Abstract
In eukaryotic cells, DNA replication is carried out by the coordinated action of three DNA polymerases (Pols), Pol α, δ, and ϵ. In this report, we describe the reconstitution of the human four-subunit Pol ϵ and characterization of its catalytic properties in comparison with Pol α and Pol δ. Human Pol ϵ holoenzyme is a monomeric complex containing stoichiometric subunit levels of p261/Pol 2, p59, p17, and p12. We show that the Pol ϵ p261 N-terminal catalytic domain is solely responsible for its ability to catalyze DNA synthesis. Importantly, human Pol (hPol) ϵ was found more processive than hPol δ in supporting proliferating cell nuclear antigen-dependent elongation of DNA chains, which is in keeping with proposed roles for hPol ϵ and hPol δ in the replication of leading and lagging strands, respectively. Furthermore, GINS, a component of the replicative helicase complex that is composed of Sld5, Psf1, Psf2, and Psf3, was shown to interact weakly with all three replicative DNA Pols (α, δ, and ϵ) and to markedly stimulate the activities of Pol α and Pol ϵ. In vivo studies indicated that siRNA-targeted depletion of hPol δ and/or hPol ϵ reduced cell cycle progression and the rate of fork progression. Under the conditions used, we noted that depletion of Pol ϵ had a more pronounced inhibitory effect on cellular DNA replication than depletion of Pol δ. We suggest that reduction in the level of Pol δ may be less deleterious because of its collision-and-release role in lagging strand synthesis.
Keywords: DNA, DNA Enzymes, DNA Polymerase, DNA Primase, DNA Replication, DNA Synthesis, siRNA
Introduction
In eukaryotes, DNA polymerases (Pols)3 α, δ, and ϵ jointly support DNA replication, and their overall roles in this process have been defined (1). The initiation of DNA replication is catalyzed by the Pol α-primase complex, composed of the Pol α complex (p180-p70) associated with the primase complex (p58-p48). Primase synthesizes short oligoribonucleotide primers (∼10–15 nt) on both DNA strands at replication origins that are elongated (to ∼30–40 nt) by Pol α. The resulting primed templates are recognized by the RFC-PCNA complex, which displaces the Pol α-primase complex and catalyzes PCNA loading onto both strands. PCNA acts as a sliding clamp and tethers Pol δ and Pol ϵ to primed templates and increases their processivity. Recent studies suggest that Pol ϵ catalyzes leading strand synthesis, whereas Pol δ supports lagging strand replication (2–5). This proposed asymmetric distribution was based on genetic observations showing that mutations of the 3′ → 5′ proofreading activities of Pol δ and Pol ϵ affected opposite DNA strands (2, 3). Moreover, budding yeast containing either the error-prone Pol δ or Pol ϵ accumulated mutations in the lagging or leading strand, respectively (3). In both budding and fission yeasts, lagging strand maturation appears to require Pol δ but not Pol ϵ (summarized in Ref. 6). Supporting evidence for the selective action of Pol δ and Pol ϵ on opposite DNA strands was derived from biochemical experiments with Xenopus extracts (4, 5). Collectively, these data support the conclusion that Pol δ is responsible for lagging strand synthesis, and Pol ϵ replicates the leading strand.
Although the mechanism contributing to the selective actions of Pol δ and Pol ϵ at the replication fork remains to be precisely defined, it is likely that factors within the replisome complex play a governing role in these processes. The initiation of eukaryotic replication is a stepwise process that is presently best understood in budding yeast (7). In this model system, replication is initiated by converting origins into prereplication complexes containing the ORC, Cdc6, Cdt1, and the Mcm2–7 complex. Prereplication complexes are activated at the G1-S phase of the cell cycle by the marked increase in the levels of the S phase cyclin-dependent kinases and the Dbf4-dependent protein kinase activities. Cyclin-dependent kinase-phosphorylated Sld2 and Sld3 mediate interactions with Dpb11, Cdc45, Pol ϵ, and GINS, which are required for the concomitant interaction of Cdc45 and GINS with the Dbf4-dependent protein kinase-activated Mcm2–7 complex. These transactions are poorly understood and probably require additional proteins (minimally including Mcm10 and Ctf4) (8–12). However, a prevailing thought is that these multifaceted protein-protein interactions result in the formation of the functional replicative helicase complex known as CMG, which contains Cdc45, Mcm2–7, and GINS, originally isolated from Drosophila (13). The CMG complex forms the core of a large macromolecular complex, the replisome progression complex (RPC), which includes a number of proteins that play roles in checkpoint regulation, nucleosome reorganization, cohesion, and DNA repair as well as replication (14). Importantly, the RPC translocates with replication forks, and this movement, as well as the integrity of the complex, is lost following the targeted degradation of a single subunit component of the CMG complex (15). To date, Pol α appears to be the only replicative polymerase associated stably with the RPC, and this association is dependent on Ctf4 (16). These findings suggest that the association of Pol δ and Pol ϵ with the RPC is likely to be weak and/or transient.
In budding yeast, it has been reported that Pol ϵ associates with the preinitiation complex prior to Pol α (17, 18). Furthermore, Pol ϵ is a component of a preloading complex (composed of Sld2, Dpb11, Pol ϵ, and GINS) (18), suggesting that it interacts with a number of critical components involved in the generation of the preinitiation complex, particularly the four-subunit GINS complex. The replication protein Dpb11 was discovered because of its ability to suppress mutations of the Pol ϵ subunit Dpb2 (p59); the Sld proteins (Sld2, Sld3, and Sld5 (a subunit of the GINS complex)) were discovered because they suppressed mutations in Dpb11 (19, 20). These findings suggest that in addition to its role as a Pol, Pol ϵ and its subunits may act as a scaffolding complex to help position other replication proteins prior to the initiation of replication.
In contrast to our understanding of the initiation of replication in budding yeast, information about this process in higher eukaryotes is less advanced. Recent reports, however, identified higher eukaryotic homologs of the budding yeast Sld2 (RecQL4) (21, 22) and Sld3 (Treslin) (23). Although the complete characterization of these proteins remains to be carried out, their action resembles that of their budding yeast homologs, suggesting that events leading to the DNA initiation may be similar in yeasts and higher eukaryotes.
In this report, we describe the reconstitution and characterization of the human four-subunit Pol ϵ complex (p261-p59-p17-p12) as well as a number of its subcomplexes using the baculovirus-insect cell system. Like the budding yeast homolog, the holocomplex of hPol ϵ is monomeric, as are all other Pol ϵ complexes examined. All Pol ϵ preparations required PCNA and RFC for DNA synthesis on a singly primed M13 template, and hPol ϵ appears to be more processive than hPol δ. In addition, both hPol ϵ and hPol δ are stimulated by the GINS complex. Collectively, the in vitro properties of hPol ϵ described here are in keeping with it acting as a processive polymerase capable of supporting leading strand replication. We also examined the role of Pol δ and Pol ϵ in cell cycle progression and fork movement by siRNA-targeted depletion. Reduction of either Pol δ or Pol ϵ by 90% caused cells to traverse the S phase more slowly than control cells. These depleted cells also exhibited reduced rates in DNA replication fork movement. Under the conditions used, Pol ϵ depletion had more pronounced inhibitory effects in DNA replication in comparison with depletion of Pol δ. These findings are consistent with the expectation that Pol ϵ plays a critical role in governing leading strand synthesis.
EXPERIMENTAL PROCEDURES
Plasmids, Enzymes, and Baculoviruses
The cDNAs expressing DNA Pol ϵ subunits p59 (GenBankTM accession number NM_002692), p17 (GenBankTM accession number BC003166), and p12 (GenBankTM accession number NM_019896) were PCR-amplified from a HeLa cDNA library and subcloned into pET28a (Novagen), pFastBac1 (Invitrogen; for baculovirus expression of untagged proteins), and pFastBacHtbFLAG. All cDNAs were sequenced to verify that no mutations were introduced during PCR and cloning. The baculoviruses were generated according to the manufacturer's instructions. The baculovirus expressing the large catalytic subunit, p261, was a generous gift from Dr. S. Linn (University of California, Berkeley, CA) (24). GINS subunits were cloned as previously described (25). hDNA Pol α-primase complex, hPol δ, hPCNA, and hRFC were prepared as indicated (26).
Purification of Human DNA Pol ϵ from Baculovirus-infected Insect Cells
Sf9 insect cells (1.15 × 109 cells, 500 ml) grown in suspension culture in Grace's medium supplemented with 10% FBS at 27 °C were infected with viruses expressing Pol ϵ subunits at multiplicities of infection of 2.5. Two days after infection, the cells were harvested by centrifugation at 600 × g for 10 min at 4 °C, washed with ice-cold PBS (yielding 9 ml of cells), and resuspended in 18 ml of Hypotonic Buffer (20 mm Hepes-NaOH, pH 7.5, 5 mm KCl, 1.5 mm MgCl2, 1 mm DTT, 1 mm phenylmethylsulfonyl fluoride (PMSF), and protease inhibitors (2 mg/ml each of aprotinin, leupeptin, and antipain and 0.1 mm benzamidine)). Following incubation on ice for 10 min, cells were lysed by Dounce homogenization (Pestle B, 20 strokes). The mixture was adjusted to 0.2 m NaCl and centrifuged at 43,000 × g for 45 min at 4 °C, and the supernatant (23 ml, 191 mg of protein) was dialyzed overnight at 4 °C in QD25 buffer (25 mm Hepes-NaOH, pH 7.5, 1 mm DTT, 1 mm EDTA, 0.1 mm EGTA, 10% glycerol, 0.01% Nonidet P-40, 25 mm NaCl, and 0.1 mm PMSF/protease inhibitors). The dialyzed material (16 ml, 155 mg of protein) was centrifuged at 43,000 × g for 10 min at 4 °C and applied to a 10-ml Q-Sepharose column (10 × 1.5 cm) pre-equilibrated with QD25 buffer. The column was washed with 10 ml of Q25 buffer, and bound proteins were eluted with 10 ml of QE300 buffer (25 mm Hepes-NaOH, pH 7.5, 1 mm DTT, 1 mm EDTA, 10% glycerol, 0.01% Nonidet P-40, 0.3 m NaCl, and protease inhibitors). Elution of bound Pol ϵ was monitored by Coomassie Blue staining, Western blotting, and DNA synthesis assays using poly(dA)4000-oligo(dT)12–18 as the primer-template (1 unit = 1 nmol of nucleotide incorporation in 30 min at 37 °C). FLAG-M2 beads (2.0 ml of 50% suspension; Sigma) were added to fractions containing Pol ϵ (50 ml, 8.82 mg of protein, 3740 units, and 424 units/mg protein) and incubated overnight at 4 °C with constant agitation. The beads were washed three times with 10 ml of FLAG buffer (25 mm Hepes-NaOH, pH 7.5, 10% glycerol, 0.15 m NaCl, 1 mm DTT, 0.01% Nonidet P-40, 1 mm EDTA, and protease inhibitors), and bound proteins were eluted with FLAG buffer supplemented with 1 mg/ml FLAG3 peptide, yielding 0.98 mg of protein (2.5 ml, 2795 units, 2852 units/mg protein). Aliquots of the FLAG peptide eluate (0.2 ml) were layered onto a 5-ml 15–40% glycerol gradient (25 mm Tris-HCl, pH 7.5, 1 mm EDTA, 0.01% Nonidet P-40, 0.4 m NaCl, 1 mm DTT, and protease inhibitors), and the mixture was centrifuged at 250,000 × g for 18 h at 4 °C. Fractions were collected from the bottom of the tube; proteins and Pol ϵ activity were monitored by 10% SDS-PAGE and the poly(dA)4000-oligo(dT)12–18 assay, respectively. The coincidental peak of protein and Pol activity from the glycerol gradient was pooled. If all of the FLAG-eluted material were subjected to the glycerol gradient sedimentation step, 0.43 mg of protein (specific activity, 7552 units/mg) would have been obtained. The fractions were aliquoted and stored at −80 °C. The hydrodynamic properties of the Pol ϵ complexes were determined as described previously (27).
