Abstract
The β-oxidation of oleic acid in Saccharomyces cerevisiae (S. cerevisiae) was studied by comparing the growth of wild-type cells on oleic acid or palmitic acid with the growth of mutants that either had a deletion in the YOR180c (DCI1) gene reported to encode Δ3,5, Δ2,4-dienoyl-CoA isomerase (dienoyl-CoA isomerase) or in the PTE1 gene encoding peroxisomal thioesterase 1. Growth of wild-type cells was indistinguishable from that of YOR180c mutant cells on either palmitic acid or oleic acid, whereas the PTE1 mutant grew slower and to a lower density on oleic acid but not on palmitic acid. The identification of 3,5-tetradecadienoic acid in the medium of wild-type cells but not in the medium of the PTE1 mutant proves the operation of the thioesterase-dependent pathway of oleate β-oxidation in S. cerevisiae. Dienoyl-CoA isomerase activity was very low in wild-type cells, fourfold higher in the YOR180c mutant, and not associated with purified Yor180c protein. These observations support the conclusion that the YOR180c gene does not encode dienoyl-CoA isomerase.
Keywords: Saccharomyces cerevisiae; peroxisomal β-oxidation; oleic acid; thioesterase-dependent pathway; Δ3,5, Δ2,4-dienoyl-CoA isomerase; Yor180c protein
1. Introduction
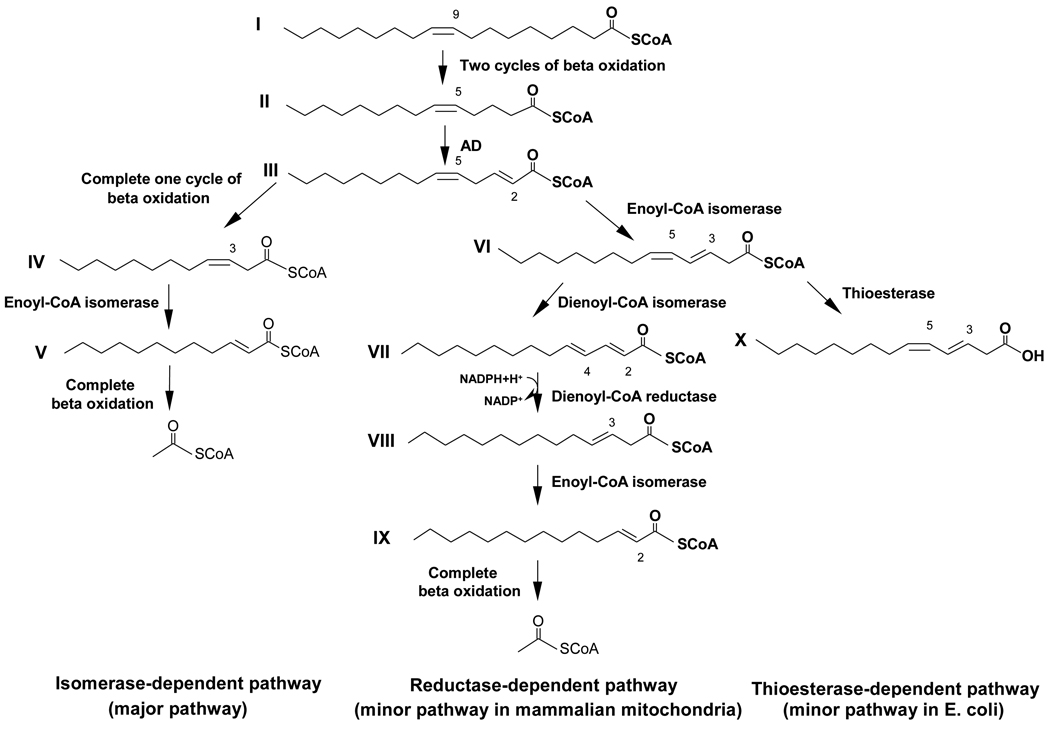
Both saturated and unsaturated fatty acids are degraded by β-oxidation. However, the breakdown of unsaturated fatty acids requires auxiliary enzymes in addition to the enzymes necessary for the β-oxidation of saturated fatty acids (reviewed in Ref. 1). The major pathway for the β-oxidation of oleic acid only requires one auxiliary enzyme named Δ3,Δ2-enoyl-CoA isomerase (enoyl-CoA isomerase)1 and therefore was named isomerase-dependent pathway (see Scheme 1) [2]. Enoyl-CoA isomerase catalyzes the positional and stereochemical isomerization of the pre-existing cis double bond of oleic acid after oleoyl-CoA has been chain shortened by three passes through the β-oxidation spiral (reviewed in Ref. 1). This simple isomerization facilitates the further β-oxidation of oleic acid by converting 3-cis-dodecenoyl-CoA (IV; all roman numerals refer to compounds shown in Scheme 1), an intermediate that cannot be degraded by β-oxidation, to 2-trans-dodecenoyl-CoA (V) that is a substrate of β-oxidation. However, the presence of enoyl-CoA isomerase creates a problem because it isomerizes a fraction of 2-trans,5-cis-tetradecadienoyl-CoA (III), an intermediate of oleate β-oxidation, to 3,5-cis-tetradecadienoyl-CoA (VI), which has two conjugated double bonds that prevent its further β-oxidation [3]. Different strategies have evolved to bypass this metabolic block. In mammalian mitochondria, Δ3,5,Δ2,4-dienoyl-CoA isomerase (dienoyl-CoA isomerase) is expressed, which shifts the two double bonds of 3,5-cis-tetradecadienoyl-CoA (VI) to yield 2-trans,4-trans-tetradecadienoyl-CoA (VII) [4,5]. The latter intermediate is reduced by 2,4-dienoyl-CoA reductase to 3-trans-enoyl-CoA (VIII) that is isomerized to 2-trans-teradecenoyl-CoA (IX) before it is completely degraded by β-oxidation [2]. This minor route of β-oxidation is referred to as the reductase-dependent pathway (see Scheme 1) even though dienoyl-CoA isomerase is the enzyme that is unique to this process while both of the other auxiliary enzymes, 2,4-dienoyl-CoA reductase and enoyl-CoA isomerase, have essential functions in other pathways of β-oxidation. E. coli has adopted a different strategy that relies on the hydrolysis of 3,5-cis-tetradecadienoyl-CoA (VI) to yield 3,5-tetradecadienoic acid (X), which is released from the cell [6]. This minor pathway of oleate β-oxidation is referred to as the thioesterase-dependent pathway (see Scheme 1).
Scheme 1.
Pathways of oleoyl-CoA β-oxidation. Emphasized are the reactions that are catalyzed by auxiliary enzymes needed for the removal of the preexisting double bond. Abbreviation: AD, acyl-CoA dehydrogenase.
This study was performed to determine which of the minor pathways is operative in Saccharomyces cerevisiae (S. cerevisiae). Does this unicellular eukaryotic organism use the simple strategy of E. coli or does it rely on the more elaborate reductase-dependent pathway of higher eukaryotes, which facilitates the complete degradation of oleic acid? It has been reported that dienoyl-CoA isomerase is present in S. cerevisiae and is encoded by the YOR180c gene that also is named DCI1 [7]. However the same authors reported that this enzyme is not required for growth of S. cerevisiae on oleic acid [7].
