Abstract
Sulfur-containing aroma compounds are key contributors to the flavor of a diverse range of foods and beverages. The tropical fruit characters of Vitis vinifera L. cv. Sauvignon blanc wines are attributed to the presence of the aromatic thiols 3-mercaptohexan-1-ol (3MH), 3-mercaptohexan-1-ol-acetate, and 4-mercapto-4-methylpentan-2-one (4MMP). These volatile thiols are found in small amounts in grape juice and are formed from nonvolatile cysteinylated precursors during fermentation. In this study, we overexpressed a Saccharomyces cerevisiae gene, STR3, which led to an increase in 3MH release during fermentation of a V. vinifera L. cv. Sauvignon blanc juice. Characterization of the enzymatic properties of Str3p confirmed it to be a pyridoxal-5′-phosphate-dependent cystathionine β-lyase, and we demonstrated that this enzyme was able to cleave the cysteinylated precursors of 3MH and 4MMP to release the free thiols. These data provide direct evidence for a yeast enzyme able to release aromatic thiols in vitro that can be applied in the development of self-cloned yeast to enhance wine flavor.
INTRODUCTION
Aromatic thiols are potent aroma compounds, with a sensory perception threshold range in the parts per trillion. They are found in a wide range of foods (39), including animal products such as yoghurt and cheese, fruits, vegetables, tea, coffee, and alcoholic beverages (2, 4, 33, 35, 38). Of particular interest in wine fermentation are the aromatic volatile thiols, 4-mercapto-4-methylpentan-2-one (4MMP), 3-mercaptohexan-1-ol (3MH), and 3-mercaptohexyl acetate. These compounds impart flavors such as “grapefruit,” “passion fruit,” and “boxwood” and are major contributors to the varietal character of Vitis vinifera L. cv. Sauvignon blanc white wines (17). Cysteine-S-conjugated bound forms of these free aromatic thiols are present in grape juice and are transformed into flavor-active thiols during fermentation by yeast (30). The carbon-sulfur (CS) β-lyase activity that is necessary for transformation of cysteine-S-conjugated forms of 3MH and 4MMP into free thiols was first inferred from cell extracts of Eubacterium limosum and Allium sativum (34). Since then, the potential for enhanced thiol release in grape juice has been demonstrated by the constitutive expression of the Escherichia coli tnaA gene, a tryptophanase with strong CS β-lyase activity (29). Thus far, there is no direct evidence of such yeast-derived enzymatic activity releasing aromatic thiols under oenological conditions although some candidate genes have been suggested based on a gene deletion approach (15, 31). The release of aromatic thiols by other microorganisms has been linked to the activity of cystathionine β- and γ-lyases, for example, Lactobacillus casei and Lactobacillus lactis in cheese production (16, 20), Staphylococcus haemolyticus in the release of human body odor (37), and Streptococcus anginosus in mouth malodor (42).
Apart from its potential role in aromatic thiol release, cystathionine β-lyase (CBL; EC 4.4.1.8) is involved in the biosynthesis of methionine. CBLs catalyze the conversion of cystathionine into homocysteine in an α,β-elimination reaction, which in a later step is converted to methionine (32). This reaction is dependent on the cofactor, pyridoxal-5′-phosphate (PLP), a derivative of vitamin B6. Some bacterial CBLs have been extensively characterized since the methionine biosynthetic pathway is absent in mammals and, thus, is an important antibiotic target (8, 9). In contrast, the enzymatic characterization of eukaryotic CBLs is more limited, with the exception of two plant CBLs from Arabidopsis thaliana (23) and Spinacia oleracea chloroplasts (28), for which the A. thaliana crystal structure has been solved (5). In yeasts, the Schizosaccharomyces pombe STR3 gene product has been shown to have activity toward cystathionine (9). Such activity can be attributed to the Saccharomyces cerevisiae STR3 homologue, YGL184C, as a strain containing a null mutant was unable to grow on glutathione or cystathionine as a sole sulfur source (12).
In this study, we purified the S. cerevisiae gene product, Str3p, and confirmed its activity as a CBL. Furthermore, we provide direct evidence that a purified form of this yeast enzyme has activity toward cysteine-S-conjugated precursors of the aromatic thiols 3MH and 4MMP. When the STR3 gene is overexpressed in a commercial wine yeast used to ferment V. vinifera L. cv. Sauvignon blanc grape must, an increase in 3MH release is detected. These data provide the basis on which a self-cloning approach could be adopted to improve the sensory properties of white wine.
MATERIALS AND METHODS
Similarities to the primary protein sequence of Str3p were identified using LALIGN (http://www.ch.embnet.org/software/LALIGN_form.html).
Chemicals.
All reagents were of analytical grade and purchased either from Amresco or Sigma-Aldrich unless otherwise stated. KH2PO4, K2HPO4, KCl, and glycerol were obtained from Chem-Supply (Adelaide, Australia). 4MMP and S-4-(4-methylpentan-2-one)-l-cysteine (Cys-4MMP) were synthesized as described by Howell et al. (14), and [2H10]4MMP and 3MH were synthesized as described by Kotseridis et al. (18). The synthesis of [2H10]3MH is described in Pardon et al. (22). S-3-(Hexan-1-ol)-l-cysteine (Cys-3MH), as a mixture of diastereoisomers, was prepared using the procedure of Wakabayashi et al. (40).
Microbial strains, medium, and culture conditions.
Chemically competent E. coli DH5α and BL21(DE3) cells (New England BioLabs) were used for amplification of plasmid DNA and protein expression, respectively. Growth and selection were carried out in Luria-Bertani medium supplemented with 30 μg/ml kanamycin. Commercial wine yeast strain VIN 13 (Anchor Yeast, Cape Town, South Africa) was used as a host strain for expression of the STR3 gene cassette. The commercial wine yeast strain EC-1118 (Lallemand, Canada) was used as a source of genomic DNA.
Yeast strains were cultivated at 30°C in either a rich medium, YPD (containing 1% yeast extract, 2% peptone, and 2% glucose), or a synthetic dropout medium, SCD (containing 2% glucose and 0.67% yeast nitrogen base without amino acids [Difco, Detroit, MI]). For the selection of sulfometuron methyl (SMM)-resistant yeast transformants, SCD medium was supplemented with 50 μg/ml SMM (Dupont, Wilmington, DE) dissolved in N-N-dimethylformamide. Solid medium contained 2% agar (Difco).
DNA constructs.
