the septum rather than on the same side at the time of cell division (as depicted in Figure 6B and C).
Figure 6.
Schematic of DNA transport by FtsK. (A, B) Limiting steps for chromosome dimer resolution in the hypothesis in which FtsK transports DNA through a pore formed by its N-terminal domain: transfer of the last segment of the circular double-stranded chromosome (A) and formation of a recombination synapse between dif sites carried by a dimer when they are positioned on either side of a closed septum at cell division (B). (C) The assembly of several FtsK complexes in an incompletely formed septum ensures synapsis of dif sites carried by a dimer and the transfer of circular DNA molecules. (D) The inertia of chromosomes drives the C-terminal domains of active FtsK molecules towards the side of the division plan from which DNA is pumped until the maximal extension of the linker is reached. To avoid clutter, only one of the six linkers attaching FtsK hexamers to the internal membrane of the cell is drawn.
Note that the idea that FtsK transports DNA across an incompletely formed septum does not exclude the possibility that a pore created by FstKN might be involved in another cellular process, such as membrane fusion at the end of cell division as suggested for SpoIIIE (Liu et al, 2006).
Generalization to other proteins of the FtsK family
In the generally admitted model for the transport of DNA by SpoIIIE during B. subtilis sporulation and by the Tra proteins during the conjugation of various streptomycetes plasmids, DNA transfer occurs across pores created by the transmembrane segments of these proteins (Grohmann et al, 2003; Liu et al, 2006; Burton et al, 2007). Our observation that a different mechanism has been adopted in E. coli for the transport of DNA during chromosome dimer resolution could be explained by the difference in the biological functions of FtsK, SpoIIIE and the Tra proteins and/or by the difference in the architecture of the cell walls of the Gram− E. coli bacterium and the Gram+ B. subtilis and streptomycetes bacteria.
However, a model in which SpoIIIE transports DNA before membrane fusion, across an incompletely formed septum, is compatible with the observation that cytoplasmic proteins and membrane dyes do not diffuse freely between the mother cell and the prespore compartments during B. subtilis sporulation. Actually, reduction of the opening during septum constriction and molecular crowding at the site of division, which involves more than a dozen proteins, could diminish the rate of diffusion of membrane dyes between these compartments. In addition, formation of the SpoIIIE motors at the periphery of the incompletely formed sporulating septum might participate in blocking the diffusion of proteins between the mother cell and prespore compartments, which could partially explain why in its absence the two compartments are no more isolated (Liu et al, 2006).
Transport of chromosomes across an incompletely formed septum is also compatible with the observation that active SpoIIIE molecules are only found on the side of the septum from which DNA is exported (Sharp and Pogliano, 2002a, 2002b; Becker and Pogliano, 2007): because of the expected inertia of chromosomes, the C-terminal domains forming the SpoIIIE engines will initially move away from the septum after loading (as depicted in Figure 6D). This movement should persist until the linker arms are fully extended. As DNA translocation by SpoIIIE is oriented by KOPS-like motifs (Ptacin et al, 2008), the C-terminal domains should end up on the side from which DNA is exported (Figure 6D).
Finally, the idea that the Streptomycetes Tra proteins transport plasmids across a pre-existing opening between contacting mycelial tips could explain how double-stranded DNA is transferred from both the donor to the recipient and vice versa without any requirement for a relaxase and a type IV secretion system (Possoz et al, 2001; Grohmann et al, 2003), in contrast to classical conjugation (Llosa et al, 2002).
Coordination between chromosome segregation and cell division
The notion that FtsK and SpoIIIE could transport DNA through an incompletely formed septum fits with the previously accumulated evidences implicating SpoIIIE in membrane fusion during sporulation (Sharp and Pogliano, 1999, 2003; Liu et al, 2006). Indeed, the role played by SpoIIIE in membrane fusion could serve as a mechanism to delay cell fission until SpoIIIE has cleared the septum from DNA. In E. coli, the long FtsK linker might enable it to function at an early stage during septum constriction, which would leave more time for DNA transport before septum closure. However, FtsK could also be implicated in a mechanism that would delay membrane fusion in the presence of un-segregated DNA at the septum. In favour of this hypothesis, pre-division figures with fully constricted septa accumulate in cells harbouring large inversions of the E. coli chromosome (Lesterlin et al, 2008), in which the activity of FtsK is required to re-distribute large amounts of chromosomal DNA in the future daughter cells at each cell cycle. Furthermore, this late cell division arrest is lost when the activity of the C-terminal domain of FtsK is affected (Lesterlin et al, 2008). Thus, it will be important to determine the role played by the C-terminal and linker domains of FtsK in cell division, how this might lead to a delay in membrane fusion in the presence of DNA, and if such a mechanism is conserved in bacteria in which the linker of FtsK is shorter than in E. coli.