2. Materials and methods
2.1. Materials
CoASH, NADH, acetyl-CoA, buturyl-CoA, hexanoyl-CoA, decanoyl-CoA, dodecanoyl-CoA, tetradecanoyl-CoA, palmitoyl-CoA, and oleoyl-CoA were purchased from Life Science Resources (Milwaukee, WI). Sigma was the source of acyl-CoA oxidase from Arthrobacter species, L-tryptophan, L-leucine, L-histidine, uracil, adenine, D-maltose, D-sorbitol, yeast nitrogen base without amino acids and most standard biochemicals. Zymolyase-100T from Arthrobacter luteus was obtained from MP Biomedicals, LLC and oleic acid from Fisher Scientific. Sep-Pak C18 cartridges used for concentrating acyl-CoAs and µBondapak C18 columns (30 cm × 3.9 mm) for high-performance liquid chromatography (HPLC) were purchased from Waters Associates. Dye reagent for protein assays, polyacrylamide ready gels, and the materials for immunobloting including the goat anti rabit IgG conjugated with alkaline phosphatase were bought from Bio-Rad Laboratories. Antibodies against maltose binding protein (MBP) and all materials for subcloning, including restriction enzymes and amylose agarose, were purchased from New England Biolabs. Rabbit antiserum against yeast Ehd3 protein was raised by Pocono Rabbit Farm and Laboratory (Canadensis, PA). Oligonucleotides were synthesized by Integrated DNA Technologies, Inc.
2.2. Synthesis of substrates
3,5-Octadienoyl-CoA, 3,5-dodecadienoyl-CoA, and 3,5-tetradecadienoyl-CoA were prepared from the corresponding 5-enoyl-CoAs by dehydrogenation with acyl-CoA oxidase as described [6,8]. 2,5-Tetradecadienoyl-CoA [2], 2-octenoyl-CoA [9], 2-tetradecenoyl-CoA [9], 3-trans-octenoyl-CoA [3], 3-trans-tetradecenoyl-CoA [3], and 3-ketooctanoyl-CoA [10] were synthesized by published procedures. All acyl-CoA preparations were purified by HPLC, and their concentrations were determined spectrophotometrically by measuring CoASH with Ellman’s reagent [11] after quantitatively cleaving the thioester bond with NH2OH at pH 7.0 [9].
2.3. S. cerevisiae strains and growth conditions
Strains of S. cerevisiae used in this study are listed in Table 1 (see references therein). Mutants PTE1Δ and PTE1Δ/YOR180cΔ were created by using a system that allows repeated URA3 selections in the construction of multiple disrupted genes [14]. For this purpose the 3.8-kb BamHI/BglII fragment of pNKY274 (14) was blunt-ended and inserted into the PTE1 gene. A linear NotI/BamHI fragment was then excised and used for transformation into W3031A and YOR180cΔ cells. Selection of URA-auxotrophs was performed using 5-fluoroorotic acid (5-FOA) as described [14]. Selected clones were screened for correct excision of the URA3 gene from the pte1::hisG-URA3-hisG fragment by PCR analysis of total DNA extracted from the URA-auxotrophs.
Table 1.
S. cerevisiae strains used in this study
| Strain | Genotype | Ref. |
|---|---|---|
| W3031A | MATa leu2 ura3 trp1 ade2 his3 | 12 |
| YOR180cΔ | MATa leu2 ura3 trp1 ade2 his3 dci1::HIS3 | 13 |
| PTE1Δ | MATa leu2 ura3 tes1::his G trp1 ade2 his3 | This study |
| YOR180cΔ/PTE1Δ | MATa leu2 ura3 tes1::his G trp1 ade2 his3 dci1::HIS3 | This study |
| PTE1Δ PRS306 | MATa leu2 ura3[pRS306-URA3] tes1::his G trp1 ade2 his3 | This study |
| PTE1Δ PRS316 | MATa leu2 ura3[pRS316-URA3] tes1::his G trp1 ade2 his3 | This study |
| PTE1Δ PRS306hDCI1POX1 | MATa leu2 ura3[pRS306-POX1-hDCI-URA3] tes1::his G trp1 ade2 his3 | This study |
| PTE1Δ PRS316hDCI1POX1 | MATa leu2 ura3[pRS316-POX1-hDCI-URA3] tes1::his G trp1 ade2 his3 | This study |
A human fibroblast cDNA library (kindly provided by Dr. A. Chen, Mount Sinai School of Medicine, New York, NY) was used to amplify the open reading frame (ORF) of human dienoyl-CoA isomerase (hDCI) by polymerase chain reaction (PCR). The hDCI ORF was then fused with the POX1 promoter using PCR and the resulting cassette was subcloned into yeast integrative (pRS306) and centromeric (pRS316) vectors to obtain pRS306hDCIPOX1 and pRS316DCIPOX1, respectively. These constructs were used to transform PTE1 deletion strain creating PTE1ΔpRS306hDCIPOX1 and PTE1ΔpRS316hDCIPOX1 strains. Wild-type and PTE1Δ strains transformed with pRS306 or pRS316 were created to serve as controls.
S. cerevisiae cells were grown overnight at 30 °C with agitation in 10 ml of medium containing 1% yeast extract, 2% peptone, 2% dextrose (YPD). The next day, cells were inoculated in fresh YPD solution at 1:10 dilution and allowed to grow at 30 °C with agitation for 6 hours. Cells were isolated by centrifugation at 2000 × g for 10 min at 4 °C (general procedure for isolating S. cerevisiae cells by centrifugation) and re-suspended in 1 liter of 1% yeast extract, 2% peptone, 3% glycerol, 0.2% Tween 40, 0.125% oleic acid (YPGO) or minimum medium containing 0.2% yeast extract, 0.67% yeast nitrogen base without amino acids, 0.2% oleic acid, 0.05% Tween 40, and auxotrophic supplements added to 20 µg/ml (40 µg/ml in the case of leucine). All growth media also contained ampicillin (100 µg/ml). Cells were allowed to grow for 18 hours in YPGO or 96 hours in minimum medium at 30 °C with vigorous aeration after which time they were harvested by centrifugation. Cell pellets were washed twice with YPGO or minimum medium and stored at –80 °C.
2.4. Growth curves
The growth of wild-type S. cerevisiae was compared with the growths of mutant cells in liquid cultures as decribed [15]. Briefly, each strain was inoculated in 5 ml of YPD followed by overnight growth at 30 °C with agitation. Cells were harvested by centrifugation and re-suspended in minimum medium containing 0.2% yeast extract, 0.67% yeast nitrogen base without amino acids, 0.1% Tween 40, and 0.125% oleic acid or 0.125% palmitic acid, supplemented with amino acids and nucleotides as required for each yeast strain. Ampicillin (10 µg;/ml) was added every 24 hours. Cells were allowed to grow overnight, then were isolated by centrifugation, washed with water, and re-suspended in 50 ml of the same medium at an initial absorbance of 0.001 at 600 nm. Growth was assessed by removing 1ml aliquots, followed by centrifugation, washing cells with water, and re-suspending them in water before measuring the absorbance at 600 nm. Aliquots were taken at 24 hours intervals for 120–144 hours. Absorbancies measured at time 0 were subtracted from the absorbancies measured thereafter. A light microscope was used to check the cultures for contaminating organisms such as bacteria.