Subcomplexes of Pol ϵ were expressed in Sf9 cells infected with viruses containing the indicated subunits and purified as described for the isolation of the holocomplex. After glycerol gradient centrifugation, protein yields of 0.67, 0.43, 0.30, 0.18, and 0.37 mg were obtained of the complexes p261-p59, p261-p12, p261-p17, p261FL, and p261N (aa 1–1305), respectively, from 0.5 liter of Sf9. The p59 subunit, which binds to the C terminus of p261, enhanced both the solubility and activity of the p261 subunit, as did truncation of p261 subunit to the p261N product.
Isolation of Saccharomyces cerevisiae Pol ϵ
The four-subunit S. cerevisiae Pol ϵ was isolated as described by Chilkova et al. (28). Plasmids expressing these subunits were generously supplied by Dr. E. Johansson (Umea University, Sweden).
In Vitro Transcription/Translation and Immunoprecipitation
Coupled in vitro transcription/translation (IVTT) reactions followed by antibody precipitations were carried out to determine subunit interactions and assembly of the holoenzyme. Following the manufacturer instructions for the Promega TNT Quick in vitro transcription/translation system, three pET plasmids (0.25 mg), each expressing a different Pol ϵ subunit, were incubated with 40 μl of TNT Quick rabbit reticulocyte lysate supplemented with 30 μCi of [35S]methionine and 1 μg of a purified FLAG-tagged DNA pol ϵ subunit that was not expressed by the pET plasmids. Reaction mixtures (50 μl) were incubated for 1.5 h at 30 °C followed by the addition of 50 units of DNase I (Roche Applied Science). After an additional 10-min incubation, reactions were divided into two 20-μl aliquots, and 30 μl of 50% FLAG-M2 beads were added to each aliquot. To one aliquot, 2.5 μl of 20 mg/ml FLAG3 peptide was added and labeled as “(+) peptide.” One hour after incubation at 4 °C with constant agitation, beads were washed three times with 0.5 ml of immunoprecipitation buffer (25 mm Tris-HCl, pH 7.5, 10% glycerol, 5 mm EDTA, 1 mm DTT, 0.15 m NaCl, and 0.2% Nonidet P-40) supplemented with 1% BSA and then twice with immunoprecipitation buffer alone; the beads were then resuspended in 1× SDS loading buffer and boiled for 5 min. Proteins were subjected to 10% SDS-PAGE followed by autoradiography to detect the radiolabeled Pol ϵ subunit adsorbed to the FLAG beads.
Purification of hGINS from Baculovirus-infected Insect Cells
A suspension culture of Sf9 cells (250 ml, 1.2 × 105 cells) was infected with viruses expressing hGINS subunits (Sld5, Psf2, Psf1, and Psf3). The Sld5 subunit was expressed with a FLAG tag. Two days after infection, cells were centrifuged at 600 × g at 4 °C for 10 min and washed with ice-cold PBS. Cells (5 ml) were resuspended in 20 ml of Hypotonic Buffer supplemented with 20 mm Hepes-NaOH, pH 8.0, incubated on ice for 10 min, and lysed by Dounce homogenization (Pestle B, 20 strokes). The lysate was adjusted to 0.5 m potassium glutamate and centrifuged at 43,000 × g for 45 min at 4 °C; the supernatant (27.5 ml; 227 mg of protein) was adjusted to 0.5% Nonidet P-40 and mixed with FLAG-M2 beads (1 ml, Sigma); and the mixture was incubated for 16 h at 4 °C with constant agitation. The beads were then washed with 14 ml of wash buffer (25 mm Hepes-KOH, pH 8.0, 10% glycerol, 0.5 m potassium glutamate, 0.5 mm DTT, 0.05% Nonidet P-40, PMSF, and protease inhibitors) for 15 min at 4 °C and then centrifuged at 600 × g for 2 min at 4 °C. The collected beads, following three washes with wash buffer, were eluted with wash buffer supplemented with 1 mg/ml FLAG3 peptide, yielding 2.3 mg of protein (2 ml). An aliquot (0.2 ml, 1.84 mg/ml) of the FLAG3 peptide-eluted material was layered onto 10 tubes containing a 5-ml 15–40% glycerol gradient (25 mm Tris-HCl, pH 7.5, 1 mm EDTA, 0.01% Nonidet P-40, 0.15 m potassium acetate, 1 mm DTT, and protease inhibitors) and centrifuged at 250,000 × g for 16 h at 4 °C. After centrifugation, 0.15-ml fractions were collected, and 20-μl samples were separated in 10% SDS-PAGE followed by Coomassie Blue staining. The stoichiometric GINS complex peaked in the aldolase region (150 kDa); these fractions were pooled (>90% homogeneous, 1.2 ml, 3.08 mg of total protein), aliquoted, and stored at −80 °C.
Pull-down Assays to Measure Interactions between GST-GINS and Pols
High Five insect cells (3 × 107 cells/T-150 flask) were grown as a monolayer in Grace's medium supplemented with 10% heat-inactivated FBS at 27 °C. Cells were infected with viruses expressing Pol α-primase subunits (or those of Pol δ or Pol ϵ) and the subunits of GINS at a multiplicity of infection of 2.5 for each virus. The Sld5 subunit of GINS contained a Precission (GE Healthcare) cleavable GST tag. Two days after infection, cells (0.2 ml) were collected, washed with ice-cold PBS, and resuspended in 1 ml of Hypotonic Buffer. The cells were incubated on ice for 10 min and lysed by Dounce homogenization (20 strokes). The sodium glutamate concentration of lysates was adjusted to 0.3 m, and extracts were centrifuged at 43,000 × g for 1 h at 4 °C. Nonidet P-40 was added to the supernatant to a final concentration of 1%. Lysates were divided into two identical aliquots (1 mg, 0.1 ml) and supplemented with DNase I (50 μg) and Benzonase (500 units) to completely digest contaminating DNA (as well as chromatin). Precission protease (10 units), which cleaved the GST tag from Sld5, was added to one aliquot and labeled “(−) control.” More than 95% of GST-Sld5 was digested by the protease (data not shown). Glutathione-Sepharose (50 μl; Amersham Biosciences) was added, and the mixtures were incubated at 4 °C for 16 h. The glutathione beads were centrifuged at 600 × g for 30 s at 4 °C, and the bound material was washed three times with 500 μl of wash buffer (25 mm Hepes-NaOH, pH 7.5, 10% glycerol, 0.3 m sodium glutamate, 0.5 mm DTT, 0.05% Nonidet P-40, 1 mm PMSF, and protease inhibitors). Bound proteins were eluted with 50 μl of 1× SDS loading buffer, and 20 μl of the eluted proteins were subjected 10% SDS-PAGE analyses. Proteins were visualized by Western blotting using specific antibodies to detect Pol and GINS subunits.
DNA Replication Assays
Poly(dA)-Oligo(dT) Assay
Reaction mixture (10 μl) containing 20 mm Tris-HCl, pH 7.5, 0.2 mm DTT, 100 μg/ml BSA, 50 μm [α32-P]dTTP (3400 cpm/pmol), 10 mm magnesium acetate, 2 mm ATP, 0.2 m sodium glutamate (not added with Pol δ), 6 nm RFC, 100 nm hPCNA, 350 nm Escherichia coli SSB, 25 μm (as nucleotides) poly(dA)4000-oligo(dT)12–18 (20:1), and variable levels of Pols were incubated for 20 min at 37 °C, and nucleotide incorporation was measured.
Singly Primed M13 Assay
Reaction mixtures (10 μl) contained 20 mm Tris-HCl, pH 7.5, 150 μg/ml BSA, 1 mm DTT, 10 mm magnesium acetate, 2 mm ATP, 20 μm [α-32P]dATP (1–2 × 104 cpm/pmol), 120 μm each of dGTP, dCTP, and dTTP, 10 nm hRFC, 100 nm PCNA, and a level of Pols as indicated. Following 20 min at 37 °C, aliquots were used to measure nucleotide incorporation and the size of DNA products following alkaline-agarose electrophoresis and autoradiography (or phosphorimaging).
RNA Interference Protocol, Flow Cytometry, and DNA Fiber Analysis
HeLa cells (ATCC) were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum at 37 °C and 5% CO2. Duplex small interfering RNAs (siRNAs) (21 bp) with 3′-dU overhangs targeting hPol ϵ (p261 subunit), hPol δ (p125 subunit), and a random sequence (AAT TCT CCG AAC GTG TCA CGT), which acted as a negative control, were synthesized by Dharmacon (Boulder, CO). siRNA sequences targeting hPol ϵ were GCG AGG AAC AGG CGA AAU A (siRNA ϵ#05), GGA GGA GGG UGC UUC GUA U (siRNA ϵ#06), GGA CAG GCG UUA CGA GUU C (siRNA ϵ#07), and CUC GGA AGC UGG AAG AUU A (siRNA ϵ#08); siRNA sequences targeting hPol δ were AGU UGG AGA UUG ACC AUU A (siRNA δ#06), CGA GAG AGC AUG UUU GGG U (siRNA δ#07), GCA AAG GCA UCU UCC CUG A (siRNA δ#08), and GCA CAG AAA CUG GGC CUG A (siRNA δ#09). Transfections were carried out as described previously (26). siRNAs were introduced into cells at a final concentration of 50 nm. Flow cytometry and DNA fiber analyses 48 h post-transfection were as described (26). For synchronization, cells were treated with 2 mm thymidine for 16 h and then washed in PBS, grown in fresh medium for 9 h, and incubated for an additional 16 h in 2 mm thymidine. siRNAs were added before the first thymidine arrest. Cells were released after the second arrest into complete medium containing 40 ng/ml nocodazole for the length of time indicated.
