Summary
Structures of the E. coli 70S ribosome show how the large and small subunits rotate to facilitate protein synthesis.
Protein biosynthesis on the ribosome requires repeated cycles of ratcheting, which couples rotation of the two ribosomal subunits with respect to each other and swiveling of the head domain of the small subunit. However, the molecular basis for how the two ribosomal subunits rearrange contacts with each other during ratcheting while remaining stably associated is not known. Here we describe x-ray crystal structures of the intact Escherichia coli ribosome, either in the apo form (3.5 Å resolution) or with one (4.0 Å res) or two (4.0 Å res) anticodon stem-loop tRNA mimics bound, that reveal intermediate states of intersubunit rotation. In the structures, the interface between the small and large ribosomal subunits rearranges in discrete steps along the ratcheting pathway. Positioning of the head domain of the small subunit is controlled by interactions with the large subunit and with the tRNA bound in the peptidyl-tRNA site. The intermediates observed here provide insight into how tRNAs move into the hybrid state of binding that precedes the final steps of mRNA and tRNA translocation.
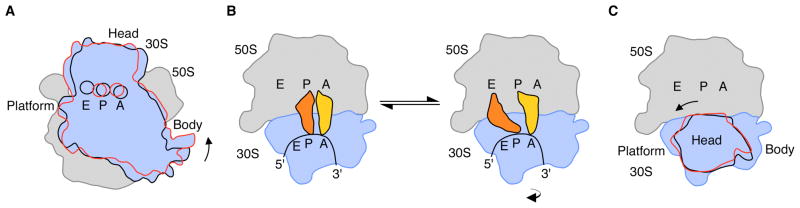
Protein biosynthesis requires many large-scale rearrangements in the ribosome as each amino acid is added to a growing polypeptide chain. Positioning of tRNA on the ribosome is proposed to occur through a ratcheting mechanism. Central to this mechanism is a rotation of the small ribosomal subunit relative to the large subunit (Fig. 1A) (1, 2) that occurs in all stages of translation–initiation, elongation, termination, and ribosome recycling (1)–and is targeted by clinically useful antibiotics (3, 4). For example after each peptide bond is formed, an ~8° intersubunit rotation results in tRNAs bound in the aminoacyl-tRNA and peptidyl-tRNA binding sites (A site and P site, respectively) moving into the P site and exit-tRNA site (E site) on the large ribosomal subunit (Fig. 1B). From this hybrid state of tRNA binding (Fig. 1B) (1, 5), the tRNAs are then translocated to the P site and E site on the small subunit.
Fig. 1.
Rotated states of the ribosome. (A) View of the bacterial 70S ribosome, composed of the small (30S) ribosomal subunit and the large (50S) ribosomal subunit. The small subunit of the ribosome (blue) can rotate from a starting conformation seen in post-initiation and termination states (12, 13, 18) (state R0, black outline) to a fully rotated conformation seen in elongation, termination and recycling steps of translation (state RF, red outline) (1, 15–17). 30S features include: Head, Platform, Body. The 50S subunit is shown in grey. Letters indicate the positions of the aminoacyl (A), peptidyl (P), and exit (E) tRNA binding sites. (B) Schematic of tRNA binding states on the ribosome. In the transition of the ribosome to the fully rotated state, tRNAs shift from binding in the A/A and P/P sites (30S subunit/50S subunit, respectively) to occupy hybrid binding sites (A/P and P/E for 30S/50S sites). The view of the ribosome is rotated 90° from that in A. (C) Rotations of the head domain of the small ribosomal subunit. Letters indicate the locations of the aminoacyl (A), peptidyl (P), and exit (E) tRNA binding sites on the large subunit. In state R0 (black), the head domain is centered over the P site (~0° rotation). Rotations of the head domain towards the E site of up to 14° (red) have been observed (1, 6, 7). The 5′ to 3′ direction of mRNA, which threads around the neck region of the 30S subunit, is also indicated.