2.5. Molecular cloning, expression, and purification of proteins from S. cerevisiae
The preparations of the plasmids pMBP-YOR180c [16] and pMBP-PTE1 [15] have been described. In order to express enoyl-CoA isomerase and the Ehd3 protein from S. cerevisiae in E. coli in the form of MBP-fusion proteins, the ORFs of both ECI1 and EHD3, respectively, were amplified by PCR using S. cerevisiae genomic DNA as a template. The complete ECI1 ORF was amplified by using the following primers 5’-AAAGTCGACAATGTCGCAAGAAATTAGGCAAAATGAG-3’ and 5’-TTTGCGGCCGCTCATAAACGATGCTTCCTTTGTTTCGAG-3’. The complete EHD3 ORF was amplified by using the following primers 5’-AAAGTCGACAATGCTCAGAAATACGCTAAAATGT-3’ and 5’-TTTGCGGCCGCTTATTTCCATCTTAAGCCATCGTTAAC-3’. The PCR products from each reaction were digested with SalI and NotI (SalI and NotI cleavage sites in the primers are underlined) and inserted into pMBP-PTE1 restricted with SalI and NotI. The resulting plasmids, named pMBP-ECI and pMBP-EHD3, were used to transform E. coli cells. Cells bearing either pMBP-YOR180c, pMBP-PTE1, pMBP-ECI, or pMBP-EHD3 were grown overnight at 37 °C in 50 ml of LB medium containing ampicillin (100 µg/ml). Forty ml of these cultures were diluted into 1 l of 2YT medium supplemented with 0.2% glucose and ampicillin (100 µg/ml). These cultures were grown at 37 °C until the A600 reached 0.4, at which time isopropyl-thio-β-D-galactopyranoside was added to a final concentration of 0.3 mM to induce protein expression. The induced cultures were incubated at 37 °C for 4 hours, 30 °C for 16 hours or 18 °C for 20 hours. After these periods, cells were harvested by centrifugation at 5000 × g for 10 min at 4 °C. Cell pellets were washed with 2YT medium and stored at –80 °C.
Six g of E.coli cell paste were suspended in 12 ml of amylose column binding buffer (20 mM Tris-HCl (pH 7.5) 200 mM NaCl, 1 mM EDTA, and 10 mM 2-mercaptoethanol). Cell suspensions were sonicated 6-times for 20 s each at intervals of 3 min to keep the temperature of the suspensions at 4 °C. The soluble extracts were separated from debris by centrifugation at 25,000 × g for 1 hour. MBP-Yor180c protein, MBP-Pte1p, MBP-Eci1p, and MBP-Ehd3 protein were purified by one-step affinity chromatography. The soluble extracts containing fusion proteins were diluted to 50 ml with amylose column binding buffer and loaded onto a 20 ml amylose agarose column at a rate of 1ml/min. The column was washed with 10 column volumes of binding buffer, and bound proteins were eluted with 1 column volume of elution buffer (column binding buffer containing 10 mM maltose). Following sodium dodecyl sulfate polyacrylamide gel analysis, homogenates of non-induced and induced cells, soluble extracts and insoluble fractions of cell homogenates, and column eluates were subjected to immunoblotting using an antibody to MBP to detect and determine the levels of MBP-fusion proteins. Purified proteins were concentrated and stored at –80 °C.
2.6. Enzyme and protein assays
Thioesterase was assayed spectrophotometrically at 412 nm. The assay mixture contained 0.1 M KPi (pH 8.0), 10 mM 5,5’-dithiobis(2-nitrobenzoic acid), 30 µM acyl-CoA, and 0.67 µg of purified MBP-Pte1p or soluble extracts to give equivalent absorbance changes. An extinction coefficient of 13,600 M−1 cm−1 was used to calculate rates. Dienoyl-CoA isomerase was assayed spectrophotometrically by measuring the increase in absorbance at 300 nm as described [4]. An extinction coefficient of 28,000 M−1 cm−1 was used to calculate rates. Enoyl-CoA hydratase was assayed by following the decrease in absorbance at 263 nm due to the hydration of the double bond of 2-trans-octenoyl-CoA [9]. L-3-Hydroxyacyl-CoA dehydrogenase was assayed by measuring the decrease in absorbance at 340 nm due to the dehydrogenation of NADH in the presence of 3-ketooctanoyl-CoA and enzyme [17]. Enoyl-CoA isomerase was measured with 3-trans-octenoyl-CoA as substrate by a coupled assay that measures the increase in absorbances at 340 nm due to the formation of NADH [17]. 3-Ketoacyl-CoA thiolase was assayed by measuring the decrease in absorbance at 303 nm due to the disappearance of the Mg2+-enolate complex of 3ketooctanoyl-CoA [18]. Extinction coefficients of 6700, 6220 and 6700 M−1 cm−1 were used to calculate the activities of the hydratase, dehydrogenase, and thiolase, respectively. One unit of enzyme activity is defined as the amount of enzyme that converts 1 µmol of substrate to product in 1 min.
2.7. Isolation of fatty acids from growth media
Wild-type and mutant S. cerevisiae cells were grown in minimum medium (for detailed conditions see under S. cerevisiae strains and growth conditions) that contained either oleic acid or palmitic acid as carbon source and ampicillin (100 µg/ml). After cells were removed by centrifugation, 500 ml of acidified growth medium were extracted four times with 100 ml of ether each. The organic phase was extracted with aqueous sodium bicarbonate, which after acidification with 2N H2SO4 was extracted four times with 8 ml of ether each. The combined ether extracts were dried over anhydrous sodium sulfate. After removal of the drying agent by filtration and ether by evaporation, the residual material was methylated by reacting it with 2 ml of BCl3-methanol (12%, w/w) for 10 minutes at 60 °C. After cooling the reaction mixture on ice, 1 ml of H2O and 1 ml of hexane were added. The organic layer was removed and dried over anhydrous sodium sulfate. After removal of sodium sulfate by filtration and ether by evaporation, the residual material containing methyl esters of acidic compounds present in the growth medium was dissolved in 50 µl of ethanol. Aliquots of 1 µl of the fatty acid methyl esters were injected at 250°C into a gas chromatograph (Shimadzu Scientific Instrument, model GC-17A) interphased with a mass spectrometer (QP-5000) and equipped with a capillary column (30m; inner diameter 0.25 mm; film thickness, 0.25 µm; EC-5; Alltech Associates Inc., Deerfield, IL). The oven temperature was raised from 100 to 230°C at 5°C/min, to 300°C at 20°C/min, and then held constant for 6 min. The mass spectrophotometer served as a detector and was operated at 280°C.