Materials and methods
Strains, growth conditions and plasmid constructions
All strains used in this study are AB1157 derivatives. They were obtained by P1 transduction and are described in Supplementary Table 1. Bacteria were grown at 37°C in Lennox-Lysogenic Broth. The following antibiotics (μg/ml) were added as appropriate: chloramphenicol (30), spectinomycin (50), kanamycin (50), ampicillin (100) and tetracyclin (15). The FtsK chimaeras were constructed by translational fusion of ZapA, ZipA or FtsW with derivatives of FtsK in which restriction sites had been introduced by PCR at the end of the cell division module (Spe1, aa 211), just before the 50 amino acid region implicated in hexamer stabilization (Spe1, aa 179) or the chromosome segregation module (XhoI, aa 818). This led to the addition of one (S) or two (LE) amino acid residues at positions 179, 210 or 818 of FtsK. Earlier studies indicated that these additions had no consequences on Xer recombination activation, septum localization and chromosome dimer resolution (Aussel et al, 2002; Yates et al, 2003; Bigot et al, 2004). The ftsK alleles were expressed from a pSC101 derivative carrying the resistance to spectinomycin under the native ftsK promoter, in translational fusion or not with fluorescent proteins. They were also placed on pBAD expression vectors carrying the resistance to ampicillin.
Microscopy
Cells were grown to OD600=0.2–0.3 at 30°C in LB, washed twice in M9 medium and spread on a microscope slide on a layer of M9 medium containing 1% agarose. A DM6000-B Microscope (Leica; Wetzlar, Germany) coupled to a Coolsnap HQ CCD camera (Photometrics; Tucson, AZ) was used to image cells. Images were analysed using the Metamorph software.
Growth competition assay
As described in Bigot et al (2004). Briefly, a 1:1 mixture of two strains was grown in serial culture in LB at 37°C and their relative frequencies determined by plating every 20 generations on appropriate media. The ftsK alleles were expressed from a pSC101 vector. The rates at which ftsA* ftsK− and ftsKLC− strains overtake their Δdif derivative correspond to the efficiencies with which the ftsK allele express sustain chromosome dimer resolution.
In vivo plasmid resolution assay
As described in Yates et al (2003). Briefly, cells were transformed with the pBAD FtsK chimaera expression vectors and a recombination substrate (pFX142; pSC101 derivative carrying the resistance to spectinomycin). At least 20 colonies were re-suspended, diluted a 100-fold in LB and grown for 4 h in 0.2% arabinose to induce FtsK chimaera production. Plasmid DNA was extracted by using a Qiagen miniprep kit and analysed by electrophoresis through a 1% agarose gel in 1 × TAE. Resolution efficiency was computed as the amount of replicative, supercoiled circle product over the amount of supercoiled substrate, which was quantitated using Sybgreen staining (Molecular probe) on a Typhoon fluorescence scanner (GE Healthcare).
qPCR analysis of chromosomal dif-cassette excision
As described in Kennedy et al (2008). Briefly, an AB1157 ftsKATP− dif-km-dif strain was transformed by pSC101 vectors expressing either ftsK, ftsKLC, ftsW∷ftsKLC or zipA∷ftsKLC under an inductible promoter. The strains were grown overnight in LB supplemented with 0.2% of glucoce, diluted in fresh LB to an OD of 0.05. After one generation, 0.2% of arabinose was added. Cultures were continually diluted with pre-warmed fresh media to maintain logarithmic growth. Aliquots were taken at several time points to measure OD at 600 nm and to purify genomic DNA. Genomic DNA was restricted with HindIII. qPCR was then performed on a LigthCycler 2.0 (Roche Scientific, Nutley, NJ) as recommanded. The primers used for qPCR were as follows: forward X, 5-TGACCGCCAACGACTGGATTC; reverse Y, 5-TTAATCGCGGCCTCGAGCAAG; reverse Z, 5-GCGACAGACACTGCGCTCTTAG.
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Table 1
Review Process File
Acknowledgments
We thank M Poidevin, J Jamin, C Lesterlin, B Michel, C Possoz, and P Stragier for critical reading and/or helpful discussions. We thank W Margolin for the kind gift of ftsA*-carrying strains. This work was supported by the Agence Nationale pour la Recherche (ANR-05-BLAN-0060) and (ANR-09-BLAN-0258-01); the Fondation pour la Recherche Médicale (Equipe 2007); the European Molecular Biology Organization (YIP 2006); the Centre National pour la Recherche Scientifique (ATIP+).
