Background: Disulfide bond formation regulates nuclear localization of transcription factor Yap1.
Results: Levels of Yap1-binding protein Ybp1 control cytoplasmic oxidative protein folding.
Conclusion: Oxidative stress response limited by levels of protein folding cofactor rather than its transcriptional regulator target.
Significance: Oxidative protein folding is the key step in oxidative stress response in fungi.
Keywords: Nuclear Translocation, Oxidative Stress, Protein Folding, Reactive Oxygen Species (ROS), Transcription Factors, Transcription Regulation
Abstract
The Saccharomyces cerevisiae transcription factor Yap1 is a central determinant of oxidative stress tolerance. This protein is found primarily in the cytoplasm in the absence of oxidative stress but, upon exposure to oxidants, rapidly translocates to the nucleus and activates expression of target genes. Although both diamide and H2O2 have been used to impose oxidative stress on cells, these different oxidants trigger Yap1 nuclear localization in distinctly different ways. Diamide appears to oxidize particular cysteine residues on Yap1, leading to inhibition of association of Yap1 with the nuclear exportin Crm1. Crm1 would normally transport Yap1 out of the nucleus. H2O2 activation of Yap1 nuclear localization requires the participation of the glutathione peroxidase Gpx3 and the Yap1-binding protein Ybp1. H2O2 exposure triggers formation of a dual disulfide bonded Yap1 that is catalyzed by the presence of Gpx3 and Ybp1. In the current study, we have determined that two distinct pools of Yap1 exist in the cell. These pools are designated by the level of Ybp1. Ybp1 interacts directly with Yap1 and these proteins form a stable complex in vivo. Genetic and biochemical experiments indicate that Ybp1 is rate-limiting for Yap1 oxidative folding during H2O2 stress. The fungal pathogen Candida glabrata expresses a protein homologous to Ybp1 called CgYbp1. Overproduction of CgYbp1 elevated H2O2 tolerance in this pathogen indicating that the determinative role of Ybp1 in setting the level of H2O2 resistance has been evolutionarily conserved.
Introduction
Oxidative stress is experienced by all cells growing in an aerobic environment. The inevitability of this environmental challenge has led to the elaboration of a wide range of strategies to deal with the production of potentially damaging reactive oxygen species (ROS).3 A central theme among the responses to ROS is the transcriptional reprogramming of cells to express new proteins that can act to detoxify these compounds. Control of the regulated response to elevated ROS levels is an important feature of all cells but especially in microorganisms because a key feature of the innate immune response in animals involves the generation of a bolus of ROS to eliminate these invading microbes (reviewed in Refs. 1 and 2).
The yeast Saccharomyces cerevisiae has served as a model eukaryotic cell for many fundamental processes and oxidative stress is no exception (3, 4). The transcription factor Yap1 is a central component in the oxidative stress response in this yeast (5, 6). Loss of YAP1 elicits an oxidant-hypersensitive phenotype and prevents ROS-induced gene expression. Regulation of Yap1 occurs in large part through control of the nuclear localization of this protein. In the absence of oxidative stress, Yap1 enters the nucleus at a relatively low basal rate compared with its rapid nuclear export via the exportin protein Crm1 (7, 8). Treatment with oxidants (like diamide or diethylmaleate) that act to directly modify cysteine residues present in the C-terminal cysteine-rich domain (C-CRD) of Yap1 blocks the ability of the protein to associate with Crm1. In turn, the oxidized form of Yap1 accumulates in the nucleus and activates expression of particular target genes (9).
Interestingly, the response of Yap1 to oxidative stress initiated by H2O2 exposure is more complex. H2O2 challenge of cells causes a covalent intermediate to be formed between the glutathione peroxidase Gpx3 and a cysteine (Cys-598) in the C-CRD (10) in a manner dependent on the presence of the Yap1-binding protein Ybp1 (11, 12). Next, disulfide bonds are formed between two cysteines present in the C-CRD with two cysteines present in the N-terminal cysteine-rich domain with the concomitant release of Gpx3 (10, 13, 14). These disulfide bonds link cysteines 303 with 598 as well as Cys-310 with Cys-629. This uniquely folded form of Yap1 is also unable to interact with Crm1 and critically can recruit the transcriptional Mediator component Rox3 to target promoters important in the response to H2O2-induced oxidative stress (15). These observations illustrate the additional components and added complexity of the response to H2O2 stress.
The role of Ybp1 in the H2O2-induced folding of Yap1 is not well understood. Ybp1 is not required for diamide resistance but is crucial for H2O2 tolerance (11, 12). We found that overproduction of Ybp1 but notably not Yap1 strongly elevated H2O2 resistance indicating that Ybp1 represented a limiting component in H2O2 resistance. Strikingly, overproduction of Ybp1 caused a diamide-hypersensitive phenotype compared with wild-type cells. Elevated dosage of Ybp1 also increased association of Ybp1 and Yap1 in vivo as well as accelerating H2O2-induced oxidative folding of Yap1 and target gene induction. Biochemical experiments indicated that Yap1 and Ybp1 directly associate in vitro. These data support the view that Yap1 exists in two different pools defined by association with Ybp1. The Yap1·Ybp1 complex is involved in the response to H2O2 but not diamide although non-Ybp1-associated Yap1 mediates diamide resistance but is less important in H2O2 tolerance. The levels of Ybp1 control the distribution of Yap1 between these two pools and the response to oxidative stress.
EXPERIMENTAL PROCEDURES
Yeast Strains and Methods
Strains used in this study are listed in Table 1. Yeast cells were grown either in rich medium (YPD: 1% yeast extract, 2% peptone, 2% dextrose) for non-selective growth or in complete synthetic medium lacking appropriate amino acids. S. cerevisiae strains were grown at 30 °C, whereas Candida glabrata strains were grown at 37 °C. To generate a S. cerevisiae yeast strain overproducing Ybp1, a TDH3::kanMX2 promoter cassette with flanking YBP1 sequence was amplified from plasmid pYM-N14 using primers F-YBP1-pYMN14 and R-YBP1-pYMN14. This cassette was transformed into a strain from the Open Biosystems TAP tag collection (BY4742 YBP1-TAP::HIS3) yielding SLS1. The genomic structure was confirmed by PCR using ScYBP1-up-F and ScYBP1-R primers. The YAP1 open reading frame (ORF) was disrupted in BY4742 YBP1-TAP::HIS3 and SLS1 (TDH3::YBP1-TAP::HIS3) using PCR-mediated gene disruption with the kanMX2 module with primers ScYAP1500upF and ScYAP1500downR, to yield SLS3 (YBP1-TAP::HIS3 yap1Δ::kanMX2). All of these disruptions were confirmed by PCR using KAN-C and ScYAP1500downR primers. To delete the CgYBP1 gene in C. glabrata, a fragment corresponding to 500 bp upstream and downstream of the CgYBP1 ORF was amplified and cloned in the YEp352 vector. The NotI fragment containing the hygromycin cassette from the pAG32 plasmid (16) was inserted into this plasmid in place of the CgYBP1 coding sequences, yielding pSL66. The KpnI-SphI fragment of pSL66 plasmid was then transformed in C. glabrata 40F1 strain and screened for hygromycin resistance. To construct a C. glabrata 40F1 strain carrying a C-terminal-tagged CgYBP1-1X HA allele under control of either the native or the strong CgPGK1 promoter, the plasmid pKGE51 was cut within the CgYBP1 ORF by enzymes ClaI and BstEII, whereas plasmid pKGE52 was cut within the CgPGK1 promoter by AvrII. The linearized plasmids were then transformed in the C. glabrata 40F1 strain to generate KGS90 (CgYBP1-1X HA URA3) and KGS91 (CgPGK1-CgYBP1-1X HA URA3). S. cerevisiae transformation was carried out by the lithium acetate technique of Ito et al. (17) and C. glabrata transformation was performed as described by Ueno et al. (18).
TABLE 1.
