Abstract
Background
Prognosis and survival are significant concerns for individuals with heart failure (HF). In order to better understand the pathophysiology of HF prognosis, the association between 2,366,858 single nucleotide polymorphisms (SNPs) and all-cause mortality was evaluated among individuals with incident HF from four community-based prospective cohorts: the Atherosclerosis Risk in Communities Study, the Cardiovascular Health Study, the Framingham Heart Study, and the Rotterdam Study.
Methods and Results
Participants were 2,526 individuals of European ancestry and 466 individuals of African ancestry who suffered an incident HF event during follow-up in the respective cohorts. Within each study, the association between genetic variants and time to mortality among individuals with HF was assessed by Cox proportional hazards models that included adjustment for sex and age at the time of the HF event. Prospective fixed-effect meta-analyses were conducted for the four study populations of European ancestry (N=1,645 deaths) and for the two populations of African ancestry (N=281 deaths). Genome-wide significance was set at P=5.0×10-7. Meta-analytic findings among individuals of European ancestry revealed one genome-wide significant locus on chromosome 3p22 in an intron of CKLF-like MARVEL transmembrane domain containing 7 (CMTM7, p = 3.2×10-7). Eight additional loci in individuals of European ancestry and four loci in individuals of African ancestry were identified by high-signal SNPs (p < 1.0×10-5), but did not meet genome-wide significance.
Conclusions
This study identified a novel locus associated with all-cause mortality among individuals of European ancestry with HF. This finding warrants additional investigation, including replication, in other studies of HF.
Keywords: heart failure, all-cause mortality, genetics, genome-wide variation
Introduction
Heart failure (HF) is a relatively common chronic condition characterized by the inability of the heart to efficiently pump blood and represents a significant public health burden, affecting nearly 6 million Americans. Prognosis and survival are a significant concern: after HF is diagnosed, 1 in 5 die within one year1, often by sudden cardiac death which occurs 6 to 9 times more often than in the general population.1,2
The public health burden of HF and its associated mortality is likely to intensify due to the aging population and an increase in the prevalence of HF risk factors, such as coronary artery disease, obesity and diabetes. The Cohorts for Heart and Aging Research in Genome Epidemiology (CHARGE) consortium includes the Atherosclerosis Risk in Communities (ARIC) Study, the Cardiovascular Health Study (CHS), the Framingham Heart Study (FHS), the Rotterdam Study (RS) and the National Institute of Aging's Iceland Age, Gene/Environment Susceptibility (AGES) Study.3 The CHARGE consortium provided an opportunity to evaluate genetic factors associated with the prognosis of HF, and the association between genome-wide variation and all-cause mortality was investigated among 2,526 incident HF cases of European ancestry and 466 incident HF cases of African ancestry. 4
Methods
Design and Study Population
The design for this study is a cohort of individuals with incident HF nested within four of the population-based studies (ARIC, CHS, FHS and RS) of the CHARGE consortium. A detailed description of each cohort study design has been published elsewhere.5-9 Selection of eligible incident HF cases (with available genome-wide scan data) and methodology for genotyping and genotype imputation are published.4 HF cases were identified during 1987-2004 for ARIC, 1989-2007 for CHS, 1988-2006 for FHS and 1990-2006 for RS. Details regarding HF diagnostic criteria are shown in supplemental Table S1.
Briefly, eligible participants for these analyses were individuals of European or African ancestry who suffered an incident HF event during follow-up and had genome-wide association data. Identification of incident HF events relied on multiple sources of clinical and administrative data. The approach was similar across the four studies and is described in more detail in previously published reports.4,10,11 A total of 2,526 incident HF events were identified from 20,926 participants of European ancestry from all four cohorts over a weighted average of 11.5 years of follow-up.4 A total of 466 HF events were identified over an average of 13.7 years among 2,895 individuals of African ancestry from ARIC and CHS.4 Each study received IRB approval and all participants provided written informed consent for the use of their DNA for research.
Genotyping and Imputation
For the individuals of European ancestry, each study independently imputed their genotype data to approximately 2.4 million autosomal single nucleotide polymorphisms (SNPs) identified in HapMap CEU samples. Imputed genotypes were not determined in ARIC individuals of African ancestry and genotypes available for analysis were restricted to successfully genotyped SNPs on the Affymetrix 6.0 chip. In CHS individuals of African ancestry, SNP data were imputed using information from HapMap YRI and CEU samples and were then restricted to the same set of SNPs evaluated in ARIC individuals of African ancestry. Imputation results were summarized as an “allele dosage” defined as the expected number of copies of the minor allele at that SNP (a fractional value between 0.0 and 2.0). A ratio of observed versus expected variance (OEV) of the dosage for each SNP was calculated within each cohort as a measure of imputation quality.
Characteristics of Participants with HF
For these analyses, measures of clinical and demographic characteristics were obtained from the clinical visit preceding HF onset. At in-person visits, clinical measures were collected (annually in CHS from 1989 through 1999, biennially in the original cohort and approximately every four years in the offspring cohort of FHS, every three years in ARIC through 1998, and every three years in RS). Measures were taken using standardized methods as specified by each study and included in-person measures of height, weight, systolic and diastolic blood pressure, and total cholesterol. Each study also identified individuals with hypertension, diabetes or myocardial infarction that occurred prior to HF diagnosis.
Mortality Ascertainment
Participants were followed prospectively with in-person visits and telephone contacts in most cohorts. Deaths were defined as all-cause mortality. In the ARIC study, deaths were ascertained through annual phone calls or through ongoing surveillance of health department death certificate files through December 31, 2004. In CHS, participant mortality was identified at 6-month surveillance contacts, National Death Index searches, and from obituaries through June 30, 2007. Additional information about cause of death was collected from death certificates, proxy interviews, and medical records.11 In the RS, deaths were ascertained through general practitioner medical records and from municipal records through January 1, 2007. All FHS participants are under continuous surveillance based on periodic Heart Study visits and biannual health history updates. Information on death (and cause of death) is obtained from hospitalization records, medical records, death certificate, family members and obituaries. All deaths are reviewed by an endpoints adjudication panel consisting of three FHS physicians. The mortality ascertainment rate for each cohort is greater than 99%.
Statistical Analyses
The association between genomic variation and time to death among individuals with incident HF was evaluated separately in each cohort by Cox proportional hazards models. The follow-up time interval was defined as the time between the date of incident HF and the date of death, last contact if lost to follow-up, or the end of follow-up, whichever came first. Each model was evaluated separately in individuals with HF of European or African ancestry, and included adjustment for sex and age at the time of the HF event. In addition, CHS adjusted for study site and FHS adjusted for generation and ancestry using principal components.12 ARIC and RS did not include additional adjustments in the analysis model. All SNPs were evaluated for association under an additive genetic model.