Footnotes
The authors declare that they have no conflict of interest.
References
- Aussel L, Barre FX, Aroyo M, Stasiak A, Stasiak AZ, Sherratt D (2002) FtsK is a DNA motor protein that activates chromosome dimer resolution by switching the catalytic state of the XerC and XerD recombinases. Cell 108: 195–205 [DOI] [PubMed] [Google Scholar]
- Barre FX (2007) FtsK and SpoIIIE: the tale of the conserved tails. Mol Microbiol 66: 1051–1055 [DOI] [PubMed] [Google Scholar]
- Barre FX, Aroyo M, Colloms SD, Helfrich A, Cornet F, Sherratt DJ (2000) FtsK functions in the processing of a Holliday junction intermediate during bacterial chromosome segregation. Genes Dev 14: 2976–2988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bath J, Wu LJ, Errington J, Wang JC (2000) Role of bacillus subtilis SpoIIIE in DNA transport across the mother cell-prespore division septum. Science 290: 995–997 [DOI] [PubMed] [Google Scholar]
- Becker EC, Pogliano K (2007) Cell-specific SpoIIIE assembly and DNA translocation polarity are dictated by chromosome orientation. Mol Microbiol 66: 1066–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begg KJ, Dewar SJ, Donachie WD (1995) A new Escherichia coli cell division gene, ftsK. J Bacteriol 177: 6211–6222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt TG, de Boer PA (2005) SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over Chromosomes in E. coli. Mol Cell 18: 555–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigot S, Corre J, Louarn J, Cornet F, Barre FX (2004) FtsK activities in Xer recombination, DNA mobilization and cell division involve overlapping and separate domains of the protein. Mol Microbiol 54: 876–886 [DOI] [PubMed] [Google Scholar]
- Bigot S, Sivanathan V, Possoz C, Barre FX, Cornet F (2007) FtsK, a literate chromosome segregation machine. Mol Microbiol 64: 1434–1441 [DOI] [PubMed] [Google Scholar]
- Burton BM, Marquis KA, Sullivan NL, Rapoport TA, Rudner DZ (2007) The ATPase SpoIIIE transports DNA across fused septal membranes during sporulation in Bacillus subtilis. Cell 131: 1301–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez AA, Farewell A, Nannmark U, Nystrom T (1997) A mutation in the ftsK gene of Escherichia coli affects cell-cell separation, stationary-phase survival, stress adaptation, and expression of the gene encoding the stress protein UspA. J Bacteriol 179: 5878–5883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorazi R, Dewar SJ (2000) Membrane topology of the N-terminus of the escherichia coli FtsK division protein. FEBS Lett 478: 13–18 [DOI] [PubMed] [Google Scholar]
- Draper GC, McLennan N, Begg K, Masters M, Donachie WD (1998) Only the N-terminal domain of FtsK functions in cell division. J Bacteriol 180: 4621–4627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errington J, Bath J, Wu LJ (2001) DNA transport in bacteria. Nat Rev Mol Cell Biol 2: 538–545 [DOI] [PubMed] [Google Scholar]
- Geissler B, Margolin W (2005) Evidence for functional overlap among multiple bacterial cell division proteins: compensating for the loss of FtsK. Mol Microbiol 58: 596–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler B, Shiomi D, Margolin W (2007) The ftsA* gain-of-function allele of Escherichia coli and its effects on the stability and dynamics of the Z ring. Microbiology 153: 814–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grohmann E, Muth G, Espinosa M (2003) Conjugative plasmid transfer in gram-positive bacteria. Microbiol Mol Biol Rev 67: 277–301, table of contents [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harry E, Monahan L, Thompson L (2006) Bacterial cell division: the mechanism and its precison. Int Rev Cytol 253: 27–94 [DOI] [PubMed] [Google Scholar]
- Kennedy SP, Chevalier F, Barre FX (2008) Delayed activation of Xer recombination at dif by FtsK during septum assembly in Escherichia coli. Mol Microbiol 68: 1018–1028 [DOI] [PubMed] [Google Scholar]
- Lesterlin C, Pages C, Dubarry N, Dasgupta S, Cornet F (2008) Asymmetry of chromosome Replichores renders the DNA translocase activity of FtsK essential for cell division and cell shape maintenance in Escherichia coli. PLoS Genet 4: e1000288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Draper GC, Donachie WD (1998) FtsK is a bifunctional protein involved in cell division and chromosome localization in Escherichia coli. Mol Microbiol 29: 893–903 [DOI] [PubMed] [Google Scholar]