RESULTS
Reconstitution of the Four-subunit hPol ϵ Complex
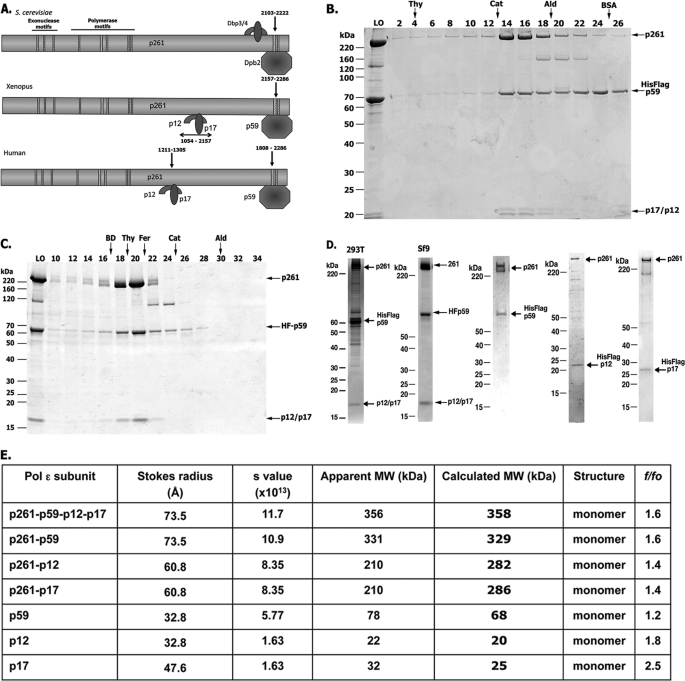
Pol ϵ is essential for replication and cell growth. It is highly conserved in yeast, Xenopus, and humans and contains a large catalytic subunit and three small subunits. The large catalytic subunit, Pol 2, is divided into two regions, the N-terminal catalytic (aa 1–1305) and the C-terminal non-catalytic domains (aa 1306–2286). The N-terminal catalytic region includes the conserved Pol and exonuclease motifs, whereas the C-terminal non-catalytic region contains binding sites for the small subunits (Fig. 1A). Whereas the binding site of Dpb2 maps to the C-terminal end of Pol 2 (29, 30), the sites at which the Dpb3 and Dpb4 subunits bind to Pol 2 differ among species. In yeast, Dpb3-Dpb4 was reported to bind near the C-terminal end of Pol 2 (29), whereas the Xenopus homologs bind to the middle portion, distal from the C terminus (30). In order to examine the biochemical properties of hPol ϵ, we reconstituted the polymerase complexes by co-expressing various combinations of Pol ϵ subunits or truncated forms of p261 in High Five insect cells. The purified complexes exhibited >90% homogeneity, as judged by glycerol gradient sedimentation (Fig. 1B) and Sepharose 6 sizing chromatography (Fig. 1C). The peak protein fractions of Pol ϵ contained stoichiometric levels of the four subunits and were active in supporting DNA synthesis (data not shown). Analysis of the hydrodynamic properties of the hPol ϵ complex showed that it is monomeric and has a moderately elongated shape (f/fo = 1.56) (Fig. 1E). These findings are consistent with those reported for Pol ϵ isolated from yeast (28) and Xenopus (30). In addition to the holocomplex, we also isolated subcomplexes of human Pol 2 (p261) that lacked one or all of the other subunits. Although the FLAG-tagged hp261 subunit was expressed and purified from insect cells, its yield and activity were low (compared with other Pol ϵ complexes), the protein tended to aggregate (shown by its distribution following glycerol gradient sedimentation), and its polymerase activity was unstable to freezing and thawing. The association of the p59 (Dpb2 subunit) stabilized the polymerase activity associated with the complex and increased the protein yield. The p261-p59 dimeric complex contained stoichiometric levels of each subunit and was detected as a single peak following size exclusion chromatography and glycerol gradient sedimentation (Fig. 1D, lane 3). Other subcomplexes isolated included p12 (Dpb4)-p261, p17 (Dpb3)-p261, and p12-p17-p261 (devoid of p59) (Fig. 1D, lanes 4 and 5) (data not shown). The hydrodynamic properties of these subcomplexes indicated that they were all monomeric and contained stoichiometric levels of the indicated subunits (Fig. 1E). The smaller subunits, p17 and p12, had f/fo ratios close to 2 (Fig. 1E), suggesting extended conformations.
FIGURE 1.
Structure and purification of hPol ϵ. A, comparison of the motifs present in the large catalytic subunit and the interaction sites of the small subunits (yeast, Xenopus, and human). The hatched rectangles indicate the conserved exonuclease and polymerase motifs located at the N-terminal half of the catalytic subunit. B, glycerol gradient sedimentation of the four-subunit hPol ϵ. FLAG-immunoprecipitated hPol ϵ (270 μg) was sedimented through 5-ml 15–35% glycerol gradients for 18 h at 250,000 × g at 4 °C. Collected fractions were subjected to 4–20% polyacrylamide gradient-SDS gel electrophoresis and Coomassie staining. LO, material loaded onto the gradient (2 μg of protein). The size markers are indicated. C, elution profile of FLAG-purified hPol ϵ following Superose 6 chromatography. hPol ϵ (270 μg) was applied to a Superose 6 10/300 GL column equilibrated with 25 mm HEPES-NaOH, pH 7.5, 10% glycerol, 0.15 m NaCl, 1 mm DTT, 1 mm EDTA, 0.05% Nonidet P-40, and protease inhibitors. The column was developed with this buffer at a rate of 250 μl/min at 4 °C, and fractions (0.5 ml) were collected. Aliquots (10 μl) were subjected to 4–20% polyacrylamide gradient-SDS gel and Coomassie staining. D, SDS-PAGE analysis of four-subunit hPol ϵ and various subcomplexes. hPol ϵ, purified as described under “Experimental Procedures,” was subjected to 4–20% polyacrylamide gradient-SDS gel electrophoresis followed by Coomassie staining. The purified proteins and amount of protein analyzed were as follows: FLAG-p59·p261·p17·p12 (3 μg, isolated from 293T cells stably expressing FLAG-p59) (lane 1), FLAG-p59·p261·p17·p12 (2 μg) (lane 2), FLAG-p59·p261 (1.75 μg) (lane 3), FLAG-p12·p261 (0.5 μg) (lane 4), and FLAG-p17·p261 (0.5 μg) (lane 5). The positions of various hPol ϵ subunits are indicated. E, complexes, subcomplexes, and subunits indicated were subjected to glycerol gradient sedimentation (to evaluate their s values) and Superose 6 (or Superdex 200) gel filtration (to obtain their Stokes radii) (Å). Their apparent molecular weight was estimated according to Siegel and Monty (27).
Mapping of p59, p17, and p12 Binding Sites on p261
The Pol ϵ p261 large subunit interacts directly with each of the three subunits (p59, p17, and p12) as evidenced by formation of subcomplexes (p261-p59, p261-p12, and p261-p17) in infected insect cells (Fig. 1D, lanes 3–5). To map the site where the p59 subunit bound, N- and C-terminal truncated derivatives of p261 were expressed by IVTT in the presence of the purified FLAG-p59 subunit. The mixture was then immunoprecipitated with FLAG antibody, the bound material was subjected to SDS-PAGE analysis, and 35S-labeled p261 derivatives were detected by autoradiography (Fig. 2A). The smallest p261 derivative that interacted with the FLAG-p59 subunit was C10 (aa 1747–2286); removal of the C-terminal 347 aa from p261 (N12) resulted in the loss of the p261-p59 interaction. The zinc finger motif of human Pol 2 is composed of four cysteines that are coordinated with a zinc ion (aa 2158–2238). To determine the zinc finger motifs required for the p59-p261 interaction, we mutated the first two cysteines in one of the two zinc finger motifs (ZF1 or ZF2) or both cysteines (ZF1 + ZF2). The wild type, ZF1, ZF2, or ZF1 + ZF2 derivative of p261-C was co-expressed with T7-tagged p59 in an IVTT reaction mixture, followed by immunoprecipitation with T7-specific antibodies (Fig. 2B). None of the zinc finger mutants interacted with T7-p59, whereas the wild-type p261-C did. We conclude that the integrity of p261 zinc finger motifs 1 and 2 is required for the p261-p59 interaction, thus confirming the observations made with yeast (29) and Xenopus Pol ϵ (30).
FIGURE 2.
Mapping of the p59 interaction site on p261. A, schematic diagram of truncated human Pol 2 mutant constructs used to map the p59 interaction site on the p261 subunit. Truncated p261 derivatives used in this study were N2 (aa 1–302), N6 (aa 1–899), p261-N (aa 1–1305), N12 (aa 1–1939), C10 (aa 1747–2286), C7 (aa 1211–2286), and p261-C (aa 1196–2286). All derivatives were expressed using IVTT reactions carried out in the presence of 1 μg of purified FLAG-tagged p59 and 35S-labeled methionine. After incubation, FLAG-p59 was immunoprecipitated (IP) with FLAG antibody coupled to agarose beads that were washed three times with FLAG buffer (25 mm Tris-HCl, pH 7.5, 10% glycerol, 5 mm EDTA, 1 mm DTT, 0.15 m NaCl, and 0.2% Nonidet P-40). Radiolabeled p261 derivatives bound by FLAG-tagged p59 were eluted in SDS sample loading buffer and subjected to 10% SDS-PAGE followed by autoradiography. The FLAG immunoprecipitations were carried out either in the presence (+F) or absence (−F) of 1 mg/ml FLAG peptide. B, diagram of the two conserved zinc finger motifs present at the C terminus of p261 (top). p261-C containing the double cysteine to alanine mutations in either of the zinc finger motifs ZF1m (C2158A/C2161A) or ZF2m (C2221A/C2224A) and the four cysteine mutations (ZF1m + ZF2m), were co-expressed with T7-tagged p59 in IVTT reactions followed by T7 immunoprecipitation, SDS-PAGE, and autoradiography. The p261-C subunit contained the wild-type zinc finger motifs. PI lanes, immunoprecipitations were carried out with control IgG; T7 lanes, immunoprecipitations were carried out with T7-specific antibody. C, mapping of the p12-p17 interaction site on the p261 subunit. Wild-type and truncated p261 derivatives were expressed in IVTT reactions in the presence of 1 mg of purified FLAG-p12·p17 complex. The p261 bound by FLAG-p12·p17 was FLAG-immunoprecipitated as described in A.
Cryoelectron microscopy suggested that the three small subunits (p59/Dpb2, p12/Dbp3, and p17/Dpb4) form a complex at the C terminus of Pol 2 that enhanced the binding of Pol ϵ to the DNA template (31). In contrast, experiments in Xenopus demonstrated that Dbp3 + Dbp4 and Dpb2 have distinct binding sites in Pol 2 (30). To map the binding site(s) of the hp12 and hp17 subunits on p261, various N- and C-terminal truncated p261 derivatives were expressed using IVTT in the presence of purified FLAG-p12 and/or p17 subunits, followed by immunoprecipitation with FLAG antibody (Fig. 2C). The FLAG-p12·p17 complex interacted with p261-N and C7 derivatives, indicating that the p12-p17 binding site on p261 maps between aa 1211 and 1305. To verify that aa 1211–1305 is the region to which p12 and/or p17 bind, we expressed a p261 derivative (p261 (Δp12-p17)) using IVTT in the presence of purified FLAG-p12·p17 heterodimer. FLAG-p12·p17 interactions with 35S-labeled p261-FL and p261-N (both containing aa 1121–1305) were detected, whereas interactions between p261 (Δp12-p17) or N6, which lacks aa 1121–1305, were not observed. These findings indicate that aa 1121–1305 of p261 are required to support the binding of p12-p17 heterodimer.