Strain list
| Strain | Description | Source |
|---|---|---|
| SEY6210 | MATα leu2–3, 112 ura3–52 his3-Δ200 trp1-Δ901 lys2–801 suc2-Δ9 Mel− | Scott Emr |
| SM13 | MATα leu2–3,112 ura3–52 his3-Δ200 trp1-Δ901 lys2–801 suc2-Δ9 Mel− yap1-Δ2::hisG | 20 |
| BY4742 | MATα his3-Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | Open Biosystems |
| BY4742 ybp1Δ | MATα his3-Δ1 leu2Δ0 lys2Δ0 ura3Δ0 ybp1Δ::kanMX2 | Open Biosystems |
| BY4742 YBP1-TAP | MATα his3-Δ1 leu2Δ0 lys2Δ0 ura3Δ0 YBP1-TAP::HIS3MX6 | Open Biosystems |
| SLS1 | MATα his3-Δ1 leu2Δ0 lys2Δ0 ura3Δ0 kanMX2::TDH3 YBP1-TAP::HIS3MX6 | This study |
| SLS3 | MATα his3-Δ1 leu2Δ0 lys2Δ0 ura3Δ0 YBP1-TAP::HIS3MX6 yap1Δ::kanMX2 | This study |
| Cg 40F1 | Cgura3Δ(−85 + 932)::Tn903NeoR | David Soll |
| SLS5 | Cgura3Δ(−85 + 932)::Tn903NeoR ybp1Δ::hphMX4 | This study |
| KGS90 | 40F1 CgYBP1–1X HA URA3 | This study |
| KGS91 | 40F1 CgPGK1-YBP1–1X HA URA3 | This study |
Plasmids
Plasmids used in this study are listed in Table 2. To express a GST-Ybp1 fusion protein, a BamHI-SalI fragment corresponding to ScYBP1 or CgYBP1 ORF was ligated into the BamHI and SalI sites of pGEX-6P1 (Amersham Biosciences). To construct a mutant Yap1 lacking a NLS, a PCR fragment carrying a truncation from residues 7 to 61 in the Yap1 N terminus was amplified using primers Yap1-NLS-For (CCAGATCATATGAAACAACATGACTTTTTCAAGTCTGCC) and Yap1-NLS-Rev (GCGGCCCGATTTTGGGCAGTCCTCTTCTGCTTAGTTTCAGGATCCAATTTGTACAATTCATCCATACCATGGG). This PCR fragment was then recombined in plasmid pNT13 (19) to generate a mutant GFP-Yap1 derivative lacking the nuclear localization signal. To generate plasmids carrying C-terminal 1× HA-tagged alleles of the C. glabrata YBP1 ORF, primers were used to amplify CgYBP1 ORF from genomic DNA isolated from wild-type C. glabrata strain 40F1. Cg-PGK YBP1 F1 and Cg-PGK YBP1 R1 to amplify 2-kb ORF without promoter or using primers CgYBP1-promoter-F1 and Cg-PGK YBP1 R1 to amplify a 2.5-kb cassette containing both the YBP1 ORF along and its native promoter. These PCR cassettes were then recombined in plasmid pJS20 (Cg PGK-PSD1–1XHA) excised with BamHI-SalI to yield pRS426 Cg YBP1-1XHA and pRS426 Cg PGK-YBP1-1× HA plasmids. A plasmid expressing Gpx3 in bacteria was constructed by inserting the GPX3 ORF as a BamHI fragment into pET28b(+) at the BamHI site to generate pKGE60. Expression of C629A Yap1 in bacteria was accomplished by use of a pET28b(+)-C629AYap1 construct. This plasmid was generated by ligating a KpnI-NotI fragment isolated from plasmid pRS316-C629A YAP1 (20) into the pJAW139 plasmid excised with the same enzymes, generating pKGE61.
TABLE 2.
Plasmid list
| Plasmid | Description | Source |
|---|---|---|
| pNT13 | pRS316 GFP-YAP1 | 19 |
| pSC99 | pSEYC102 TRX2-lacZ | 19 |
| YEp352-YAP1 | YEp352-YAP1 | Lab Plasmid |
| pJAW139 | pET28b(+)- YAP1 (63–650 amino acids) | Lab Plasmid |
| pKGE2 | pRS315 PGK1-HA-YBP1 | 11 |
| pKGE3 | pRS315 PGK1-HA-YBP2 | 11 |
| pKGE4 | pRS315 PGK1-HA-GPX3 | 11 |
| pKGE50 | GFP-ΔNLS-YAP1 in pRS316 | This study |
| pKGE51 | pRS426 URA3 Cg YBP1–1XHA | This study |
| pKGE52 | pRS426 CgPGK-YBP1–1XHA | This study |
| pSLS | pGEX-6P1-GST-CgYBP1 | This study |
| pKGE60 | pET28b(+)-GPX3 | This study |
| pKGE61 | pET28b(+)-C629A YAP1 | This study |
Western Blot Analysis
Cells grown in 10 ml to mid-log phase were harvested, washed, and lysed by either glass bead lysis in the presence of TCA or in buffer containing 300 mm sorbitol, 100 mm NaCl, 5 mm MgCl2, 10 mm Tris (pH 7.4), and protease inhibitors. Cell lysates were cleared and the Bradford protein assay (Bio-Rad) was used to determine the protein concentration. Equal portions of protein from each sample were run on a SDS-polyacrylamide gel. The proteins were transferred to nitrocellulose membrane, blocked with 5% nonfat dry milk in phosphate-buffered saline, and probed with polyclonal anti-Yap1 antibody (20), polyclonal anti-TAP antibody (Open Biosystems), or monoclonal anti-HA antibodies (Covance). Horseradish peroxidase-conjugated secondary antibody and the ECL kit (Pierce) were used to visualize immunoreactive protein.
β-Galactosidase Assays
Wild-type cells were transformed with low-copy number plasmids corresponding to the empty pRS315 vector or pRS315-PGK1-YBP1 plasmid along with the TRX2-lacZ gene fusion construct. Transformants were grown to mid-log phase in selective media and then either left untreated or induced by adding oxidants at the indicated concentrations for an additional 1 h. An equal number of cells from each sample was harvested at various time points, washed, and assayed for β-galactosidase activity as described (21). All assays were carried out in triplicate and represented at least three independent trials.
Co-immunoprecipitation
All immunoprecipitation assays were performed using lysed spheroplasts. To obtain spheroplasts, 300 ml of cells were harvested at A600 ∼ 0.8 and washed with 10 ml of solution 1 (1 m sorbitol, 10 mm MgCl2, 30 mm DTT, 100 μg/ml of phenylmethlysulfonyl fluoride, 50 mm K2HPO4) and followed by centrifugation. The pellet was resuspended in 10 ml of solution 2 (1 m sorbitol, 10 mm MgCl2, 30 mm DTT, 100 μg/ml of phenylmethlysulfonyl fluoride, 50 mm K2HPO4, 25 mm sodium succinate, pH 5.5) containing oxalyticase and incubated at 30 °C for 1 h. The resulting spheroplasts were pelleted by centrifugation at 5,000 × g for 20 min at 4 °C. Protein lysates were prepared by lysing these spheroplasts with lysis buffer (1% Triton X-100, 0.15 m NaCl, 50 mm Tris-HCl, pH 7.2) containing 1× protease inhibitor mixture, followed by addition of 2 mm EDTA, 200 μm sodium vanadate, 50 mm sodium fluoride, and 1 mm DTT. Lysis was performed by shaking cell suspensions on a Tomy shaker in the presence of glass beads for 5 min at 4 °C. Protein extracts were clarified by centrifuging at 14,000 × g for 5 min at 4 °C. For immunoprecipitation, washed protein A beads were incubated with cell lysates for 2 h at 4 °C, then anti-TAP antibody was added to the lysates and incubated at 4 °C overnight. This mixture was centrifuged at 14,000 × g for 5 min at 4 °C, and 50 μl of supernatant was saved as a control. After washing the beads three times with PBS (pH 7.0) containing 1 mm DTT, immunoprecipitated proteins were recovered by adding 3× Laemmli dye (0.125 m Tris, pH 6.8, 4% SDS, 20% glycerol, 10% mercaptoethanol, 2% bromphenol blue dye). These precipitates along with the inputs and other control proteins were then loaded on an 8% polyacrylamide gel and analyzed by Western blotting using a polyclonal anti-Yap1 antibody.