- Liu NJ, Dutton RJ, Pogliano K (2006) Evidence that the SpoIIIE DNA translocase participates in membrane fusion during cytokinesis and engulfment. Mol Microbiol 59: 1097–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llosa M, Gomis-Ruth FX, Coll M, de la Cruz Fd F (2002) Bacterial conjugation: a two-step mechanism for DNA transport. Mol Microbiol 45: 1–8 [DOI] [PubMed] [Google Scholar]
- Massey TH, Mercogliano CP, Yates J, Sherratt DJ, Lowe J (2006) Double-stranded DNA translocation: structure and mechanism of hexameric FtsK. Mol Cell 23: 457–469 [DOI] [PubMed] [Google Scholar]
- Perals K, Capiaux H, Vincourt JB, Louarn JM, Sherratt DJ, Cornet F (2001) Interplay between recombination, cell division and chromosome structure during chromosome dimer resolution in Escherichia coli. Mol Microbiol 39: 904–913 [DOI] [PubMed] [Google Scholar]
- Possoz C, Ribard C, Gagnat J, Pernodet JL, Guerineau M (2001) The integrative element pSAM2 from Streptomyces: kinetics and mode of conjugal transfer. Mol Microbiol 42: 159–166 [DOI] [PubMed] [Google Scholar]
- Ptacin JL, Nollmann M, Becker EC, Cozzarelli NR, Pogliano K, Bustamante C (2008) Sequence-directed DNA export guides chromosome translocation during sporulation in Bacillus subtilis. Nat Struct Mol Biol 15: 485–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh OA, Bigot S, Barre FX, Allemand JF (2005) Analysis of DNA supercoil induction by FtsK indicates translocation without groove-tracking. Nat Struct Mol Biol 12: 436–440 [DOI] [PubMed] [Google Scholar]
- Saleh OA, Perals C, Barre FX, Allemand JF (2004) Fast, DNA-sequence independent translocation by FtsK in a single-molecule experiment. EMBO J 23: 2430–2439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp MD, Pogliano K (1999) An in vivo membrane fusion assay implicates SpoIIIE in the final stages of engulfment during Bacillus subtilis sporulation. Proc Natl Acad Sci USA 96: 14553–14558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp MD, Pogliano K (2002a) MinCD-dependent regulation of the polarity of SpoIIIE assembly and DNA transfer. EMBO J 21: 6267–6274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp MD, Pogliano K (2002b) Role of cell-specific SpoIIIE assembly in polarity of DNA transfer. Science 295: 137–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp MD, Pogliano K (2003) The membrane domain of SpoIIIE is required for membrane fusion during Bacillus subtilis sporulation. J Bacteriol 185: 2005–2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner WW, Kuempel PL (1998) Sister chromatid exchange frequencies in Escherichia coli analyzed by recombination at the dif resolvase site. J Bacteriol 180: 6269–6275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Val M-E, Kennedy SP, El karoui M, Bonné L, Chevalier F, Barre F-X (2008) FtsK-dependent dimer resolution on multiple chromosomes in the pathogen Vibrio cholerae. PLoS Genet 4: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Lutkenhaus J (1998) FtsK is an essential cell division protein that is localized to the septum and induced as part of the SOS response. Mol Microbiol 29: 731–740 [DOI] [PubMed] [Google Scholar]
- Wu LJ, Errington J (1994) Bacillus subtilis spoIIIE protein required for DNA segregation during asymmetric cell division. Science 264: 572–575 [DOI] [PubMed] [Google Scholar]
- Wu LJ, Errington J (2004) Coordination of cell division and chromosome segregation by a nucleoid occlusion protein in Bacillus subtilis. Cell 117: 915–925 [DOI] [PubMed] [Google Scholar]
- Wu LJ, Lewis PJ, Allmansberger R, Hauser PM, Errington J (1995) A conjugation-like mechanism for prespore chromosome partitioning during sporulation in Bacillus subtilis. Genes Dev 9: 1316–1326 [DOI] [PubMed] [Google Scholar]
- Yates J, Aroyo M, Sherratt DJ, Barre FX (2003) Species specificity in the activation of Xer recombination at dif by FtsK. Mol Microbiol 49: 241–249 [DOI] [PubMed] [Google Scholar]
- Yu XC, Tran AH, Sun Q, Margolin W (1998) Localization of cell division protein FtsK to the Escherichia coli septum and identification of a potential N-terminal targeting domain. J Bacteriol 180: 1296–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Table 1
Review Process File