3. Results
3.1. The minor pathway of oleate β-oxidation in S. cerevisiae
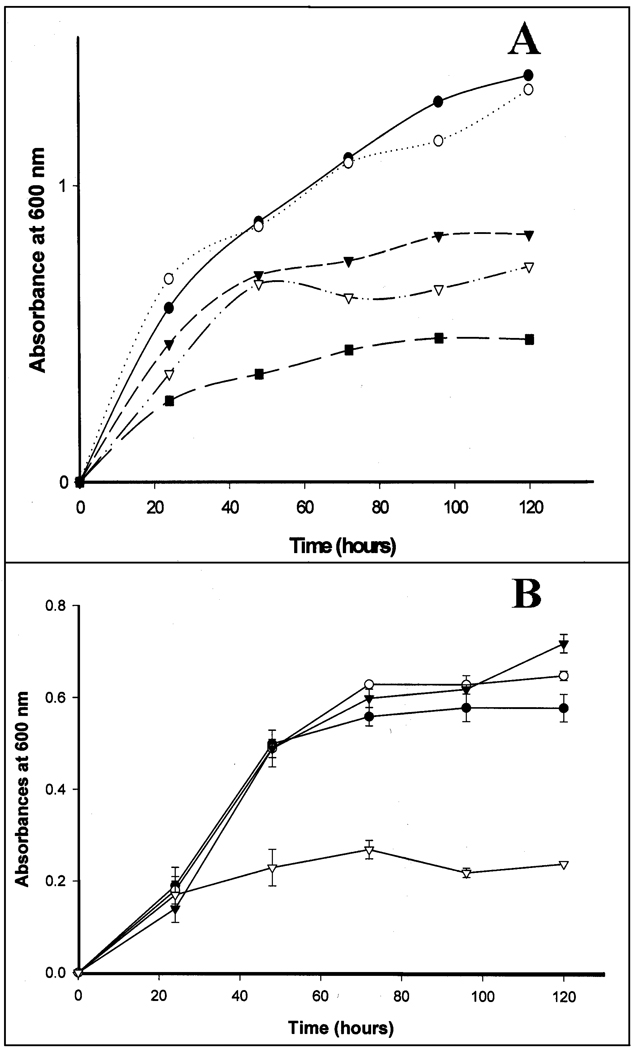
3,5-Tetradecadienoyl-CoA (compound VI in Scheme 1), which is formed from 2,5-tetradecadienoyl-CoA by enoyl-CoA isomerase during the β-oxidation of oleic acid, is either degraded by the reductase-dependent pathway or disposed of via the thioesterase-dependent pathway (see Scheme 1). It remains unknown which of these two pathways is operative in yeast. In an attempt to obtain information about the minor pathway of β-oxidation in S. cerevisiae, the growth of wild-type cells on oleic acid or palmitic acid was compared with the growths of S. cerevisiae mutants that either had YOR180c or PTE1, or both genes deleted. The former gene has been reported to encode dienoyl-CoA isomerase [7] while the PTE1 gene encodes peroxisomal acyl-CoA thioesterase 1 (Pte1p) [15]. As is apparent from the data shown in Fig. 1, growth of S. cerevisiae on either oleic acid or palmitic acid was not affected by the deletion of the YOR180c gene. The absence of a growth defect indicates that either the reductase-dependent pathway does not operate in this organism or that the YOR180c gene does not encode dienoyl-CoA isomerase. In contrast, deletion of the PTE1 gene impaired the growth on oleic acid but not on palmitic acid (see Fig. 1A&B). This observation is in agreement with the reported growth defect of a PTE1 mutant when grown on oleic acid [15]. It seems that Pte1p is important for the β-oxidation of oleic acid in S. cerevisiae but not for the degradation of palmitic acid.
Fig. 1.
Growth of wild-type and mutant cells of S. cerevisiae on oleic acid or palmitic acid. A, growth on oleic acid. (●) Wild-type cells; (○) YOR180c mutant; (▼) PTE1 mutant; (∇) YOR180c1/PTE1 double mutant; (■) wild-type cells in the absence of oleic acid. B, growth on palmitic acid. (●) Wild-type cells; (○) YOR180c mutant; (▼) PTE1 mutant; (∇) wild-type cells in the absence of palmitic acid.
3.2. Peroxisomal thioesterase 1 functions in oleate β-oxidation in S. cerevisiae
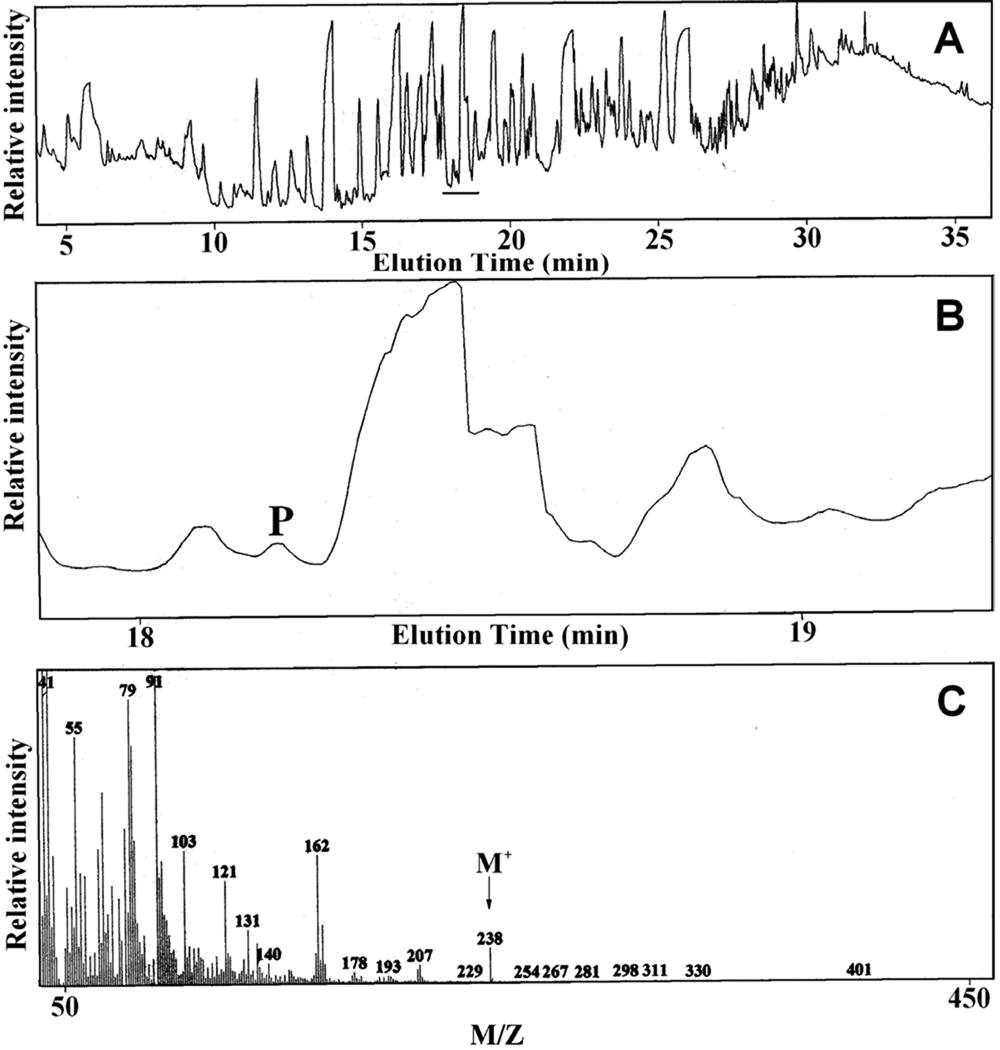
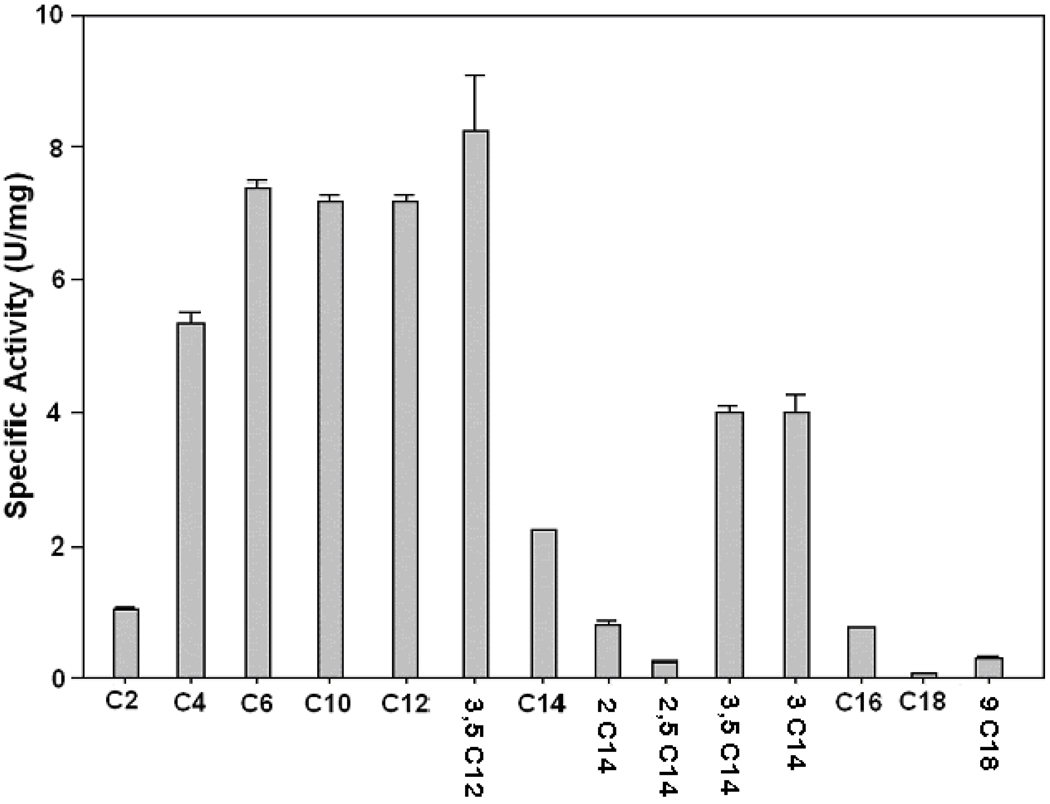
Because the deletion of the PTE1 gene impaired growth of S. cerevisiae on oleate but not on palmitate, Pte1p might be involved in the β-oxidation of unsaturated fatty acids like oleic acid but not in that of saturated fatty acids. Specifically, Pte1p may hydrolyze 3,5-cis-tetradecadienoyl-CoA that is formed during the β-oxidation of oleic acid (see Scheme 1). To test this hypothesis, the growth medium was analyzed to determine if 3,5-tetradecadienoic acid (compound X in Scheme 1) had been formed during growth of S. cerevisiae on oleic acid. For this analysis, the acidic components of the growth medium were extracted with ether, converted to methyl esters, and analyzed by gas chromatography/mass spectrometry. The gas chromatogram of this fraction shown in Fig. 2A is indicative of a large number of acidic compounds in the growth medium. A search of the region between 18 min and 19 min, where methyl 3,5-tetradecadienoate would be eluted, revealed the presence of a compound (marked P in Fig. 2, Panel B) that yielded a molecular ion with a mass/charge ratio (M/Z) of 238 (Fig. 2, Panel C) as expected of methyl 3,5-tetradecadienoate. Even though fraction P contained some impurities, its mass spectrum (Fig. 2, Panel C) was similar to the spectrum