Standard procedures for the isolation and manipulation of DNA were used as described in Ausubel et al. (1). The pDLG42-CSL1 plasmid (29) served as a template to amplify the E. coli tryptophanase (tnaA) gene using Phusion High-Fidelity DNA Polymerase (Finnzymes). For E. coli expression of proteins with a C-terminal six-histidine tag, we first cloned the tnaA gene into the pET-24(+) vector (Merck Biosciences) with the primers pET24-TnA-FWD (5′-TCTAGGATCCAAAATAAGGAGGAAAAACATATGAAGGATTATGTAATGGAAAACTTTAAAC-3′) and pET24-TnA-REV (5′-GTGCTCGAGAACTTCTTTCAGTTTTGCGGTGAAG-3′) using BamHI and XhoI restriction sites (underlined in both primers). The subsequent construct, designated pET-T, had an E. coli Shine Dalgarno sequence (in boldface) (24) to ensure efficient translation and an NdeI restriction site. The S. cerevisiae STR3 gene was amplified from genomic DNA from the wine yeast strain EC-1118 with primers pET24-STR3-FWD (5′-TCTACATATGCCGATCAAGAGATTAGATACA-3′) and pET24-STR3-REV (5′-GTGCTCGAGCAATTTCGAACTCTTAATATTCAATTCTGA-3′). The STR3 coding region (1,398 bp) was cloned using the NdeI and XhoI restriction sites (underlined in both primers) in the pET-T plasmid, thus yielding the pET-STR3 construct.
The pDLG42-PGK1 plasmid was constructed by cloning the 1.8-kb HindIII fragment released from the pHVXII plasmid containing the phosphoglycerate kinase I gene (PGK1) constitutive promoter (PGK1P) and terminator (PGK1T) cassettes, into the HindIII site of the yeast single-copy integrating plasmid pDLG42. This plasmid contains the ILV2 (SMR1-410) marker gene, which confers resistance to SMM. To clone STR3 into the pDLG42-pGK1 plasmid, the gene was amplified by PCR using pET-STR3 as a template and primers STR3-XhoI-FWD (5′-GACTCTCGAGATGCCGATCAAGAGATTAGATAC-3′), and STR3-XhoI-REV (5′-TAGCCTCGAGTTACAATTTCGAACTCTTAATATTC-3′), which were engineered to introduce an XhoI restriction site (underlined in both primers). The PCR product was cloned into the XhoI site of the pDLG42-PGK1 plasmid between the PGK1P and PGK1T sequences. The resulting plasmid, pDLG42-PGK1-STR3, was linearized with ApaI and transformed into VIN 13. Transformants were selected in SCD-SMM medium, genomic DNA was isolated, and the integration of the STR3 expression cassette into the genome was confirmed by PCR. This transformant was designated VIN 13 (STR3). The integrity of all expression constructs used in this study was confirmed by DNA sequencing using the Australian Genome Research Facility, Brisbane, Australia.
Protein expression and purification.
Transformants were grown in 250 ml of Luria-Bertani medium to the log phase (optical density at 600 nm [OD600] of 0.6 to 0.8) from an overnight culture supplemented with 30 μg/ml kanamycin. Expression of recombinant protein was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 18°C. After 16 h, the bacterial cells were harvested by centrifugation and washed twice with 20 mM Tris, pH 7.5, 130 mM KCl, 10% (wt/vol) glycerol, and 10 mM EDTA. About 1.5 g of cell pellets was resuspended in cell lysis buffer (50 mM Tris, pH 8.0, 100 mM KCl, 10% [wt/vol] glycerol, 0.25 mM Triton X-100, 10 mM imidazole, 100 μM PLP, and protease inhibitors 1 mM phenyl-methanesulfonyl fluoride and 5 mM ε-amino-n-caproic acid) and lysed by four sequential passes through a precooled cell disruptor (8,000 to 15,000 lb/in2) (EmulsiFlex-05 Homogenizer; Aventis). All subsequent steps were carried out at 4°C. The lysates were centrifuged at 45,000 × g for 1 h in a Sorvall SW32 rotor and filtered through a 0.22-μm-pore-size (Millipore) filter. The resulting cleared lysate was then exposed to nickel affinity chromatography using a 4-ml Ni-Sepharose 6 Fast Flow matrix according the manufacturer's instructions (GE Lifesciences). Protein was purified by washing with 5 column volumes of NiA buffer containing 20 mM Tris, pH 7.4, 250 mM KCl, 100 μM PLP, and 10 mM imidazole and a subsequent wash with 5 column volumes of NiA wash buffer containing 100 mM imidazole for Str3p and 70 mM for tryptophanase. Bound protein was then eluted in NiA buffer containing 500 mM imidazole for Str3p and 250 mM imidazole for tryptophanase. Protein was separated by electrophoresis on NuPAGE 10% Bis-Tris SDS-PAGE gels with morpholinepropanesulfonic acid (MOPS)-SDS running buffer (Invitrogen) and stained with Imperial stain (Thermo Scientific). The molecular weight of monomeric Str3p was determined from the Precision Plus molecular weight marker (Bio-Rad).
Dialysis, protein concentration, and estimation.
Purified proteins were dialyzed overnight at 4°C with continuous stirring in 1 liter of buffer A (20 mM Tris, pH 8.0, 500 mM KCl, 20 mM EDTA, 8% [wt/vol] glycerol, 100 μM PLP) for Str3p and with 20 mM potassium phosphate, pH 7.0, and 10 μM PLP for tryptophanase. Dialyzed enzymes were concentrated at 4°C with Vivaspin 15R 10,000-molecular-weight-cutoff (MWCO) concentrators (Sartorius Stedim Biotech) to 10 to 12 mg/ml. Protein concentration was determined using a Bio-Rad protein assay with a standard of bovine serum albumin. Purified Str3p was then snap frozen and kept at −80°C.
Size exclusion chromatography.
Size exclusion chromatography was carried out with a Superdex 200HR 10/30 analytical column (GE Lifesciences) using an ÄKTA Explorer 100 fast protein liquid chromatograph (FPLC; Pharmacia/GE Lifesciences). The flow rate was 0.3 ml/min. Prior to sample loading, columns were equilibrated with buffer A. The column was calibrated with proteins of known molecular weights (Sigma-Aldrich) to produce standard curves.
MALDI-TOF MS.
Twenty-five micrograms of purified Str3p was resolved by SDS-PAGE and subjected to a trypsin digestion and matrix-assisted laser desorption ionization–time of flight mass spectroscopy (MALDI-TOF MS) by the Australian Proteome Analysis Facility Ltd., Sydney, Australia. Data were submitted to the database search program Mascot (Matrix Science Ltd., London, United Kingdom).