Across the four study populations of European ancestry and separately the two populations of African ancestry, fixed-effect meta-analyses combined β-coefficients and study-specific p-values using genomic control lambdas to adjust for remaining population substructure. The meta-analysis produced an overall hazards ratio (HR), standard error and a p-value. Prior to meta-analysis, a filter was applied to SNPs with a β-coefficient greater than 5.0 or less than -5.0 to avoid numerical instabilities. Meta-analyses were performed using MetABEL software (http://mga.bionet.nsc.ru/∼yurii/ABEL/)13 and METAL (http://www.sph.umich.edu/csg/abecasis/Metal/index.html) for confirmation. To avoid inaccuracies in the asymptotic assumptions of Cox models when the number of events carrying the variant allele was small (i.e., <70 individuals with an endpoint and carrying the minor allele), SNPs were excluded from further consideration when the post-meta-analysis population-size-weighted minor allele frequency (MAF) was ≤0.03 in individuals of European ancestry and ≤0.135 in individuals of African ancestry.
For each ancestry group, a quantile-quantile (Q-Q) plot was generated from Cox proportional hazards models and shows the distribution of the observed and expected p-values for all SNPs. Also, meta-analyzed p-values were plotted according to location across the 22 autosomes. An a priori threshold of 5.0×10-7 was used to indicate genome-wide significance in the analysis of individuals of European and African ancestry.14 High-signal SNPs had a p-value smaller than 1.0×10-5, but did not meet genome-wide significance. If more than one significant or high-signal SNP clustered at a locus, the SNP with the smallest p-value was reported as the locus marker. Regional plots showing detailed linkage disequilibrium (LD) and gene location information and forest plots were created for genome-wide significant markers. Additionally, top hits from the meta-analysis in the European and African ancestry populations were compared. The minor allele frequencies, hazard ratios and p-values for these markers are reported for each ancestral population.
Results
Among the 2,526 participants of European ancestry with incident HF, a total of 1,645 deaths from all causes occurred at a weighted average of 3.6 years. A total of 281 deaths occurred among the 466 individuals with incident HF of African ancestry at a weighted average of 3.4 years. Baseline characteristics of cohort participants with incident HF are shown in Table 1. The weighted average age at diagnosis of HF was 77.3 years (range from 44 to 101) among individuals of European ancestry and 70.0 years (range from 46 to 96) among individuals of African ancestry.
Table 1. Characteristics of individuals with incident HF stratified by study and ancestral group.
| ARIC | CHS | RS | FHS | |||
|---|---|---|---|---|---|---|
| Characteristics | EA | AA | EA | AA | EA | EA |
| Individuals with HF, n | 691 | 331 | 838 | 135 | 748 | 249 |
| Mean age at HF event, yrs | 67.3 | 65.6 | 82.4 | 80.9 | 79.6 | 80.7 |
| Age range, yrs | 47-81 | 46-81 | 66-98 | 67-96 | 57-101 | 44-99 |
| Male, % | 60.1 | 40.5 | 56.3 | 66.7 | 48.8 | 47.8 |
| Body mass index, kg/m2 | 29.5 | 31.5 | 27.0 | 29.7 | 27.0 | 28.5 |
| Systolic blood pressure, mmHg | 132 | 140 | 141 | 144 | 146 | 143 |
| Diastolic blood pressure, mmHg | 70 | 77 | 69 | 73 | 74 | 70 |
| Total cholesterol, mg/dL | 203 | 209 | 201 | 210 | 252 | 193 |
| Hypertensive*, % | 58.5 | 80.0 | 73.5 | 85.9 | 44.7 | 76.3 |
| Diabetic*, % | 28.7 | 53.3 | 15.6 | 31.9 | 17.4 | 17.7 |
| Myocardial infarction†, % | 25.2 | 20.2 | 0.0 | 0.0 | 28.1 | 27.7 |
| Current smoker, % | 28.6 | 29.9 | 8.8 | 14.1 | 23.1 | 10.4 |
| Mean follow-up, yrs | 3.9 | 3.4 | 3.5 | 3.3 | 3.9 | 1.8 |
| One-year mortality rates‡ | 0.12 | 0.17 | 0.20 | 0.20 | 0.19 | 0.34 |
| All-cause deaths, n | 327 | 191 | 606 | 90 | 556 | 156 |
EA = European ancestry; AA = African ancestry; BMI = body mass index.
Self-reported history of treated hypertension and diabetes was determined in CHS, RS and FHS. Measurements of blood pressure and blood glucose were also used for the diagnosis of hypertension and diabetes in FHS using standardized criteria. In the ARIC study, the definition of hypertension included use of antihypertensive medication and measured blood pressure (systolic blood pressure ≥ 140 mm Hg, diastolic blood pressure ≥ 90 mm Hg), and diabetes was defined by a fasting glucose level ≥126 mg/dl, a nonfasting glucose level ≥200 mg/dl, or a self-reported diagnosis of diabetes or use of diabetic medication.
Myocardial infarction that occurs prior to HF diagnosis
among individuals with heart failure
Participants of European Ancestry
Figure 1 shows the Q-Q plot of p-values derived from the approximately 2.4 million tests performed in participants of European ancestry plotted against the expected distribution. Except for some of the very lowest p-values, the observed and expected distributions of p-values were similar. The genomic inflation lambda values for the individual cohorts were 0.95 for CHS, 0.99 for RS, 1.00 for ARIC and 1.09 for FHS. The meta-analysis genomic inflation lambda value for those of European ancestry was equal to 1.0, suggesting minimal population stratification or other technical artifacts. Figure 2 presents p-values for the approximately 2.4 million SNPs organized by chromosome and genomic position. As shown in Table 2, one SNP (rs12638540, p-value = 3.21×10-7, HR=1.53, genome-wide confidence interval 1.01-2.31, MAF of 0.04) on chromosome 3p22 exceeded the pre-specified genome-wide significance threshold. This SNP resides in an intron of CKLF-like MARVEL transmembrane domain containing 7 (CMTM7), and was genotyped in RS and FHS and imputed with high quality in ARIC (OEV=0.981) and CHS (OEV=0.921). Figure 3 presents detailed information about LD structure and p-values for other SNPs in this region, and a forest plot of the meta-analysis findings is shown in Figure 4.