In addition to intersubunit rotation, ratcheting also involves a nearly orthogonal rotation of the head domain of the small ribosomal subunit (Fig. 1C) that plays a role in controlling the position of tRNAs within the ribosome (1, 6, 7). As with intersubunit rotation, movement of the head domain is a target for clinically useful antibiotics (8). Swiveling of the head domain relative to the body of the small subunit may also be required for the intrinsic helicase activity of the ribosome in unwinding secondary structure in mRNA (8, 9). Rotations of up to 14° allow the head domain to change its position by 20 Å or more at the ribosomal subunit interface, or the width of a tRNA substrate (7).
The molecular basis for how the ribosomal subunits rotate with respect to each other while remaining stably associated remains unknown (1, 10). Furthermore, the precise timing of movements of the small subunit head domain during ratcheting are not clear, as the head domain can move independently of the body and platform domains of the small subunit (1, 6, 7). Here we present three x-ray crystal structures of the E. coli 70S ribosome that reveal intermediates along the ratcheting pathway. These structures show that the ribosome can rearrange the interface between the ribosomal subunits in discrete steps, and suggest how these rearrangements may direct tRNAs into hybrid states of binding essential for mRNA and tRNA translocation.
Using new crystal forms, we determined x-ray crystal structures of the E. coli 70S ribosome in the absence of ligands, with mRNA and an anticodon stem-loop mimic of tRNAMetf (ASLMetf) bound in the P site of the small (30S) subunit, or with mRNA and two ASL mimics of tRNAPhe (ASLPhe) bound in the A and P sites of the 30S subunit (11) (3.5 Å, 4.0 Å and 4.0 Å resolution, respectively, Tables S1 and S2). One of the two copies of the ribosome in each crystal structure resembles previously determined high-resolution structures of the ribosome (7, 12). Notably, in the other copies of the ribosome in each structure, the small subunit of the ribosome is rotated to an intermediate position with respect to the large subunit when compared to structures of the ribosome determined previously. In the new conformation, the small subunit is rotated by 3°–6° relative to its position in a post-initiation state of the ribosome, in which initiator tRNA is bound in the P site (defined here as state R0, Fig. 2A, 2B, Table S3) (12, 13), and 2°–3° relative to the ribosome with tRNAs bound in the P/P and E/E sites (here denoted state R1, Fig. 2C, Table S3) (14). In ribosomes in which tRNA occupies a hybrid P/E binding site (1, 15–17) (here called state RF, Fig. 1A, 1B) the small subunit is rotated by an additional 2°–4° relative to the rotational state described here (Fig. 2D, Table S3), which we term state R2. The ribosome can therefore adopt at least four stable states of intersubunit rotation, R0, R1, R2 and RF.
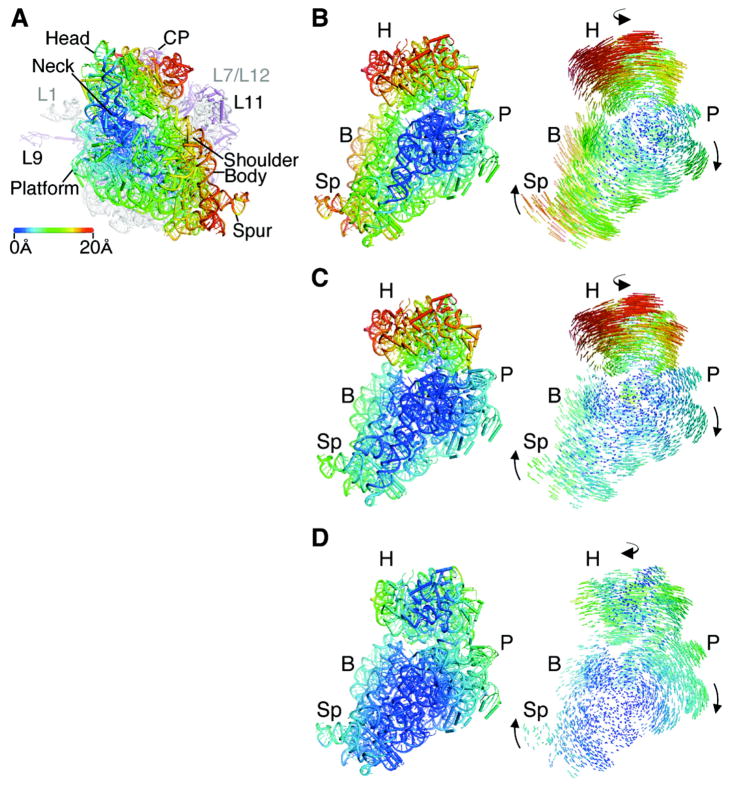
Fig. 2.