GST Interaction Assays
To prepare protein extracts, yeast cells were grown to mid-log phase, and 10 mm sodium azide and 10 mm potassium fluoride were directly added to the culture. After chilling on ice for 5 min, cells were harvested and resuspended with 4 m sorbitol buffer containing 1× protease inhibitor mixture (Roche Applied Science) and 2 mm DTT. After lysing the cells on a Tomy shaker in the presence of glass beads at 4 °C for 45 min, the extract was clarified by centrifugation. The resulting cell extract was pre-cleared by rotation with the washed glutathione-Sepharose 4B beads for 1 h at 4 °C, followed by centrifugation. To prepare crude extracts from cells expressing recombinant Yap1 proteins, a pET28b+ plasmid expressing Yap1 amino acid residues 63–650 (pJAW139) or a similar clone expressing the C629A form of Yap1 were transformed into BL21/DE3 bacterial cells. These transformants were induced with isopropyl 1-thio-β-d-galactopyranoside (Research Products International) and protein extracts were prepared. To decrease nonspecific interaction, the crude extract was pre-cleared by incubating for 1 h with glutathione beads, onto which recombinant GST had been bound, followed by centrifugation. GST fusion proteins were expressed and purified from bacterial lysates as described (22). The GST fusion proteins were incubated with washed glutathione-Sepharose 4B beads for 1 h. These beads were further incubated with the pre-cleared extracts for 3 h at 4 °C and then spun down at 13,000 × g for 5 min and washed three times with C buffer (25 mm HEPES, pH 7.6, 10% glycerol, 50 mm KCl, 0.1 mm EDTA) containing 2 mm DTT. The bound proteins were eluted by 50 μl of PBS, 50 mm reduced glutathione (pH 7.2). Samples were then loaded on 10% polyacrylamide gel and analyzed by Western blotting using anti-TAP (1:1000, Open Biosystems) or anti-polyclonal Yap1 antibodies (1:1500).
The experiments using Gpx3 were carried out by loading the GSH-Sepharose beads with GST-Ybp1 and Yap1 as above. Extracts from either pET28b(+)- or pKGE60 (Gpx3)-containing cells were prepared as described above. Protein extracts were split into equal fractions and then incubated at room temperature for 10 min in the presence or absence of oxidants (1 mm of either diamide or H2O2). These extracts were then passed over the Yap1-containing beads, eluates were collected and precipitated with TCA. In some cases, the proteins remaining after treatment with the extracts were eluted by addition of reduced GSH, followed by TCA precipitation. Precipitates were washed with acetone and analyzed by Western blotting using anti-Yap1 and anti-GST antisera.
Analysis of Yap1 Oxidation Status
To monitor status of Yap1 oxidation, we followed the method described earlier (13). After exposure to 1 mm H2O2 for the indicated time periods, 50 ml of cells were collected and washed with 20% TCA. The pellets were then resuspended in 12.5% TCA, glass beads were added and vortexed vigorously in a Tomy shaker for 10 min. After breakage, the extracts were collected and beads were washed again with 12.5% TCA. Beads and debris were removed by centrifugation, the supernatant was collected and pooled with the initial extract. Proteins were pelleted by high speed centrifugation and washed with ice-cold acetone twice, followed by air drying for 20 min. Dried pellets were then dissolved in 75 mm iodoacetamide, 1% SDS, 100 mm Tris-HCl (pH 8), 1 mm EDTA, complete protease inhibitors (Roche Applied Science), and incubated at 25 °C for 15 min. Samples were then subjected to dialysis against 10 mm Tris-HCl (pH 8), 50 mm NaCl, 10 mm MgCl2 for 4 h. Protein extracts were boiled in 3× Laemmli buffer (without β-mercaptoethanol) and separated by nonreducing SDS-PAGE. The separated proteins were then transferred to nitrocellulose membrane and immunoblotted using anti-Yap1 serum.
Fluorescence Microscopy
To examine the localization of Yap1-GFP fusion proteins, cells were grown to an A600 of ∼0.6 and first stained with 4′,6′-diamidino-2-phenylindole (DAPI) for 1 h to visualize the nuclei and mitochondria. Cells were then exposed to hydrogen peroxide or diamide for 20 min. Equal amounts of cells were spun down, washed, and resuspended in 50 μl of water. 8 μl of concentrated culture were visualized using an Olympus BX60 microscope. Images were captured on a Hamamatsu digital camera.
RESULTS
Ybp1 Is Limiting for H2O2 Resistance in S. cerevisiae
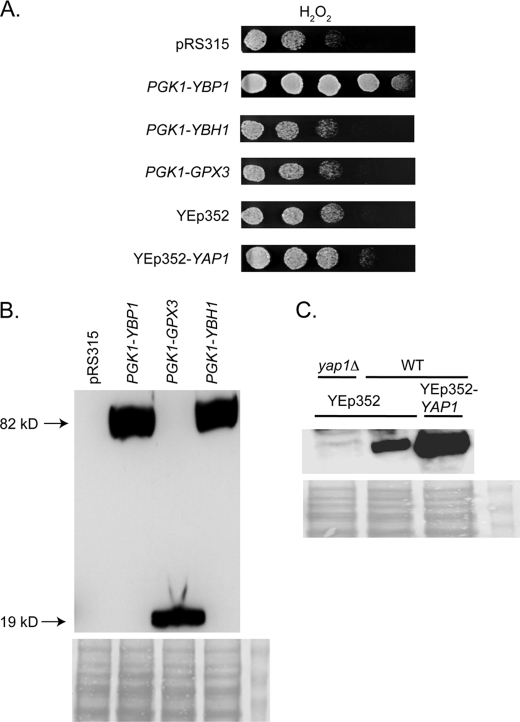
Given that several factors are required for Yap1-mediated H2O2 tolerance, we wanted to determine which of these might be limiting for this oxidative stress phenotype. Wild-type cells were transformed with plasmids that placed the various components involved in the Yap1-mediated response to H2O2 under control of the strong constitutive PGK1 promoter. Transformants were then tested for the ability to grow on medium containing H2O2 (Fig. 1A).
FIGURE 1.
Ybp1 is limiting for H2O2 tolerance. A, a low-copy number vector plasmid (pRS315) or this same plasmid containing the strong promoter from the glycolytic gene PGK1 driving YBP1, YBH1 (YBP1 homologue involved in centromere function (11, 12, 32)), or GPX3 were introduced into wild-type (SEY6210) S. cerevisiae cells. Addtionally, a high-copy number vector plasmid (YEp352) or this same plasmid containing the YAP1 gene were similarly introduced into SEY6210 cells. Transformants were grown to mid-log phase, placed on medium containing H2O2, and photographed after incubation at 30 °C. B, whole cell protein extracts were prepared from transformants described above containing the indicated expression plasmids. Equal aliquots of extracts were separated on SDS-PAGE and analyzed by Western blotting with an anti-HA antibody. Each expression plasmid contains two HA epitopes fused to the N terminus of the PGK1-driven protein. Molecular mass of the recombinant proteins is indicated in kDa on the left side of the panel. Ponceau S staining of total protein is shown at the bottom of panels B and C to confirm equal loading. C, either wild-type (WT:SEY6210) or isogenic yap1Δ cells transformed with the indicated high-copy number plasmids were grown to mid-log phase, protein extracts were prepared and analyzed by Western blotting using anti-Yap1 antibody (20).
Transformants expressing Ybp1 under control of the strong PGK1 promoter exhibited robust resistance to H2O2. Similar transformants producing driving Gpx3 or the Ybp1 homologue Ybh1 from the PGK1 promoter control did not attain similar increased H2O2 resistance. Expression of all these components were comparably high (Fig. 1B) and we verified that Yap1 was overproduced relative to the chromosomal copy (Fig. 1C). Failure to increase H2O2 resistance could not be explained by poor expression.
Strikingly, overproduction of Yap1 itself failed to significantly increase H2O2 resistance although steady-state levels of Yap1 were at least 5 times above those seen in wild-type cells. These data argued that Ybp1 levels were limiting for production of Yap1-dependent resistance to H2O2 challenge.
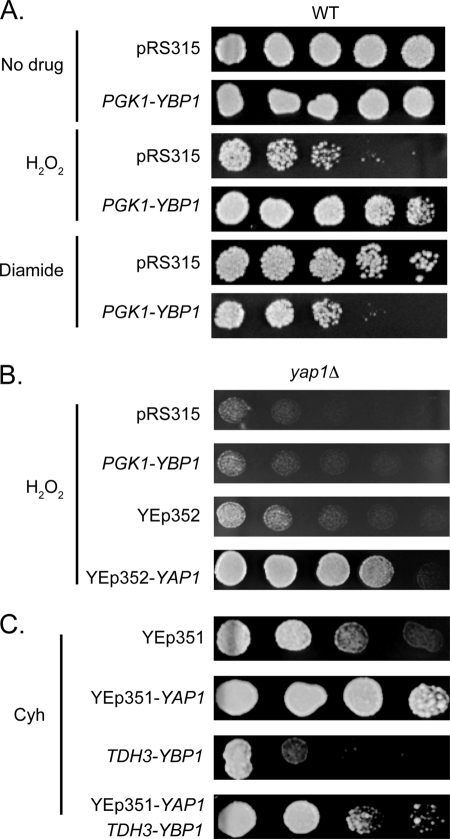
Because Ybp1 overproduction led a dramatic increase in H2O2 tolerance, these same cells were evaluated for their resistance to the oxidant diamide. Our expectation was that Ybp1 overproduction would have negligible influence on diamide resistance because loss of the YBP1 gene has been shown to specifically depress H2O2 but not diamide tolerance (11, 12). Wild-type cells transformed with the low-copy number vector plasmid or this same plasmid containing the PGK1-YBP1 fusion gene were grown to mid-log phase and then placed on medium containing H2O2, diamide, or lacking any oxidant. These transformants were allowed to grow and then photographed (Fig. 2A).