We also examined whether the p12, p17, and p59 subunits formed complexes in the absence of p261. Expression of p59, p12, and p17 using IVTT followed by immunoprecipitation with specific antibodies revealed stable interactions between p12 and p17, but neither of these subunits interacted with p59 (data not shown).
Catalytic Activity of Pol ϵ Preparations
All hPol ϵ derivatives containing the polymerase domains supported DNA-dependent dNMP incorporation. As summarized in Table 1, the turnover rate, using poly(dA)4000-oligo(dT)12–18 as the template-primer, varied between 50 and 90 pmol of nucleotide at 37 °C. Comparison of hPol δ and hPol ϵ revealed that Pol δ was 2.4-fold more active than Pol ϵ; under the same conditions, the four-subunit S. cerevisiae Pol ϵ was ∼10-fold more active than the human homolog. In the presence of the poly(dA)4000-oligo(dT)12–18 template-primer, Pol ϵ preparations (including the yeast) were stimulated ∼6-fold by RFC and PCNA (data not shown). These findings suggest that under the conditions described, the Pol ϵ large subunit is solely responsible for the catalytic activity of the holoenzyme. The activity of the four-subunit hPol ϵ, stored as glycerol gradient fractions at −80 °C, was stable for at least 1 year following repeated freezing and thawing. The p261-FL preparation, however, was relatively unstable, and its activity was reduced (>50%) following freezing and thawing (2–3 times).
TABLE 1.
Specific activity of various Pol preparations
Reaction mixtures (10 μl) contained 20 mm Tris-HCl, pH 7.5, 0.2 mm DTT, 100 μg/ml BSA, 50 μm [α−32P]dTTP (3400 cpm/pmol), 2 mm ATP, 0.2 m sodium glutamate (not added with Pol δ), 6 nm hRFC, 100 nm hPCNA, 350 nm E. coli SSB, 25 μm (as nucleotides), poly(dA)4000-oligo(dT)12–18 (20:1). Variable levels of Pols were incubated for 20 min at 37 °C, and nucleotide incorporation was measured.
| Protein preparation | Specific activity | Turnovera |
|---|---|---|
| μmol/h/mg protein | pmol incorporated/min/pmol protein | |
| S. cerevisiae Pol ϵ | 120 | 690 |
| hPol δ | 51 | 212 |
| p261N | 29 | 48.3 |
| p261-FL | 14 | 60.1 |
| p261-p59 | 12 | 88.5 |
| 4S hPol ϵ (four-subunit) | 10 | 72 |
| p261-p12-p17 | 14 | 68 |
a Turnover refers to pmol of nucleotide incorporated/min/pmol of protein. S. cerevisiae Pol ϵ assays were carried out using S. cerevisiae RFC/S. cerevisiae PCNA in place of the human homologs.
Role of PCNA and RFC
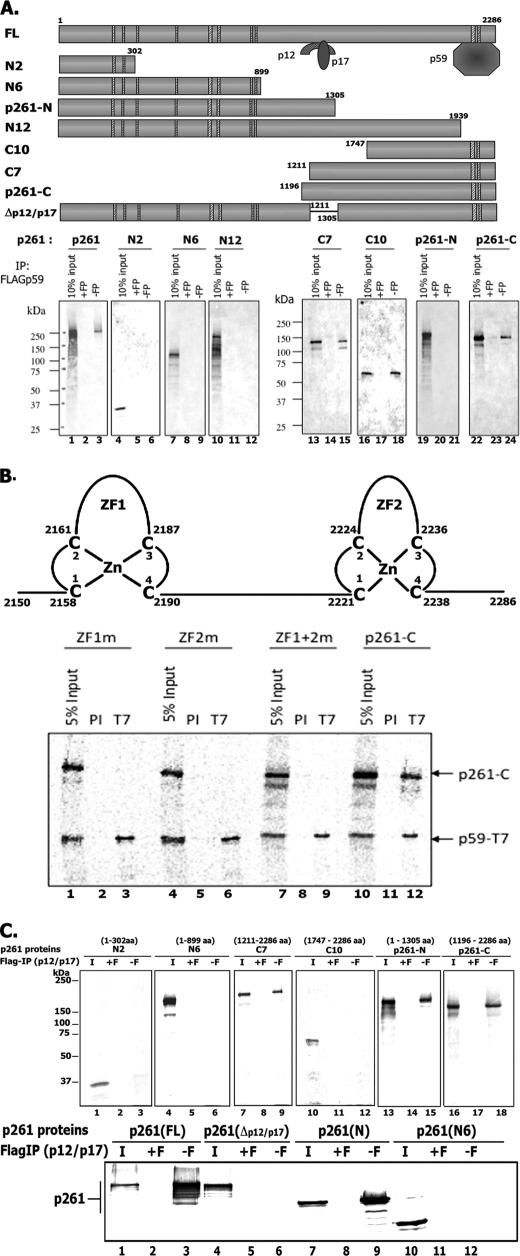
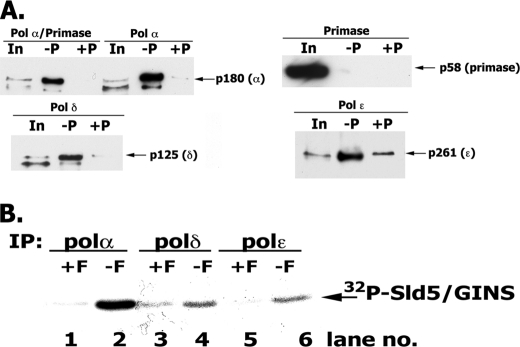
All hPol ϵ preparations, including the four-subunit holoenzyme, p261N, p261, and p261-p59, catalyzed DNA synthesis on singly primed M13, and this activity required RFC and PCNA (Fig. 3A). The amounts of PCNA required to support DNA synthesis with the four-subunit Pol ϵ and p261N preparations were almost identical (KD ∼2.4 and 3.5 nm, respectively) and similar to that observed with Pol δ (Fig. 3B). Direct PCNA interactions with hPol ϵ, the p261N derivative, as well as the p59 and p12 subunits were observed. However, efforts to detect a PCNA interaction site within the catalytically active p261N derivative required for DNA synthesis failed (supplemental Fig. 1). Because the levels of PCNA required to activate the p261-N derivative and the holocomplex were identical, it is likely that the PCNA binding sites present in the p59 and p12 subunits do not contribute significantly to the activation of the four-subunit complex. In the presence of excess PCNA, relatively low RFC levels were required for DNA synthesis with either Pol ϵ (four-subunit) or Pol δ (Table 2).
FIGURE 3.
Influence of hPCNA and hRFC on various hPol ϵ preparations. A, influence of PCNA. Reaction mixtures (10 μl) contained 20 mm Tris-HCl, pH 7.5, 0.2 mm DTT, 200 μg/μl BSA, 10 mm magnesium acetate, 35 μm [α-32P]dATP (18,280 cpm/pmol), 2 mm ATP, 130 μm each of dCTP, dGTP, and dTTP, 0.2 m sodium glutamate, 1 nm singly primed M13, 350 nm E. coli SSB, 10 nm p261-N, 10.3 nm p261-FL, 11.2 nm p261 p59 complex or 11 nm four-subunit hPol ϵ (where indicated), 6 nm hRFC, and 100 nm PCNA. Mixtures were incubated for 30 min at 37 °C, and aliquots were used to measure nucleotide incorporation and size of DNA products following 1% alkaline-agarose gel separation and autoradiography. B, influence of PCNA levels on DNA synthesis using singly primed M13 DNA catalyzed by hPol δ, hPol ϵ, and p261-N. Reaction mixtures were as described in A (except that the level of glutamate was reduced to 30 mm) with the indicated levels of PCNA and either the four-subunit Pol ϵ, p261-N, or hPol δ preparation.
TABLE 2.
Influence of hRFC on hPol δ and hPol ϵ
Reaction mixtures were as described in the legend to Fig. 3A with 50 fmol of hPCNA and 50 fmol of the four-subunit Pol ϵ and 80 fmol of Pol δ where indicated.
| RFC added | hPol ϵ | hPol δ |
|---|---|---|
| nm | pmol nt incorporated/20 min | pmol nt incorporated/20 min |
| 5.6 | 23.7 | 26.7 |
| 0.56 | 13.4 | 11.2 |
| 0.11 | 3.2 | 3.36 |
| None | 0.47 | 0.58 |
Comparison of the Processivity of DNA Synthesis Catalyzed by hPol ϵ and hPol δ
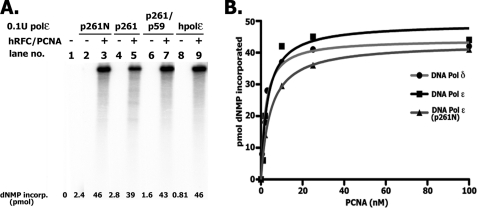
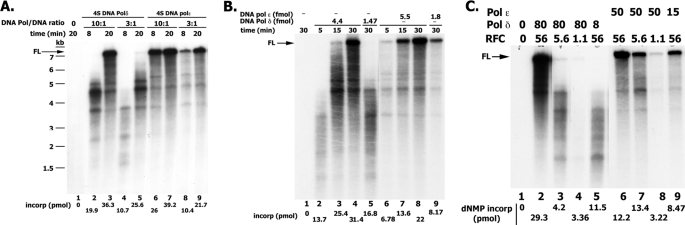
When the rate of DNA synthesis catalyzed by hPol ϵ in the presence of singly primed M13 and RFC/PCNA was examined, the level of nucleotide incorporation increased with increasing levels of Pol ϵ, as expected. However, the size distribution of products formed was similar to that observed at low and high levels of enzyme. Such properties are characteristic of a processive polymerase and differ from those expected for a distributive enzyme. These findings, as well as side-by-side experiments carried out with Pol ϵ and Pol δ are summarized in Fig. 4. As shown in Fig. 4A, variations in the amount of Pol δ added to a fixed level of singly primed M13 DNA altered the length and amount of the DNA products formed. In the presence of hPol δ at molar ratios to DNA of ∼3 and 10, the sizes of DNA products formed were ∼3.0 and 4.5 kb after 8 min of incubation (compare lanes 4 and 2), which increased to 5–6 kb and full length (∼7 kb) after 20 min at 37 °C (compare lanes 5 and 3), respectively. At these molar ratios of Pol ϵ/DNA, full-length DNA products were detected at all times (lanes 6–9) regardless of the ratio of enzyme to DNA and the amount of nucleotide incorporated. We examined whether these differences depended on the preloading of RFC and PCNA or template-primer used. Hence, experiments were carried out in which reactions were preincubated with RFC and PCNA to load the clamp/clamp loader. The Pols (at different molar ratios to DNA) were then added, and the rate of DNA synthesis and size of DNA products were determined. As shown (Fig. 4B), hPol ϵ acted more processively then hPol δ using these altered conditions. At lower enzyme/DNA ratios (1.4:1 for Pol δ and 1.8:1 for Pol ϵ), shown in Fig. 4B (lanes 5 and 12), processivity differences between the Pols were detected. Experiments with singly primed φx174 DNA, used in place of singly primed M13 DNA, as well as alterations in the order of the addition of reagents also showed that hPol ϵ acted more processively than hPol δ.