Structure of the apo-70S ribosome in an intermediate state of intersubunit rotation, state R2. (A) Comparison of the ribosome in state R2 with the ribosome in state R0 (12, 13), with the 50S subunit serving as reference (1). Arrows indicate the direction of movement in the transition from state R0 to state R2. The distance changes in 30S subunit positions are color-coded in Å units, as shown, in this and the subsequent panels. Ribosomal RNA and proteins in the 50S subunit are colored grey and magenta, respectively. 30S features include: Head, Neck, Platform, Body, Shoulder, and Spur. 50S features include: L1, protein L1/rRNA arm; CP, central protuberance; L11, protein L11/rRNA arm, L9, protein L9. The approximate location of proteins L7/L12 and L1, not observed in the structure, are noted in grey. (B) Comparison of the ribosome in state R2 to the ribosome in state R0, viewed from the perspective of the 50S subunit. Difference vectors between phosphorous and Cα atoms are shown to the right, with arrows indicating the direction of the change. (C) Comparison of the ribosome in state R2 to the ribosome in state R1 (14). (D) Comparison of the ribosome in state R2 to the ribosome in the fully rotated state RF (15, 16).
In state R2, the central contacts or “bridges” between the ribosomal subunits (1, 7) are nearly indistinguishable from those observed in ribosomes in states R0 and R1 (7, 12, 14, 18–20) (Fig. 3A). These bridges include contacts between ribosomal RNA elements in the small and large subunits (16S rRNA and 23S rRNA, respectively) that are near the tRNA binding sites. The largest change in the central bridges occurs in bridge B2a, adjacent to the mRNA decoding site (Fig. 3A). In this region, nucleotide A1913 in 23S rRNA and nucleotides A1492-A1493 in helix h44 of 16S rRNA adopt different conformations depending on the tRNA occupancy of the A site, as observed in other structures (7, 12, 14). To maintain contacts in bridges at the center of the interface (Fig. 3A) during subunit rotation, helix h44 in the small subunit bends near 16S rRNA helix h14 (Fig. 3B).
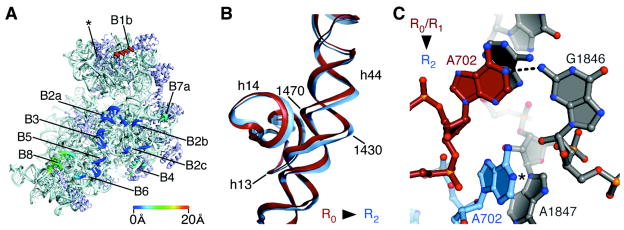
Fig. 3.
Contacts, or “bridges”, between the ribosomal subunits in the apo-70S ribosome in state R2. (A) The position of bridges in state R2 compared to those in state R0 (12, 13). Bridge numbering is the same as in (7). The direction of view and color coding are the same as in Fig. 2C. Bridge B1a (asterisk), includes the A-site Finger (H38 in 23S rRNA) which spans the subunit interface parallel to the A and P sites (29). This contact is not visible in the present structures due to disorder at the end of H38 in both states of the ribosome. (B) Bend in rRNA helix h44 in 16S rRNA that accommodates rotated state R2. Nucleotides and 16S rRNA helices are marked. The view is the same as in Fig. 2B. (C) Bridge B7a in state R2 compared to that in states R0 (12, 18–20) and R1 (14). Nucleotide A702 in 16S rRNA in the 30S subunit (light blue) and nucleotides in H68 of 23S rRNA in the 50S subunit (grey) are shown for state R2. Nucleotide A702 in state R0 or R1 is shown in red. The N1 position of A702 that would be methylated by dimethylsulfate is marked (5).