Figure 1.
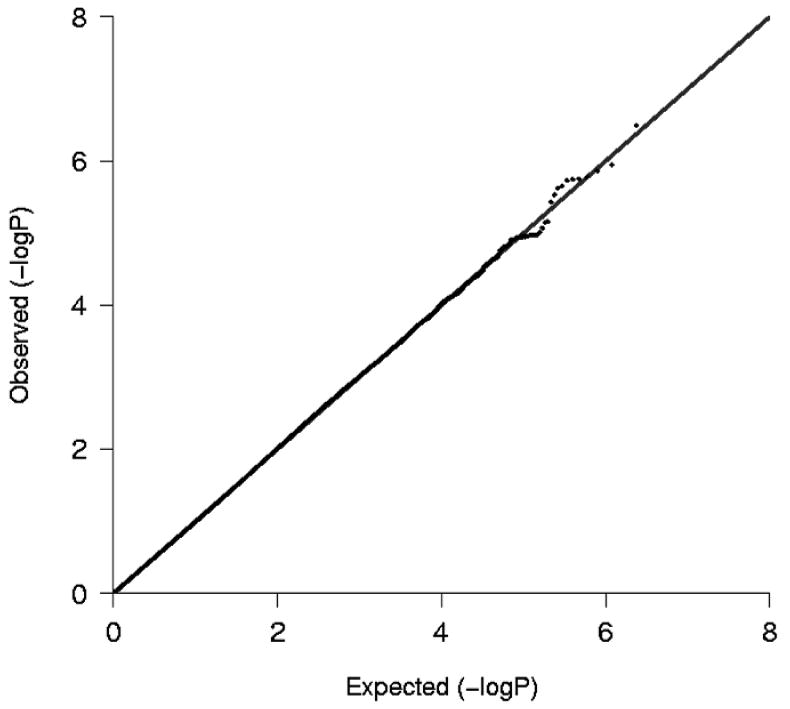
Plot of the expected and observed -log P-values from analyses of participants of European ancestry.
Figure 2.
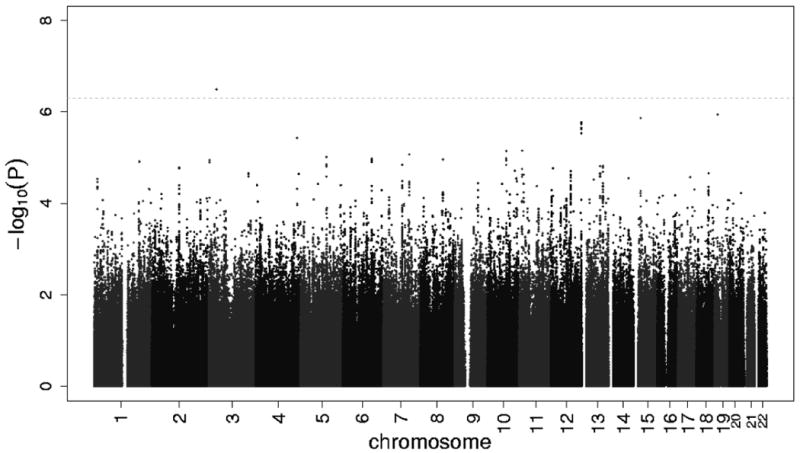
Among participants of European ancestry, the -log P-value of the additive genetic model for each SNP with a study size-weighted average minor allele frequency of greater than or equal to 3%, according to location in the 22 autosomal chromosomes. Horizontal line indicates the 5.0×10-7 threshold of genome-wide significance.
Table 2. Description of all SNPs by chromosome and position with meta-analysis P-values less than 1.0×10-5.
| Population | RS number | Chr | Position | Variant* | MAF | P-value | HR (GWAS CI) | Closest Gene |
|---|---|---|---|---|---|---|---|---|
| EA | rs12638540 | 3 | 32463538 | A/G | 0.043 | 3.21 ×10-7 | 1.53 (1.01-2.31) | CMTM7 (intron) |
| EA | rs4528684 | 19 | 14212574 | C/T | 0.094 | 1.13 ×10-6 | 1.42 (0.99 – 2.03) | 34.6 kb from LPHN1 |
| EA | rs2125623 | 15 | 29616999 | C/T | 0.285 | 1.37 ×10-6 | 0.81 (0.65 – 1.01) | OTUD7A (intron) |
| EA | rs7965445 | 12 | 130428856 | G/A | 0.098 | 1.68 ×10-6 | 1.30 (0.99 – 1.72) | 11.9 kb from LOC338797 |
| EA | rs12422521 | 12 | 130426315 | T/C | 0.099 | 1.78 ×10-6 | 1.30 (0.99 – 1.72) | 9.4 kb from LOC338797 |
| EA | rs12422973 | 12 | 130430537 | G/C | 0.097 | 1.80 ×10-6 | 1.30 (0.99 – 1.72) | 13.6 kb from LOC338797 |
| EA | rs7299374 | 12 | 130428360 | G/T | 0.099 | 1.86 ×10-6 | 1.30 (0.99 – 1.72) | 11.4 kb from LOC338797 |
| EA | rs7977251 | 12 | 130436978 | C/T | 0.095 | 2.22 ×10-6 | 1.30 (0.98 – 1.73) | 20.0 kb from LOC338797 |
| EA | rs7976504 | 12 | 130437027 | G/A | 0.095 | 2.38 ×10-6 | 1.31 (0.98 – 1.73) | 20.1 kb from LOC338797 |
| EA | rs6486658 | 12 | 130432243 | C/T | 0.097 | 2.94 ×10-6 | 1.29 (0.98 – 1.70) | 15.3 kb from LOC338797 |
| EA | rs7687921 | 4 | 177096020 | A/T | 0.039 | 3.74 ×10-6 | 1.77 (0.95 – 3.28)** | GPM6A (intron) |
| EA | rs7120489 | 11 | 12425973 | G/A | 0.069 | 6.94 ×10-6 | 1.35 (0.96 – 1.90) | PARVA (intron) |
| EA | rs4979906 | 10 | 79114759 | A/G | 0.186 | 7.13 ×10-6 | 1.23 (0.98 – 1.55) | 46.6 kb from KCNMA1 |
| EA | rs17159640 | 7 | 111873569 | A/T | 0.048 | 8.52 ×10-6 | 1.59 (0.94 – 2.68) | IFRD1 (intron) |
| EA | rs9885413 | 5 | 110204027 | G/T | 0.065 | 9.76 ×10-6 | 1.34 (0.96 – 1.87) | 77.6 kb from SLC25A46 |
| AA | rs6868223 | 5 | 33672351 | G/A | 0.338 | 1.78 ×10-6 | 1.58 (0.98 – 2.56) | ADAMTS12 (intron) |
| AA | rs12733856 | 1 | 219618162 | C/A | 0.382 | 2.86 ×10-6 | 1.56 (0.97 – 2.51) | 42.0 kb from LOC400804 |
| AA | rs8017423 | 14 | 89749663 | T/C | 0.376 | 7.02 ×10-6 | 0.61 (0.36 – 1.06) | 27.7 kb from KCNK13 |
| AA | rs769044 | 14 | 89740891 | A/G | 0.372 | 9.52 ×10-6 | 0.62 (0.36 – 1.07) | 18.9 kb from KCNK13 |
| AA | rs6573013 | 14 | 54805626 | G/T | 0.141 | 9.64 ×10-6 | 1.74 (0.93 – 3.26) | 2.2 kb from FBXO34 |
EA = European ancestry; AA = African ancestry; Chr = chromosome; MAF = estimated minor allele frequency; HR = hazard ratio for the minor allele from the additive genetic model; GWAS CI = confidence interval for 5.0×10-7;
major/minor allele;
Only in CHS, RS and FHS
Figure 3.