FIGURE 2.
Overproduction of Ybp1 inhibits diamide and cycloheximide resistance. A, wild-type cells were transformed with the indicated plasmids. Transformants were tested as above for the ability to tolerate H2O2 or diamide. B, a strain lacking Yap1 (yap1Δ) was transformed with the indicated plasmids. Transformants were placed on rich medium containing H2O2 to evaluate their relative resistance to this oxidant. C, isogenic wild-type cells or cells overexpressing Ybp1 (TDH3-YBP1) were transformed with an empty high-copy number vector plasmid (YEp351) or the same plasmid containing the wild-type YAP1 gene (YEp351-YAP1). Transformants were grown to mid-log phase and then placed on rich medium containing a gradient of cycloheximide (Cyh).
Surprisingly, we found that Ybp1 overproduction caused a reduction in diamide resistance while still increasing H2O2 tolerance. These data indicate that high levels of Ybp1 cause an increase in H2O2 resistance with a concomitant decrease in diamide tolerance. To ensure that the observed increase in H2O2 exhibited the expected Yap1 dependence, the PGK1-YBP1 plasmid was introduced into a yap1Δ strain along with several control plasmids. Transformants were tested for the ability to grow in the presence of H2O2 (Fig. 2B). The absence of Yap1 completely blocked the ability of the PGK1-YBP1 fusion gene to elevate H2O2 resistance above transformants containing the pRS315 vector alone. Only returning the YAP1 gene to this strain was sufficient to elevate H2O2 resistance. The ability of Ybp1 overproduction to increase H2O2 resistance was strictly dependent on the presence of Yap1.
Expression of Yap1 has also been demonstrated to markedly elevate drug resistance (23–25). Overproduction of Yap1 elevated cycloheximide resistance in wild-type cells (Fig. 2C). However, elevated expression of Ybp1 lowered the ability of cells to tolerate cycloheximide as seen above for resistance to the oxidant diamide. These data argue that the interaction of Ybp1 and Yap1 has similar negative effects on diamide and drug resistance but very different consequences to H2O2 tolerance.
Elevated Levels of Ybp1 Differentially Influence Yap1-mediated Response to Oxidants
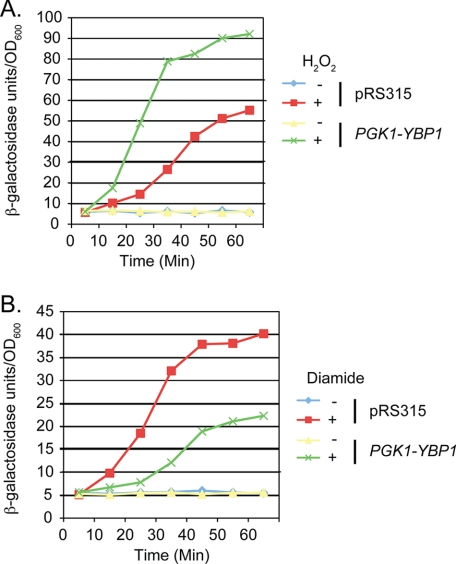
Previous studies (19, 26) have established that Yap1 activation of transcription of the thioredoxin-encoding TRX2 gene is critical for H2O2 resistance. To determine whether overproduction of Ybp1 influenced TRX2 inducible expression, a low-copy number plasmid containing a TRX2-lacZ fusion gene was introduced into wild-type cells along with the pRS315 vector containing or lacking the PGK1-YBP1 fusion gene. Transformants were grown to early log phase and then analyzed for their induction in response to challenge with either H2O2 or diamide (Fig. 3).
FIGURE 3.
Elevated Ybp1 levels exhibit differential effects on inducible gene expression. A, wild-type cells were transformed with a low-copy number plasmid carrying a TRX2-lacZ reporter gene along with a vector plasmid (pRS315) or the same vector containing a fusion gene overproducing Ybp1 (PGK1-YBP1). Transformants were grown to early log phase and challenged with H2O2 or left untreated. At the indicated time points, samples were withdrawn and TRX2-dependent β-galactosidase activity was measured. B, the same protocol as in A was followed except oxidative stress was imposed with the addition of diamide.
Overproduction of Ybp1 caused TRX2 induction in the presence of H2O2 to occur both more rapidly and to a greater extent than in wild-type cells. Overproduction of Ybp1 caused the TRX2-lacZ reporter gene to reach 50 units/A600 of β-galactosidase activity in 20 min, whereas wild-type cells required 50 min to reach this level of induction. At the last time point assayed (60 min), TRX2 expression in the presence of PGK1-YBP1 was over 90 units/A600, whereas wild-type cells only exhibited 55 units/A600.
Carrying out this same experiment in the presence of diamide as the inducing oxidant produced a distinctly different behavior (Fig. 3B). Wild-type cells rapidly elevated TRX2 expression to nearly 40 units/A600 after 45 min of diamide induction, whereas cells overproducing Ybp1 only increased to 20 units/A600. As seen for the resistance phenotypes, increased Ybp1 expression led to a striking increase in TRX2-lacZ expression upon H2O2 challenge but inhibited expression of this same reporter gene when cells were stressed with diamide.
Mechanism of Ybp1 Influence on Yap1 Activation by H2O2 and Diamide
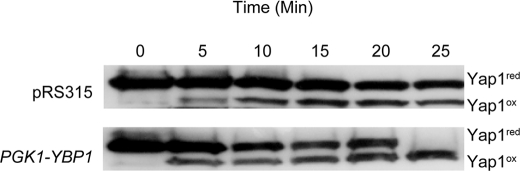
The work of Morgan and colleagues (12) provided evidence that Ybp1 was required for oxidative folding of Yap1 upon peroxide stress. Because overproduction of Ybp1 elevated both resistance to H2O2 and accelerated the induction of TRX2 by this oxidant, we tested if oxidative folding of Yap1 was similarly enhanced. Earlier work established that the H2O2-oxidized form of Yap1 migrates more rapidly through nonreducing SDS-PAGE than does the reduced form of Yap1 (13). A wild-type strain was transformed with either a vector plasmid or a similar plasmid driving YBP1 from the PGK1 promoter. Transformants were challenged with H2O2 and protein extracts were prepared in the absence of reductants. Equal aliquots of these extracts were then separated by nonreducing SDS-PAGE, transferred to nitrocellulose, and probed with an anti-Yap1 polyclonal antibody (Fig. 4).
FIGURE 4.
Wild-type cells transformed with the indicated plasmids were grown to log phase. Transformants were exposed to H2O2 and samples were withdrawn at the indicated times. Protein extracts were prepared and equal aliquots were resolved on SDS-PAGE under nonreducing conditions as described (13). Proteins were transferred to nitrocellulose and analyzed by Western blotting using the anti-Yap1 antibody. The positions of the reduced form of Yap1 (Yap1red) and the H2O2-folded form of Yap1 (Yap1ox) are indicated on the right side.
After 25 min of H2O2 stress, the only detectable form of Yap1 corresponded to the fully oxidized species when Ybp1 was produced from the PGK1 promoter. At the same time point in the presence of wild-type levels of Ybp1, the majority of Yap1 was still in the reduced form although the oxidized form was clearly detectable. These data illustrate the stimulation of H2O2-induced folding in the presence of elevated dosage of Ybp1.