By contrast, key bridges between the platform of the 30S subunit and the 50S subunit (B4, B7a) are shifted half way to their position in the RF state (1, 15, 16) (Fig. 2, 3A). In the structures observed here, the platform of the 30S subunit rotates about the helical axis of the top of helix h44 in 16S rRNA. The rotation exposes nucleotide A702 in 16S rRNA to solvent, whereas it is buried in the minor groove of H68 in 23S rRNA in high-resolution structures of states R0 and R1 (Fig. 3C, Fig. S1) (12, 14, 18–20). In 16S rRNA, nucleotide A702 is protected from chemical probes when tRNAs are bound in the A/A and P/P sites (5). However, when tRNAs occupy hybrid binding sites (A/P and P/E, Fig. 1B), nucleotide A702 becomes exposed to chemical probes (5), and bridge B7a is rearranged (1, 11, 15, 16). Exposure of A702 to solvent suggest that the conformation of the ribosome in state R2 is at least part way to the fully rotated state (RF) that accommodates tRNA binding in hybrid A/P and P/E sites (Fig 1B). Notably, apart from protein S15 in bridge B4 which is also shifted half way to its position in state RF (1, 11, 15, 16) (Fig. S2), most of the remainder of the 30S platform does not make direct contact with the large subunit (Fig. 3A). Limited contacts likely allow large shifts in the position of the interface in this region.
Contacts between the 30S subunit head domain and the 50S subunit have been shown to adopt many different configurations. In states R0 and R1, protein L5 in the central protuberance of the 50S subunit contacts the N-terminal lobe of protein S13 in the 30S head domain when it is centered over the 30S P site (Bridge B1b, Fig. 1C, Fig. 4A) (1, 12, 14). In the RF state of the ribosome the head domain of the 30S subunit is shifted such that protein S13 forms a key interaction between its long central α-helix and protein L5 in the large subunit (1, 21, 22). Notably, in the structure of the ribosome in state R2 with no bound ligands, the key interaction between proteins L5 and S13 (21, 22) is essentially indistinguishable from that in the fully rotated ribosome (1, 15, 16) (Fig. 4B). To enable the contact, the head of the small subunit is swiveled towards the E site by 11° relative to its conformation when the head domain is aligned with the 30S subunit P site (14) (Fig. 1C, Fig. 4A), whereas the head domain is swiveled by only 5° in the RF state when tRNAs are bound in the hybrid A/P and P/E sites (Fig. 1B, Fig. 4B) (1, 11, 15, 16). CryoEM reconstructions of the yeast ribosome in the RF state reveal that tRNA bound in the P/E site remains associated with the head domain of the small subunit as the head domain adopts an “open” configuration required for tRNA transit to the small subunit E site (1). Using the head domain of the small subunit as a guide, we modeled full-length A/P and P/E tRNAs (11, 16) in the ribosome in state R2. Based on the model, tRNAs could bind in the hybrid A/P and P/E sites in the R2 state due to swiveling of the head domain of the small subunit by ~11°, as observed in the apo-70S structure (Fig. 4C).
