Abstract
WNK1 and WNK4 encode two members of the WNK serine-threonine kinase subfamily. Greater WNK1 expression associates with higher BP. A combination of promoters, enhancers, repressors, and insulators regulate WNK1 expression, but whether microRNAs also modulate WNK1 expression is unknown. Here, computational analysis revealed the presence of a target sequence for miR-192 and miR-215 at the same site in the 3′ untranslated region of the ubiquitous L- and the kidney-specific KS-WNK1. We functionally validated this target sequence by transient transfection and reporter assays. Although we observed expression of both miRs along the distal nephron, only miR-192 regulated endogenous WNK1 ex vivo. Furthermore, a potassium load, sodium depletion, and aldosterone infusion each significantly reduced miR-192 expression in the kidney. Taken together, these results suggest a miR-driven mechanism of gene regulation by aldosterone and a role for miR-192 in the regulation of sodium and potassium balance in the kidney.
Micro-RNAs (miRs) are small noncoding RNAs (21–22nt) known to regulate gene expression at the post-transcriptional level by inhibition of translation,1 degradation, or storage of mRNA.2 In mammals, most of the studies described a role for miRs in embryonic development or pathologic states, such as oncogenesis, cardiovascular, or infectious diseases.3 Only a few studies showed that miRs could also be involved in adult physiologic mechanisms. miR-155 plays a role in hematopoïesis and immunity,4 whereas miR-375 regulates insulin secretion.5
Some miRs are specifically expressed in one or some tissues,6 suggesting a specific role in these tissues. Quantitative RT-PCR6 and microarray7 studies both identified miR-192, miR-194, and miR-215 as specifically expressed in the kidney. Microarray analysis also identified miR-204 and miR-216 as being enriched in the kidney. Moreover, some miRs were preferentially expressed in the renal cortex or medulla.8 However, little is known about the role of miRs in normal renal physiology and regulation of electrolytes balance. miR-192 has been implicated in the development of diabetic nephropathy.9 The inactivation of Dicer in developing renal structures10 or in podocytes11,12 demonstrated a requirement for miRs in kidney development and in the maintenance of podocyte structure and functions.
WNK1 and WNK4, two members of the WNK (With No lysine [K]) serine-threonine kinase subfamily,13 are essential for the coordinated regulation of electrolytes transport in the kidney.14 Mutations in both genes lead to familial hyperkaliemic hypertension (FHHt), a rare mendelian form of human hypertension,15 also known as pseudohypoaldosteronism type 2 or Gordon syndrome. Mutations identified in the WNK1 gene are large deletions within the first intron, leading to overexpression of WNK1 in leukocytes.15 The characterization of transgenic mice with intron 1 deletion and reporter genes allowed us to show that this overexpression corresponds to an abnormal expression of the kidney-specific isoform of WNK1 (KS-WNK1) in extra-renal tissues.16 The overexpression of the long isoform (L-WNK1) in the distal convoluted tubule (DCT) also observed in this model may explain the renal phenotype observed in FHHt. The physiologic importance of the finely tuned regulation of WNK1 expression has also been demonstrated in WNK1 knockout mice. Heterozygous WNK1+/− mice with a 50% reduction in WNK1 expression present with decreased BP, whereas homozygous embryos died in utero.17,18
The regulation of WNK1 transcription is complex.16,19 Proximal promoters give rise to ubiquitously expressed L-WNK1 isoforms containing a complete kinase domain. A third promoter, rP, located upstream of exon 4a, controls the expression of the KS-WNK1 isoform, lacking the major part of the kinase domain and expressed specifically in the DCT and the connecting tubule (CNT). In addition, the 3′ end of the WNK1 gene contains two polyadenylation (polyA) sites, separated by 1.6 kb.16 Finally, WNK1 transcription is regulated by changes in dietary or hormonal conditions such as changes in sodium and potassium intake or aldosterone administration.18,20
Here, we further study the regulation of WNK1 expression. Computational analysis revealed that a target sequence for miR-192 and miR-215 is present in the 3′ untranslated region (3′UTR) of WNK1. We show that these miRs, expressed in the distal nephron, efficiently target WNK1 3′UTR in vitro, whereas only miR-192 is able to regulate WNK1 expression ex vivo. We also found that miR-192 expression in the kidney is regulated by sodium and potassium intake and aldosterone, in a way opposite to WNK1. These results suggest that miR-192 could be involved in the regulation of a physiologic process, that is, sodium and potassium balance.
Results
Identification of miR-192 and miR-215 Target Sites in WNK1 3′UTR
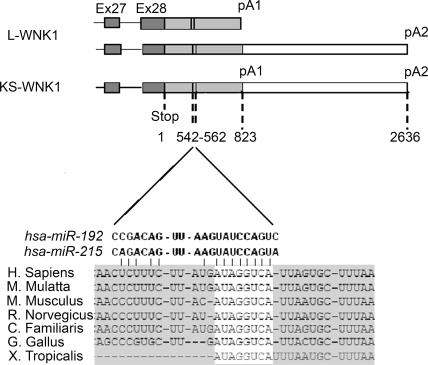
Two polyA sites are present in the 3′UTR of WNK1.16 Multitissue Northern blot analysis suggest that L-WNK1 isoforms contain both the proximal and distal polyA tails whereas KS-WNK1 contains only the distal one (reference 21; Figure 1). We therefore focused on the first part of WNK1 3′UTR, common to both isoforms. To explore systematically the computational involvement of miRs in targeting WNK1, we used a framework of tools and data integration (Henrion-Caude, personal communication). We used four prediction programs (miRanda, DianamicroT, Pictar, and TargetScan), based on the complementarity between the miR and its target and cross-species conservation of the target sequence. Of the 18 miRs target sites predicted by at least two algorithms (Supplemental Table S1), miR-192 and miR-215 appeared to be the best candidates because they are specifically expressed in the kidney.6 Both miRs share the same target sequence, located 542 bp downstream of the stop codon (Figure 1).