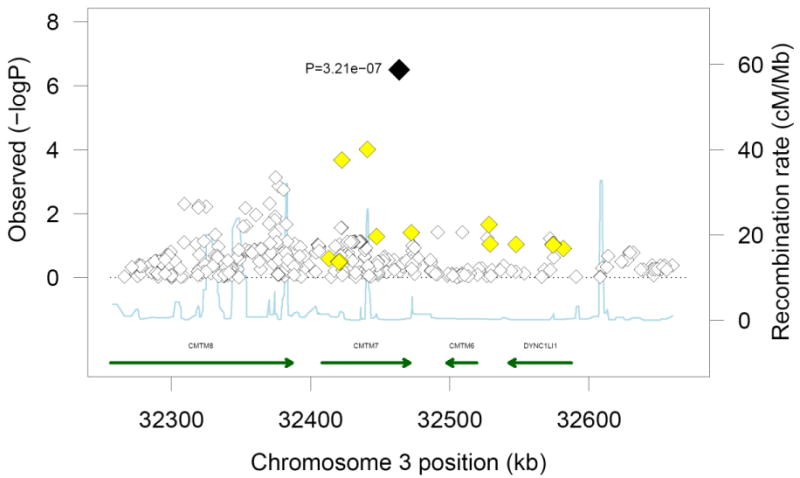
Regional association plot for the genome-wide significant marker, rs12638540, in individuals of European ancestry. The top marker is presented in black font and neighboring variants are presented in different colors based on linkage disequilibrium: yellow: r2 ≥ 0.2 and < 0.5; and white: r2 < 0.2. There were no SNPs in the area with LD ≥ 0.5 with the top marker.
Figure 4.
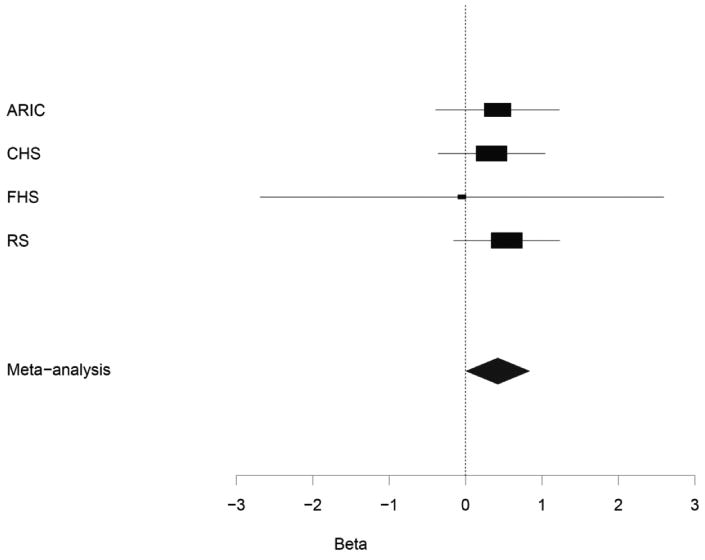
Meta-analysis and study-specific hazards ratios and confidence intervals (α=5.0×10-7) for rs12638540 associated with all-cause mortality in individuals of European ancestry.
In addition to the one SNP exceeding genome-wide significance, fourteen additional high-signal SNPs were identified with p-values less than 1.0×10-5 that marked eight chromosomal regions of interest. Table 2 provides detail about those loci identified in individuals of European ancestry. Of these eight loci, the OTU domain containing 7A (OTUD7A), glycoprotein M6A (GPM6A), alpha parvin (PARVA) and interferon-related developmental regulator 1 (IFRD1) genes were identified by a single intronic SNP. An intergenic SNP on chromosome 19 was 34.6 kb from the latrophilin 1 (LPHN1) gene. An intergenic SNP on chromosome 10 was 46.6 kb from the potassium large conductance calcium-activated channel, subfamily M, alpha member 1 (KCNMA1) gene, and an intergenic SNP on chromosome 5 was 77.6 kb from the solute carrier family 25, member 46 (SLC25A46) gene. An intergenic region on chromosome 12 was also identified. Cohort-specific results for the index markers for each locus are provided in supplemental Table S2.
Participants of African Ancestry
Figure 5 shows the Q-Q plot of p-values derived from the 560,383 tests performed in participants of African ancestry. Except for some of the very lowest p-values, the observed and expected distributions of p-values were similar. Figure 6 presents all p-values organized by chromosome and genomic position. In individuals of African ancestry with incident HF, no SNP exceeded the pre-specified genome-wide significance threshold for an association with mortality. Five high-signal SNPs were identified with p-values less than 1.0×10-5 that marked four loci of interest (Table 2). The smallest p-value among individuals of African ancestry was for an intronic SNP in the ADAM metallopeptidase with thrombospondin type 1 motif, 12 (ADAMTS12) gene. Two SNPs were within 27.7 kb from the potassium channel, subfamily K, member 13 (KCNK13) gene and one SNP was in the promoter region of F-box protein 34 (FBXO34). An additional intergenic SNP was greater than 100 kb from a known gene on chromosome 1. Cohort-specific results are provided in supplemental Table S2.