Carbon-sulfur lyase assay.
Reactions were carried out in a total volume of 1 ml containing a final concentration of 2 μg/ml of Str3p, 50 mM phosphate buffer, pH 8.5, 20 μM PLP, 1 mM EDTA, and a 2 mM concentration of the sulfur-containing amino acid substrate. Reaction mixtures were incubated for 1 h at 37°C and kept frozen until assayed for CS lyase activity. For pH optimum tests, the reaction buffer contained 50 mM morpholineethanesulfonic acid (MES), 50 mM bis-Tris-propane (BTP), and 50 mM CAPS (N-cyclohexyl-3-aminopropanesulfonic acid), and, depending on the target pH (5.5 to 11), it was titrated with either KOH or HCl. Experiments were performed in triplicate.
The formation of the α-ketoacids pyruvate (indicator of β-lyase activity) and α-ketobutyrate (indicator of γ-lyase activity) was determined by high-performance liquid chromatography (HPLC) using an HPX-87H Aminex ion exchange column (Bio-Rad) and an Agilent Technologies 1200 series liquid chromatograph. The operating conditions were as follows: a flow rate of 0.5 ml/min, 65°C, and detection at 210 nm. The mobile phase was 5 mM H2SO4. Pyruvate and α-ketobutyrate (Sigma-Aldrich) were used as standards. For the negative control, a cleared lysate from BL21(DE3) cells transformed with an empty pET vector was treated in an identical way to cells containing pET-STR3. For kinetic analysis we used Ellman's reagent [5,5′-dithiobis-(2-nitrobenzoic acid)] to quantify free thiol groups with absorbance measured at 412 nm.
Michaelis-Menten kinetics.
Reactions were carried out as described above with 2 μg/ml purified Str3p. The data from three individual experiments were pooled and fitted to the Michaelis-Menten equation using PRISM (version 5) with R2 values of 0.91 and 0.94 for l-cystathionine and l-djenkolate, respectively. For calculation of the catalytic turnover and catalytic efficiency, we used the assumption that Str3p was purified to homogeneity.
Enzymatic reactions with Cys-4MMP and Cys-3MH.
Cleavage of Cys-4MMP and Cys-3MH was done in a total volume of 1.3 ml. E. coli tryptophanase was included as a positive control. For negative controls, reactions were performed with Str3p that was heat inactivated for 2 min at 95°C and using protein eluted from nickel affinity chromatography and extracted from induced E. coli cells transformed with an empty pET-24(+) vector. Indistinguishable concentrations of free thiol were detected in both negative controls. The conditions of the reactions were as follows: 31 μg/ml enzyme, 50 mM phosphate buffer, pH 7.0 or 7.5, 20 μM PLP, and 1 mM EDTA with 0.25 mM or 2 mM substrate, incubated at 28°C for 1 h and kept at 4°C until assayed by headspace gas chromatography (GC)/MS.
Wine fermentation.
YPD medium was inoculated with strains VIN 13, VIN 13 (STR3), and VIN 13 (CSL1). Yeast starter cultures were made in autoclaved grape juice and incubated for 48 h at 28°C to stationary phase. Frozen 2007 V. vinifera L. cv. Sauvignon blanc clarified juice (obtained from the Adelaide Hills, Australia) was thawed, thoroughly mixed, and filter sterilized using a VacuCap 60PF filter unit (0.8/0.2-μm pore diameter; Pall Life Sciences).
The basic chemical parameters of the juice were 198 g/liter sugar, 460 mg/liter yeast-assimilable nitrogen, and a pH of 3.2. A volume of 200 ml of the juice was transferred to 250-ml fermentation flasks with air locks, and the juice was inoculated at a density of 1 × 106 cells/ml from the starter cultures. The wines were fermented in triplicate at 18°C for 15 days and then cold stabilized at 4°C. The wines were then racked and kept in 100-ml glass reagent bottles at 4°C until analysis. The concentrations of sugars, ethanol, glycerol, acetic acid, malic acid, tartaric acid, and succinic acid were measured by HPLC using a Bio-Rad HPX-87H column as described above for quantitation of α-ketoacids. Low-molecular-weight sulfur compounds that are known off-odors were quantified by gas chromatography coupled with sulfur chemiluminescence detection (GC/SCD) (27).
Headspace GC/MS analysis.
An aliquot of 1 ml of the enzymatic reaction mixture (or diluted tryptophanase reaction mixture) was assayed in a total volume of 5 ml containing approximately 20 mg EDTA and 2 g NaCl in a 20-ml solid-phase microextraction vial with a magnetic crimp cap (Gerstel, Baltimore, MD). A solution containing a mixture of deuterated standards of [2H10]4MMP (9.88 μg/ml) and [2H10]3MH (14.32 μg/ml) in ethanol was added using a glass syringe (SGE; Grace Davison Discovery Sciences, Rowville, Victoria, Australia) to each of the samples. The instrumental conditions were as described in Swiegers et al. (29) with the following modification: the autosampler was fitted with an automated 2-cm divinylbenzene/carboxen/polydimethylsiloxane solid-phase microextraction fiber (Supelco, Bellefonte, PA). For quantification, mass spectra were recorded in the selective ion monitoring mode. The ions monitored were m/z 81, 96, 108, 141, and 142 for [2H10]4MMP; m/z 55, 75, 89, 99, and 132 for 4MMP; m/z 60, 62, 92, 109, and 144 for [2H10]3MH; and m/z 55, 82, 100, and 134 for 3MH. Selected fragment ions were monitored for 20 ms each. The underlined ion for each compound was the ion typically used for quantitation, having the best signal-to-noise ratio and the least interference from other components. The other ions were used as qualifiers. Analysis of 3MH at wine-like concentrations was carried out using a newly developed method (6).
RNA purification.
Approximately 1 × 107 yeast cells were harvested from fermentations by centrifugation, prepared in RNALater (Ambion), and kept at −80°C until analysis. The cells were washed by resuspension in 300 μl of ice-cold nuclease-free H2O and centrifuged at 1,500 × g at 4°C. The cells were then resuspended in 100 μl of Zymolyase buffer (50 mM Tris-HCl pH 7.5, 1 M sorbitol and 10 mM MgCl2) containing 30 mM dithiothreitol (DTT), and incubated for 15 min at room temperature to reduce any disulfide bonds. Zymolyase 20T from Arthrobacter luteus (MP Biomedicals, Aurora, OH) (20 units) was added to Zymolyase buffer containing 1 mM DTT and incubated at 30°C for 40 min.