Figure 1.
miR-192 and miR-215 target sequences localize within the 3′UTR common to both WNK1 isoforms. miR-192 and miR-215 target sequences span from position 542 to 562 relative to the stop codon, in WNK1 3′UTR. The seed sequence of miR-192 and miR-215 completely matches with the sequence extending from 554 to 562 within the 3′UTR of WNK1 (sequence highlighted in white). This sequence is conserved among species. The target sequence is located upstream of WNK1 first polyadenylation signal (pA1), which is common to both isoforms as indicated in the schematic depiction of L-WNK1 and KS-WNK1.
Independently of any conservation, targeting of this sequence, referred to as TS (for target sequence), was assessed through pattern-based22 and support vector machine-based (SVM23) methods. If only miR-215-TS duplex was thus retrieved, central loop integration indicated that miR-192 was also able to target TS.24 Finally, SVM postfitting of the predictions25 revealed targeting of TS by miR-192 but not miR-215. This systematic computational exploration was a prompt method to further characterize this TS motif (Table 1).
Table 1.
Computational analysis identified miR-192 and miR-215 as good candidates to target WNK1 3′UTR
| Methods | hsa-miR-192 | hsa-miR-215 |
|---|---|---|
| Cross-species comparison | ||
| TargetScan 5.1 | + | + |
| Miranda | + | + |
| DianamicroT 3.0 | − | + |
| miRTarget2 | + | + |
| Pattern-based (RNA 22) | − | + |
| SVM classifier-based (miTarget) | − | + |
| Central loop integrating | + | + |
| miRNA:target interaction filter | True | False |
Cross-species comparison, pattern-based, SVM classifier–based, or central loop integrating tools were used to predict the presence (+) or absence (−) of miR-192 or miR-215 binding sites in WNK1 3′UTR. Postfitting of the predictions was done according to Yang et al.25
Expression of miR-192 and miR-215 in the Distal Nephron
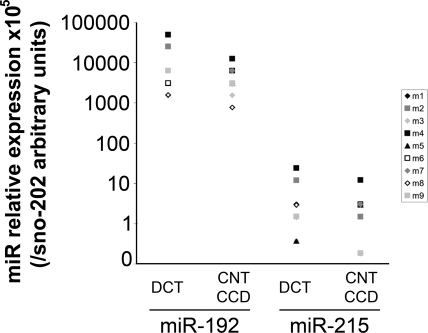
miR-192 and miR-215 were previously described as specifically expressed in the kidney.6,8 We quantified their expression by real-time PCR in DCT, CNT, and cortical collecting duct (CCD) microdissected from nine C57BL/6N adult males. miR-192 was expressed at a higher level than miR-215 but expression of each miR was detected at similar levels along the distal nephron, despite the high variability observed between individuals (Figure 2). This variability in miR expression was probably caused by the microdissection process as it was not observed in whole kidney extracts (Supplemental Figure S1).
Figure 2.
miR-192 and miR-215 are expressed in the distal nephron. Relative expression of miR-192 or miR-215 in the distal convoluted tubule, the connecting tubule, and the cortical convoluted tubule, dissected from nine C57BL/6N males. Each point represents the 2−ΔCT value obtained from one animal, with ΔCT = CtmiR − Ctsno202, and a different symbol is used for each mouse. sno202 is a tiny RNA used as a reference gene.
In Vitro Effect of miR-192 and miR-215 on WNK1 Expression
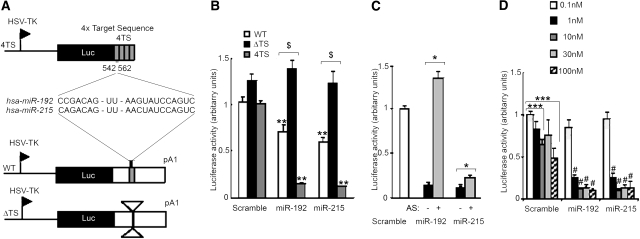
To explore the effect of miR-192 and miR-215 on the regulation of WNK1 expression, we generated three different constructs (Figure 3A). The 4TS plasmid contained four copies of a 63-bp fragment containing the TS downstream of the luciferase reporter gene. The wild-type plasmid carried the complete 3′UTR of WNK1 down to the first polyA and the ΔTS one carried the 3′UTR deleted from a 63-bp fragment containing the TS downstream of the luciferase gene. The constructs were transfected with 30 nM of either a scramble miR, miR-192, or miR-215 in Madin-Darby canine kidney (MDCK) cells. Addition of miR-192 or miR-215 led to a strong decrease in luciferase activity compared with the scramble miR when cotransfected with the 4TS reporter construct (86% for miR-192 and 89% for miR-215; Figure 3, B and C). Cotransfection of miR-192 or miR-215 with the WT construct led to a 29 or 40% significant decrease in luciferase activity, respectively. Luciferase activity was not reduced when miR-192 and miR-215 were cotransfected with the ΔTS construct (Figure 3B). Interestingly, cotransfection of the WT construct with miR-17*, an ubiquitous miR also predicted to target WNK1 3′UTR, did not inhibit luciferase activty (data not shown). To ensure that the decrease in luciferase activity was specifically due to miR-192 and miR-215, we cotransfected the 4TS construct with miR-192 or miR-215 alone or in combination with their antisense inhibitor (AS-192 or AS-215; Figure 3C). Addition of AS-192 completely reversed the effect of miR-192. The effect of miR-215 was only partially reversed by AS-215.