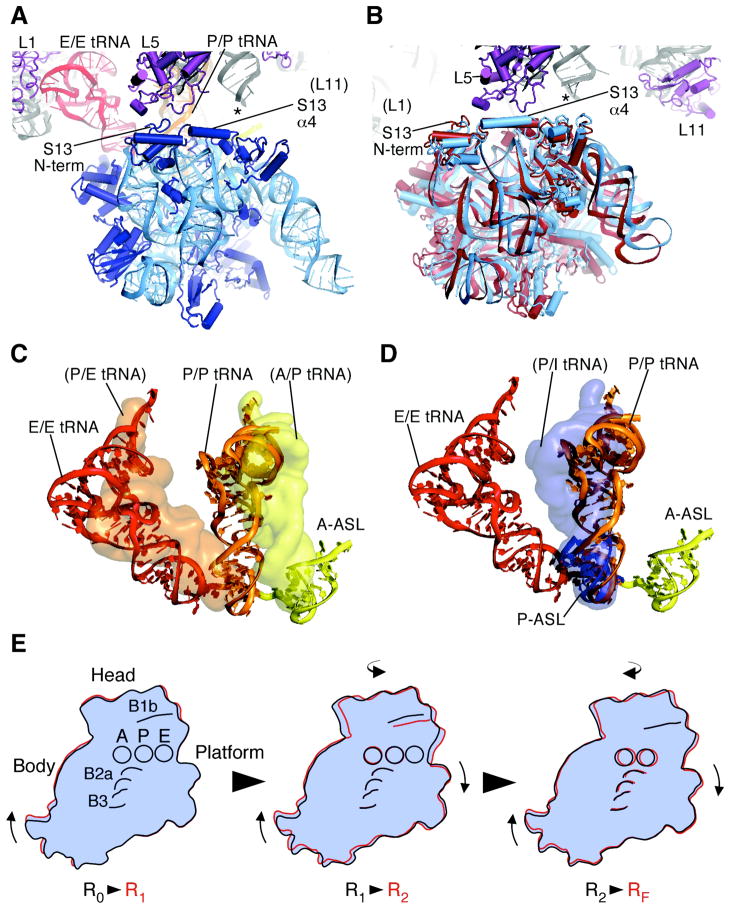
Fig. 4.
Changes in the position of the head domain in the 30S subunit in state R2. (A) Bridge B1 in ribosomes in state R1 (14). The tRNAs bound in the 30S subunit A site (yellow), and in the P/P (orange) and E/E sites (red) are shown. Domains in protein S13 in the 30S subunit head domain (blue) and protein L5 in the 50S subunit (purple) are marked. An asterisk marks the approximate location of the A-site finger (ASF) helix H38 in 23S rRNA, the tip of which is disordered in the crystal structure (14). Protein L31, not seen in E. coli 70S ribosome structures, has been removed for clarity. (B) Bridge B1 in the apo-70S ribosome in state R2 (light blue) compared to state RF (red). Domains in protein S13 in the 30S subunit head domain and protein L5 in the 50S subunit are marked. Asterisk as in panel A. (C) Position of full-length tRNAs modeled onto the apo-70S ribosome in state R2. The superposition used the head domain of the 30S subunit in the fully-rotated state RF (16) for reference (1, 11). Surfaces of the modeled tRNAs (yellow and orange) are compared to the position of tRNAs in state R1 (14), as described in panel A and shown as ribbons. (D) Position of full-length tRNA in the P site of state R1 (14) modeled onto the ribosome complexed with ASLMetf in the P site in state R2 (11), using the 30S subunit body and platform of the ribosome in state R1 as a reference. Surface of the modeled tRNA (blue) is compared to the position of the P-site ASLMetf in state R2 (blue) and tRNAs in state R1 (described above) shown as ribbons. (E) Step-wise rearrangements in the ribosome along the ratcheting pathway. The molecular envelope of the 30S subunit is shown for clarity. Domains of the 30S subunit (head, body, platform), tRNA binding sites (A, P, and E, respectively), and bridges B1b, B2a, and B3 are shown. The view is the same as in Fig. 2B. Arrows indicate the direction of movement from one state to the next.