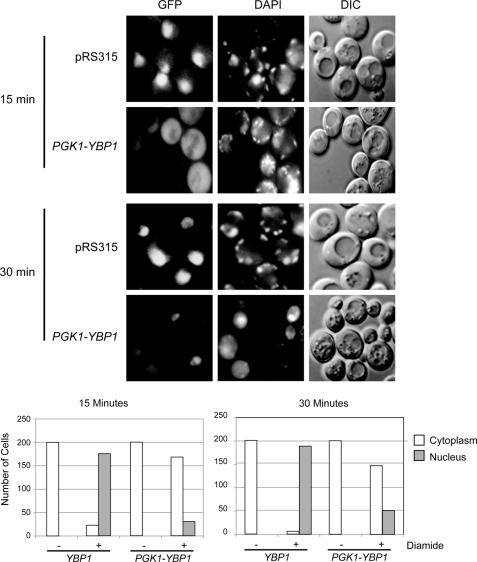
Because high level Ybp1 production inhibited both diamide induction of TRX2 expression and lowered diamide tolerance, we introduced the same pair of plasmids containing or lacking the PGK1-YBP1 fusion gene along with a GFP-Yap1-expressing plasmid into a wild-type strain. These double transformants were grown to early log phase and then treated with diamide. After 15 and 30 min of diamide exposure, these transformants were stained with DAPI and visualized by fluorescence microscopy or Nomarski optics (Fig. 5).
FIGURE 5.
Oxidative folding of Yap1 responds to Ybp1 dosage. Increased Ybp1 expression inhibits diamide-induced nuclear translocation of Yap1. Wild-type cells were transformed with a low-copy number plasmid expressing GFP-Yap1 and either a vector plasmid (pRS315) or pRS315 carrying the PGK1-YBP1 fusion gene. Transformants were grown to early log phase and then subjected to diamide stress for the times indicated on the left. Cells were withdrawn and visualized for GFP-Yap1 (GFP), DNA (DAPI), or by Nomarski optics (DIC). Quantitation of the fluorescence pattern seen in 200 representative cells is presented at the bottom of the figure.
Overproduction of Ybp1 strongly inhibited nuclear localization of GFP-Yap1 after diamide treatment. 15 min of diamide exposure was sufficient to cause nearly complete nuclear accumulation of GFP-Yap1 in the presence of wild-type levels of Ybp1 but few cells exhibited nuclear localization of the factor when PGK1-YBP1 was present. After 30 min of diamide treatment, a low level of nuclear localization was seen for GFP-Yap1 in PGK1-YBP1-containing cells but still less than that in cells containing normal Ybp1 levels.
These data suggest that whereas Ybp1 acts as a positive regulator of H2O2 resistance, as described earlier (11, 12), overproduction of Ybp1 exerts a negative influence on Yap1-mediated diamide tolerance. We hypothesized that two pools of Yap1 may exist in the cell, determined by the expression level of Ybp1. Free Yap1 (not bound to Ybp1) can mediate the response to diamide, whereas a Yap1·Ybp1 complex is responsible for gene induction during H2O2 exposure. To test this model, we directly examined the interaction of Yap1 and Ybp1 both in vitro and in vivo.
Yap1 and Ybp1 Bind Directly and Form a Stable Complex in Vivo
Previous studies found that Yap1 and Ybp1 are able to associate in vivo (11, 12). To determine whether these two proteins can directly interact, recombinant forms of each wild-type protein were expressed in bacteria as either glutathione S-transferase or polyhistidine-containing fusion proteins. A mutant form of Yap1 was also expressed in this bacterial system. C629A Yap1 was determined previously to be constitutively localized to the nucleus, irrespective of the presence of oxidants (19). C629A Yap1 was unable to respond to H2O2 challenge although this mutant protein caused cells to be hyper-resistant to diamide. The failure of C629A Yap1 to confer H2O2 resistance could be due to this mutant form being unable to interact with Ybp1 or simply being restricted to a different compartment of the cell than Ybp1. Use of this cell-free interaction assay allowed discrimination between these two different hypotheses underlying the H2O2 defect of C629A Yap1.
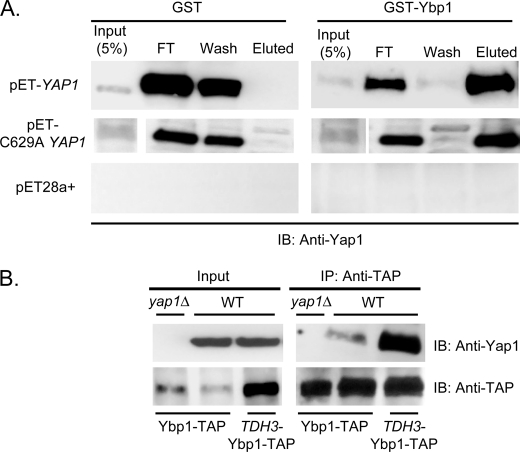
The GST-Ybp1 fusion protein or GST alone was bound to GSH-Sepharose beads. Bacterially expressed polyhistidine-Yap1 or C629A Yap1 was then passed over these beads. Unbound proteins were collected and the beads were washed extensively with buffer. Specifically bound proteins were eluted by treating the beads with reduced GSH, collected, and subjected to Western blotting using the anti-Yap1 antibody (Fig. 6A).
FIGURE 6.
Yap1 and Ybp1 interact in vitro and in vivo. A, recombinant forms of Yap1 (pET-YAP1) and Ybp1 (GST-Ybp1) were prepared in Escherichia coli and purified by standard techniques. Glutathione S-transferase (GST) or GST-Ybp1 were bound to glutathione-Sepharose beads. Next, protein lysates from cells containing an empty expression vector (pET28a+) or the same vector driving bacterial expression of Yap1 (pET-YAP1) or a constitutively nuclear mutant form (C629A YAP1) were applied. An aliquot of the lysate (Input) was blotted to estimate binding efficiency. After binding, unbound material was collected (FT) and the beads were washed with buffer (wash). Finally, bound material was eluted with reduced glutathione (Eluted). Samples of each fraction were resolved on SDS-PAGE and analyzed by Western blotting with the anti-Yap1 antibody. B, isogenic wild-type or yap1Δ cells containing the indicated YBP1-TAP fusion genes were grown to mid-log phase and protein extracts were prepared. Equal aliquots of these extracts (Input) were retained to permit confirmation of the presence of each protein. Ybp1-TAP was immunoprecipitated using the anti-TAP antibody. These immunoprecipates (IP), together with the input samples, were resolved on SDS-PAGE and analyzed by Western blotting (IB) using the indicated antibodies.
Beads with GST-Ybp1 bound efficiently captured recombinant Yap1 that could be eluted with GSH. Importantly, recombinant C629A Yap1 was able to directly associate with Ybp1 supporting the view that this mutant protein could interact with Ybp1 in vivo but their different subcellular distributions precludes this. Conversely, beads with only GST bound failed to interact with bacterially produced Yap1. This result indicates that Yap1 and Ybp1 can directly interact and no additional yeast factor is required for this complex to form.
To test if increasing the dosage of Ybp1 would lead to enhanced Yap1·Ybp1 complex formation, we constructed a strain containing a YBP1-TAP fusion gene in place of the wild-type YBP1 locus. The YBP1 promoter was then replaced with the TDH3 promoter to overproduce the Ybp1-TAP fusion protein. Additionally, we constructed a yap1Δ derivative in the presence of the YBP1-TAP fusion gene. These three strains were grown to mid-log phase, whole cell protein extracts were prepared, and the Ybp1-TAP protein was recovered by immunoprecipitation using the anti-TAP antibody. These immunoprecipitates were separated on SDS-PAGE and subjected to Western blotting analysis using either the anti-Yap1 or anti-TAP antibodies (Fig. 6B).
Elevating the expression of Ybp1-TAP led to increased Yap1 association with Ybp1. Steady-state levels of Yap1 were unaffected by the overproduction of Ybp1-TAP. These data provide further support for the idea that the level of the Yap1·Ybp1 complex represents the limiting parameter for H2O2 tolerance. Increasing the level of this complex prior to H2O2 exposure leads to an increased ability to tolerate this oxidant but decreases resistance to diamide.
Dissociation of the Yap1·Ybp1 Complex in Vitro
Our finding that Yap1 and Ybp1 can directly interact in vitro provided the possibility to examine the regulation of this binding interaction. Previous work provided important insight into the oxidative folding of Yap1. The glutathione peroxidase homologue Gpx3 is covalently linked to Yap1 in the presence of H2O2 and Ybp1 (10). Structural analysis indicated that the H2O2-oxidized form of Yap1 is likely to undergo dramatic changes in the tertiary structure of the factor because of the formation of two new disulfide bonds between the N- and C-CRD regions (14). This likely structural reorganization suggested the possibility that the Yap1·Ybp1 complex might dissociate upon H2O2 oxidation.