of synthetic methyl 3,5-tetradecadienoate previously published [6]. Most importantly, the molecular ion of 3,5-tetradecadienoate with a mass/charge ratio of 238 was not detected when the medium of wild-type S. cereviciae grown on palmitate or that of the PTE1 mutant grown on oleate was analyzed by gas chromatography/mass spectrometry. Hence, 3,5-tetradecadienoate is formed in S. cerevisiae by Pte1p during oleate β-oxidation. This conclusion prompted us to check if 3,5-tetradecadienoyl-CoA is a substrate of Pte1p. In agreement with a previous report [19], the substrate profile (see Fig. 3) proves Pte1p to be most active with short-chain and medium-chain acyl-CoAs. Most important for this study was the demonstration that 3,5-tetradecadienoyl-CoA is a good substrate of Pte1p, which hydrolyzes it almost twice as fast as myristoyl-CoA, its saturated analog. A comparison of activities observed with different substrates having the same acyl chain length leads to the conclusion that 3-enoyl-CoAs are better substrates than the corresponding saturated acyl-CoAs, whereas 2-enoyl-CoAs are hydrolyzed more slowly than their saturated analogs.
Fig. 2.
Identification of 3,5-tetradecadienoic acid in the medium of S. cerevisiae grown on oleate as the sole carbon source. A, gas chromatogram of the methyl esters of the acidic fraction extracted from the growth medium. B, region of the gas chromatogram where methyl 3,5-tetradecadienoate would be eluted. C, mass spectrum of the material labeled P in Panel B.
Fig. 3.
Substrate specificity of purified yeast peroxisomal thioesterase (Pte1p) expressed in E. coli. Acyl-CoA thioesterase activities were determined under standard conditions (see Experimental Procedures) with purified Pte1p fused to maltose binding protein (MBP) and the following substrates: C2, acetyl-CoA; C4, butyryl-CoA; C6, hexanoyl-CoA; C10, decanoyl-CoA; C12, dodecanoyl-CoA; 3,5C12, 3,5-cis-dodecadienoyl-CoA; C14, tetradecanoyl-CoA (myristoyl-CoA); 2C14, 2-trans-tetradecenoyl-CoA; 2,5C14, 2-trans-5-cis-tetradecadienoyl-CoA; 3,5C14, 3,5-cis-tetradecadienoyl-CoA; 3C14, 3-trans-tetradecenoyl-CoA; C16, hexadecanoyl-CoA (palmitoyl-CoA); C18, octadecanoyl-CoA (stearoyl-CoA); 9C18, 9-cis-octadecenoyl-CoA (oleoyl-CoA).
3.3. Dienoyl-CoA isomerase is present in S. cerevisiae but is not encoded by the YOR180c gene
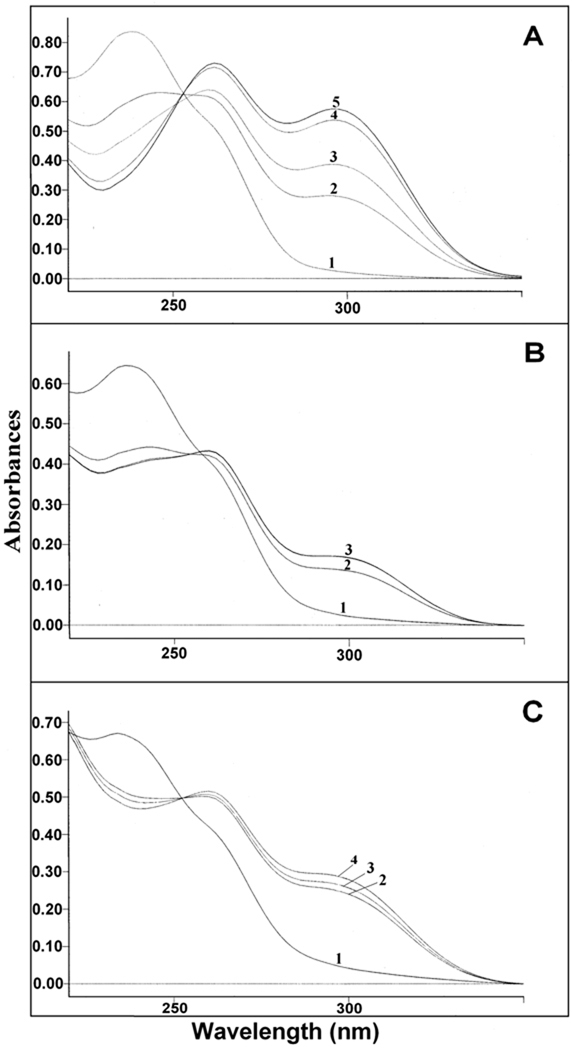
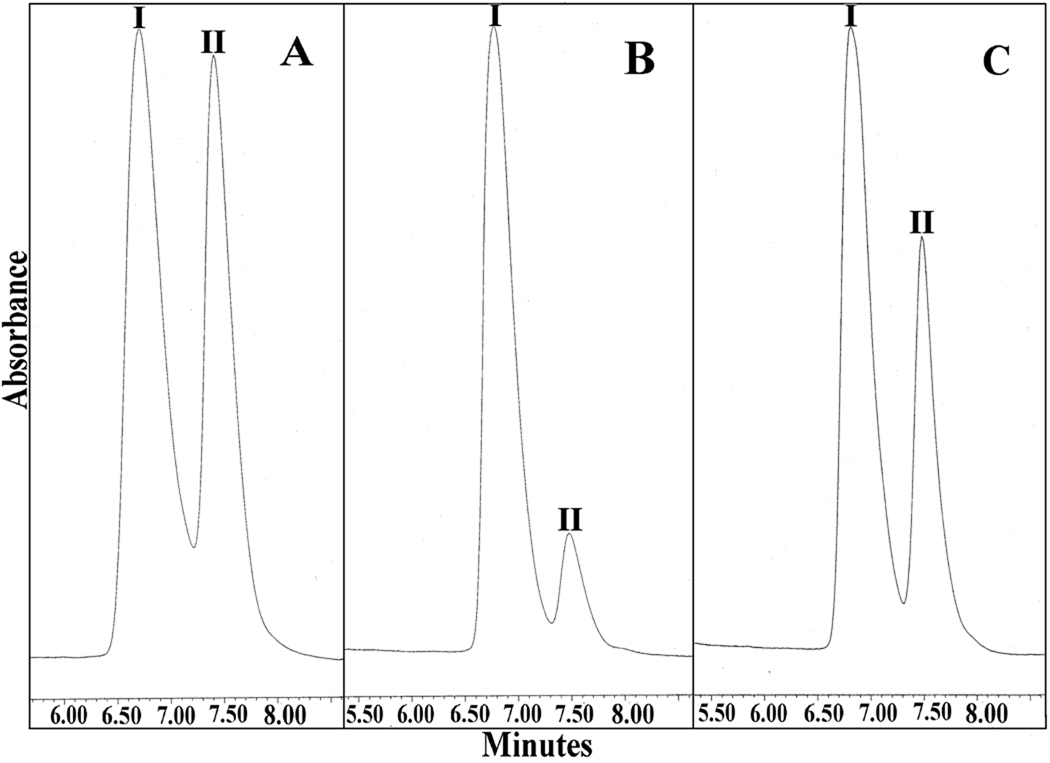
It has been reported that YOR180c, also referred to as DCI1, encodes dienoyl-CoA isomerase, but that this enzyme is not required for growth of S. cerevisiae on oleic acid [7]. In agreement with this claim is our observation that the deletion of YOR180c does not affect growth of S. cerevisiae on oleic acid (see Fig. 1A). This result prompted us to question whether dienoyl-CoA isomerase is present in S. cerevisiae and, if it is present, whether it is encoded by the YOR180c gene? When a soluble extract of wild-type S. cerevisiae cells was assayed for dienoyl-CoA isomerase, a low activity of 9.2 milliunits/mg of protein was detected (see Table 2). Most important, however, was the observation that the deletion of YOR180c did not abolish the dienoyl-CoA isomerase activity, but rather caused it to increase fourfold. Moreover, the purified Yor180c protein fused to MBP did not exhibit any dienoyl-CoA isomerase activity. This data does not agree with the claim that YOR180c