Figure 3.
miR-192 and miR-215 functionally interact with their target sequence within WNK1 3′UTR in vitro. (A) Schematic representation of the luciferase expression vectors. (B) Luciferase to β-gal chemiluminescence ratios measured when cotransfecting MDCK cells with the indicated luciferase reporter and 30 nM of either a scramble miR (Sc), miR-192, or miR-215. Cotransfection of the WT or 4TS plasmid with miR-192 or miR-215 induced a significant decrease in luciferase activity compared with the scramble (**P < 0.01). Inhibition of luciferase activity was lost in cells cotransfected with the ΔTS plasmid and any of the two miRs compared with cells cotransfected with the WT plasmid ($P < 0.0005). (C) Luciferase to ß-gal chemiluminescence ratios measured when cotransfecting MDCK cells with the 4TS reporter and 30 nM of either Sc, miR-192, or miR-215, alone or in combination with their inhibitor. The inhibition induced by the miRs was reversed by addition of their inhibitor (*P < 0.05). (D) Luciferase to ß-gal chemiluminescence ratios measured when cotransfecting MDCK cells with the 4TS reporter and increasing quantities (0.1, 1, 10, 30, or 100 nM) of either Sc, miR-192, or miR-215. Surprinsingly, luciferase activity was inhibited by cotransfection of 10 or 100 nM of Sc compared with 0.1 nM (***P < 0.005) but was significantly more suppressed by transfection of increasing concentrations of miR-192 and miR-215 (#P < 0.005 compared with the same concentration of Sc). Results are expressed as mean ± SEM, from three independent transfections realized in triplicate.
Finally, the dose-dependent effect of miR-192 and miR-215 on luciferase activity was tested. The 4TS construct was cotransfected with increasing concentrations (0.1, 1, 10, 30, and 100 nM) of either a scramble miR, miR-192, or miR-215. Whereas no significant effect was seen when 0.1 nM of miR-192 or miR-215 was used, a 74% decrease in luciferase activity was observed for a concentration as low as 1 nM and a maximal effect was obtained with 10 nM of miRs, with more than a 85% decrease in luciferase activity (Figure 3D). Surprisingly, we observed a significant 34 and 50% decrease in luciferase activity with 10 and 100 nM, respectively, of scramble miR, but the decrease obtained with miR-192 and miR-215 was significantly more pronounced at these concentrations (87 and 89% decrease for miR-192 and miR-215, respectively, at 10 nM and 89 and 86% at 100 nM). Taken together, these data suggest that miR-192 and miR-215 could regulate WNK1 expression in a dose-dependent manner through binding to their target sequence in the 3′UTR of the gene.
miR-192 Regulates L-WNK1 Protein Expression Ex Vivo
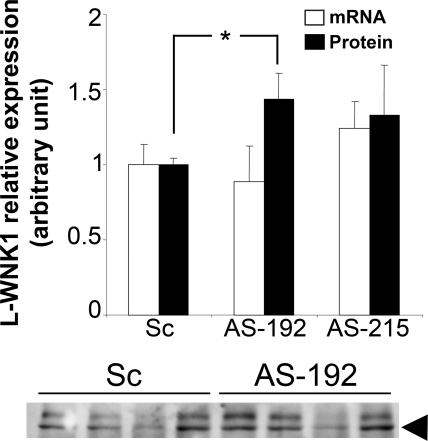
HEK293 cells, which express L-WNK1,16 miR-192, and miR-215 (data not shown), were transfected with either AS-192 or AS-215 (30 nM). Transfection of AS-192, but not AS-215, led to a significant 1.4-fold increase in L-WNK1 protein expression compared with the scramble (Figure 4A and Supplemental Figure S2) in the absence of any modification of L-WNK1 transcripts level (Figure 4B). Although modest, the degree of WNK1 stimulation by inhibition of miR-192 was comparable to what has been previously described in similar experiments.26,27 This effect was restricted to L-WNK1 as β-actin protein expression was not affected (data not shown). Endogenous miR-192 can therefore regulate L-WNK1 expression at the post-transcriptional level in HEK293 cells.
Figure 4.
miR-192 regulates WNK1 expression in HEK293 cells. 30 nM of scramble, AS-192, or AS-215 were transfected in HEK293 cells. (A) Total RNA or proteins were extracted 48 hours after transfection and the level of expression of L-WNK1 was quantified by quantitative RT-PCR or Western blot. Whereas L-WNK1 mRNA was not affected, L-WNK1 protein expression was increased by AS-192 transfection (*P < 0.05). Results are expressed as mean ± SEM from two independent transfections realized in triplicate for RNA expression and four independent transfections realized in duplicate for protein expression. (B) Representative immunoblot for L-WNK1: in HEK293 cells, a doublet is recognized by the L-WNK1 antibody. The lower band (arrowhead) corresponds to L-WNK1.
miR-192 Expression Is Regulated by Na+ or K+ Intake and Aldosterone
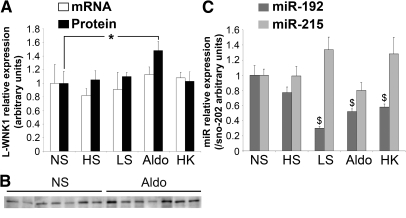
KS-WNK1 mRNA expression is stimulated by aldosterone,21,22 whereas L-WNK1 expression is unaffected. We asked if L-WNK1 expression could be regulated at the post-transcriptional level. mRNA expression of L-WNK1 and KS-WNK1 and protein expression of L-WNK1 were measured in the kidney of 16-week-old C57BL/6N mice fed with a normal, low-sodium, or high sodium diet, submitted to a potassium load or a chronic aldosterone infusion for 10 days (Supplemental Table S2). In agreement with a previous study,21 we confirmed that aldosterone and potassium load led respectively to a 1.6- and 1.5-fold increase in KS-WNK1 mRNA expression (Supplemental Figure S3) and that L-WNK1 mRNA expression is unaffected by the different challenges (Figure 5A). At the protein level, L-WNK1 was not modified by changes in Na+ or K+ intake but was significantly increased by aldosterone infusion (1.9 ± 0.17 versus 1 ± 0.12 in control animals; Figure 5B). This modest increase in L-WNK1 expression was comparable to the variation in WNK1 expression described in other studies.21,28 The expression of KS-WNK1 protein could not be evaluated in the absence of specific antibody. These results suggest that L-WNK1 expression is regulated only at the post-transcriptional level by aldosterone.