RNA was isolated using a PureLink RNA mini-kit (Invitrogen) according to the manufacturer's instructions. Briefly, lysis was carried out by adding 200 μl of lysis buffer containing 1% β-mercaptoethanol, followed by centrifugation at 16,000 × g for 2 min. An equal volume of 99% ethanol was added to the supernatant, followed by thorough vortexing. The resulting lysate (500 μl) was transferred to a PureLink RNA mini-spin column, and RNA was isolated according to the manufacturer's instructions. Purified RNA was treated with 1 unit of amplification-grade DNase I (Invitrogen) for 15 min at room temperature. The reactions were stopped by the addition of 5 mM EDTA and incubation at 65°C for 5 min. A typical yield of 500 ng of total RNA was obtained as quantified by the QUBIT quantification platform using a Quant-iT RNA assay kit and standards (Invitrogen).
Reverse transcription.
cDNA was synthesized from 100 ng of total RNA using an oligo(dT)20 primer and an Affinity Script quantitative PCR (qPCR) cDNA synthesis kit (Stratagene, Agilent Technologies). All steps in cDNA synthesis were performed according to the manufacturer's instructions.
qPCR.
Quantitative PCR was performed using a Bio-Rad CFX96 real-time detection system with Brilliant II SYBR green reagent (Agilent Technologies) and cDNA made from 2.5 ng of total RNA in a volume of 25 μl. To quantify the transcript level of the STR3 gene, data were normalized using ACT1 as a reference transcript. Primers for STR3 (STR3.FWD, 5′-TCAAACCTACCAGAACAAACAAG-3′; STR3.REV, 5′-CGTCACAGCCCATATACTCAG-3′) and ACT1 (Act.FWD, 5′-GCCAAGATAGAACCACCAATCC-3′; Act.REV, 5′-CTGATGTCGATGTCCGTAAGG-3′) were validated with efficiencies of 99.4% and 100.6%, respectively. Threshold cycle (CT) values were obtained from duplicate fermentations, and STR3 expression was normalized against the actin reference gene by the 2−ΔΔCT method and expressed relative to the native promoter at day 5.
Nucleotide sequence accession number.
The sequence of the variant STR3 gene used in this study was submitted to GenBank under accession number HQ008776.
RESULTS
Analysis of the S. cerevisiae cystathionine β-lyase STR3 sequence.
The STR3 gene from the diploid commercial wine yeast, EC-1118, displayed three heterozygous allelic variants, G244A, A411G, and T633C, compared with the S288c haploid reference strain. Of these, only the G244A variant results in an amino acid change in the protein sequence (A82T). This variant is not present in the recently published whole genome shotgun sequence of the EC-1118 strain (21), assembled as a pseudo-haploid due to the low rate of heterozygosity observed in this strain. The STR3 gene used in our study contained both G244A and T633C variants (GenBank under accession number HQ008776). Based on its primary amino acid sequence, Str3p is highly conserved among other budding yeasts, with identities of 66% for Lachancea thermotolerans and 51% for Clavispora lusitaniae. It also displays 44% identity to the fission yeast S. pombe STR3 homologue, 40% identity to an A. thaliana CBL, and 29% identity to the E. coli CBL encoded by metC. The Str3p amino acid sequence diverges from that of E. coli tryptophanase; both belong to the large group of aspartate aminotransferase fold type I enzymes (25).
Purification of S. cerevisiae Str3p.
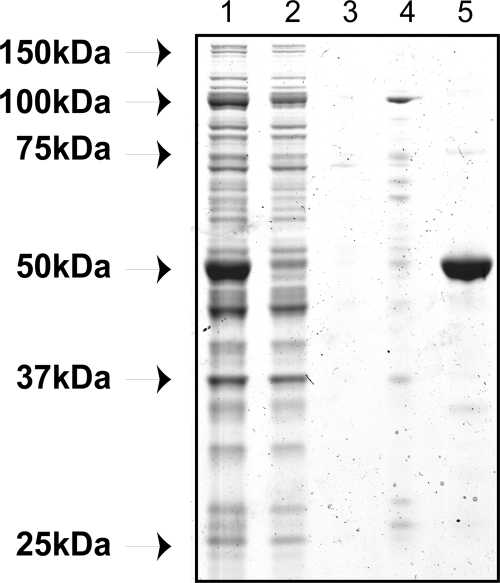
We expressed recombinant Str3p in E. coli and purified the protein in the presence of its cofactor, PLP, using Ni-nitrilotriacetic acid (NTA) chromatography to capture the C-terminal six-histidine-tagged protein (Fig. 1). Monomeric recombinant Str3p has a predicted molecular size of 53 kDa and migrated at approximately 52 kDa on an SDS-PAGE gel. A yield of 40 mg of pure protein per liter of culture was typically obtained. The buffer exchange conditions to stabilize the isolated protein and maintain its enzymatic activity included 500 mM KCl and 20 mM EDTA. By including glycerol in the buffer, less than 6% of the activity was lost in a freeze-thaw cycle, compared to the 56% loss in the presence of imidazole. MALDI-TOF MS was performed on purified Str3p, and its identity as the STR3-encoded protein was confirmed (E value of 7.9e−68).
Fig. 1.
SDS-polyacrylamide gel of nickel affinity-purified Str3p. Lane 1, cleared lysate; lane 2, flowthrough; lane 3, 10 mM imidazole wash; lane 4, 100 mM imidazole wash; lane 5, elution with 500 mM imidazole.
The size of native purified Str3p was investigated by size exclusion chromatography. A protein peak with CBL activity eluted at 13.2 ml, which corresponds to a molecular mass of 195 kDa (data not shown). This indicates that S. cerevisiae Str3p purified from E. coli forms a stable homotetramer, a finding consistent with other CBLs (7, 23, 28). PLP enzymes with cofactor bound to the active site show a characteristic absorbance at 420 to 435 nm (10, 41). We observed absorption at 428 nm together with the protein peak, suggesting that the protein was associated with PLP and purified as the holoenzyme.
Kinetic properties of purified Str3p.