encodes dienoyl-CoA isomerase. Yet a definite conclusion necessitated a more rigorous proof that the dienoyl-CoA activity detected by measuring the change in absorbance at 300 nm was due to the isomerization of 3,5-octadienoyl-CoA to 2,4-octadienoyl-CoA. For this, the isomerization reaction was studied by analyzing the spectral changes in the UV region. As shown in Fig. 4A, the isomerization of 3,5-octadienoyl-CoA to 2,4-octadienoyl-CoA catalyzed by purified rat dienoyl-CoA isomerase was associated with a decrease of the absorbance at 238 nm, an increase in absorbance at 263 nm, and the appearance on an absorbance at 300 nm. The same absorbance changes were observed with a soluble extract of wild-type S. cerevisiae (see Fig. 4B) except that they occurred at a slower rate and were smaller, most likely due to the partial hydrolysis of the substrate during the long incubation necessary for detecting the isomerization. The extract of YOR180c mutant cells yielded larger absorbance changes than the extract of wild-type cells in agreement with the several fold higher dienoyl-CoA isomerase activity observed in the mutant strain. 2,4-Octadienoyl-CoA, the product of the isomerization of 3,5-octadienoyl-CoA, was also identified by HPLC. For this analysis, the substrate and product of the reaction were separated by HPLC and identified by comparison with authentic compounds. The results shown in Fig 5A–C prove that extracts of both wild-type S. cerevisiae and strain YOR180c converted 3,5-octadienoyl-CoA to 2,4-octadienoyl-CoA, except that the latter extract yielded more product than the wild-type material when they were incubated for the same period of time and with the same amount of cellular extract. Taken together, the data prove that S. cerevisiae contains a low but definite activity of dienoyl-CoA isomerase that is not associated with the Yor180c protein.
Table 2.
Dienoyl-CoA isomerase activity in S. cerevisiae
| Sample | Specific activity |
|---|---|
| (milliunits/mg) | |
| Soluble extract of wild-type S. cerevisiae | 9.2 ± 0.2 |
| Soluble extract of S. cerevisiae strain YOR180cΔ | 39.1 ± 0.2 |
| Purified MBP-Yor180c protein | 0* |
The limit of detecting
dienoyl-CoA isomerase activity with 3,5-cis-octadienoyl-CoA as substrate was 0.46 milliunits/mg of protein.
Fig. 4.
Spectrophotometric analysis of the isomerization of 3,5-octadienoyl-CoA to 2,4-octadienoyl-CoA catalyzed by: A, purified rat dienoyl-CoA isomerase; spectrum 1 was recorded at time 0; spectra 2–5 were recorded 1 min, 2 min, 4 min, 5 min and 10 min after the addition of 20 ng of enzyme. B, extract of wild-type S. cerevisiae; spectrum 1 was recorded at time 0, spectrum 2 was recorded 5 min and spectrum 3 10 min and 20 min after the addition of 27 µg of the soluble extract. C, extract of strain YOR180c; spectrum 1 was recorded at time 0, spectra 2–4 were recorded 5 min, 10 and 20 min after the addition of 27 µg of the soluble extract.
Fig. 5.
HPLC analysis of the product formed from 3,5-octadienoyl-CoA by soluble extracts from S. cerevisiae. A, Authentic 3,5-cis-octadienoyl-CoA (I) and 2-trans-4-trans-octadienoyl-CoA (II); B, conversion of 3,5-cis-octadienoyl-CoA (I) to 2-trans-4-trans-octadienoyl-CoA (II) by 27 µg of an extract from wild-type cells; C, conversion of 3,5-cis-octadienoyl-CoA (I) to 2-trans-4-trans-octadienoyl-CoA (II) by 27 µg of a soluble extract from strain YOR180c.