Figure 5.
Aldosterone in the mouse kidney regulates expression of miR-192 and L-WNK1. Adult mice were submitted to specific Na+ or K+ dietary treatments or chronic aldosterone infusion for 10 days (NS, normal salt; HS, high salt; LS, low salt; aldo, aldosterone; HK, high potassium). Protein, total, and small RNA were extracted from the kidneys. (A) Quantification by real time RT-PCR relative to 18S showed that L-WNK1 expression was not affected by the different challenges, whereas semiquantitative immunoblotting revealed that L-WNK1 protein expression was stimulated by aldo (*P < 0.05 versus NS). (B) Representative L-WNK1 immunoblot. Results are expressed as mean ± SEM from ten animals or nine for HS. (C) Quantification by Taqman assay relative to sno202 showed that miR-192 expression was inhibited by LS, aldo, and HK ($P < 0.0005 versus NS). Results are expressed as mean ± SEM from seven (HS), eight (aldo and NS), nine (HK), or ten (LS) animals.
To determine the potential role played by miR-192 in the regulation of L-WNK1 expression, we tested if their expression could be regulated by aldosterone and K+ and Na+ intake, in a way opposite to KS-WNK1 and L-WNK1. Whereas miR-215 expression was unaffected, miR-192 expression in the kidney was significantly reduced by potassium load (0.6 ± 0.04 versus 1 ± 0.12 in controls), sodium depletion (0.3 ± 0.02 versus 1 ± 0.12), and aldosterone infusion (0.5 ± 0.04 versus 1 ± 0.12) (Figure 5C). These results suggest that miR-192 could participate in the fine-tuning of WNK1 production under these conditions.
Discussion
Little is known on the role of miRs in the kidney, especially in normal renal physiology. Two studies implicated miRs in polycystic kidney disease29 and in the development of diabetic nephropathy.9 Deletion of Dicer in various compartments of the kidney demonstrated the requirement of miRs in the establishment of renal structure and podocyte function.10 We describe here that, in addition to an already complex transcriptional regulation, the expression of WNK1 is likely to be regulated at the post-transcriptional level by miR-192, a miR expressed only in the kidney and the gastrointestinal tract.6 We also show that the expression of miR-192 is regulated by sodium/potassium intake and aldosterone in the kidney, in a way opposite to WNK1 isoforms. Computational analysis of miR-192 potential targets in the kidney also showed that, in addition to WNK1, this micro-RNA could directly regulate the expression of ion transporters and channels, such as the chloride channel CLC5 and the β1-subunit of the Na+-K+ ATPase, a sodium pump crucial for the maintenance of the transmembrane gradient at the basolateral side of the renal distal tubule.30 Taken together, our results suggest that miRs could be involved in the physiologic regulation of ion transport in the kidney.
The regulation of miRs expression by androgens,31 estrogens,32 and LH/hCG33 was previously described. We show for the first time in vivo that aldosterone could regulate miR expression. miR-192 expression was reduced in mice submitted to sodium depletion, potassium loading, or aldosterone infusion, three situations of elevated aldosterone level. Interestingly, miR-192 was implicated in the development of diabetic nephropathy through TGF-β–induced collagen expression via its binding to the 3′UTR of Smad-interacting protein 1.9 Aldosterone promotes renal fibrosis by stimulating collagen synthesis via ERK1/2 phosphorylation and TGF-β induction.34 miR-192 could thus participate in the multiple roles of aldosterone in health and diseases. The mechanisms by which aldosterone regulates miR-192 expression remain to be defined. A recent in vitro study showed that aldosterone stimulates the expression of miR-23a through the calcineurin-NFATc3 pathway in cardiomyocytes.35 The regulation of miR-192 expression has been poorly characterized. A conserved noncoding sequence containing a consensus binding site for the proto-oncogene ets-1 has been identified 2158 bp upstream of the miR-192/miR-194 -2 cluster.7 Several glucorticoid response elements are present in the 5′ region of the cluster but are also found upstream of the miR-215 gene. Interestingly, the expression of miR-192 was not modified in the colon of mice receiving an aldosterone infusion (data not shown). This result suggests that the regulation of miR-192 expression by aldosterone in the kidney could depend on tissue-specific effectors or co-factors.
Transient transfection and reporter assays showed that these miR-192 and miR-215 could repress luciferase expression by binding to their target sequence in WNK1 3′UTR. Although computational analysis by the commonly used tools would have predicted miR-215 as a better candidate, postfitting of the predictions rather indicated that miR-192 was truly targeting the TS and not miR-215 (Table 1). Interestingly, this latter search was experimentally validated by the finding that only miR-192 regulated endogenous WNK1 expression ex vivo. Our study thus highlights the importance of the experimental validation of any predicted miR targeting.