Figure 5.
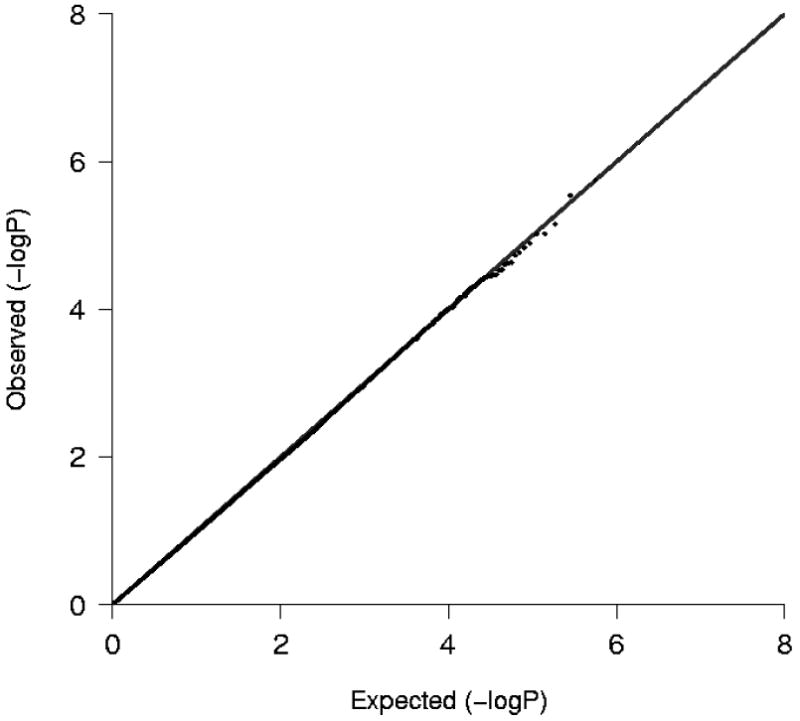
Plot of the expected and observed -log P-values from analyses of participants of African ancestry.
Figure 6.
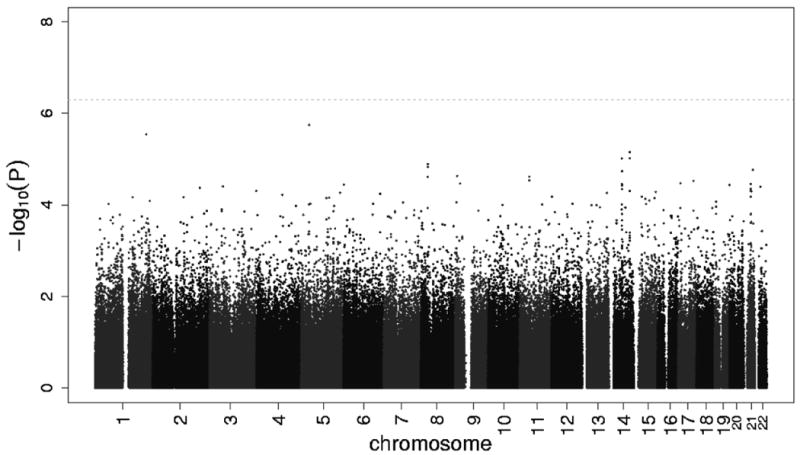
Among participants of African ancestry, the -log P-value of the additive genetic model for each SNP with a study size-weighted average minor allele frequency of greater than or equal to 13.5%, according to location in the 22 autosomal chromosomes. Horizontal line indicates the 5.0×10-7 threshold of genome-wide significance.
Cross Ancestry Comparisons
None of the loci identified in the African ancestry analysis were associated with mortality in individuals with HF of European ancestry (all p-values > 0.05). The genome-wide significant and high-signal SNPs among individuals of European ancestry were not associated with mortality among African ancestry CHS participants (all p-values >0.05). These SNPs were only imputed and evaluated for an association in CHS participants of African ancestry and were not imputed in individuals of African ancestry from the ARIC study. The minor allele frequency for rs12638540 is 0.07 in CHS individuals with heart failure of African ancestry.
Discussion
This study evaluated the association between genome-wide genetic variation and all-cause mortality among individuals with incident HF. The meta-analysis included 2,526 individuals of European ancestry with HF and revealed an intronic SNP in CMTM7 on chromosome 3 that exceeded genome-wide significance and was associated with a 53% increase in risk of death. Approximately 8% of the HF population carried this variant. Among African ancestry participants, no SNP had a p-value that exceeded genome-wide significance. Additional high-signal SNPs identified in either the European or African ancestry populations were in or close to LPHN1, OTUD7A, GPM6A, PARVA, KCNMA1, IFRD1, SLC25A46, ADAMTS12, KCNK13 and FBXO34 (p-value range between 1.10×10-6 and 9.64×10-6). Regions of the genome without known genes also identified in these analyses reside on chromosome 1q41 and 12q24. SNPs identified in GWAS for incident heart failure4 did not overlap with those identified here.
The genome-wide significant association was for CMTM7, one of several chemokine-like factor genes clustered in the same region on chromosome 3p22. The protein encoded by CMTM7 is highly expressed in leukocytes and is also expressed in the heart, but its function is unknown. A recent study indicates that chemokine receptor gene expression is upregulated among HF patients compared with controls.15 The CMTM7 chemokine-like factor may act as a chemoattractant to guide migration of cells in the heart. Some chemokines play a role as a pro-inflammatory marker in the context of an immune response or may promote angiogenesis during HF.