3.4. Does dienoyl-CoA isomerase function in oleate β-oxidation and what is the role of YOR180c?
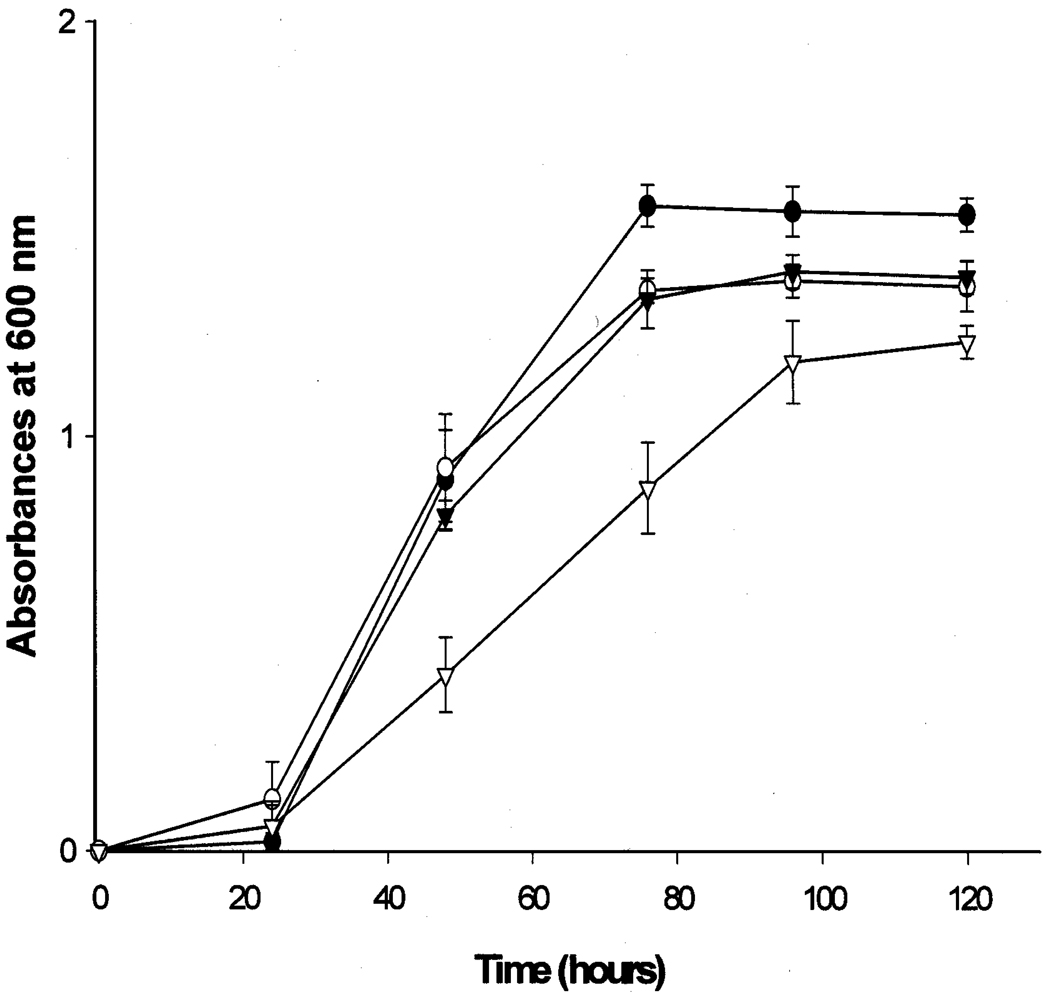
Because YOR180c does not encode dienoyl-CoA isomerase, homologous proteins were tested for dienoyl-CoA isomerase activity. Specifically, enoyl-CoA isomerase and the EHD3 gene product, a protein of unknown function, were expressed in E. coli as fusion proteins with maltose binding protein and after purification were assayed for enoyl-CoA hydratase, enoyl-CoA isomerase, and dienoyl-CoA isomerase. The former enzyme exhibited the expected enoyl-CoA isomerase activity of 21.5 units per mg of protein, but was devoid of enoyl-CoA hydratase or dienoyl-CoA isomerase activity. The Ehd3 protein did not catalyze any of the three reactions. Without a good clue about the identity of dienoyl-CoA isomerase in S. cerevisiae we did not continue attempts to identify this enzyme. However, we asked if dienoyl-CoA isomerase could function in oleate β-oxidation and perhaps could substitute for Pte1p and reverse the growth impairment observed with the PTE1 mutant (see Fig. 1A). As shown in Fig. 6, the PTE1 mutant grew almost as well as wild-type cell when transformed with a plasmid that carried cDNA encoding human dienoyl-CoA isomerase. This result suggests that both Pte1p and dienoyl-CoA isomerase can function in the β-oxidation of oleic acid, presumably by facilitating the degradation or disposal of 3,5-tetradecadienoyl-CoA.
Fig. 6.
Growth of yeast transformants on oleic acid. (●) Wild-type cells transformed with vector pRS316; (○) PTE1 mutant cells transformed with plasmid pRS306hDCIPOX1; (▼) PTE1 mutant cells transformed with plasmid pRS316hDCIPOX1; (∇) PTE1 mutant cells transformed with vector pRS306.
4. Discussion
The major aim of this study was to determine if 3,5-tetradecadienoyl-CoA, which is formed during the β-oxidation of oleic acid, is degraded via the reductase-dependent pathway or is disposed of by the thioesterase-dependent pathway. Evidence in support of the existence or operation of either pathway in S. cerevisiae had previously been obtained. The presence of the YOR180c (DCI1) gene claimed to encode dienoyl-CoA isomerase [7] agrees with the existence of the reductase-dependent pathway while an impairment of oleate β-oxidation upon deletion of the PTE1 gene [15] points to the operation of the thioesterase-dependent pathway. The involvement of the thioesterase-dependent pathway in oleate β-oxidation was proven in this study by the identification of 3,5-tetradecadienoic acid in the medium of S. cerevisiae growing on oleic acid. Additionally, the substrate specificity of Pte1p proved it to be effective in hydrolyzing 3,5-tetradecadienoyl-CoA. Hence the thioesterase-dependent pathway is operative in S. cerevisiae. However the question remained whether the reductase-dependent pathway also operates in this organism? The answer is less certain because the pathway does not produce a unique metabolite that accumulates and thereby would prove the operation of the pathway. The key enzyme of the reductase-dependent pathway is dienoyl-CoA isomerase. Although this activity is present at a very low level in a soluble extract from S. cerevisiae grown on oleate, the protein responsible for this activity has not been identified. However, this study disproves the claim that dienoyl-CoA isomerase is encoded by the YOR180c (DCI1) gene. Altogether it remains uncertain if the low dienoyl-CoA isomerase activity detected in S. cerevisiae is involved in oleate β-oxidation. This doubt increased further when it was demonstrated that the deletion of the YOR180c gene resulted in a more than fourfold increase of the wild-type dienoyl-CoA isomerase activity without stimulating the growth of S. cerevisiae on oleic acid. Moreover, 9-cis,11-trans-octadecadienoic acid (conjugated linoleic acid), which requires dienoyl-CoA isomerase for its complete β-oxidation, does not support the growth of S cerevisiae [20], possibly because the dienoyl-CoA isomerase activity of this organism is insufficient to generate the necessary energy and carbon precursors. This observation is not indicative of the operation of a reductase-dependent pathway in S cerevisiae. However, the reductase–dependent pathway might operate in peroxisomes of S cerevisiae if the activity level of dienoyl-CoA isomerase would be higher. This idea is supported by the improved growth of the PTE1 mutant after transformation with an expression plasmid for human dienoyl-CoA isomerase. Altogether, the data obtained during this investigation support the conclusion that the disposal of 3,5-tetradecadienoyl-CoA formed during oleate β-oxidation occurs via the thioesterase-dependent pathway although a small contribution by the reductase-dependent pathway cannot be ruled out.