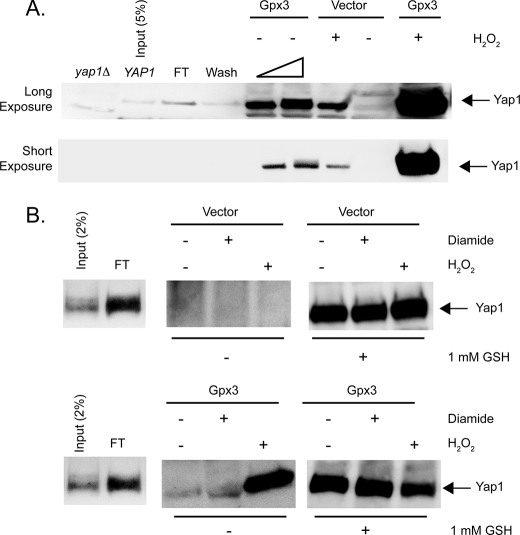
To examine this possibility, we formed the GST-Ybp1·Yap1 complex on Sepharose-GSH beads as described above. These beads represent the starting material for the subsequent assays. Whole cell protein extracts were prepared from bacterial cells containing an empty expression vector (pET28b(+)) or the same plasmid expressing S. cerevisiae Gpx3 (pET-Gpx3). These protein extracts were treated with 1 mm H2O2 or used directly with no additions. Elution of Yap1 was accomplished by the addition of appropriately prepared protein extracts to the beads. The eluate was collected from each column and concentrated by TCA precipitation. The precipitates were then subjected to Western blotting using the anti-Yap1 antibody (Fig. 7A).
FIGURE 7.
Yap1 can be released from complex formation with Ybp1 by addition of Gpx3 and H2O2. A, a bacterial lysate expressing glutathione S-transferase-Ybp1 was bound to glutathione-Sepharose beads. After washing, these beads were incubated with a protein extract prepared from cells containing a pET28a+ plasmid producing Yap1. This extract was mixed with the GSH-Sepharose bound GST-Ybp1 to form an immobilized GST·Ybp1·Yap1 complex bound to Sepharose-GSH. The beads containing the GST·Ybp1·Yap1 complex were then treated with protein lysates from bacterial cells expressing Gpx3 or containing the empty pET28b+ vector plasmid only. Where indicated, 1 mm H2O2 was added to the extracts and incubated for 10 min at room temperature. The eluate from each column was collected and concentrated by precipitation with TCA. The precipitates were washed with acetone, resuspended in 1× Laemmli buffer, and electrophoresed through SDS-PAGE. After transfer to nitrocellulose membranes, the presence of Yap1 was assessed by Western blotting using the rabbit anti-Yap1 antiserum. Protein extracts from yeast cells lacking the Yap1 protein (yap1Δ) and bacterial lysate from cells expressing Yap1 (YAP1) were analyzed as controls for the location of authentic Yap1 in this assay. A sample of the Yap1-containing extract corresponding to 5% of the total was analyzed here. FT indicates the amount of recombinant Yap1 that failed to associate with the GSH-Sepharose bound GST-Ybp1. Wash denotes the amount of Yap1 that was eluted from the column by low-salt buffer treatment. The bar of increasing width refers to the use of 20 or 40 μg of extract from Gpx3-expressing cells in these two lanes. 40 μg of extract was used in all other lanes. The top panel presents a long exposure of the same Western blot to increase the ability to detect Yap1, whereas the lower panel is a short exposure that emphasizes the dramatic increase in Yap1 release in the presence of Gpx3 and H2O2. B, beads containing GST·Ybp1·Yap1 complexes were formed as above. These beads were then incubated with protein extracts from cells containing the empty expression vector pET28b+ (Vector) (top panel) or the same plasmid expressing Gpx3 (Gpx3) (bottom panel). These incubations were carried out with no additions or with the addition of 1 mm diamide or H2O2 as indicated. Eluates were collected and blotted for the presence of Yap1 as above. After elution, the beads were then treated with 1 mm reduced GSH to elute the remaining Yap1. These GSH-dependent eluates were also concentrated by TCA precipitation and subjected to Western blot analysis to determine the level of Yap1 remaining on the column.
Addition of bacterial extracts containing Gpx3 induced a small but reproducible amount of elution of Yap1 from the beads. Importantly, incubation of these same extracts in the presence of 1 mm H2O2 released ∼10 times as much Yap1, consistent with a requirement for this oxidant in the reaction. Use of the same amount of extract from cells containing the empty expression vector led to release of very small amounts of Yap1, either in the presence or absence of H2O2 treatment.
To further examine the specificity of this in vitro reaction, the ability of diamide to influence Ybp1·Yap1 association was compared with H2O2. Reactions were performed as described above except that 1 mm diamide was used to treat extracts to determine whether any oxidant could trigger Yap1 release from this column. As can be seen in Fig. 7B, exposure of the Gpx3-containing extract to diamide failed to induce any significant Yap1 release. To confirm that equal levels of Yap1 were present in all reactions, the remaining Yap1 was eluted by treatment with reduced GSH. To ensure that release of Yap1 from the column was not accompanied by simultaneous release of GST-Ybp1, a polyclonal anti-GST antibody was used to probe the samples in Fig. 7B. No significant release of the GST fusion protein was detected upon treatment with Gpx3 and H2O2 (data not shown), consistent with the interpretation that the effect of Gpx3 and H2O2 acts on Yap1 as expected. These data support the view that the presence of Gpx3 and H2O2 triggers a change in Yap1 that in turn leads to dissociation of the Ybp1·Yap1 complex, likely by triggering the oxidative folding of Yap1. Further experiments are underway to confirm this suggestion.
Yap1 Is Folded by Ybp1 in the Cytoplasm
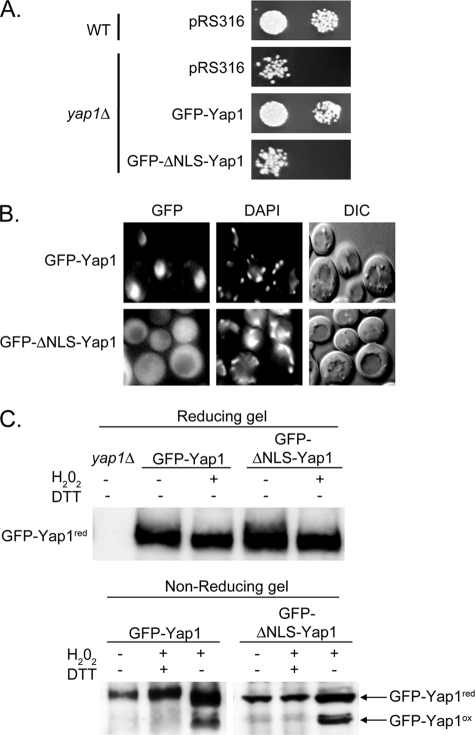
Although Ybp1 is clearly required for H2O2-induced folding of Yap1 in the cell, the compartment in which this folding takes place is unknown. To determine whether Ybp1 could fold Yap1 in the cytoplasm, a mutant form of Yap1 was constructed that lacked the previously described nuclear localization signal (NLS) of this transcription factor (27). The intent was to trap Yap1 in the cytoplasm, then challenge cells with H2O2 and assess the ability of Ybp1 to fold cytoplasmically localized Yap1. Yap1 contains a bipartite NLS in its amino terminus between residues 5 and 59. This region of the protein was removed in the context of a GFP-Yap1 fusion protein to allow visualization of the localization defect in the ΔNLS mutant protein. The GFP-ΔNLS-Yap1 protein was expressed from the native YAP1 promoter and carried on low-copy number plasmid. This mutant YAP1 allele, along with the wild-type locus, was transformed into a yap1Δ strain. Transformants were grown to mid-log phase and tested for H2O2 tolerance, nuclear localization phenotype, and ability to be folded by Ybp1 in the presence of H2O2 (Fig. 8).
FIGURE 8.
Evidence for H2O2-induced folding of Yap1 occurring in the cytoplasm. A, wild-type or yap1Δ cells were transformed with a low-copy number vector plasmid (pRS316) or the same plasmid expressing GFP-Yap1 or the NLS-deficient mutant (GFP-ΔNLS-Yap1). Transformants were placed on rich medium containing H2O2 to compare their ability to tolerate H2O2 as before. B, the transformants containing the two indicated forms of GFP-Yap1 were treated with H2O2 for 10 min and then visualized by microscopy for localization of GFP-Yap1 (GFP), organellar DNA (DAPI), or by Nomarski optics (DIC). C, protein extracts were prepared under conditions allowing detection of the H2O2-folded form of GFP-Yap1 as described above. In the top panel, these extracts were resolved using standard SDS-PAGE (Reducing gel). In the bottom panel, a nonreducing gel was used to electrophorese proteins prepared under the indicated conditions. DTT was included in some samples to fully reduce GFP-Yap1. The locations of the reduced and oxidized forms of GFP-Yap1 are indicated as before. Both Western blots were probed with anti-Yap1 antibody.