The target sequence for miR-192 and miR-215 is located upstream of the first polyA, common to all WNK1 isoforms.16 As a consequence, miR-192 could potentially regulate the expression of L- and KS-WNK1. This hypothesis is reinforced by the fact that expression of this miR is not restricted to the DCT. Aldosterone stimulates the transcription of KS-WNK1, whereas it stimulates L-WNK1 only at the post-transcriptional level. The inhibition of miR-192 expression by aldosterone could participate in the regulation of WNK1 expression by this hormone. Whether KS-WNK1 protein expression is regulated or not in a similar manner remains to be determined. miR-192 renal expression was also decreased by sodium depletion and potassium load, two situations of elevated aldosterone, whereas L-WNK1 mRNA and protein expression was unaffected. This probably reflects the complex regulation of this kinase. The absence of an increase in L-WNK1 protein and KS-WNK1 mRNA expression during salt depletion could be explained by the elevated angiotensin II level, which would inhibit WNK1. A previous study indeed reported that L-WNK1 protein expression was not regulated by potassium load.28 In addition, several studies in rats and mice suggest that potassium load and aldosterone have differential effects.36,37
Mutations in WNK1 lead to the development of FHHt. Two microRNAs were reported to play a role in the control of BP but none of them through a specific effect on sodium transport. A target site for miR-155 was found in the 3′UTR of the angiotensin II type 1 receptor (AGTR1). The 1166C minor allele, which is associated with hypertension, reduces the interaction between miR-155 and its target sequence, thus leading to an increased expression of AGTR1.38 Another polymorphism at the miR-122 target site in the 3′UTR of SLC7A1 causes endothelial dysfunction and is associated with essential hypertension.39 We searched for polymorphisms or mutations in WNK1 3′UTR in 34 unrelated FHHt patients, carrying no WNK1 or WNK4 mutations. No variant was found by direct sequencing of the 290-bp region surrounding the miR-192 site (data not shown). The polymorphism rs11571461 was found at a frequency similar to that previously described in Caucasian subjects.40 However, this polymorphism described downstream of WNK1 is actually located in the RAD52 gene.
In conclusion, although the critical role played by aldosterone in sodium and potassium handling and BP regulation has been well established, the mechanisms by which this hormone exerts its actions are still not fully elucidated. Besides its main transcriptional effects mediated by the mineralocorticoid receptor, aldosterone was also described to have rapid nongenomic effects.41 Here, we present a potential new mechanism of regulation of gene expression by aldosterone, taking place at the post-transcriptional level and mediated by microRNAs.
Concise Methods
Computational Analysis
For predictions based on conservation, four algorithms were used: TargetScan (version 5.1: http://www.targetscan.org/vert_50/), miRanda (http://microrna.sanger.ac.uk/targets/v5/ and http://www.microrna.org/microrna/getGeneForm.do), Diana microT (version 3.0: http://diana.cslab.ece.ntua.gr/microT/), and MirTarget2 (http://mirdb.org/miRDB/). We subsequently analyzed the miRNA/target-island partners as described previously22 and also used a support vector machine classifier.23 The effect of central loops was taken into account.24 Postfitting of the predictions was done as described.25
Tubules Microdissection
All animals used in this study were treated in full compliance with the French government animal welfare policy (agreement no. A 75-05-12). Microdissection of the kidney of nine 8-week-old C57BL/6N was performed in sterile conditions under a binocular microscope as described.42 Pools of tubules of each category (about 10 mm in length) were transferred to 50 μl of QIAzol Lysis Reagent (miRNeasy mini kit, Qiagen) and stored at −80°C. One, two, or three pools of DCT and CNT/CCD were collected from the same mouse kidney.
miRNA TaqMan Low-Density Arrays on Tubules
Total RNA including microRNAs was extracted using miRNeasy Mini Kit (Qiagen). Reverse transcription was done with 5 ng of RNA using the megaplex reverse transcription and preamplification pool set and TaqMan microRNA reverse transcription kit (Applied Biosystems). A 12-cycle preamplification was made using the megaplex reverse transcription and preamplification pool set and the TaqMan preamplification master mix (Applied Biosystems). The preamplification product was diluted in 75 μl of water and 1:100 was used for TaqMan low-density arrays on TaqMan rodent miRNA array A and B, using the TaqMan Universal PCR Master Mix, No AmpErase UNG, and an Applied Biosystems 7900 HT Fast real time PCR system. Results were expressed as 2−ΔCt values with ΔCT = CtmiR − Ctsno202.
Cells and Cell Culture
All cell culture reagents were purchased from Invitrogen. MDCK cells were grown in Dulbecco modified Eagle medium high glucose (4500 mg/L d-glucose) with l-glutamine and sodium pyruvate, supplemented with 1% nonessential amino acids and 10% FCS. Human embryonic kidney (HEK293) cells were grown in Dulbecco modified Eagle medium high glucose (4500 mg/L d-glucose) with l-glutamine and sodium pyruvate, supplemented with 7.5% FCS. All cell culture media were supplemented with penicillin and streptomycin.
Generation of Luciferase Reporter Constructs
The pGL3-TK vector was generated by inserting the TK promoter, isolated from the pRL-TK vector (Promega), into the BglII and HindIII sites of the pGL3-Basic Vector (Promega). To generate the WT construct, the fragment corresponding to a partial human WNK1 3′UTR starting at 236 bp and finishing at 1072 bp after WNK1 stop codon was amplified from the BAC AC004765 (gi: 4731046) by PCR. The ΔTS construct was generated by deleting from the WT construct a 63-bp fragment (tactggggtgcaactctttcttatgataggtcattagtgctttaagcaaaagatattagcagc) containing the target sequence identified for miR-192 and miR-215 (in bold) by computational analysis. The sequences upstream and downstream of the deletion were amplified by PCR from the 3′UTR construct and ligated together in pGEM-T (Promega). To generate the 4TS plasmid, the 63-bp target sequence of miR-192 and miR-215 was amplified by PCR from the BAC AC004765 and submitted to ligation. The 252-bp tetrameric ligation products were selected. The different PCR fragments were then inserted in the XbaI restriction site of the pGL3-TK vector.