Of the 12 loci that did not meet genome-wide significance, four merit further discussion. The observed association with OTUD7A in individuals of European ancestry, and the association with FBXO34 in individuals of African ancestry, suggests that these genes may play a role in HF progression and survival by contributing to inadequate degradation of ubiquitin/protein conjugates and resulting in autophagic myocyte death. A mouse model for desmin-related cardiomyopathy indicates that accumulation of polyubiquitinated proteins is correlated with hastened HF progression.16 Other studies show that ubiquitinated proteins accumulate in the cytosol of human hearts with idiopathic or ischemic cardiomyopathies.17,18 The observed association with potassium channels encoded by KCNMA1 in individuals of European ancestry and KCNK13 in individuals of African ancestry provides evidence that control of vascular tone and regulation of the contraction of cardiac smooth muscle may be important components of HF progression and survival. Substantial previous work has documented that potassium channels are associated with cardiovascular and mortality endpoints.19-24
Although there was not a consistent finding for a particular SNP across the European and African ancestry populations, genes involved in deubiquitinating activity and genes encoding potassium channels were identified in each ethnic group. It is important to note that for many loci, there was a clear difference in the MAF between ethnic groups. This difference in MAF coupled with variable patterns of LD between the populations obfuscates the interpretability of cross-ancestral comparisons. In addition, it is known that certain HF treatments affect length of survival with HF, and access to care and treatment practices are known to vary by region and ethnicity.25,26
Study Strengths and Limitations
Although this is the largest study to date of individuals with incident HF evaluated for an association between genome-wide genetic variation and all-cause mortality, there are study limitations. SNP imputation in individuals of European ancestry resulted in the ability to meta-analyze results across approximately 2.4 million SNPs. Given 2.4 million tests, one significant SNP is what would have been expected by chance for the a priori level of genome-wide significance and only one SNP exceeded this threshold in individuals of European ancestry. This finding may represent a false positive result and requires replication in other studies. Similarly, the number of high-signal loci identified in either ancestry group was far fewer than would be expected by chance given a threshold of P=1.0×10-5.
For some variants, imputation quality varied and there was reduced statistical power to detect an association for poorly imputed SNPs. Across-ancestry comparisons were limited to data from CHS individuals of African ancestry, and the overall statistical power to detect an association was limited in African ancestry participants given a total sample of 466 individuals.
Due to limited data availability across the cohorts, this study did not account for ejection fraction, treatment for heart failure, socioeconomic status, access to medical care or hospitalizations subsequent to HF diagnosis. Heterogeneity among the individuals investigated for this study may have resulted from variation in heart failure diagnostic criteria across the four studies. However, a study comparing diagnostic criteria between CHS and FHS showed that all-cause mortality rates were similar across these diagnostic classifications.27 A study evaluating heart failure diagnosis by central adjudication compared to hospital discharge diagnoses showed that mortality rates are similar.28 A limitation of this study is that given differences in diagnostic criteria, the heart failure cases may not represent the same disease. Since the four study populations likely comprise both systolic and diastolic heart failure, it is important to note that the cause of death may be markedly different between these two disease states. This study addressed all-cause mortality and there was lack of information across cohorts to adequately conduct analyses of cause-specific mortality.
Finally, the observed signals are described in the context of the nearest gene; however, it is possible that true causal variants may be in neighboring genes or intergenic regions. Additional signals may have been missed due to coverage in particular genomic regions or the lack of statistical power to detect modest effects.
Conclusions
This study investigated 2,526 incident HF cases of European ancestry from ARIC, CHS, FHS and RS and 466 incident HF cases of African ancestry from ARIC and CHS for an association between genome-wide variation and all-cause mortality. One SNP in CMTM7 was significantly associated with all-cause mortality in individuals of European ancestry with HF, suggesting a role of chemokines with survival. However, no SNP was found to be significantly associated with mortality in individuals with HF of African ancestry. Future studies of this type, especially those that address some of the limitations of this study, may identify genes that lead to an improved understanding of HF pathophysiology and treatment of the disease. These findings warrant additional investigation, including replication, in other studies of HF.
This study investigated 2,526 incident heart failure cases of European ancestry and 466 incident heart failure cases of African ancestry for an association between genome-wide variation and all-cause mortality. One variant in the CKLF-like MARVEL transmembrane domain containing 7 (CMTM7) genes was significantly associated with all-cause mortality in individuals of European ancestry with heart failure. CMTM7 is one of several chemokine-like factor genes clustered in the same region on chromosome 3p22. These results suggest that chemokines may play a role in survival of heart failure patients. No genomic variation was significantly associated with mortality in individuals with heart failure of African ancestry. Future studies of this type may identify genes that lead to an improved understanding of heart failure pathophysiology and treatment of the disease. These findings warrant additional investigation, including replication, in other studies of heart failure.
Supplementary Material
Acknowledgments
The authors thank the staff and participants of the ARIC study for their important contributions. We thank Mila Jhamai, Pascal Arp, Dr Michael J. Moorhouse, Marijn Verkerk and Sander Bervoets for their help in creating the database and Maxim Struchalin for his contributions to the imputations of the data. The authors acknowledge the essential role of the CHARGE (Cohorts for Heart and Aging Research in Genome Epidemiology) Consortium in development and support of this manuscript. CHARGE members include ARIC, CHS, FHS, RS and AGES. The funding sources had no role in the study design, analyses, or drafting of the manuscript. The National Heart, Lung, and Blood Institute reviews all manuscripts submitted for publication but it was not involved in the decision to publish.
Funding Sources: The Atherosclerosis Risk in Communities Study is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute contracts N01-HC-55015, N01-HC-55016, N01-HC-55018, N01-HC-55019, N01-HC-55020, N01-HC-55021, N01-HC-55022, R01HL087641, R01HL59367 and R01HL086694; National Human Genome Research Institute contract U01HG004402; and National Institutes of Health contract HHSN268200625226C. Infrastructure was partly supported by Grant Number UL1RR025005, a component of the National Institutes of Health and NIH Roadmap for Medical Research. This research was supported in part by the intramural research program of the NIH, National Institutes of Environmental Health Sciences. The Cardiovascular Health Study is supported by contract numbers N01-HC-85079 through N01-HC-85086, N01-HC-35129, N01 HC-15103, N01 HC-55222, N01-HC-75150, N01-HC-45133, grant numbers U01 HL080295 and R01 HL 087652 from NHLBI with additional contribution from the National Institute of Neurological Disorders and Stroke. A full list of principal CHS investigators and institutions can be found at http://www.chs-nhlbi.org/pi.htm. DNA handling and genotyping was supported in part by National Center for Research Resources grant M01RR00425 to the Cedars-Sinai General Clinical Research Center Genotyping core and National Institute of Diabetes and Digestive and Kidney Diseases grant DK063491 to the Southern California Diabetes Endocrinology Research Center. The Framingham Heart Study was supported by NHLBI (Contract No. N01-HC-25195) and its contract with Affymetrix, Inc for genotyping services (Contract No. N02-HL-6-4278). This work was also supported in part by grants from the National Heart Lung Blood Institute 2K24HL04334, R01HL077477, and R01HL093328 (all to RSV). This study is based on analyses by Framingham Heart Study investigators participating in the SNP Health Association Resource (SHARe) project. The Rotterdam Study is supported by the Erasmus Medical Center and Erasmus University Rotterdam; the Netherlands Organization for Scientific Research (NWO); the Netherlands Organization for Health Research and Development (ZonMw); the Research Institute for Diseases in the Elderly (RIDE); The Netherlands Heart Foundation (Nederlandse hartstichting); the Ministry of Health, Welfare and Sports; the Ministry of Education, Culture and Science; the European Commission (DG XII); and the Municipality of Rotterdam. The GWAS database of the Rotterdam study was funded through NWO (175.010.2005.011, 911.03.012) and RIDE. This study was supported by the Netherlands Genomics Initiative (NGI)/NWO, project number 050-60-810.