A plasmid expressing the GFP-ΔNLS-Yap1 was unable to complement the H2O2 hypersensitive allele of a yap1Δ strain (Fig. 8A). The defect caused by loss of the Yap1 NLS reduced complementation of the H2O2-sensitive phenotype by this plasmid-borne allele to a level indistinguishable from a strain lacking any functional Yap1. Consistent with the loss of complementation, the GFP-ΔNLS-Yap1 also failed to detectably accumulate in the nucleus upon treatment with H2O2 (Fig. 8B). The wild-type GFP-Yap1-expressing plasmid was able to carry out both of these functions. These results support the view that the GFP-ΔNLS-Yap1 is unlikely to enter the nucleus at a significant rate and exists primarily if not exclusively in the cytoplasm.
Having established the severity of the nuclear localization defect of GFP-ΔNLS-Yap1, the ability of this mutant form of Yap1 to be oxidatively folded was assessed. These transformants were treated with H2O2 and protein extracts were prepared under nonreducing conditions as described above. Levels of oxidatively folded Yap1 were determined by Western blotting after electrophoresis through a nonreducing SDS-PAGE (Fig. 8C).
H2O2-induced folding of the GFP-ΔNLS-Yap1 was not disturbed when compared with wild-type Yap1. This result argues that Ybp1-catalyzed folding of Yap1 can occur in the cytoplasm prior to nuclear localization of the protein during oxidative stress.
Conservation of the Role of Ybp1 in C. glabrata
The pathogenic yeast C. glabrata expresses a protein closely related to S. cerevisiae Yap1 called CgYap1 (28). Similarly, the C. glabrata genome contains a protein exhibiting striking sequence identity with Ybp1 encoded by a gene called CAGL0K06743g. We will refer to the protein product of this gene as CgYbp1. To determine whether CgYbp1 was playing a similar role in H2O2 resistance in C. glabrata, several strains of this yeast were constructed. First, a null allele lacking the entire coding sequence of CgYBP1 was generated by replacing the ORF with a hygromycin resistance cassette. To enable detection of CgYbp1 expression, a HA tag was introduced at the C terminus of the chromosomal CgYBP1 gene. Finally, this CgPGK1 promoter was inserted into the CgYbp1-HA strain in place of the wild-type CgYBP1 promoter. These strains allowed the dosage of CgYbp1 to be varied across a wide range of expression values.
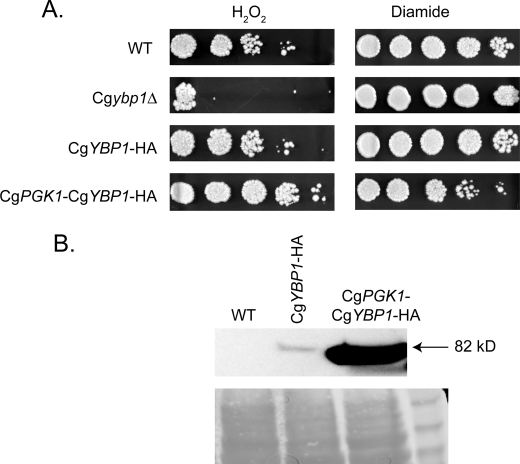
This set of strains, along with the unmodified wild-type strain, were grown to mid-log phase, placed on medium containing either H2O2 or diamide, and then allowed to grow at 37 °C (Fig. 9A). Loss of CgYbp1 from the cell resulted in a striking defect in H2O2 tolerance but had no significant influence on diamide resistance. Similar to the case in S. cerevisiae, replacing the CgYBP1 promoter with that of the CgPGK1 gene led to a pronounced increase in H2O2 resistance while causing a coupled decrease in diamide tolerance. To ensure that the CgYbp1-HA-tagged protein was being overproduced in cells containing the CgPGK1 promoter in place of the cognate wild-type promoter, protein extracts were prepared from these transformants and analyzed by Western blotting using the anti-HA antibody (Fig. 9B). CgYbp1-HA was strongly overproduced when driven from the CgPGK1 promoter compared with expression supported by the wild-type CgYBP1 promoter. These data indicate that the action of Ybp1 is conserved between S. cerevisiae and the pathogen, C. glabrata.
FIGURE 9.
Ybp1 function is conserved in C. glabrata. A, C. glabrata strains derived from the wild-type (40F1: WT) were placed on YPD medium containing the indicated oxidants. These strains included an isogenic Cgybp1Δ mutant, a strain containing an integrated CgYBP1-HA allele (CgYBP1-HA), or a strain expressing the Ybp1-HA-tagged protein from the CgPGK1 promoter (CgPGK1-CgYBP1-HA). B, whole cell protein extracts were prepared from the strains described above. Equal aliquots of protein were resolved on SDS-PAGE and analyzed by Western blotting using anti-HA antibody. The top panel presents the Western blotting data with the estimated size of CgYbp1-HA indicated on the right. The bottom panel is the same blot stained with Ponceau S prior to Western analysis. Size standards are shown in the most right lane.
DISCUSSION
Our finding that Yap1 exists in two distinct pools adds a level of fine control to the redox response controlled by this transcription factor. We divide these pools into the cycling or Ybp1-free pool along with the Ybp1-bound pool. The cycling pool represents Yap1 that moves from the cytoplasm to the nucleus and back. This pool is responsible for diamide resistance and represents the major fraction of Yap1 in the cell. The Ybp1-bound pool of Yap1 is present as a stable subpopulation and corresponds to the minor fraction of Yap1. The Ybp1-bound pool mediates the response to H2O2 challenge. These two pools can be selectively activated in an oxidant-dependent fashion to induce genes relevant to detoxification of a given oxidative stress.
Our data also suggests very different behaviors of these different pools in the presence of different oxidants. For example, although the Ybp1-bound pool represents the minority of Yap1, overexpression of Ybp1 leads to near complete conversion of Yap1 to the H2O2-folded form in the presence of this oxidant (Fig. 4). We suggest that the initial Ybp1-bound Yap1 is folded and then released from Ybp1. This free Ybp1 can then recruit more Yap1 from the cycling pool to continue production of the H2O2-folded form. This ultimately will convert all Yap1 into the H2O2-folded form and is likely at the root of eliciting diamide hypersensitivity upon Ybp1 overproduction. Elevated levels of Ybp1 cause progressively more Yap1 to be associated with Ybp1. This in turn lowers the level of the cycling pool of Yap1, leading to a diminished ability to mount a transcriptional response to diamide stress.
Conversely, diamide challenge of cells causes nuclear recruitment of the cycling pool of Yap1 but is unlikely to extract Yap1 from the Ybp1-bound pool. This assertion is supported by the observation that overproduction of Ybp1 caused diamide hypersensitivity. This is most consistent with the notion that the Ybp1·Yap1 complex remains stable even during diamide challenge.
The data reported in Fig. 6 provide the first demonstration that Yap1 and Ybp1 are likely to directly interact in the cell. We and others have been able to document this interaction via the use of two hybrid approaches (11, 12). However, our efforts to further localize the interaction motifs of these two proteins were unsuccessful, although both GST pulldown and two hybrid strategies were employed (data not shown). Small modifications of the Ybp1 protein sequence eliminated any detectable interaction with Yap1. We suggest that interaction of Yap1 and Ybp1 is likely to involve multiple, low-affinity interactions of these two proteins. The large rearrangement that occurs in the Yap1 structure upon H2O2-induced folding (14) is likely to trigger release of the folded protein from its Ybp1 partner. This suggestion is fully consistent with the release of Yap1 from the Ybp1·Yap1 complex immobilized on Sepharose-GSH beads shown in Fig. 7 upon the addition of Gpx3 and H2O2. Evidence in support of this notion comes from the observation that, whereas Yap1 clearly accumulates in the nucleus upon H2O2 stress (13, 19), Ybp1 does not (11, 12). It is important to recognize that further work is required to confirm that our in vitro reaction faithfully recapitulates H2O2-induced Yap1 folding. Direct biochemical confirmation such as done before (29) will be required to validate that the correct disulfide bonds are being produced in our in vitro reaction system.