Transient Transfections and Reporter Gene Assays
MDCK cells in 12-well dishes were transfected with 1 μg of each luciferase plasmid using cationic liposomes (Lipofectamine 2000; Invitrogen) and 0.1 to 100 nM of scramble, precursor miR-192, or precursor miR-215 (Applied Biosystems) for the dose-dependent response experiments, or 30 nM of scramble, precursor miR-192, precursor miR-215, anti—miR-192 antisense (AS-192), or anti–miR-215 antisense (AS-215; Applied Biosystems) for the functional experiments. Cells were co-transfected with 200 ng of pCH11043 as a control for transfection efficiency. Forty-eight hours after transfection, luciferase and β-galactosidase activities were measured in a GLOMAX Microplate Luminometer (Promega), with a chemiluminescence reporter assay system (dual light luciferase assay system; Applied Biosystems). Luciferase activity was normalized to β-galactosidase activity.
L-WNK1 Real-Time RT-PCR
HEK cells were cultured in 12-well dishes and transfected with 0 or 30 nM of scramble, AS-192, or AS-125. Forty-eight hours after transfection, total RNA was extracted using the miRNeasy Mini Kit (Qiagen) and 2 μg was converted to cDNA by reverse transcription using the Superscript II reverse transcriptase and 1-μg random primers (Invitrogen). L-WNK1 and human 18S expression was measured using qPCR MasterMix Plus with fluorescein (Eurogentec). The primer sequences were given elsewhere.16,44 L-WNK1 expression was normalized to 18S.
Animal Treatment
The treated groups and control groups were handled in parallel during a 10-day period.
NaCl Depletion and NaCl or KCl Loading.
Each group consisted of ten 16-week-old C57BL/6N male mice. All of the animals had free access to water. Mice were fed ad libitum with a semisynthetic diet (Scientific Food and Engineering, Augis, France) containing 0.3, 0.03, or 3% NaCl for control, NaCl-depleted, or NaCl-loaded animals, respectively. The KCl-loaded animals were given 2.4% KCl in the drinking water.
Aldosterone Chronic Infusion.
Mice were given 60 μg/kg per day of aldosterone via minipump (Charles River). At the end of the experimental period, animals were killed after anesthesia by peritoneal injection of ketamine and xylazine (0.1 and 0.01 mg/g body wt, respectively). Excised kidneys were cut into 5-mm slices. To prepare whole kidney protein extract, one kidney slice was placed into 200-μl ice-cold isolation buffer (250 mM sucrose and 20 mM Tris-HEPES [pH 7.4]), containing protease and phosphatase inhibitors (PhosphoSTOP Phopshatase Inhibitor Cocktail Tablet and Complete Protease Inhibitor Cocktail Tablet, Roche Diagnostics), and homogenized in an Ultraturrax homogenizer at maximum speed for 3 × 10 s. The homogenates were centrifuged at 4000g for 15 minutes at 4°C and the supernatant was collected.
miRNA Real-Time PCR
Total RNA, including miRs, was extracted from one slice of mouse kidney or from HEK293 cells using the miRNeasy Mini Kit (Qiagen). miR-192 and miR-215 expression was analyzed by TaqMan miRNA assays according to the manufacturer's protocol (Applied Biosystems) and normalized to the expression of the control tiny RNA sno-202.
Immunoblotting
Immunoblotting procedures was performed as described previously45 using an antibody directed against the N-terminal domain of L-WNK1 (Novus Biological Ltd., 1:500). Coomassie blue–stained polyacrylamide gels were used to control equality of protein loading for each series. Densitometric values were normalized to the mean for the control group in a given experiment, which was defined as 100%.
Statistical Analysis
Data are expressed as mean ± SEM. Statistical comparisons between two groups were made by the Wilcoxon test and between multiple groups by ANOVA. A value of P < 0.05 was considered significant.
Disclosures
None.
Supplementary Material
Acknowledgments
This work was supported by INSERM, ANR-05-MRAR-010-01, the EUNEFRON European Network, and the Fondation Leducq Transatlantic Network on Hypertension. E.E.M. received a fellowship from the MESR and X.-O.Z. received a fellowship from INSERM. We thank S. Harrap, C. Büsst, S. Bergaya, and J. Favier for their critical reading of the manuscript.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
Supplemental information for this article is available online at http://www.jasn.org/.