FIGURE 4.
Rate of chain elongation reaction catalyzed by Pol δ and Pol ϵ. A, influence of the ratio of Pol ϵ to DNA on length of DNA products formed. Reaction mixtures (10 μl) containing 20 mm Tris-HCl, pH 7.5, 150 μg/ml BSA, 10 mm magnesium acetate, 1 mm DTT, 0.1 mm EDTA, 30 mm potassium glutamate, 1.5 mm ATP, 30 μm [α-32P]dATP (23,230 cpm/pmol), 130 μm each of dCTP, dTTP, and dGTP, 0.73 nm singly primed M13, 70 nm E. coli SSB, 5.6 nm RFC, 50 nm PCNA, and either 7.3 or 2.19 nm hPol δ or the four-subunit hPol ϵ (as indicated) were incubated at 37 °C for the time specified. Reactions were halted with EDTA (10 mm final), and aliquots were removed to determine the level of nucleotide incorporation and size of DNA products following alkaline-agarose electrophoresis. B, influence of Pol/DNA ratio following preincubation of RFC/PCNA with DNA. Reactions were as described in A and contained 1 nm singly primed M13 DNA and the indicated levels of hPol δ and hPol ϵ. Reaction mixtures lacking Pols were preincubated for 3 min at 37 °C to preload RFC and PCNA onto DNA, after which the Pols were added. Mixtures were then incubated for the time indicated at 37 °C. C, influence of RFC levels on the rate and size of DNA synthesized with hPol ϵ and hPol δ. Reaction mixtures, as described in A, with the indicated levels of RFC and hPols, were incubated for 20 min at 37 °C.
Similar processivity differences were detected when the elongation of singly primed M13 was limited by the level of RFC rather than the polymerase/DNA ratio. As shown in Fig. 4C, the length of DNA products formed by hPol δ decreased markedly as the level of hRFC was reduced. In contrast, full-length M13-DNA products were detected with hPol ϵ even at the lowest level of RFC added. It should be noted that the amount of dNTP incorporated with either Pol was drastically affected by RFC levels, in keeping with its requirement for DNA synthesis. Collectively, these data indicate that under the different conditions used, hPol δ was less processive in its action than hPol ϵ. Previous studies also suggested that the hPol δ was distributive in its action and underwent repeated dissociation-association events during its elongation cycle (32, 33). These findings differ substantially from those reported for S. cerevisiae Pol δ, which was shown to be highly processive (34). The significance of these findings is discussed in more detail below. All experiments described in Fig. 4 were carried out with the E. coli SSB as the DNA-binding protein; identical results were obtained when RPA was used in place of E. coli SSB, although the activity with Pol ϵ was reduced ∼15–30%. Qualitatively, both the rate of elongation of primed DNA templates and the size of the products formed were the same with either DNA-binding protein (data not shown).
Assembly of the GINS Complex
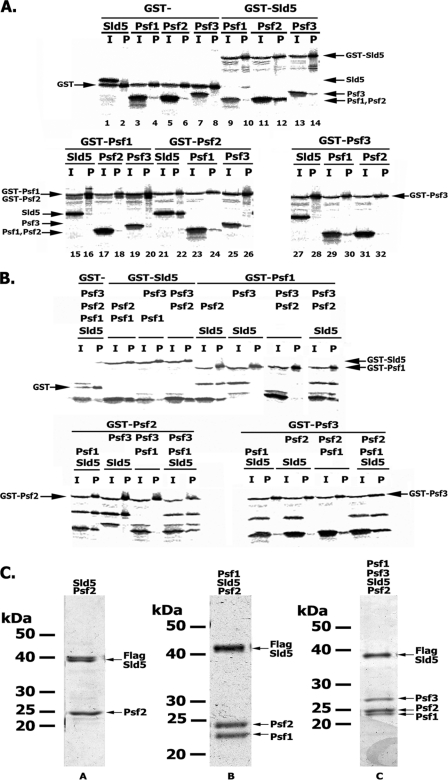
In yeast, genetic screens for gene products that interacted with Dpb11 identified the highly conserved four-subunit GINS complex (composed of Sld5, Psf3, Psf2, and Psf1), which is essential for DNA replication in vivo (35) and in vitro (36). Crystal structures of the hGINS revealed a tightly packed four-subunit complex showing multiple subunit interactions (25, 37, 38). As we reported previously, the four-subunit GINS complex exhibited a central cavity with dimensions that could accommodate ssDNA (25). Although GINS binds ssDNA weakly (39), it is not clear whether this activity contributes to this property. Although the crystal structures show subunit interactions, the data do not indicate how the complex assembles or predict the existence of subcomplexes. We examined the subunit interactions and assembly of GINS by carrying out IVTT reactions followed by GST pull-downs. In these experiments, one subunit of the complex was expressed with a GST tag along with one, two, or three of the other subunits. After protein synthesis, mixtures were subject to GST pull-downs; after washing and release of the bound 35S-labeled proteins, mixtures were subject to SDS-PAGE and autoradiography (Fig. 5A). This analysis revealed that only Sld5 and Psf2 formed a stable dimer (Fig. 5A, lanes 12 and 22), although weak interactions between Psf3 and Psf1 were observed when GST-Psf3 was used (Fig. 5A, lane 20 versus lane 30). Moreover, Sld5 formed a stable complex with Psf1 and Psf2 (Fig. 5B, lanes 2, 8, and 16) but not with Psf1 and Psf3 (Fig. 5B, lanes 4, 10, and 24). We detected the formation of low levels of the GST-Psf3·Psf2·Psf1 trimer (lane 28); however, the trimer was only detected with GST-Psf3 and not with GST-Psf1 or GST-Psf2 (Fig. 5B, lanes 12 and 20). The four-subunit GINS complex formed regardless of which subunit was fused with GST (Fig. 5B, lanes 14, 22, and 30). Together, these data suggest the order of GINS assembly, beginning with the formation of an Sld5-Psf2 dimer, which is then bound by Psf1 and followed by Psf3 interaction with the Sld5-Psf2-Psf1 trimer to complete the assembly.
FIGURE 5.
Assembly of the four-subunit hGINS complex. A and B, two- and three-subunit interactions. Various subunit combinations of the hGINS complex with one subunit tagged with GST were expressed using the IVTT system followed by GST pull-down. The material bound to the beads was eluted with SDS loading buffer and subjected to 12% SDS-PAGE, and the 35S-labeled proteins were detected by autoradiography. The proteins expressed in each reaction are indicated above the lanes. I, 10% input; P, GST pull-down step. C, in vivo formation and isolation of GINS complexes. Four-subunit hGINS and subcomplexes were isolated from Sf9-infected cells as described under “Experimental Procedures” and subjected to 12% SDS-PAGE followed by Coomassie staining. The specific complexes loaded in each lane are indicated above each lane; the amount of protein loaded was as follows: His-FLAG-Sld5·Psf2 (0.3 μg), His-FLAG-Sld5·Psf2·Psf1 (0.7 μg), and His-FLAG-Sld5·Psf2·Psf1·Psf3 (0.5 μg).
Based on the IVTT and GST pull-down experiments, we expressed and purified the GINS holo- and subcomplexes from baculovirus-infected insect cells. In keeping with our IVTT experiments, we purified to >90% homogeneity the two-subunit complex (FLAG-Sld5·Psf2), the three-subunit complex (FLAG-Sld5·Psf2·Psf1), and the holocomplex (FLAG-Sld5·Psf2·Psf3·Psf1) (Fig. 5C). Densitometric scans of the Coomassie-stained SDS-polyacrylamide gel of the glycerol gradient peak fractions indicated that these complexes contained stoichiometric amounts of each subunit. Efforts to isolate the individual GINS subunits have been unsuccessful due to insolubility or aggregation.
Interaction of hGINS with Pols
S. cerevisiae GINS was shown to interact with S. cerevisiae Pol ϵ/Dpb2, Sld2, and Dpb11 to form a preloading complex that is thought to load GINS, Pol ϵ, and perhaps other components of the replication machinery at replication origins (18). We examined the interactions between the Pols and GINS in insect cells infected with baculoviruses expressing the different subunits of Pol α-primase (or Pol δ or Pol ϵ) and GINS (including a GST-tagged Sld5). Pol α, δ, and ϵ co-eluted with GST-GINS (Fig. 6A, lanes 2, 5, 11, and 14). Despite nonspecific binding observed with p261 (Pol ϵ), p261 bound to GST-GINS at least 20 times higher than that observed in the input (1%) and the negative control (Fig. 6A, compare lane 13 versus lane 14 and lane 15 versus lane 14). Under the conditions used, we did not detect hGINS binding to the primase subunits (p48/p58), indicating that GINS interacted with the p180/p68 subunits of Pol α, in contrast to previous reports (40). However, when analyzed by glycerol gradient sedimentation, GINS and Pols dissociated during the 16-h centrifugation (data not shown), suggesting that the GINS-Pol interactions are unstable.
FIGURE 6.
Interaction of hGINS with replication Pols. A, interaction of GINS and Pols detected following infection of High Five insect cells. High Five insect cells were infected with viruses expressing the indicated FLAG-tagged Pols and the four GINS subunits (including GST-Sld5). After infection, cells were lysed, and the GST-GINS (and associated proteins) was pulled-down with glutathione-agarose beads as described under “Experimental Procedures.” GST-precipitated material was subjected to 4–20% polyacrylamide gradient gel/SDS electrophoresis and Western blotting, and bands were visualized with Pol-specific antibodies. In, 1% of input; −P, GST pull-down carried out without Precission protease treatment; +P, GST pull-downs carried out following Precission protease treatment. B, in vitro interaction of GINS and Pols. Reaction mixtures (100 μl) containing 3 pmol of FLAG-tagged Pol α-primase complex (or Pol δ or Pol ϵ) and 5 pmol of 32P-labeled GINS (1454 cpm/fmol) were incubated in Binding Buffer (50 mm HEPES, pH 7.5, 0.05% Nonidet P-40, 100 mm NaCl, 0.1 mg/ml BSA, and protease inhibitors) for 16 h at 4 °C. After incubation, reaction mixtures were divided into two equal aliquots, 15 μl of FLAG-agarose beads were added to both, and FLAG peptide (final concentration, 1 mg/ml) was added to one aliquot (+F) that served as a negative control. Pols were immunoprecipitated (IP) with FLAG antibody for 2 h at 4 °C, and the beads were washed three times with 0.5 ml of Binding Buffer. The bound proteins were eluted by boiling the beads in 15 μl of 1× SDS loading buffer, and the proteins were resolved in 12% SDS-PAGE; GINS was detected by autoradiography and phosphorimaging.