Notably, contacts between the ribosome and the ASL portion of tRNA may oppose movement of tRNAs into the hybrid state of binding. The ASL portion of P-site tRNA has been proposed to stabilize the 30S subunit head domain centered over the P site (0° rotation, Table S3) and prevent frameshifting (12). ASL binding to the ribosome in state R2 also centers the head domain of the small subunit over the P site (11), as seen in state R0 (12). This is true regardless of whether an ASL is bound in the A site or not (11). In state R2, centering of the head domain over the P site would leave the acceptor end of P-site tRNA positioned part way between the P and E sites on the large subunit (Fig. 4D), presumably an unstable configuration except for translation initiation (23). To counterbalance the stabilizing effect of the ASL on the positioning of the head domain, bridge B1b between proteins S13 and L5 and contacts between the large subunit and the elbow and acceptor ends of tRNA (1, 24–27) may favor 30S subunit head rotation and P-site tRNA movement into the hybrid P/E site in states R2 and RF, in agreement with the in vivo importance of the α-helical region of S13 involved in the bridge (21, 22) and the requirement of full-length P-site tRNA for translocation to occur (25). Also consistent with our structural model of hybrid state formation, single-molecule FRET experiments have shown that ribosomes lacking substrates or complexed with a P-site ASLfMet exhibit spontaneous subunit rotation, but not with the same rate or efficiency as when full-length P-site tRNA is bound (10).
The structures of the E. coli 70S ribosome presented here reveal that in addition to the known R0, R1 and RF states, the ribosome can adopt an intermediate state of subunit rotation, state R2. This state would likely occur transiently, before the ribosome adopts a fully rotated conformation (1, 10, 16). Based on the structures presented above, we propose that during ratcheting, which combines intersubunit rotation and rotation of the small subunit head domain, key bridges between the ribosomal subunits rearrange in a step-wise manner. Ratcheting likely begins with the 30S subunit body, continuing with the 30S platform and head domains, and completes with rearrangement of the central bridges (Fig. 4E, Movie S1, S2, Table S3). Such a step-wise rearrangement would assist the ribosome in making large shifts at the interface without fully destabilizing the subunit interface. In addition, the multiple conformations of the head domain of the 30S subunit would help to position tRNAs on the ribosome during ratcheting (1, 6, 7). Intriguingly, single-molecule FRET studies have shown that P-site tRNA fluctuates between the P/P and P/E sites at a faster rate than intersubunit rotation (10, 26). The ability of tRNAs to adopt a hybrid state of binding in state R2 may in part explain the different rates between these processes. Fluctuation of tRNAs between the P/P and P/E states would not require the full extent of intersubunit rotation, but could occur in state R2 due to swiveling of the 30S subunit head domain and the inherent flexibility of P-site tRNA (14, 28). High-resolution structures of the ribosome in different rotated states with intact tRNAs will be required to complete our molecular understanding of the large-scale conformational rearrangements in the ribosome that allow ratcheting and protein synthesis to occur.
Supplementary Material
Acknowledgments
We thank K. Frankel, S. Classen, and G. Meigs for help with data measurement at the SIBYLS and 8.3.1 beamlines at the Advanced Light Source (ALS), R. Kanagalaghatta, D. Neau and F. Murphy for help with data measurement at ID-24 at the Advanced Photon Source (APS), P. Afonine and P. Adams for help with Phenix refinement, S.-H. Kim, P. Adams and J. Holton for useful crystallographic discussions, and J. Frank for providing the cryo-EM reconstructions of the E. coli 70S ribosome. We also thank J. Doudna, S. Blanchard, H. Noller, A. Korostelev, and D. Ermolenko for helpful discussions and comments on the manuscript. Atomic coordinates and structure factors are deposited in the Protein Data Bank (accession codes 3I1M, 3I1N, 3I1O, 3I1P, 3I1Q, 3I1R, 3I1S, 3I1T, 3I1Z, 3I20, 3I21, 3I22). This work was funded by the National Institutes of Health (GM65050 to J.H.D.C., NCI grant CA92584 for the SIBYLS and 8.3.1 beamlines at the ALS, and NRCC grant RR-15301 for the Northeastern Collaborative Access Team beamlines at 24-ID at the APS) and by the Department of Energy (DE-AC03 76SF00098 for the SIBYLS and 8.3.1 beamlines at the ALS, and DE-AC02-06CH11357 for the APS).