References
- 1. Filipowicz W, Bhattacharyya SN, Sonenberg N: Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat Rev Genet 9: 102–114, 2008 [DOI] [PubMed] [Google Scholar]
- 2. Rana TM: Illuminating the silence: Understanding the structure and function of small RNAs. Nat Rev Mol Cell Biol 8: 23–36, 2007 [DOI] [PubMed] [Google Scholar]
- 3. Boyd SD: Everything you wanted to know about small RNA but were afraid to ask. Lab Invest 88: 569–578, 2008 [DOI] [PubMed] [Google Scholar]
- 4. Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, van Dongen S, Grocock RJ, Das PP, Miska EA, Vetrie D, Okkenhaug K, Enright AJ, Dougan G, Turner M, Bradley A: Requirement of bic/microRNA-155 for normal immune function. Science 316: 608–611, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P, Stoffel M: A pancreatic islet-specific microRNA regulates insulin secretion. Nature 432: 226–230, 2004 [DOI] [PubMed] [Google Scholar]
- 6. Liang Y, Ridzon D, Wong L, Chen C: Characterization of microRNA expression profiles in normal human tissues. BMC Genomics 8: 166, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sun Y, Koo S, White N, Peralta E, Esau C, Dean NM, Perera RJ: Development of a micro-array to detect human and mouse microRNAs and characterization of expression in human organs. Nucleic Acids Res 32: e188, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tian Z, Greene AS, Pietrusz JL, Matus IR, Liang M: MicroRNA-target pairs in the rat kidney identified by microRNA microarray, proteomic, and bioinformatic analysis. Genome Res 18: 404–411, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kato M, Zhang J, Wang M, Lanting L, Yuan H, Rossi JJ, Natarajan R: MicroRNA-192 in diabetic kidney glomeruli and its function in TGF-beta-induced collagen expression via inhibition of E-box repressors. Proc Natl Acad Sci U S A 104: 3432–3437, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pastorelli LM, Wells S, Fray M, Smith A, Hough T, Harfe BD, McManus MT, Smith L, Woolf AS, Cheeseman M, Greenfield A: Genetic analyses reveal a requirement for Dicer1 in the mouse urogenital tract. Mamm Genome 20: 140–151, 2009 [DOI] [PubMed] [Google Scholar]
- 11. Harvey SJ, Jarad G, Cunningham J, Goldberg S, Schermer B, Harfe BD, McManus MT, Benzing T, Miner JH: Podocyte-specific deletion of dicer alters cytoskeletal dynamics and causes glomerular disease. J Am Soc Nephrol 19: 2150–2158, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shi S, Yu L, Chiu C, Sun Y, Chen J, Khitrov G, Merkenschlager M, Holzman LB, Zhang W, Mundel P, Bottinger EP: Podocyte-selective deletion of dicer induces proteinuria and glomerulosclerosis. J Am Soc Nephrol 19: 2159–2169, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Huang CL, Cha SK, Wang HR, Xie J, Cobb MH: WNKs: Protein kinases with a unique kinase domain. Exp Mol Med 39: 565–573, 2007 [DOI] [PubMed] [Google Scholar]
- 14. McCormick JA, Yang CL, Ellison DH: WNK kinases and renal sodium transport in health and disease: An integrated view. Hypertension 51: 588–596, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wilson FH, Disse-Nicodeme S, Choate KA, Ishikawa K, Nelson-Williams C, Desitter I, Gunel M, Milford DV, Lipkin GW, Achard JM, Feely MP, Dussol B, Berland Y, Unwin RJ, Mayan H, Simon DB, Farfel Z, Jeunemaitre X, Lifton RP: Human hypertension caused by mutations in WNK kinases. Science 293: 1107–1112, 2001 [DOI] [PubMed] [Google Scholar]
- 16. Delaloy C, Lu J, Houot AM, Disse-Nicodeme S, Gasc JM, Corvol P, Jeunemaitre X: Multiple promoters in the WNK1 gene: One controls expression of a kidney-specific kinase-defective isoform. Mol Cell Biol 23: 9208–9221, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xie J, Wu T, Xu K, Huang IK, Cleaver O, Huang CL: Endothelial-specific expression of WNK1 kinase is essential for angiogenesis and heart development in mice. Am J Pathol 175: 1315–1327, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zambrowicz BP, Abuin A, Ramirez-Solis R, Richter LJ, Piggott J, BeltrandelRio H, Buxton EC, Edwards J, Finch RA, Friddle CJ, Gupta A, Hansen G, Hu Y, Huang W, Jaing C, Key BW, Jr., Kipp P, Kohlhauff B, Ma ZQ, Markesich D, Payne R, Potter DG, Qian N, Shaw J, Schrick J, Shi ZZ, Sparks MJ, Van Sligtenhorst I, Vogel P, Walke W, Xu N, Zhu Q, Person C, Sands AT: Wnk1 kinase deficiency lowers blood pressure in mice: A gene-trap screen to identify potential targets for therapeutic intervention. Proc Natl Acad Sci U S A 100: 14109–14114, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xu BE, Min X, Stippec S, Lee BH, Goldsmith EJ, Cobb MH: Regulation of WNK1 by an autoinhibitory domain and autophosphorylation. J Biol Chem 277: 48456–48462, 2002 [DOI] [PubMed] [Google Scholar]
- 20. Naray-Fejes-Toth A, Snyder PM, Fejes-Toth G: The kidney-specific WNK1 isoform is induced by aldosterone and stimulates epithelial sodium channel-mediated Na+ transport. Proc Natl Acad Sci U S A 101: 17434–17439, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. O'Reilly M, Marshall E, Macgillivray T, Mittal M, Xue W, Kenyon CJ, Brown RW: Dietary electrolyte-driven responses in the renal WNK kinase pathway in vivo. J Am Soc Nephrol 17: 2402–2413, 2006 [DOI] [PubMed] [Google Scholar]
- 22. Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I: A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell 126: 1203–1217, 2006 [DOI] [PubMed] [Google Scholar]