We confirmed and quantified the weak interactions between Pols and GINS using co-immunoprecipitation involving purified FLAG-tagged Pols and radiolabeled GINS ([32P]Sld5-Psf2-Psf3-Psf1). As shown in Fig. 6B, incubation of Pols (3 pmol) with [32P]GINS (5 pmol) resulted in formation of Pol-[α-32P]GINS (∼0.05 fmol) or Pol δ/Pol ϵ-[32P]GINS (∼0.004 fmol), respectively. These weak interactions were not augmented significantly by altering reaction conditions that included increasing the amount of GINS, prolonging the incubation time (2–16 h), decreasing the salt concentration, or varying incubation temperatures (4, 0, and 37 °C) (data not shown).
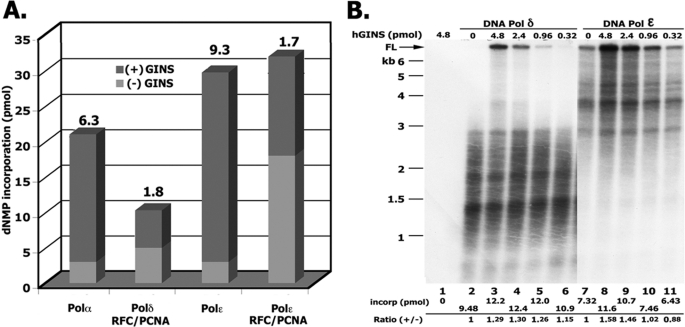
The findings that hGINS interacted weakly with the replicative Pols (Fig. 6) prompted us to examine whether these interactions affected their polymerase activities. We first examined these effects using a primed oligonucleotide substrate, a 90-mer containing a (TG)20 single strand region (26) that supported DNA synthesis catalyzed by Pol α as well as Pol ϵ in the absence of RFC, PCNA, and a DNA-binding protein (Fig. 7A). As shown, hGINS stimulated Pol ϵ activity ∼9-fold. Under the conditions used, maximal stimulation was detected at ∼480 nm GINS with a KD of ∼150 nm (data not presented). When these reactions were supplemented with RFC and PCNA, DNA synthesis catalyzed by Pol ϵ was increased ∼7-fold. hGINS increased the PCNA-stimulated DNA synthesis by Pol ϵ only 1.7-fold. The addition of RPA or E. coli SSB did not affect the level of DNA synthesis. Identical experiments with hPol δ showed that it supported significant DNA synthesis only in the presence of RFC and PCNA. Supplementation of Pol δ reactions with hGINS increased the level of nucleotide incorporation ∼2-fold. Pol α activity was stimulated ∼6-fold by GINS (KD ∼200 nm) (data not shown).
FIGURE 7.
Influence of hGINS on replicative Pols. A, reactions with an oligonucleotide primer-template. Reaction mixtures (20 μl) contained 40 μm HEPES-NaOH buffer, pH 7.5, 100 μg/ml BSA, 0.75 mm DTT, 10 mm magnesium acetate, 17.5 μm [α-32P]dATP (9157 cpm/pmol), 75 μm dCTP, 125 μm oligonucleotide primer-template 90-mer (TG)20 as template (26), 50 mm NaCl, and, where specified, a 0.3 nm concentration of the two-subunit (p180-p70) Pol α complex, 4 nm four-subunit Pol ϵ, 9 nm Pol δ, 5.6 nm RFC, 50 nm PCNA, and 1 mm ATP. After 30 min at 37 °C, aliquots were removed to measure DNA synthesis. B, stimulation of Pol δ and Pol ϵ holoenzyme activity by GINS using singly primed M13. Reaction mixtures (10 μl) contained 20 mm Tris-HCl, pH 7.5, 150 μg/ml BSA, 1 mm DTT, 10 mm magnesium acetate, 2 mm ATP, 20 μm [α-32P]dATP (17,750 cpm/pmol), 120 μm each of dCTP, dGTP, and dTTP, 30 mm sodium glutamate, 1 nm singly primed M13, 400 nm E. coli SSB, 5.6 nm RFC, 50 nm PCNA, and varying levels of the four-subunit hGINS complex. Mixtures were incubated for 3 min at 37 °C and then supplemented with 2.2 nm Pol δ or 2.2 nm Pol ϵ, as indicated. Following incubation for 10 min at 37 °C, aliquots were removed to measure DNA synthesis and the size of labeled DNA products formed.
In order to evaluate the effects of hGINS on DNA synthesis catalyzed by Pol ϵ and Pol δ on a more physiologically relevant template, elongation of singly primed M13 was examined. With this template, DNA synthesis catalyzed by these Pols is totally dependent on RFC, PCNA, and a single-stranded DNA-binding protein (E. coli SSB or RPA) (Fig. 3). As shown in Fig. 7B, in the presence of either Pol δ or Pol ϵ, hGINS stimulated nucleotide incorporation marginally (1.3–1.6-fold). Under the conditions used, the length of DNA products formed by Pol δ were extended to ∼3 kb (Fig. 7B, lane 2), and a small percentage of this material was elongated substantially by hGINS in a dose-dependent manner (lanes 3–6). Reaction with Pol ϵ were also stimulated by hGINS, although this effect differed quantitatively from that observed with Pol δ. As shown in Fig. 7B, full-length 7-kb DNA products were formed by Pol ϵ, and the stimulatory effects of GINS were evident among products of all sizes. As observed with Pol δ, this stimulation increased in the presence of a higher level of GINS.
It was reported previously that hGINS stimulated Pol α activity weakly and interacted with the DNA primase subunits (40). A GINS homolog has been identified in the archaea Sulfolobus solfataricus. In this organism, the S. solfataricus GINS complex consists of a dimer of a heterodimer (41), which was shown to interact with S. solfataricus Mcm and the S. solfataricus DNA primase heterodimer, although functional effects of these interactions were not reported. As shown in Fig. 6, we failed to detect interactions between GINS and the primase subunits; nor did we detect any stimulation of primase activity (data not shown). It should be noted that the two-subunit Pol α complex (p180-p70) preparation was used in the experiments reported in Fig. 7. GINS stimulated the elongation of a primed oligonucleotide catalyzed by the four-subunit Pol α complex (p180-p70-p55-p45) as well (supplemental Fig. 2). We also investigated whether GINS stimulated Pol α in the SV40 T-antigen-SV40 origin-dependent replication reaction (supplemental Fig. 3). Under the conditions used, hGINS activated both the SV40-T antigen-dependent monopolymerase (with Pol α-primase as the only Pol present) and dipolymerase reactions (Pol α-primase plus the Pol δ holoenzyme). We also noted that the stimulation of Pol α and Pol ϵ was significantly greater with the four-subunit hGINS complex than with the three-subunit (devoid of Psf3) or the two-subunit (containing only Sld5-Psf2) complexes (supplemental Fig. 4 and supplemental Table 1). Based on these findings, we suggest that hGINS may affect the association of the replicative Pols with the replicative helicase. By stimulating the polymerization activity of replicative Pols, hGINS may play roles in preventing fork collapse as well as coordinating the actions of the replicative Pols and fork unwinding/movement.
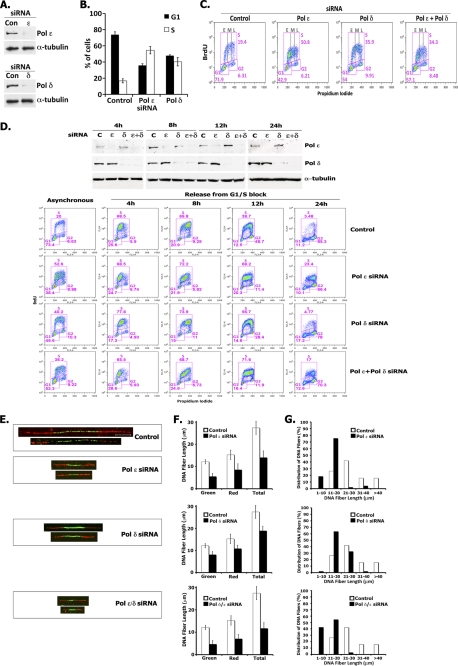
Depletion of hPol ϵ and hPol δ Slows S Phase Progression
To evaluate the role of hPol ϵ and hPol δ in cellular DNA replication, we depleted the large catalytic subunit of hPol ϵ (p261) and hPol δ (p125) in HeLa cells by 93 and 90%, respectively, using siRNAs against their ORFs (Fig. 8A). Asynchronous populations of cells depleted of either hPol ϵ or hPol δ displayed a marked increase in the amount of S phase cells (Fig. 8B). Similar results were obtained with other siRNAs targeting different regions of hPol ϵ or hPol δ (supplemental Fig. 5), establishing that reduction of either polymerase increases S phase population. As shown in Fig. 8C, we observed a ≥2-fold increase in the S phase population in Pol ϵ- or Pol δ-depleted cells compared with control cells (50.8% of S phase cells in hPol ϵ siRNA-treated and 35.9% of S phase cells in hPol δ siRNA-treated cells versus 19.4% of S phase cells in control siRNA-treated cells). Whereas cells depleted of hPol δ distributed equally throughout early, middle and late S phases (indicated as E, M, and L, respectively, in Fig. 8C), the majority of cells depleted of hPol ϵ accumulated in early S phase. The simultaneous depletion of both hPol ϵ and hPol δ showed a distribution similar to that observed for cells depleted of hPol ϵ alone (i.e. increase in number of cells in early S). To determine whether the accumulation of cells in S phase was due to a cell cycle arrest or to the slow progression of cells through S phase, we monitored S phase progression in synchronized HeLa cells (Fig. 8D). The majority of untargeted cells, halted in G1/S by a double thymidine block followed by their release in nocodazole-containing medium, progressed through S and accumulated in mitosis before 24 h, with less than 5% of cells still in S phase. hPol ϵ-depleted cells progressed much more slowly than the control cells, with 25% of cells still in S phase at the 24 h time point. Surprisingly, Pol δ-depleted cells progressed more slowly through S phase but entered mitosis, whereas cells that were simultaneously depleted of hPol ϵ and hPol δ showed delayed S phase progression similar to cells depleted of hPol ϵ alone (17% of cells still in S phase at 24 h). We then examined whether the slower S phase progression in depleted cells was due to slower fork movement. For this purpose, cells were sequentially pulsed for 20 min each with IdU and CldU and lysed, and DNA fibers were spread on slides and incubated with primary antibodies specific to IdU and CldU and fluorescence-labeled secondary antibodies. DNA fibers containing green IdU fluorescent label flanked on each side with red CldU fluorescent label represent DNA molecules formed by bidirectional movement of replication forks. Representative DNA fibers isolated from control, hPol ϵ-depleted, hPol δ-depleted, and hPol ϵ/δ-doubly depleted cells are shown in Fig. 8E. Both the green IdU label, representing DNA synthesis from replication origins, and the red CldU label were significantly shorter in hPol ϵ-depleted, hPol δ-depleted, and hPol ϵ/δ-doubly depleted cells than fibers isolated from siRNA control cells, indicating that decreased levels of these Pols slowed fork movement. The average length of DNA fibers from hPol ϵ-depleted, hPol δ-depleted, hPol ϵ/δ-doubly depleted, and control cells was 14.4, 18.9, 11.7, and 27.4 μm, respectively (Fig. 8F). Based on measurements of >100 individual DNA fibers isolated from each sample, the majority of DNA fibers isolated from control cells were 21–30 μm in length, whereas the majority of the fibers from hPol ϵ-depleted cells were 11–20 μm (2-fold reduction in rate); ∼60% of the fibers from hPol δ-depleted cells were also 11–20 μm in length, and ∼30% were 21–30 μm. The reduction in fork movement was even more dramatic in cells depleted of both hPol ϵ and hPol δ; ∼50% of the fibers were 11–20 μm, and ∼40% were 1–10 μm in length (∼2–30-fold reduction) (Fig. 8G). These findings indicate that depletion of either replicative Pol significantly reduced fork progression.