Previous analyses were not able to clearly define localization of Ybp1 in the cell (11, 12). Our finding that a form of Yap1 that is unable to be delivered to the nucleus can be folded by Ybp1 in the presence of H2O2 confirms the ability of this regulatory step to occur in the cytoplasm. When cells initially experience oxidative stress, one of the first compartments to acquire an oxidizing potential will be the cytoplasm. Positioning the Yap1·Ybp1 complex in this compartment would allow oxidant sensing to occur relatively early. This is crucial because Yap1 does not directly detoxify oxidants but instead must move to the nucleus and induce gene expression. Coupling this geographical positioning to the high H2O2 reactivity of the Gpx3 sensor protein (discussed in Ref. 3) helps trigger Yap1 induction early in the onset of oxidative stress.
Mutant forms of Yap1 that constitutively localize in the nucleus confer hyper-resistance to diamide but fail to normally complement the H2O2 hypersensitivity of a yap1Δ strain (13, 19, 20, 30). A striking feature of these mutants is the range of different changes that produce the same phenotypic behavior: constitutive nuclear localization, diamide hyper-resistance, and H2O2 hypersensitivity. These mutations include small and large deletions as well as a variety of point mutations in various residues. Some of these mutants retain the various cysteine residues known to be required for proper oxidative folding of Yap1. A simplifying model for the consistent behavior of this wide range of mutation types is suggested by all of these mutants lacking normal Crm1 association. This would explain their nuclear localization. Combined with our finding that Ybp1 can fold Yap1 in the cytoplasm and the ability of C629A Yap1 to interact with Ybp1, these data suggest that the defect in H2O2-triggered folding in these C-terminal Yap1 mutant derivatives might be caused by separation of the cytoplasmic folding machinery from the constitutively nuclear Yap1.
Although Yap1 is clearly a key player in the response to oxidative stress, it is important to note that early identification of the YAP1 gene was driven by its role in drug resistance (23). Overproduction of Yap1 strongly elevated resistance to drugs such as cycloheximide and sulfometuron methyl, neither of which is believed to have a significant contribution from oxidative stress in terms of their toxicity. This is most consistent with an increase in the cycling pool of Yap1 leading to an elevation in expression of drug resistance genes in a fashion similar to the observed increase in resistance to thiol-reactive compounds like diethylmaleate or diamide. Evidence that this same cycling pool is also required for drug resistance is provided by the observation that increased Ybp1 levels also diminished cycloheximide resistance in a manner similar to that seen for diamide tolerance (Fig. 2C). This observation is also consistent with our suggestion that these two pools of Yap1, defined by Ybp1, carry out different roles in the cell.
Homologues of Ybp1 can be found in Candida albicans as well as in S. cerevisiae and C. glabrata. It is interesting to note that Schizosaccharomyces pombe has a Yap1 homologue called Pap1 but no recognizable Ybp1-like protein encoded by its genome. Regulation of Pap1 and Yap1 is very similar, but S. pombe surprisingly lacks this important modulator of H2O2-induced folding. Pap1 is translocated to the nucleus upon H2O2 stress and contains analogous cysteine residues to those of Yap1 (31). The different environments faced by S. cerevisiae and its Candida relatives compared with those encountered by S. pombe may have inspired these different solutions to the problem of oxidative folding of Yap1.
Acknowledgments
We thank Rob Piper and Michael Henry for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grant GM49825.
- ROS
- reactive oxygen species
- N- or C-CRD
- N- or C-terminal cysteine-rich domain
- NLS
- nuclear localization signal.
REFERENCES
- 1. Iles K. E., Forman H. J. (2002) Immunol. Res. 26, 95–105 [DOI] [PubMed] [Google Scholar]
- 2. Nauseef W. M. (2007) Immunol. Rev. 219, 88–102 [DOI] [PubMed] [Google Scholar]
- 3. Toledano M. B., Delaunay A., Monceau L., Tacnet F. (2004) Trends Biochem. Sci. 29, 351–357 [DOI] [PubMed] [Google Scholar]
- 4. Herrero E., Ros J., Bellí G., Cabiscol E. (2008) Biochim. Biophys. Acta 1780, 1217–1235 [DOI] [PubMed] [Google Scholar]
- 5. Moye-Rowley W. S. (2003) Eukaryot. Cell 2, 381–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rodrigues-Pousada C., Menezes R. A., Pimentel C. (2010) Yeast 27, 245–258 [DOI] [PubMed] [Google Scholar]
- 7. Kuge S., Toda T., Iizuka N., Nomoto A. (1998) Genes Cells 3, 521–532 [DOI] [PubMed] [Google Scholar]
- 8. Yan C., Lee L. H., Davis L. I. (1998) EMBO J. 17, 7416–7429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kuge S., Jones N., Nomoto A. (1997) EMBO J. 16, 1710–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Delaunay A., Pflieger D., Barrault M. B., Vinh J., Toledano M. B. (2002) Cell 111, 471–481 [DOI] [PubMed] [Google Scholar]
- 11. Gulshan K., Rovinsky S. A., Moye-Rowley W. S. (2004) Eukaryot. Cell 3, 318–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Veal E. A., Ross S. J., Malakasi P., Peacock E., Morgan B. A. (2003) J. Biol. Chem. 278, 30896–30904 [DOI] [PubMed] [Google Scholar]
- 13. Delaunay A., Isnard A. D., Toledano M. B. (2000) EMBO J. 19, 5157–5166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wood M. J., Storz G., Tjandra N. (2004) Nature 430, 917–921 [DOI] [PubMed] [Google Scholar]
- 15. Gulshan K., Rovinsky S. A., Coleman S. T., Moye-Rowley W. S. (2005) J. Biol. Chem. 280, 40524–40533 [DOI] [PubMed] [Google Scholar]
- 16. Goldstein A. L., McCusker J. H. (1999) Yeast 15, 1541–1553 [DOI] [PubMed] [Google Scholar]
- 17. Ito H., Fukuda Y., Murata K., Kimura A. (1983) J. Bacteriol. 153, 163–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ueno K., Uno J., Nakayama H., Sasamoto K., Mikami Y., Chibana H. (2007) Eukaryot. Cell 6, 1239–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Coleman S. T., Epping E. A., Steggerda S. M., Moye-Rowley W. S. (1999) Mol. Cell. Biol. 19, 8302–8313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wemmie J. A., Steggerda S. M., Moye-Rowley W. S. (1997) J. Biol. Chem. 272, 7908–7914 [DOI] [PubMed] [Google Scholar]
- 21. Guarente L. (1983) Methods Enzymol. 101, 181–191 [DOI] [PubMed] [Google Scholar]
- 22. Guan K. L., Dixon J. E. (1991) Anal. Biochem. 192, 262–267 [DOI] [PubMed] [Google Scholar]
- 23. Leppert G., McDevitt R., Falco S. C., Van Dyk T. K., Ficke M. B., Golin J. (1990) Genetics 125, 13–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wu A., Wemmie J. A., Edgington N. P., Goebl M., Guevara J. L., Moye-Rowley W. S. (1993) J. Biol. Chem. 268, 18850–18858 [PubMed] [Google Scholar]
- 25. Bossier P., Fernandes L., Rocha D., Rodrigues-Pousada C. (1993) J. Biol. Chem. 268, 23640–23645 [PubMed] [Google Scholar]
- 26. Kuge S., Jones N. (1994) EMBO J. 13, 655–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Isoyama T., Murayama A., Nomoto A., Kuge S. (2001) J. Biol. Chem. 276, 21863–21869 [DOI] [PubMed] [Google Scholar]
- 28. Chen K. H., Miyazaki T., Tsai H. F., Bennett J. E. (2007) Gene 386, 63–72 [DOI] [PubMed] [Google Scholar]
- 29. Okazaki S., Tachibana T., Naganuma A., Mano N., Kuge S. (2007) Mol. Cell 27, 675–688 [DOI] [PubMed] [Google Scholar]
- 30. Takeuchi T., Miyahara K., Hirata D., Miyakawa T. (1997) FEBS Lett. 416, 339–343 [DOI] [PubMed] [Google Scholar]
- 31. Toone W. M., Kuge S., Samuels M., Morgan B. A., Toda T., Jones N. (1998) Genes Dev. 12, 1453–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ohkuni K., Abdulle R., Tong A. H., Boone C., Kitagawa K. (2008) PLoS One 3, e1617 [DOI] [PMC free article] [PubMed] [Google Scholar]