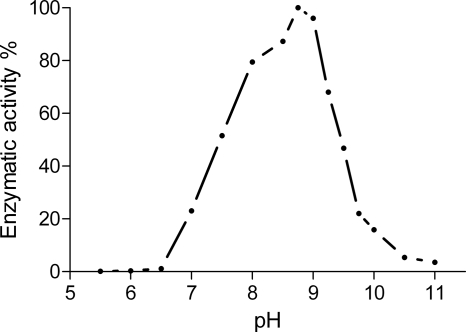
The effect of pH on recombinant Str3p activity was investigated using its physiological substrate, l-cystathionine. Enzyme activity was measured by the formation of pyruvate, one of the end products of the α,β-elimination (β-lyase) reaction. The recombinant enzyme displayed a bell-shaped pH rate profile, with an optimum activity at pH 8.75 (Fig. 2), and no significant activity was observed below pH 7 or above pH 10.5.
Fig. 2.
The activity of Str3p toward l-cystathionine was measured by detection of pyruvate by HPLC. The activity was normalized against a blank, and the result is expressed as a percentage of the maximal activity at pH 8.75. Data shown are the mean of three experiments. The standard deviation did not exceed 10% of any of the values.
Several sulfur-containing amino acids were assayed as potential substrates for the Str3p enzyme (Table 1). The nonprotein amino acid l-djenkolate was the most effective substrate for Str3p, preferred even above its physiological substrate, l-cystathionine. To a lesser extent, l-cystine, S-methyl-l-cysteine, and S-ethyl-l-cysteine acted as substrates, and a residual activity was also detected using l-cysteine as a substrate. No α-ketobutyrate was detected with any of the substrates susceptible for γ-lyase activity, confirming that Str3p has only β-lyase activity. Reactions involving recombinant Str3p with the two substrates l-cystathionine and l-djenkolate obeyed Michaelis-Menten kinetics. The catalytic turnover (Kcat) of Str3p was slightly higher for l-djenkolate than for l-cystathionine (1.27 versus 0.91 s−1), while a 2-fold binding preference for l-cystathionine compared to l-djenkolate was observed (Km of 96 versus 178 μM). Consequently, the catalytic efficiency, Kcat/Km, was higher for l-cystathionine than for l-djenkolate.
Table 1.
Substrate specificity of purified Str3p
| Substrate | Relative activity (%)a |
|---|---|
| l-Cystathionineb | 100 ± 10.9 |
| l-Djenkolate | 154 ± 11.0 |
| l-Cystine | 22.0 ± 2.1 |
| S-Ethyl-l-cysteine | 9.0 ± 0.7 |
| S-Methyl-l-cysteine | 7.4 ± 0.3 |
| l-Cysteine | 1.0 ± 0.3 |
| l-Methionine | 0.0 |
The formation of pyruvate was detected by HPLC and expressed relative to the activity with l-cystathionine ± standard deviations. Data are from triplicate experiments.
Specific activity toward l-cystathionine was 1,258 ± 138 μmol/min/mg of protein.
Enzymatic release of 3MH and 4MMP from their cysteine-S-conjugate precursors.
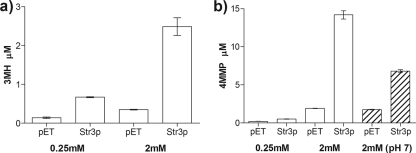
The volatile thiols 3MH and 4MMP are influential odorants for a wide range of wines (36). Since we have demonstrated that Str3p displays a broad specificity toward cysteine-S-conjugates, we asked whether Str3p would also release the aromatic thiols 3MH and 4MMP from their respective cysteinylated precursors, Cys-3MH and Cys-4MMP. S. cerevisiae Str3p was able to release 12.3 μM 4MMP and 2.1 μM 3MH from 2 mM concentrations of their respective precursors when reactions were conducted with 31 μg/ml purified enzyme (Fig. 3). This side activity of Str3p against Cys-4MMP and Cys-3MH corresponds to 1.3% and 0.2% of the E. coli tryptophanase activity, respectively, and approximately 0.6 to 0.1% of the specific activity toward its physiological substrate, l-cystathionine. In addition, a reaction at pH 7.0 released 47% of the 4MMP released by Str3p at pH 7.5 (Fig. 3B). This reduced activity at pH 7.0 is consistent with the pH rate profile of Str3p with l-cystathionine as a substrate (Fig. 2). The amount of 3MH and 4MMP formed was dependent on the concentration of precursor for both enzymes. Cys-3MH was, however, the most effective substrate at a concentration of 0.25 mM (0.54 versus 0.30 μM free 3MH and 4MMP, respectively). This led us to investigate the influence of Str3p in 3MH release under fermentation conditions.
Fig. 3.
GC/MS quantification of enzymatic reactions with purified Str3p. The release of 3MH and 4MMP was quantified with headspace GC/MS in reaction mixtures incubated with 0.25 mM or 2 mM cysteine-S-conjugated precursor. Experiments were carried out with 31 μg/ml purified Str3p at 28°C and at pH 7.5 to minimize hydrolysis of the 4MMP precursor (26). Data shown are the means of triplicate experiments ± standard deviations of Str3p reactions and empty vector controls (pET), which were significantly different (P < 0.01) for both substrates. An additional negative control, using heat-inactivated Str3p, was indistinguishable from the empty vector control (P = 0.225 for 3MH and P = 0.442 for 4MMP). We observed a strong correlation (R2 = 0.95) between thiol and pyruvate formation for both Cys-3MH and Cys-4MMP (data not shown). Hatched bars show results of an experiment carried out at pH 7.0.
Volatile thiol release during fermentation of a V. vinifera L. cv. Sauvignon blanc grape must.
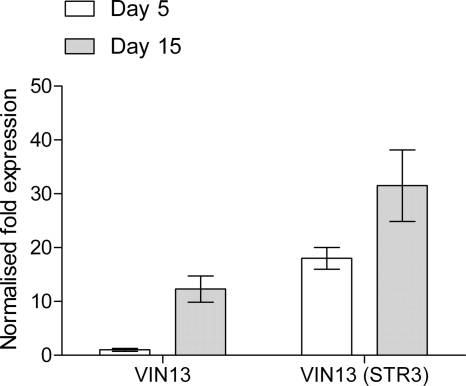
Since Str3p displayed a modest side activity toward the cysteinylated aroma precursors, we tested whether overexpression of the STR3 gene in the wine yeast VIN 13 could increase the release of the aromatic thiol 3MH under oenological conditions. The modified strain, VIN 13 (STR3), was used to ferment a V. vinifera L. cv. Sauvignon blanc grape juice, alongside the VIN 13 (CSL1) strain previously engineered to express the E. coli tryptophanase gene tnaA (29). The level of the STR3 mRNA transcript was monitored during fermentation by reverse transcription-quantitative PCR (RT-qPCR) at day 5 (when 60% of the sugar was consumed) and at day 15 (at the end of fermentation). In the VIN 13 control strain, the level of STR3 transcript under the control of its native promoter increased 12.3-fold between day 5 and day 15 of fermentation (Fig. 4). Compared with the control strain, STR3 transcript levels in the modified strain, VIN 13 (STR3), were 18-fold higher at day 5 but only 50% higher by the end of fermentation. The STR3 overexpression strain displayed sustained induction during the course of fermentation, whereas STR3 under the control of the native promoter was highly expressed toward the end of fermentation.