FIGURE 8.
Depletion of hPol ϵ or hPol δ slows down S phase progression. A, siRNA depletion of Pol δ or Pol ϵ. HeLa cells were transfected with control (Con) siRNA or specific siRNAs targeting Pol ϵ or Pol δ (siRNAs used for depletion of Pol ϵ and Pol δ were #08 and #06, respectively; supplemental Fig. 5). The levels of the large subunits of Pol ϵ and Pol δ were measured by immunoblotting 48 h after transfection. To quantitate the extent of depletion of Pol ϵ and Pol δ, the protein levels were adjusted using the α-tubulin loading control and quantified relative to the protein level present in the control sample. B and C, siRNA depletion of Pol δ or Pol ϵ leads to accumulation of cells in S phase. HeLa cells were transfected with control siRNA or with siRNAs targeting either Pol ϵ or Pol δ. After 48 h, cells were incubated with BrdU for 90 min, stained with BrdU-FITC antibody and propidium iodide (PI), and analyzed by flow cytometry. The bar graph (B) shows the percentage of S phase (BrdU-positive) cells versus G1 phase cells present in each sample. Plots (C) show BrdU incorporation (y axis), DNA content (x axis), and cell cycle distribution (described for B) of HeLa cells following siRNA treatment. The distribution of cells present in early (E), middle (M), and late (L) S phase is also indicated. D, Pol ϵ-depleted cells progress more slowly through S phase. HeLa cells were transfected with control siRNA, Pol ϵ siRNA, Pol δ siRNA, or a combination of these siRNAs 4 h before synchronization by double thymidine block. Arrested cells were released into nocodazole-containing medium, harvested at the times indicated following BrdU treatment for 90 min, and analyzed for protein depletion by immunoblotting (top panel) and by flow cytometry (lower panels) as described in A. The BrdU plots show the profiles of samples treated with the indicated Pol ϵ/Pol δ siRNAs compared with control siRNA samples at the indicated cell cycle stage: asynchronous cells, 4 h (early-middle S), 8 h (late S), 12 h (G2), and 24 h (mitosis). E–G, replication fork progression analyses. HeLa cells transfected with Pol ϵ or Pol δ siRNA alone and with both siRNAs were incubated for 20 min with IdU followed by 20 min with CldU and then subjected to replication fork movement analysis. Individual replicating forks were visualized by immunofluorescence of the incorporated halogenated nucleotides present in isolated DNA fibers, as described under “Experimental Procedures.” E, images of fibers. The bar in the fiber image of the control sample corresponds to 10 μm. The mean DNA fiber length was calculated by measuring at least 100 individual fibers in each experiment, and the results were plotted (F). The data from one representative experiment are plotted as percentage of DNA fibers possessing the specified length indicated (G). Error bars, S.E.
DISCUSSION
Here we have described the isolation and characterization of the four-subunit hPol ϵ complex and showed that its subunits are present in stoichiometric levels and that the holocomplex is a monomer. These findings indicate that both Pol ϵ and Pol δ (28, 42–44) are monomeric complexes, suggesting that each could act on an opposite strand during replisome translocation. We determined the sites within the large catalytic Pol 2 subunit (p261) that interact with the small subunits (p59, p12, and p17). The p59 (Dpb2) binds to the two C-terminal zinc finger motifs in p261 (located between aa 2150 and 2186), whereas the p12 (Dpb3) and p17 (Dpb4) subunits interact within the Pol 2 region spanning aa 1211–1305. Similar findings were reported for the four-subunit Xenopus Pol ϵ (30). Based on studies with the monomeric S. cerevisiae Pol ϵ complex (28), the binding of S. cerevisiae Dpb2 (77 kDa) to S. cerevisiae Pol 2 also occurs through the conserved zinc finger domains at the C terminus of Pol 2 (29). However, the S. cerevisiae Dpb3 (23 kDa) and S. cerevisiae Dpb4 (22 kDa) subunits were reported to bind close to the Dpb2 binding site (29).
As described above, all hPol ϵ derivatives containing the N-terminal catalytic domains possessed polymerase activity that, under the conditions used, was unaffected by the presence or absence of the other subunits. Although it is clear that the hPol ϵ p261 N terminus is solely responsible for its catalytic activity, small subunits (p59, p12, and p17) probably play roles in DNA replication. In S. cerevisiae, both Pol 2 and Dpb2 subunits are essential for viability, whereas Dpb3 and Dpb4 are not. In keeping with these findings, the Xenopus p261-p59 complex supports in vitro Xenopus DNA replication, whereas the p261 subunit alone or complexed with the p12 and p17 subunits did not (30). It may be that the Dpb2 subunit plays critical roles when Pol ϵ is associated with the replication fork by interacting with other replisome components. Although Dpb3 and Dpb4 are not essential in budding yeast, phenotypic changes have been detected following their deletion (45, 46). Dpb3 and Dpb4 were shown to possess histone fold motifs, and in S. cerevisiae and human cells (47), Dpb4 was shown to be a component of the CHRAC chromatin remodeling complex (48).
In the presence of RFC, PCNA, and singly primed M13, the rates of nucleotide incorporation catalyzed by hPol δ and hPol ϵ were not markedly different. Important differences, however, were noted in their elongation properties. Under the conditions used, hPol δ was less processive than Pol ϵ, indicating that during elongation of DNA chains, Pol δ dissociated more readily than Pol ϵ. Elongation reactions with Pol ϵ led to the synthesis of full-length products even at relatively low enzyme/DNA molar ratios. These findings differ from previous studies with the S. cerevisiae Pol ϵ; however, these studies were carried out in reactions in which the Pol ϵ/DNA ratios were extremely low, possibly contributing to the dissociation and low processivity of S. cerevisiae Pol ϵ (31). Our findings that hPol δ is a poorly processive enzyme, although in agreement with earlier reports (32, 33), differ substantially from the properties described for the highly processive S. cerevisiae Pol δ. The S. cerevisiae Pol δ is a three-subunit complex (49), whereas the hPol δ (as well as the Schizosaccharomyces pombe enzyme) is a four-subunit complex (44, 50). It is unlikely that this contributes to their marked processivity differences because the two-subunit S. cerevisiae Pol δ complex (p125-p31), devoid of the non-essential p32 subunit, was as processive as the three-subunit complex (34). Although the processivity measurements described here suggest that hPol δ could act efficiently as the lagging strand Pol and Pol ϵ as the leading strand Pol, this arrangement is likely to be influenced by additional factors, particularly the replicative helicase working in front of these replicative Pols. In S. cerevisiae, the only replicative Pol shown to be stably linked to the RPC was Pol α, and this association required Ctf4 (16). Although a substantial amount of in vivo data (both in yeast and higher eukaryotes) suggest that Ctf4 stabilizes the Pol α subunit (16, 51), we previously found that Ctf4 interacted weakly with all three replicative Pols and markedly stimulated Pol α and Pol ϵ (26). We have carried out similar studies with GINS, a component of the CMG complex, the replicative helicase. Similar to findings with hCtf4, hGINS interacted weakly with all replicative Pols (best with Pol α) and strongly stimulated Pol α (but not DNA primase) and Pol ϵ activities. Maximal stimulation required the four-subunit GINS complex because the two (Sld5-Psf2) and three (Sld5-Psf2-Psf1) subcomplexes were less effective. Although we detected functional interactions between GINS and Pol α and Pol ϵ, the physiological significance of these findings is unclear. Because GINS is a critical component of the CMG complex, functional interaction studies between the CMG complex and the replicative Pols may yield more relevant results. The influence of other replisome components Mcm10 and Mrc1 (claspin in higher eukaryotes) on the replicative Pols has been examined. Mcm10 was shown to interact strongly with the catalytic subunit of Pol α (as well as to stimulate its Pol activity (52)) and Mrc1 was reported to bind to Pol ϵ (53), suggesting that replisome-associated proteins are likely to play multiple roles during fork movement.
In addition to replication, Pol ϵ and its subunits play other roles. In S. cerevisiae, it plays a role in the formation of the preinitiation complex and is loaded onto chromatin prior to Pol α (17). It is not clear whether the Pol ϵ loaded at this stage participates in subsequent DNA synthesis. In yeast, the C-terminal end of Pol ϵ plays critical roles in supporting checkpoint activation and viability of cells deleted of the catalytic domains of the Pol 2 subunit. In S. pombe, it was shown that the checkpoint proteins, which are normally not essential, became essential under these conditions (54). In both yeasts, deletion of the Pol 2 catalytic region markedly reduced the growth rate of cells. It was also reported that in fission yeast, co-expression of the catalytic N- and C-terminal domains of Pol 2 in trans and the p59 subunit increased cell viability and rendered the checkpoint proteins non-essential (54). A possible explanation for these observations could be that the N- and C-terminal regions of Pol ϵ interact to form a complex. However, our efforts to detect in vitro interactions between the C- and N-terminal hPol ϵ fragments were unsuccessful. Interactions between these purified components were not observed following immunoprecipitation or sizing column chromatography (data not presented). It may be that in vivo other components, which we did not supply in vitro, are required for such an interaction.
In higher eukaryotes, replication origins are poorly defined. It has been estimated that somatic cells contain ∼1 origin/150 kbp of DNA (55), The size of the human genome is estimated to be ∼2.9 × 106 kbp (56), suggesting that a human cell may contain ∼2 × 104 origins. Using quantitative Western blot analyses, specifically of the large catalytic subunits, we determined that each HeLa cell contains ∼3 × 106 molecules of hPol ϵ and somewhat lower levels of hPol δ (0.5 × 106 molecules; supplemental Fig. 6). These findings suggest that each cell contains ∼40–100 molecules of each Pol per origin. For comparative purposes, Escherichia coli contains 1 origin and ∼10–20 molecules of Pol III holoenzyme/cell (56).