- 23. Kim SK, Nam JW, Rhee JK, Lee WJ, Zhang BT: miTarget: MicroRNA target gene prediction using a support vector machine. BMC Bioinformatics 7: 411, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ye W, Lv Q, Wong CK, Hu S, Fu C, Hua Z, Cai G, Li G, Yang BB, Zhang Y: The effect of central loops in miRNA:MRE duplexes on the efficiency of miRNA-mediated gene regulation. PLoS One 3: e1719, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yang Y, Wang YP, Li KB: MiRTif: A support vector machine-based microRNA target interaction filter. BMC Bioinformatics 9 [Suppl 12]: S4, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Harris TA, Yamakuchi M, Ferlito M, Mendell JT, Lowenstein CJ: MicroRNA-126 regulates endothelial expression of vascular cell adhesion molecule 1. Proc Natl Acad Sci U S A 105: 1516–1521, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ji R, Cheng Y, Yue J, Yang J, Liu X, Chen H, Dean DB, Zhang C: MicroRNA expression signature and antisense-mediated depletion reveal an essential role of MicroRNA in vascular neointimal lesion formation. Circ Res 100: 1579–1588, 2007 [DOI] [PubMed] [Google Scholar]
- 28. Wade JB, Fang L, Liu J, Li D, Yang CL, Subramanya AR, Maouyo D, Mason A, Ellison DH, Welling PA: WNK1 kinase isoform switch regulates renal potassium excretion. Proc Natl Acad Sci U S A 103: 8558–8563, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pandey P, Brors B, Srivastava PK, Bott A, Boehn SN, Groene HJ, Gretz N: Microarray-based approach identifies microRNAs and their target functional patterns in polycystic kidney disease. BMC Genomics 9: 624, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kaplan JH: Biochemistry of Na,K-ATPase. Annu Rev Biochem 71: 511–535, 2002 [DOI] [PubMed] [Google Scholar]
- 31. Shi XB, Xue L, Yang J, Ma AH, Zhao J, Xu M, Tepper CG, Evans CP, Kung HJ, DeVere White RW: An androgen-regulated miRNA suppresses Bak1 expression and induces androgen-independent growth of prostate cancer cells. Proc Natl Acad Sci U S A 104: 19983–19988, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dai R, Phillips RA, Zhang Y, Khan D, Crasta O, Ahmed SA: Suppression of LPS-induced Interferon-gamma and nitric oxide in splenic lymphocytes by select estrogen-regulated microRNAs: A novel mechanism of immune modulation. Blood 112: 4591–4597, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fiedler SD, Carletti MZ, Hong X, Christenson LK: Hormonal regulation of MicroRNA expression in periovulatory mouse mural granulosa cells. Biol Reprod 79: 1030–1037, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Han JS, Choi BS, Yang CW, Kim YS: Aldosterone-induced TGF-beta1 expression is regulated by mitogen-activated protein kinases and activator protein-1 in mesangial cells. J Korean Med Sci 24 [Suppl]: S195–S203, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lin Z, Murtaza I, Wang K, Jiao J, Gao J, Li PF: miR-23a functions downstream of NFATc3 to regulate cardiac hypertrophy. Proc Natl Acad Sci U S A 106: 12103–12108, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stanton B, Pan L, Deetjen H, Guckian V, Giebisch G: Independent effects of aldosterone and potassium on induction of potassium adaptation in rat kidney. J Clin Invest 79: 198–206, 1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wald H, Popovtzer MM, Garty H: Differential regulation of CHIF mRNA by potassium intake and aldosterone. Am J Physiol 272: F617–F623, 1997 [DOI] [PubMed] [Google Scholar]
- 38. Sethupathy P, Borel C, Gagnebin M, Grant GR, Deutsch S, Elton TS, Hatzigeorgiou AG, Antonarakis SE: Human microRNA-155 on chromosome 21 differentially interacts with its polymorphic target in the AGTR1 3′ untranslated region: A mechanism for functional single-nucleotide polymorphisms related to phenotypes. Am J Hum Genet 81: 405–413, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yang Z, Kaye DM: Mechanistic insights into the link between a polymorphism of the 3′UTR of the SLC7A1 gene and hypertension. Hum Mutat 30: 328–333, 2009 [DOI] [PubMed] [Google Scholar]
- 40. Newhouse S, Farrall M, Wallace C, Hoti M, Burke B, Howard P, Onipinla A, Lee K, Shaw-Hawkins S, Dobson R, Brown M, Samani NJ, Dominiczak AF, Connell JM, Lathrop GM, Kooner J, Chambers J, Elliott P, Clarke R, Collins R, Laan M, Org E, Juhanson P, Veldre G, Viigimaa M, Eyheramendy S, Cappuccio FP, Ji C, Iacone R, Strazzullo P, Kumari M, Marmot M, Brunner E, Caulfield M, Munroe PB: Polymorphisms in the WNK1 gene are associated with blood pressure variation and urinary potassium excretion. PLoS One 4: e5003, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Grossmann C, Gekle M: New aspects of rapid aldosterone signaling. Mol Cell Endocrinol 308: 53–62, 2009 [DOI] [PubMed] [Google Scholar]
- 42. Farman N, Vandewalle A, Bonvalet JP: Autoradiographic determination of dexamethasone binding sites along the rabbit nephron. Am J Physiol 244: F325–F334, 1983 [DOI] [PubMed] [Google Scholar]
- 43. Norton PA, Coffin JM: Bacterial beta-galactosidase as a marker of Rous sarcoma virus gene expression and replication. Mol Cell Biol 5: 281–290, 1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Patberg KW, Obreztchikova MN, Giardina SF, Symes AJ, Plotnikov AN, Qu J, Chandra P, McKinnon D, Liou SR, Rybin AV, Shlapakova I, Danilo P, Jr., Yang J, Rosen MR: The cAMP response element binding protein modulates expression of the transient outward current: Implications for cardiac memory. Cardiovasc Res 68: 259–267, 2005 [DOI] [PubMed] [Google Scholar]
- 45. Quentin F, Eladari D, Frische S, Cambillau M, Nielsen S, Alper SL, Paillard M, Chambrey R: Regulation of the Cl-/HCO3- exchanger AE2 in rat thick ascending limb of Henle's loop in response to changes in acid-base and sodium balance. J Am Soc Nephrol 15: 2988–2997, 2004 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.