Fig. 4.
Quantitative RT-PCR of the STR3 mRNA level at day 5 and day 15 during fermentation with the commercial wine yeast, VIN 13, and a strain modified to overexpress STR3, VIN 13 (STR3). Data shown are the means of four data points from duplicate fermentations ± standard errors of the means.
All fermentations proceeded at similar rates and were dry (less than 2 g/liter sugar) after 15 days. Other chemical parameters (pH, glycerol, ethanol, and organic acid production) were analyzed after fermentation with the different strains and found to be almost identical (Table 2), including concentrations of other key low-molecular-weight volatile sulfur compounds known to adversely affect wine flavor.
Table 2.
Basic composition of the V. vinifera L. cv. Sauvignon blanc wines made using the modified VIN 13 (STR3), VIN 13 (CSL1), and the control VIN 13 strains
| Detection method and component | Amount of component in straina: |
||
|---|---|---|---|
| VIN 13 | VIN 13 (STR3) | VIN 13 (CSL1) | |
| HPLC | |||
| Alcohol (% [vol/vol]) | 11.5 ± 0.3 | 11.6 ± 0.2 | 11.4 ± 0.3 |
| Residual sugar (g/liter) | 0.1 ± 0.0 | 0.4 ± 0.2 | 0.0 ± 0.0 |
| Acetic acid (g/liter) | 0.05 ± 0.01 | 0.06 ± 0.01 | 0.03 ± 0.005 |
| Glycerol (g/liter) | 4.6 ± 0.13 | 4.4 ± 0.03 | 4.5 ± 0.31 |
| Malic acid (g/liter) | 2.6 ± 0.02 | 2.5 ± 0.01 | 2.5 ± 0.05 |
| Tartaric acid (g/liter) | 1.8 ± 0.01 | 1.8 ± 0.01 | 1.8 ± 0.12 |
| Succinic acid (g/liter) | 2.0 ± 0.07 | 2.0 ± 0.04 | 2.0 ± 0.14 |
| GC/SCD | |||
| Hydrogen sulfide (μg/liter) | 1.2 ± 0.2 | 1.2 ± 0.3 | 1.6 ± 1.1 |
| Methanethiol (μg/liter) | 4.1 ± 1.8 | 5.0 ± 1.1 | 4.6 ± 2.4 |
| Ethanethiol (μg/liter) | ND | ND | ND |
| Dimethyl sulfide (μg/liter) | 11.4 ± 6.0 | 10.7 ± 3.2 | 10.6 ± 5.7 |
| Carbon disulfide (μg/liter) | 0.3 ± 0.1 | 0.4 ± 0.1 | 0.5 ± 0.2 |
| Diethyl sulfide (μg/liter) | 0.3 ± 0.0 | 0.3 ± 0.0 | 0.2 ± 0.0 |
| Methyl thioacetate (μg/liter) | 3.2 ± 0.8 | 4.6 ± 2.6 | 2.9 ± 1.0 |
| Dimethyl disulfide (μg/liter) | ND | ND | ND |
| Ethyl thioacetate (μg/liter) | 0.3 ± 0.4 | 0.5 ± 0.6 | 0.5 ± 0.5 |
| Diethyl disulfide (μg/liter) | ND | ND | ND |
Data are from triplicate fermentations ± standard deviations. ND, not detected.
The VIN 13 (CSL1) strain released substantial amounts of 3MH, 9.5 times more than the control VIN 13 strain, consistent with the levels described previously in a synthetic medium spiked with the Cys-3MH precursor (29). The 3MH concentration after fermentation with the VIN 13 (STR3) strain was 27% (278 ng/liter) higher than in fermentation with the VIN 13 control strain (P = 0.014) (Table 3).
Table 3.
Production of 3MH in a V. vinifera L. cv. Sauvignon blanc grape must using the modified VIN 13 (STR3), VIN 13 (CSL1), and the control VIN 13 strains
| Strain | 3MH (ng/liter)a | Pb |
|---|---|---|
| VIN 13 | 1,084 ± 66 | |
| VIN 13 (STR3) | 1,362 ± 84 | 0.014 |
| VIN 13 (CSL1) | 10,268 ± 548 | 0.001 |
Data shown are the means of triplicate fermentations ± standard deviations quantified by headspace GC/MS.
Determined by a one-tailed student t test.
DISCUSSION
The biochemical properties of the S. cerevisiae STR3 gene product have not previously been characterized. Its function has been inferred by gene disruption, yielding a yeast strain that could not grow on cystathionine as the sole sulfur source (3, 12). In this study, we purified Str3p in order to determine some of its biochemical properties. We confirmed that Str3p is a CBL, with the highest catalytic efficiency for l-cystathionine. In accordance with previously characterized CBL enzymes, Str3p forms a stable tetrameric enzyme consisting of four PLP-bound subunits.
As observed for enzymes of this class from A. thaliana, S. oleracea, and E. coli, Str3p also displayed broad substrate specificity, including some activity toward the cysteine-S-conjugate substrates S-ethyl-l-cysteine and S-methyl-l-cysteine. The latter substrate, together with S-methyl-l-cysteine sulfoxide, occurs in high concentrations in Brassica and Allium vegetables. Characteristic flavors of these vegetables are partly derived through enzymatic degradation of these amino acids by CS lyases when their tissue is disrupted (11).
The in vitro incubation of purified Str3p with cysteine-S-conjugates of 3MH and 4MMP confirmed our hypothesis that the enzyme has a residual cysteine-S-conjugate β-lyase activity and was able to cleave these substrates to release the corresponding aromatic thiols. The reactions occurred in a concentration-dependent manner, and Str3p displayed a preference for Cys-3MH at low substrate concentrations and for Cys-4MMP at high concentrations. To our knowledge, this is the first direct evidence of a purified yeast enzyme displaying a CS β-lyase activity necessary to cleave the cysteine-S-conjugates of 4MMP and 3MH.