References
- 1.Frank J, Gao H, Sengupta J, Gao N, Taylor DJ. Proc Natl Acad Sci U S A. 2007;104:19671–8. doi: 10.1073/pnas.0708517104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Horan LH, Noller HF. Proc Natl Acad Sci U S A. 2007;104:4881–5. doi: 10.1073/pnas.0700762104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johansen SK, Maus CE, Plikaytis BB, Douthwaite S. Mol Cell. 2006;23:173–82. doi: 10.1016/j.molcel.2006.05.044. [DOI] [PubMed] [Google Scholar]
- 4.Ermolenko DN, et al. Nat Struct Mol Biol. 2007;14:493–7. doi: 10.1038/nsmb1243. [DOI] [PubMed] [Google Scholar]
- 5.Moazed D, Noller HF. Nature. 1989;342:142–8. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- 6.Spahn CM, et al. EMBO J. 2004;23:1008–19. doi: 10.1038/sj.emboj.7600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schuwirth BS, et al. Science. 2005;310:827–34. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 8.Borovinskaya MA, Shoji S, Holton JM, Fredrick K, Cate JH. ACS Chem Biol. 2007;2:545–52. doi: 10.1021/cb700100n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takyar S, Hickerson RP, Noller HF. Cell. 2005;120:49–58. doi: 10.1016/j.cell.2004.11.042. [DOI] [PubMed] [Google Scholar]
- 10.Cornish PV, Ermolenko DN, Noller HF, Ha T. Mol Cell. 2008;30:578–88. doi: 10.1016/j.molcel.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Materials and Methods are available as supporting material on Science Online.
- 12.Berk V, Zhang W, Pai RD, Cate JH. Proc Natl Acad Sci U S A. 2006;103:15830–4. doi: 10.1073/pnas.0607541103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gabashvili IS, et al. Cell. 2000;100:537–49. doi: 10.1016/s0092-8674(00)80690-x. [DOI] [PubMed] [Google Scholar]
- 14.Selmer M, et al. Science. 2006;313:1935–42. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 15.Connell SR, et al. Mol Cell. 2007;25:751–64. doi: 10.1016/j.molcel.2007.01.027. [DOI] [PubMed] [Google Scholar]
- 16.Agirrezabala X, et al. Mol Cell. 2008;32:190–7. doi: 10.1016/j.molcel.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Julian P, et al. Proc Natl Acad Sci U S A. 2008;105:16924–7. doi: 10.1073/pnas.0809587105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laurberg M, et al. Nature. 2008;454:852–7. doi: 10.1038/nature07115. [DOI] [PubMed] [Google Scholar]
- 19.Weixlbaumer A, et al. Science. 2008;322:953–6. doi: 10.1126/science.1164840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Korostelev A, et al. Proc Natl Acad Sci U S A. 2008;105:19684–9. doi: 10.1073/pnas.0810953105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cukras AR, Green R. J Mol Biol. 2005;349:47–59. doi: 10.1016/j.jmb.2005.03.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoang L, Fredrick K, Noller HF. Proc Natl Acad Sci U S A. 2004;101:12439–43. doi: 10.1073/pnas.0405227101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Allen GS, Zavialov A, Gursky R, Ehrenberg M, Frank J. Cell. 2005;121:703–12. doi: 10.1016/j.cell.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 24.Lill R, Robertson JM, Wintermeyer W. EMBO J. 1989;8:3933–8. doi: 10.1002/j.1460-2075.1989.tb08574.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joseph S, Noller HF. EMBO J. 1998;17:3478–83. doi: 10.1093/emboj/17.12.3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Munro JB, Altman RB, O’Connor N, Blanchard SC. Mol Cell. 2007;25:505–17. doi: 10.1016/j.molcel.2007.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pan D, Kirillov SV, Cooperman BS. Mol Cell. 2007;25:519–29. doi: 10.1016/j.molcel.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Korostelev A, Trakhanov S, Laurberg M, Noller HF. Cell. 2006;126:1065–77. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]
- 29.Valle M, et al. Cell. 2003;114:123–34. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.