Footnotes
Atherosclerosis Risk in Communities Study: Univ of TX at Houston Health Sci Ctr, Houston, TX (ACM, EB, JTW); Dept of Med (LRL, PPC), Biostatistics (DC), & Epidemiology (WR) Univ of NC at Chapel Hill, Chapel Hill, NC; Nat Inst of Envtl Health Sci (NIEHS), Rsrch Triangle Park, NC (LRL), Univ of MN School of Public Health, Minneapolis, MN (AF); Dept of Med, Univ of MS Med Ctr (ERF) and the Nat Heart, Lung, and Blood Inst Jackson Heart Study (ERF), TX Heart Inst (JTW)
Rotterdam Study: Dept of Epidemiology (JFF, AD, YSA, BHS, AH, CMvD, JCMW) and Internal Med (FR, AGU), Erasmus MC, Rotterdam, the Netherlands; Member of the Netherlands Consortium for Healthy Aging (JFF, JCMW)
Framingham Heart Study: Dept of Biostatistics, Boston Univ School of Public Health (LAC, SD, YAW), Dept of Med and Preventive Med, Boston Univ School of Med (EJB, RSV), Boston Univ and NHLBI's Framingham Heart Study (LAC, EJB, COD, DL, RSV), Dept of Epidemiology, Boston Univ School of Public Health (RSV), Nat Heart Lung Blood Inst (COD, DL), Mass Gen Hospital, Harvard Med School (COD, TJW).
Cardiovascular Health Study: Dept of Epidemiology (NLS, BMP), Med (NLG, JCB, BMP), Biostatistics (TL, KMR), and Health Services (BMP) Univ of Washington, Seattle, WA; Seattle Epidemiologic Research and Information Ctr of the Dept of Veterans Affairs Office of Rsrch and Dev, Seattle, WA (NLS); Group Health Ctr for Health Studies, Group Health, Seattle, WA (BMP); Med Genetics Inst, Cedars-Sinai Med Ctr, Los Angeles, CA (TH, JIR).
Conflict of Interest Disclosures: None
References
- 1.American Heart Association Statistics Committee and Stroke Statistics Subcommittee Heart Disease and Stroke Statistics-2009 Update. Circulation. 2009;119:e1–e161. doi: 10.1161/CIRCULATIONAHA.108.191259. [DOI] [PubMed] [Google Scholar]
- 2.Thom T, Kannel W, Silbershatz H, D'Agostino R. Cardiovascular diseases in the United States and prevention approaches. In: Fuster V, Alexander R, O'Rourke R, editors. Hurst's The Heart, Arteries and Veins. 10th. New York, NY: McGraw-Hill; 2001. pp. 3–17. [Google Scholar]
- 3.Psaty B, O'Donnell C, Gudnason V, Lunetta K, Folsom A, Rotter J, Uitterlinden A, Harris T, Witteman J, Boerwinkle E, Consortium obotC Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium: design of prospective meta-analyses of genome-wide association studies from five cohorts. Circulation: Cardiovascular Genetics. 2009;2:73–80. doi: 10.1161/CIRCGENETICS.108.829747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith N, Felix J, Morrison A, Demissie S, Glazer N, Heiss G, Cupples A, Dehghan A, Lumley T, Rosamond W, Lieb W, Rivadeneira F, Bis J, Folsom A, Benjamin E, Aulchenko Y, Haritunians T, Couper D, Murabito J, Stricker B, Gottdiener J, Chang P, Wang T, Harris T, Rice K, Loehr L, Gudnason V, Hofman A, Heckbert S, Shahar E, Donnell CO, Uitterlinden A, Rotter J, Fox E, Levy D, Duijn Cv, Psaty B, Witteman J, Boerwinkle E, Vasan R. The association of genome-wide genetic variation with incident heart failure in adults of European and African ancestry: the CHARGE Consortium. Co-submitted to Circulation: Cardiovascular Genetics. 2009 doi: 10.1161/CIRCGENETICS.109.895763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.ARIC Investigators The Atherosclerosis Risk in Communities (ARIC) Study: design and objectives. American Journal of Epidemiology. 1989;129:687–702. [PubMed] [Google Scholar]
- 6.Fried L, Borhani N, Enright P, Furberg C, Gardin J, Kronmal R, Kuller L, Manolio T, Mittelmark M, Newman A, et al. The Cardiovascular Health Study: design and rationale. Annals of Epidemiology. 1991;1:263–276. doi: 10.1016/1047-2797(91)90005-w. [DOI] [PubMed] [Google Scholar]
- 7.Dawber T, Meadors G, Moore F. Epidemiological approaches to heart disease: the Framingham Study. Am J Public Health Nations Health. 1951;41:279–281. doi: 10.2105/ajph.41.3.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feinleib M, Kannel W, Garrison R, McNamara P, Castelli W. The Framingham Offspring Study. Design and preliminary data. Prevention Medicine. 1975;4:518–525. doi: 10.1016/0091-7435(75)90037-7. [DOI] [PubMed] [Google Scholar]
- 9.Hofman A, Breteler M, van Duijn C, Janssen H, Krestin G, Kuipers E, Stricker B, Tiemeier H, Uitterlinden A, Vingerling J, Witteman J. The Rotterdam Study: 2010 objectives and design update. European Journal of Epidemiology. 2009;24:553–572. doi: 10.1007/s10654-009-9386-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.White A, Folsom A, Chambless L, Sharret A, Yang K, Conwill D, Higgins M, Williams O, Tyroler H. Community surveillance of coronary heart disease in the Atherosclerosis Risk in Communities (ARIC) Study: Methods and initial two years' experience. Journal of Clinical Epidemiology. 1996;49:223–233. doi: 10.1016/0895-4356(95)00041-0. [DOI] [PubMed] [Google Scholar]
- 11.Ives D, Fitzpatrick A, Bild D, Psaty B, Kuller L, Crowley P, Cruise R, Theroux S. Surveillance and ascertainment of cardiovascular events: The Cardiovascular Health Study. Annals of Epidemiology. 1995;5:278–285. doi: 10.1016/1047-2797(94)00093-9. [DOI] [PubMed] [Google Scholar]
- 12.Price A, Patterson N, Plenge R, Weinblatt M, Shadick N, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nature Genetics. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 13.Aulchenko Y, Ripke S, Isaacs A, Duijn Cv. GenABEL: an R library for genome-wide association analyses. Bioinformatics. 2007;23:1294–1296. doi: 10.1093/bioinformatics/btm108. [DOI] [PubMed] [Google Scholar]