Previous studies based on gene disruption have identified several yeast genes (IRC7, CYS3, and BNA3) that may contribute to the release of 3MH and/or 4MMP (13, 15, 31). The contribution of each was, however, unclear since the effect of the deletions on thiol release was strongly dependent on the experimental conditions used, and some results were contradictory. Interestingly, IRC7, which was suggested to encode the main enzymatic activity involved in the release of 4MMP during fermentation (31), is annotated as a putative CBL based on sequence similarity. This activity has never been demonstrated experimentally; nonetheless, yeast CBLs, apart from their physiological function of cleaving cystathionine to yield homocysteine for methionine biosynthesis, could be harnessed to drive the formation of aromatic thiols during beverage fermentation. We therefore integrated an additional copy of the gene under the control of the constitutive promoter, PGK1P, in the commercial yeast strain VIN 13. Enhanced 3MH released by the VIN 13 (STR3) strain represents the first proof of concept that a yeast-derived gene can be used in place of the CSL1 construct (29) to harness latent flavor potential during wine fermentation. STR3 may not encode the optimal enzyme for this purpose since, in addition to its side activity against aromatic thiol precursors, the STR3 gene is among a group of genes transcriptionally upregulated during fermentation (19). Nonetheless, since the VIN 13 (STR3) strain was able to release 278 ng/liter more 3MH than a control strain and since 3MH has a sensory detection threshold of 60 ng/liter (33), this increase illustrates the potential for CBLs to modulate wine aroma. Although the results of the in vitro experiment indicate that enzymatic activity of Str3p is directly responsible for the increase in 3MH during wine fermentation, we cannot rule out that Str3p overexpression could affect expression of other genes in sulfur amino acid metabolism. However, overexpression of Str3p did not affect the concentrations of other low-molecular-weight volatile sulfur compounds known to adversely affect wine flavor.
In conclusion, we have demonstrated that a yeast enzyme, Str3p, is a CBL with side activity toward cysteine-S-conjugated thiols and that expression of STR3 can be manipulated in wine yeast to effectively alter the composition of volatile thiols in a wine fermentation. In vitro characterization of other yeast enzymes with putative cysteine-S-conjugate β-lyase activity, in conjunction with structural bioinformatics, represents a path forward to improve our understanding of volatile thiol release during fermentation and develop optimal self-cloned yeast strains for enhanced wine flavor.
ACKNOWLEDGMENTS
We thank the Department of Wine Innovation at Chr-Hansen A/S, Denmark, and Hentie Swiegers for financial support. Research at the Australian Wine Research Institute is supported by Australia's grape growers and winemakers through their investment body, The Grape and Wine Research Development Corporation, with matching funding from the Australian Government.
We also acknowledge Drew Sutton at Flinders University for assistance with size exclusion chromatography and Paul Chambers for critical reading of the manuscript.
Footnotes
Published ahead of print on 8 April 2011.
REFERENCES
- 1. Ausubel F. M., Brent R., Kingston R. E. (ed.). 1994. Current protocols in molecular biology. Wiley, New York, NY [Google Scholar]
- 2. Bailly S., Jerkovic V., Marchand-Brynaert J., Collin S. 2006. Aroma extraction dilution analysis of Sauternes wines. Key role of polyfunctional thiols. J. Agric. Food Chem. 54:7227–7234 [DOI] [PubMed] [Google Scholar]
- 3. Barreto L., Garcerá A., Jansson K., Sunnerhagen P., Herrero E. 2006. A peroxisomal glutathione transferase of Saccharomyces cerevisiae is functionally related to sulfur amino acid metabolism. Eukaryot. Cell 5:1748–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bouchilloux P., Darriet P., Henry R., Lavigne-Cruège V., Dubourdieu D. 1998. Identification of volatile and powerful odorous thiols in Bordeaux red wine varieties. J. Agric. Food Chem. 46:3095–3099 [Google Scholar]
- 5. Breitinger U., et al. 2001. The three-dimensional structure of cystathionine β-lyase from Arabidopsis and its substrate specificity. Plant Physiol. 126:631–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Capone D. L., Sefton M. A., Jeffery D. W. 1 April 2011, posting date Application of a modified method for 3-mercaptohexan-1-ol determination to investigate the relationship between free thiol and related conjugates in grape juice and wine. J. Agric. Food Chem. doi: 10.1021/jf200116q [DOI] [PubMed] [Google Scholar]
- 7. Clausen T., Huber R., Laber B., Pohlenz H. D., Messerschmidt A. 1996. Crystal structure of the pyridoxal-5′-phosphate dependent cystathionine β-lyase from Escherichia coli at 1.83 Å. J. Mol. Biol. 262:202–224 [DOI] [PubMed] [Google Scholar]
- 8. Ejim L. J., et al. 2007. Inhibitors of bacterial cystathionine β-lyase: leads for new antimicrobial agents and probes of enzyme structure and function. J. Med. Chem. 50:755–764 [DOI] [PubMed] [Google Scholar]
- 9. Ejim L. J., et al. 2004. Cystathionine β-lyase is important for virulence of Salmonella enterica serovar Typhimurium. Infect. Immun. 72:3310–3314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gentry-Weeks C. R., Keith J. M., Thompson J. 1993. Toxicity of Bordetella avium β-cystathionase toward MC3T3-E1 osteogenic cells. J. Biol. Chem. 268:7298–7314 [PubMed] [Google Scholar]
- 11. Hamamoto A., Mazelis M. 1986. The C-S Lyases of Higher Plants: Isolation and properties of homogeneous cystine lyase from Broccoli (Brassica oleracea var. botrytis) buds. Plant Physiol. 80:702–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hansen J., Johannesen P. F. 2000. Cysteine is essential for transcriptional regulation of the sulfur assimilation genes in Saccharomyces cerevisiae. Mol. Gen. Genet. 263:535–542 [DOI] [PubMed] [Google Scholar]
- 13. Harsch M. 2009. Identification of yeast genes involved in Sauvignon Blanc aroma development. Ph.D. thesis University of Auckland, Auckland, New Zealand [Google Scholar]