Another aim of this investigation was to determine whether the Yor180c protein is a dienoyl-CoA isomerase. This evaluation of the Yor180c protein was prompted by the claim that it is a dienoyl-CoA isomerase, which is not required for growth of S. cerevisiae on oleic acid [7] even though it would be needed for the complete degradation of 3,5-tetradecadienoyl-CoA formed during oleate β-oxidation. The evidence obtained in this study supports the conclusion that the Yor180c protein is not a dienoyl-CoA isomerase. Firstly, the deletion of the YOR180c gene did not eliminate the dienoyl-CoA isomerase activity and secondly, the purified fusion protein of Yor180c protein and maltose binding protein did not have any dienoyl-CoA isomerase activity. This result raised the question as to which protein might be responsible for the dienoyl-CoA isomerase activity in S. cerevisiae? It is conceivable that an enzyme that has a binding site for an acyl-CoA and that catalyzes a proton transfer at the α-carbon of the acyl chain might also exhibit low dienoyl-CoA isomerase activity. A candidate protein was enoyl-CoA isomerase, which, however, did not exhibit any dienoyl-CoA isomerase activity. When Ehd3 protein, a protein of unknown function but with some homology to enoyl-CoA isomerase, was found to be devoid of dienoyl-CoA isomerase activity, we abandoned our attempts to identify the protein responsible for the low dienoyl-CoA isomerase activity of S. cerevisiae.
Acknowledgement
This work was supported by U.S. Public Health Service Grant GM008168 from the National Institute of General Medical Sciences and by U.S. Public Health Service Grant RR03060 to Research Centers of Minority Institutions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Abreviations used are: Enoyl-CoA isomerase, Δ3,Δ2-enoyl-CoA isomerase; dienoyl-CoA isomerase, Δ3,5,Δ2,4-dienoyl-CoA isomerase; S. cerevisiae, Saccharomyces cerevisiae; HPLC, high-performance liquid chromatography; MBP, maltose binding protein; 5-FOA, 5-fluoroorotic acid; ORF, open reading frame; hDCI, human dienoyl-CoA isomerase; PCR, polymerase chain reaction; Pte1p, peroxisomal acyl-CoA thioesterase 1;
References
- 1.Kunau W-H, Dommes V, Schulz H. β-Oxidation of fatty acids in mitochondria, peroxisomes, and bacteria: A century of continued progress. Prog. Lipid Res. 1995;34:267–342. doi: 10.1016/0163-7827(95)00011-9. [DOI] [PubMed] [Google Scholar]
- 2.Ren Y, Schulz H. Metabolic functions of the two pathways of oleate β-oxidation. Double bond metabolism during the β-oxidation of oleic acid in rat heart mitochondria. J. Biol. Chem. 2003;278:111–116. doi: 10.1074/jbc.M209261200. [DOI] [PubMed] [Google Scholar]
- 3.Zhang D, Yu W, Geisbrecht BV, Gould SJ, Sprecher H, Schulz H. Functional characterization of Δ3,Δ2-enoyl-CoA isomerases from rat liver. J. Biol. Chem. 2002;277:9127–9132. doi: 10.1074/jbc.M112228200. [DOI] [PubMed] [Google Scholar]
- 4.Luo MJ, Smeland TE, Shoukry K, Schulz H. Δ3,5,Δ2,4-Dienoyl-CoA isomerase from rat liver mitochondria. J. Biol. Chem. 1994;269:2384–2388. [PubMed] [Google Scholar]
- 5.Zhang D, Liang X, He X-Y, Alipui OD, Yang S-Y, Schulz H. Δ3,5,Δ2,4-Dienoyl-CoA isomerase is a multifunctional isomerase. A structural and mechanistic study. J. Biol. Chem. 2001;276:13622–13627. doi: 10.1074/jbc.M011315200. [DOI] [PubMed] [Google Scholar]
- 6.Ren Y, Aguirre J, Ntamack AG, Chu CH, Schulz H. An alternative pathway of oleate β-oxidation in Escherichia coli involving the hydrolysis of a dead end intermediate by a thioesterase. J. Biol. Chem. 2004;279:11042–11050. doi: 10.1074/jbc.M310032200. [DOI] [PubMed] [Google Scholar]
- 7.Gurvitz A, Mursula AM, Yagi AI, Hartig A, Ruis H, Rottensteiner H, Hiltunen JK. Alternatives to the isomerase-dependent pathway for the β-oxidation of oleic acid are dispensable in Saccharomyces cerevisiae. J. Biol. Chem. 1999;274:24514–24521. doi: 10.1074/jbc.274.35.24514. [DOI] [PubMed] [Google Scholar]
- 8.Shoukry K, Schulz H. Significance of the reductase-dependent pathway for the β-oxidation of unsaturated fatty acids with odd-numbered double bonds. J. Biol. Chem. 1998;273:6892–6899. doi: 10.1074/jbc.273.12.6892. [DOI] [PubMed] [Google Scholar]
- 9.Fong JC, Schulz H. Short-chain and long-chain enoyl-CoA hydratases from pig heart muscle. Methods Enzymol. 1981;71:390–398. doi: 10.1016/0076-6879(81)71049-8. [DOI] [PubMed] [Google Scholar]
- 10.Thorpe C. A method for the preparation of 3-ketoacyl-CoA derivatives. Anal. Biochem. 1986;155:391–394. doi: 10.1016/0003-2697(86)90452-5. [DOI] [PubMed] [Google Scholar]
- 11.Ellman GL. Tissue sulfhydryl groups. Arch. Biochem. Biophys. 1959;82:70–77. doi: 10.1016/0003-9861(59)90090-6. [DOI] [PubMed] [Google Scholar]
- 12.Thomas BJ, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- 13.Karpichev IV, Small GM. Evidence for a novel pathway for the targeting of a Saccharomyces cerevisiae peroxisomal protein belonging to the isomerase/hydratase family. J. Cell Sci. 2000;113:533–544. doi: 10.1242/jcs.113.3.533. [DOI] [PubMed] [Google Scholar]
- 14.Alani E, Cao L, Kleckner N. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiple disrupted yeast strains. Genetics. 1987;116:541–545. doi: 10.1534/genetics.112.541.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones JM, Nau K, Geraghty MT, Erdman R, Gould SJ. Identification of peroxisomal acyl-CoA thioesterases in yeast and humans. J. Biol. Chem. 1999;274:9216–9223. doi: 10.1074/jbc.274.14.9216. [DOI] [PubMed] [Google Scholar]
- 16.Geisbrecht BV, Schulz K, Nau K, Geraghty MT, Schulz H, Erdmann R, Gould SJ. Preliminary characterization of Yor180c: Identification of a novel peroxisomal protein of Saccharomyces cerevisiae involved in fatty acid metabolism. Biochem. Biophys. Res. Commun. 1999;260:28–34. doi: 10.1006/bbrc.1999.0860. [DOI] [PubMed] [Google Scholar]
- 17.Binstock JF, Schulz H. Fatty acid oxidation complex from Escherichia coli. Methods Enzymol. 1981;71:403–411. doi: 10.1016/0076-6879(81)71051-6. [DOI] [PubMed] [Google Scholar]
- 18.Schulz H, Staack H. 3-Ketoacyl-CoA thiolase with broad chain length specificity from pig heart muscle. Methods Enzymol. 1981;71:398–403. doi: 10.1016/0076-6879(81)71050-4. [DOI] [PubMed] [Google Scholar]
- 19.Maeda I, Delessert S, Hasegawa S, Seto Y, Zuber S, Poirier Y. The peroxisomal acyl-CoA thioesterase Pte1p from Saccharomyces cerevisiae is required for efficient degradation of short straight chain and branched chain fatty acids. J. Biol. Chem. 2006;281:11729–11735. doi: 10.1074/jbc.M511762200. [DOI] [PubMed] [Google Scholar]
- 20.Gurvitz A, Hamilton B, Ruis H, Hartig A, Hiltunen K. Degradation of conjugated linoleic acid isomers in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta. 2001;1533:81–85. doi: 10.1016/s1388-1981(01)00148-2. [DOI] [PubMed] [Google Scholar]