- 14.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cappuzzello C, Napolitano M, Arcelli D, Melillo G, Melchionna R, DiVito L, Carlini D, Silvestri L, Brugaletta S, Liuzzo G, Crea F, Capogrossi M. Gene expression profiles in peripheral blood mononuclear cells of chronic heart failure patients. Physiological Genomics. 2009 doi: 10.1152/physiolgenomics.90364.2008. Epub. [DOI] [PubMed] [Google Scholar]
- 16.Tannous P, Zhu H, Johnstone J, Shelton J, Rajasekaran N, Benjamin I, Nguyen L, Gerard R, Levine B, Rothermel B, Hill J. Autophagy is an adaptive response in desmin-related cardiomyopathy. Proceedings of the National Academy of Sciences, USA. 2008;105:9745–9750. doi: 10.1073/pnas.0706802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heling A, Zimmermann R, Kostin S, Maeno Y, Hein S, Devaux B, Bauer E, Klovekorn W, Schlepper M, Schaper W, Schaper J. Increased expression of cytoskeletal, linkage, and extracellular proteins in failing human myocardium. Circulation Research. 2000;86:846–853. doi: 10.1161/01.res.86.8.846. [DOI] [PubMed] [Google Scholar]
- 18.Kostin S, Pool L, Elsasser A, Hein S, Drexler H, Arnon E, Hayakawa Y, Zimmermann R, Bauer E, Klovekorn W, Schaper J. Myocytes die by multiple mechanisms in failing human hearts. Circulation Research. 2003;92:715–724. doi: 10.1161/01.RES.0000067471.95890.5C. [DOI] [PubMed] [Google Scholar]
- 19.Tanaka Y, Meera P, Song M, Knaus H, Toro L. Molecular constituents of maxi KCa channels in human coronary smooth muscle: predominant alpha + beta subunit complexes. Journal of Physiology (London) 1997;502:545–547. doi: 10.1111/j.1469-7793.1997.545bj.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marijic J, Li QX, Song M, Nishimaru K, Stefani E, Toro L. Decreased expression of voltage- and Ca2+-activated K+ channels in coronary smooth muscle during aging. Circulation Research. 2001;88:210–216. doi: 10.1161/01.res.88.2.210. [DOI] [PubMed] [Google Scholar]
- 21.Tomas M, Vazquez E, Fernandez-Fernandez J, Subirana I, Plata C, Heras M, Vila J, Marrugat J, Valverde M, Senti M. Genetic variation in the KCNMA1 potassium channel alpha subunit as risk factor for severe essential hypertension and myocardial infarction. Journal of Hypertension. 2008;26:2147–2153. doi: 10.1097/HJH.0b013e32831103d8. [DOI] [PubMed] [Google Scholar]
- 22.Arnett D, Tang W, Rao D, Devereux R, Claas S, Kraemer R, Broeckel U. Genome-wide association study identifies single-nucleotide polymorphism in KCNB1 associated with left ventricular mass in humans: The HyperGEN Study. BMC Medical Genetics. 2009;10:43. doi: 10.1186/1471-2350-10-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newton-Cheh C, Eijgelsheim M, Rice K, Bakker Pd, Yin X, Estrada K, Bis J, Marciante K, Rivadeneira F, Noseworthy P, Sotoodehnia N, Smith N, Rotter J, Kors J, Witteman J, Hofman A, Heckbert S, O'Donnell C, Uitterlinden A, Psaty B, Lumley T, Larson M, Stricker B. Common variants at ten loci influence QT interval duration in the QTGEN Study. Nature Genetics. 2009;41:399–406. doi: 10.1038/ng.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pfeufer A, Sanna S, Arking D, Muller M, Gateva V, Fuchsberger C, Ehret G, Orru M, Pattaro C, Kottgen A, Perz S, Usala G, Barbalic M, Li M, Putz B, Scuteri A, Prineas R, Sinner M, Gieger C, Najjar S, Kao W, Muhleisen T, Dei M, Happle C, Mohlenkamp S, Crisponi L, Erbel R, Jockel K, Naitza S, Steinbeck G, Marroni F, Hicks A, Lakatta E, Muller-Myhsok B, Pramstaller P, Wichmann H, Schlessinger D, Boerwinkle E, Meitinger T, Uda M, Coresh J, Kaab S, Abecasis G, Chakravarti A. Common variants at ten loci modulate the QT interval duration in the QTSCD Study. Nature Genetics. 2009;41:407–414. doi: 10.1038/ng.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shah B, Hernandez A, Liang L, Al-Khatib S, Yancy C, Fonarow G, Peterson E. Hospital variation and characteristics of implantable cardioverter-defibrillator use in patients with heart failure. Data from the GWTG-HF (Get With The Guidelines - Heart Failure) Registry. Journal of the American College of Cardiology. 2009;53:416–422. doi: 10.1016/j.jacc.2008.09.045. [DOI] [PubMed] [Google Scholar]
- 26.Forman D, Cannon C, Hernandez A, Liang L, Yancy C, Fonarow G. Influence of age on the management of heart failure: findings from Get With the Guidelines - Heart Failure (GWTG-HF) American Heart Journal. 2009;157:1010–1017. doi: 10.1016/j.ahj.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 27.Schellenbaum G, Rea T, Heckbert S, Smith N, Lumley T, Roger V, Taylor H, Kitzman D, Levy D, Psaty B. Survival associated with two sets of diagnostic criteria for congestive heart failure. American Journal of Epidemiology. 2004;160:628–635. doi: 10.1093/aje/kwh268. [DOI] [PubMed] [Google Scholar]
- 28.Schellenbaum G, Heckbert S, Smith N, Rea T, Lumley T, Kitzman D, Roger V, Taylor H, Psaty B. Congestive heart failure incidence and prognosis: case identification using central adjudication versus hospital discharge diagnoses. Annals of Epidemiology. 2006;16:115–122. doi: 10.1016/j.annepidem.2005.02.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