- 14. Howell K. S., et al. 2004. Variation in 4-mercapto-4-methyl-pentan-2-one release by Saccharomyces cerevisiae commercial wine strains. FEMS Microbiol. Lett. 240:125–129 [DOI] [PubMed] [Google Scholar]
- 15. Howell K. S., et al. 2005. Genetic determinants of volatile-thiol release by Saccharomyces cerevisiae during wine fermentation. Appl. Environ. Microbiol. 71:5420–5426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Irmler S., Raboud S., Beisert B., Rauhut D., Berthoud H. 2008. Cloning and characterization of two Lactobacillus casei genes encoding a cystathionine lyase. Appl. Environ. Microbiol. 74:99–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. King E. S., Osidacz P., Curtin C., Bastian S., Francis I. L. Assessing desirable levels of sensory properties in Sauvignon blanc wines—consumer preferences and contribution of key aroma compounds. Austr. J. Grape Wine Res., in press [Google Scholar]
- 18. Kotseridis Y., Ray J. L., Augier C., Baumes R. 2000. Quantitative determination of sulfur containing wine odorants at sub-ppb levels. 1. Synthesis of the deuterated analogues. J. Agric. Food Chem. 48:5819–5823 [DOI] [PubMed] [Google Scholar]
- 19. Marks V. D., et al. 2008. Dynamics of the yeast transcriptome during wine fermentation reveals a novel fermentation stress response. FEMS Yeast Res. 8:35–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Martínez-Cuesta M. C., et al. 2006. YtjE from Lactococcus lactis IL1403 is a C-S lyase with α,γ-elimination activity toward methionine. Appl. Environ. Microbiol. 72:4878–4884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Novo M., et al. 2009. Eukaryote-to-eukaryote gene transfer events revealed by the genome sequence of the wine yeast Saccharomyces cerevisiae EC1118. Proc. Natl. Acad. Sci. U. S. A. 106:16333–16338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pardon K. H., et al. 2008. Synthesis of the individual diastereomers of the cysteine conjugate of 3-mercaptohexanol (3-MH). J. Agric. Food Chem. 56:3758–3763 [DOI] [PubMed] [Google Scholar]
- 23. Ravanel S., Job D., Douce R. 1996. Purification and properties of cystathionine β-lyase from Arabidopsis thaliana overexpressed in Escherichia coli. Biochem. J. 320:383–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ringquist S., et al. 1992. Translation initiation in Escherichia coli: sequences within the ribosome-binding site. Mol. Microbiol. 6:1219–1229 [DOI] [PubMed] [Google Scholar]
- 25. Schneider G., Käck H., Lindqvist Y. 2000. The manifold of vitamin B6 dependent enzymes. Structure 8:R1–R6 [DOI] [PubMed] [Google Scholar]
- 26. Shinkaruk S., et al. 2008. Surprising structural lability of a cysteine-S-conjugate precursor of 4-methyl-4-sulfanylpentan-2-one, a varietal aroma in wine of Vitis vinifera L. cv. Sauvignon blanc. Chem. Biodivers. 5:793–810 [DOI] [PubMed] [Google Scholar]
- 27. Siebert E. T., Solomon M. R., Pollnitz A. P., Jeffery D. W. 2010. Selective determination of volatile sulfur compounds in wine by gas chromatography with sulphur chemiluminescence detection. J. Agric. Food Chem. 58:9454–9462 [DOI] [PubMed] [Google Scholar]
- 28. Staton A. L., Mazelis M. 1991. The C-S lyases of higher plants: homogeneous beta-cystathionase of spinach leaves. Arch. Biochem. Biophys. 290:46–50 [DOI] [PubMed] [Google Scholar]
- 29. Swiegers J. H., et al. 2007. Engineering volatile thiol release in Saccharomyces cerevisiae for improved wine aroma. Yeast 24:561–574 [DOI] [PubMed] [Google Scholar]
- 30. Swiegers J. H., et al. 2009. The influence of yeast on the aroma of Sauvignon blanc wine. Food Microbiol. 26:204–211 [DOI] [PubMed] [Google Scholar]
- 31. Thibon C., et al. 2008. Nitrogen catabolic repression controls the release of volatile thiols by Saccharomyces cerevisiae during wine fermentation. FEMS Yeast Res. 8:1076–1086 [DOI] [PubMed] [Google Scholar]
- 32. Thomas D., Surdin-Kerjan Y. 1997. Metabolism of sulfur amino acids in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 61:503–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tominaga T., Furrer A., Henry R., Dubourdieu D. 1998. Identification of new volatile thiols in the aroma of Vitis vinifera L. var. Sauvignon blanc wines. Flavour Fragrance J. 13:159–162 [Google Scholar]
- 34. Tominaga T., Peyrot des Gachons C., Dubourdieu D. 1998. A new type of flavor precursors in Vitis vinifera L. cv. Sauvignon blanc: S-cysteine conjugates. J. Agric. Food Chem. 46:5215–5219 [DOI] [PubMed] [Google Scholar]
- 35. Tominaga T., Guimbertau G., Dubourdieu D. 2003. Role of certain volatile thiols in the bouquet of aged champagne wines. J. Agric. Food Chem. 51:1016–1020 [DOI] [PubMed] [Google Scholar]
- 36. Tominaga T., Masneuf I., Dubourdieu D. 2004. Powerful aromatic volatile thiols in wines made from several Vitis vinifera grape varieties and their releasing mechanism. ACS Symp. Ser. Am Chem. Soc. 871:314–337 [Google Scholar]
- 37. Troccaz M., Benattia F., Borchard G., Clark A. J. 2008. Properties of recombinant Staphylococcus haemolyticus cystathionine β-lyase (metC) and its potential role in the generation of volatile thiols in axillary malodor. Chem. Biodivers. 5:2372–2385 [DOI] [PubMed] [Google Scholar]
- 38. Vermeulen C., Lejeune I., Tran T. T. H., Collin S. 2006. Occurrence of polyfunctional thiols in fresh lager beers. J. Agric. Food Chem. 54:5061–5068 [DOI] [PubMed] [Google Scholar]
- 39. Vermeulen C., Gijs L., Collin S. 2005. Sensorial contribution and formation pathways of thiols in foods: a review. Food Rev. Int. 21:69–145 [Google Scholar]
- 40. Wakabayashi H., Wakabayashi M., Eisenreich W., Engel K. 2004. Stereochemical course of the generation of 3-mercaptohexanal and 3-mercaptohexanol by β-lyase-catalyzed cleavage of cysteine conjugates. J. Agric. Food Chem. 52:110–116 [DOI] [PubMed] [Google Scholar]
- 41. Wu W., Morris D. R. 1973. Biosynthetic arginine decarboxylase from Escherichia coli. Subunit interactions and the role of magnesium ion. J. Biol. Chem. 248:1696–1699 [PubMed] [Google Scholar]
- 42. Yoshida Y., Negishi M., Amano A., Oho T., Nakano Y. 2003. Differences in the βC-S lyase activities of viridans group streptococci. Biochem. Biophys. Res. Commun. 300:55–60 [DOI] [PubMed] [Google Scholar]