We depleted HeLa cells of Pol δ (p125 subunit), Pol ϵ (p261 subunit), and both Pol δ and Pol ϵ and examined their effects on cell cycle progression and fork movement. Although siRNA-targeted depletion reduced the levels of the Pols by 90%, based on the above measurements, approximately ∼105 molecules of Pol ϵ and Pol δ/cell were still present. Under these conditions, we noted that reduction of Pol ϵ levels had a more pronounced effect on cell cycle progression and fork movement than depletion of Pol δ. As expected, the simultaneous depletion of both Pols resulted in more severe effects, although cell cycle progression was similar to that observed following depletion of Pol ϵ alone. We speculate that decreased levels of Pol δ may have less of a biological effect than decreased levels of Pol ϵ. This notion is based on the action of Pol δ as a lagging strand Pol. Because ∼103 Okazaki fragments are produced for each Pol δ present in mammalian cells (57), the lagging strand Pol must dissociate rapidly and transfer to a new lagging strand primer site, possibly leading to the recycling of Pol δ. Recent studies demonstrated that the highly processive S. cerevisiae Pol δ undergoes this release and transfer cycle when the Pol encounters a downstream duplex after completing the elongation of an Okazaki fragment (34). Although the hPol δ acts less processively than the S. cerevisiae Pol δ, the size of Okazaki fragments in eukaryotes is relatively short (∼250 bp), and a collision-release cycle may permit a catalytic role for Pol δ. It is likely that replication of leading strand by Pol ϵ requires substantially longer association of this Pol with the elongating strand.
In both yeasts, Pol ϵ mutants with deleted polymerase domains are viable, suggesting that another polymerase can support replication of the leading strand in lieu of an active Pol ϵ (54, 58). The likely candidate for this role is Pol δ. We investigated whether transfection of plasmids expressing the C terminus of hPol ϵ rescued the slow S phase progression and fork movement observed in cells depleted of Pol ϵ. We noted no changes in the rate of S phase progression or fork movement following transfection of plasmids expressing of the C-terminal Pol ϵ region (data not shown). Although these are negative results, we think it unlikely that the poorly processive hPol δ, in contrast to the highly processive S. cerevisiae Pol δ, would replace Polϵ as the leading strand Pol in HeLa cells. Collectively, our in vivo studies indicate that both Pols are required for normal cell cycle and fork progression.
Supplementary Material
Acknowledgment
We thank the Molecular Cytology Facility at Memorial Sloan Kettering Cancer Center for help with the microscope imaging.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–6 and Table 1.
- Pol
- polymerase
- RFC
- replication factor C
- PCNA
- proliferating cell nuclear antigen
- RPC
- replisome progression complex
- hPol
- hPCNA, hRFC, and hGINS, human Pol, PCNA, RFC, and GINS, respectively
- aa
- amino acids
- IVTT
- in vitro transcription/translation
- nt
- nucleotides
- IdU
- iododeoxyuridine
- CldU
- chlorodeoxyuridine.
REFERENCES
- 1. Johansson E., Macneill S. A. (2010) Trends Biochem. Sci. 35, 339–347 [DOI] [PubMed] [Google Scholar]
- 2. Pursell Z. F., Isoz I., Lundström E. B., Johansson E., Kunkel T. A. (2007) Science 317, 127–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nick McElhinny S. A., Gordenin D. A., Stith C. M., Burgers P. M., Kunkel T. A. (2008) Mol. Cell 30, 137–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fukui T., Yamauchi K., Muroya T., Akiyama M., Maki H., Sugino A., Waga S. (2004) Genes Cells 9, 179–191 [DOI] [PubMed] [Google Scholar]
- 5. Waga S., Masuda T., Takisawa H., Sugino A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 4978–4983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kang H. Y., Choi E., Bae S. H., Lee K. H., Gim B. S., Kim H. D., Park C., MacNeill S. A., Seo Y. S. (2000) Genetics 155, 1055–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Remus D., Diffley J. F. (2009) Curr. Opin. Cell Biol. 21, 771–777 [DOI] [PubMed] [Google Scholar]
- 8. Wang J., Wu R., Lu Y., Liang C. (2010) Biochem. Biophys. Res. Commun. 395, 336–341 [DOI] [PubMed] [Google Scholar]
- 9. Im J. S., Ki S. H., Farina A., Jung D. S., Hurwitz J., Lee J. K. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 15628–15632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sawyer S. L., Cheng I. H., Chai W., Tye B. K. (2004) J. Mol. Biol. 340, 195–202 [DOI] [PubMed] [Google Scholar]
- 11. Lee J. K., Seo Y. S., Hurwitz J. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 2334–2339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wohlschlegel J. A., Dhar S. K., Prokhorova T. A., Dutta A., Walter J. C. (2002) Mol. Cell 9, 233–240 [DOI] [PubMed] [Google Scholar]
- 13. Moyer S. E., Lewis P. W., Botchan M. R. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 10236–10241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gambus A., Jones R. C., Sanchez-Diaz A., Kanemaki M., van Deursen F., Edmondson R. D., Labib K. (2006) Nat. Cell Biol. 8, 358–366 [DOI] [PubMed] [Google Scholar]
- 15. Kanemaki M., Labib K. (2006) EMBO J. 25, 1753–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gambus A., van Deursen F., Polychronopoulos D., Foltman M., Jones R. C., Edmondson R. D., Calzada A., Labib K. (2009) EMBO J. 28, 2992–3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Aparicio O. M., Weinstein D. M., Bell S. P. (1997) Cell 91, 59–69 [DOI] [PubMed] [Google Scholar]
- 18. Muramatsu S., Hirai K., Tak Y. S., Kamimura Y., Araki H. (2010) Genes Dev. 24, 602–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Araki H., Leem S. H., Phongdara A., Sugino A. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 11791–11795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kamimura Y., Masumoto H., Sugino A., Araki H. (1998) Mol. Cell Biol. 18, 6102–6109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sangrithi M. N., Bernal J. A., Madine M., Philpott A., Lee J., Dunphy W. G., Venkitaraman A. R. (2005) Cell 121, 887–898 [DOI] [PubMed] [Google Scholar]
- 22. Wu J., Capp C., Feng L., Hsieh T. S. (2008) Dev. Biol. 323, 130–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kumagai A., Shevchenko A., Shevchenko A., Dunphy W. G. (2010) Cell 140, 349–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li Y., Asahara H., Patel V. S., Zhou S., Linn S. (1997) J. Biol. Chem. 272, 32337–32344 [DOI] [PubMed] [Google Scholar]
- 25. Chang Y. P., Wang G., Bermudez V., Hurwitz J., Chen X. S. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 12685–12690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bermudez V. P., Farina A., Tappin I., Hurwitz J. (2010) J. Biol. Chem. 285, 9493–9505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Siegel L. M., Monty K. J. (1966) Biochim. Biophys. Acta 112, 346–362 [DOI] [PubMed] [Google Scholar]
- 28. Chilkova O., Jonsson B. H., Johansson E. (2003) J. Biol. Chem. 278, 14082–14086 [DOI] [PubMed] [Google Scholar]
- 29. Dua R., Levy D. L., Campbell J. L. (1999) J. Biol. Chem. 274, 22283–22288 [DOI] [PubMed] [Google Scholar]
- 30. Shikata K., Sasa-Masuda T., Okuno Y., Waga S., Sugino A. (2006) BMC Biochem. 7, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Asturias F. J., Cheung I. K., Sabouri N., Chilkova O., Wepplo D., Johansson E. (2006) Nat. Struct. Mol. Biol. 13, 35–43 [DOI] [PubMed] [Google Scholar]
- 32. Masuda Y., Suzuki M., Piao J., Gu Y., Tsurimoto T., Kamiya K. (2007) Nucleic Acids Res. 35, 6904–6916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Podust V. N., Podust L. M., Müller F., Hübscher U. (1995) Biochemistry 34, 5003–5010 [DOI] [PubMed] [Google Scholar]
- 34. Langston L. D., O'Donnell M. (2008) J. Biol. Chem. 283, 29522–29531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Takayama Y., Kamimura Y., Okawa M., Muramatsu S., Sugino A., Araki H. (2003) Genes Dev. 17, 1153–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kubota Y., Takase Y., Komori Y., Hashimoto Y., Arata T., Kamimura Y., Araki H., Takisawa H. (2003) Genes Dev. 17, 1141–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Choi J. M., Lim H. S., Kim J. J., Song O. K., Cho Y. (2007) Genes Dev. 21, 1316–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kamada K., Kubota Y., Arata T., Shindo Y., Hanaoka F. (2007) Nat. Struct. Mol. Biol. 14, 388–396 [DOI] [PubMed] [Google Scholar]
- 39. Yoshimochi T., Fujikane R., Kawanami M., Matsunaga F., Ishino Y. (2008) J. Biol. Chem. 283, 1601–1609 [DOI] [PubMed] [Google Scholar]
- 40. De Falco M., Ferrari E., De Felice M., Rossi M., Hübscher U., Pisani F. M. (2007) EMBO Rep. 8, 99–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Marinsek N., Barry E. R., Makarova K. S., Dionne I., Koonin E. V., Bell S. D. (2006) EMBO Rep. 7, 539–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Johansson E., Majka J., Burgers P. M. (2001) J. Biol. Chem. 276, 43824–43828 [DOI] [PubMed] [Google Scholar]
- 43. Bermudez V. P., MacNeill S. A., Tappin I., Hurwitz J. (2002) J. Biol. Chem. 277, 36853–36862 [DOI] [PubMed] [Google Scholar]
- 44. Xie B., Mazloum N., Liu L., Rahmeh A., Li H., Lee M. Y. (2002) Biochemistry 41, 13133–13142 [DOI] [PubMed] [Google Scholar]
- 45. Araki H., Hamatake R. K., Morrison A., Johnson A. L., Johnston L. H., Sugino A. (1991) Nucleic Acids Res. 19, 4867–4872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ohya T., Maki S., Kawasaki Y., Sugino A. (2000) Nucleic Acids Res. 28, 3846–3852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li Y., Pursell Z. F., Linn S. (2000) J. Biol. Chem. 275, 23247–23252 [DOI] [PubMed] [Google Scholar]
- 48. Iida T., Araki H. (2004) Mol. Cell Biol. 24, 217–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gerik K. J., Li X., Pautz A., Burgers P. M. (1998) J. Biol. Chem. 273, 19747–19755 [DOI] [PubMed] [Google Scholar]
- 50. Zuo S., Bermudez V., Zhang G., Kelman Z., Hurwitz J. (2000) J. Biol. Chem. 275, 5153–5162 [DOI] [PubMed] [Google Scholar]
- 51. Zhu W., Ukomadu C., Jha S., Senga T., Dhar S. K., Wohlschlegel J. A., Nutt L. K., Kornbluth S., Dutta A. (2007) Genes Dev. 21, 2288–2299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Fien K., Cho Y. S., Lee J. K., Raychaudhuri S., Tappin I., Hurwitz J. (2004) J. Biol. Chem. 279, 16144–16153 [DOI] [PubMed] [Google Scholar]
- 53. Lou H., Komata M., Katou Y., Guan Z., Reis C. C., Budd M., Shirahige K., Campbell J. L. (2008) Mol. Cell 32, 106–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Feng W., D'Urso G. (2001) Mol. Cell Biol. 21, 4495–4504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Errico A., Costanzo V. (2010) EMBO Rep. 11, 270–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kornberg A., Baker T. (2005) DNA Replication, 2nd Ed., University Science Books, Herndon, VA [Google Scholar]
- 57. Burgers P. M. (2009) J. Biol. Chem. 284, 4041–4045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kesti T., Flick K., Keränen S., Syväoja J. E., Wittenberg C. (1999) Mol. Cell 3, 679–685 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.