Abstract
The fission yeast Schizosaccharomyces pombe exhibits invasive growth and nonsexual flocculation in response to nitrogen limitation. Gsf2, a flocculin of fission yeast, is required not only for nonsexual flocculation but also for invasive growth through the recognition of galactose residues on cell surface glycoconjugates. We found that pyruvylation negatively regulates nonsexual flocculation by capping the galactose residues of N-linked galactomannan. We investigated whether pyruvylation also regulates invasive growth. The pvg4+ gene originally was isolated as a multicopy suppressor of a pvg4 mutant defective in the pyruvylation of N-linked oligosaccharides. However, we did not detect a defect in cell surface pyruvylation in the pvg4/mbx2 deletion mutant, as assessed by alcian blue staining and a Q-Sepharose binding assay. Instead, the deletion prevented invasive growth under conditions of low nitrogen and high glucose, and it reduced the adhesion and flocculation of otherwise flocculent mutants by reducing gsf2+ expression. mbx2+-overexpressing strains exhibited nonsexual and calcium-dependent aggregation, which was inhibited in the presence of galactose but mediated by the induction of gsf2+. These findings indicate that Mbx2 mediates invasive growth and flocculation via the transcriptional activation of gsf2+ in fission yeast. In addition, we found that fission yeast Mbx2 induces the nonsexual flocculation of budding yeast by the activation of FLO1.
INTRODUCTION
Yeasts propagate in a unicellular fashion by budding or by binary fission. However, many types of yeast can switch growth modes, changing from unicellular growth to filamentous branching multicellular hyphae. This transition can be induced by a wide variety of environmental changes, ranging from pH to the nature of the carbon source. Many species of dimorphic yeasts are pathogenic toward humans and plants in hyphal form (19, 32). Invasive filamentous growth has been reported in Schizosaccharomyces pombe, and S. pombe cells grow as long and branched invasive structures under conditions of low nitrogen and high glucose (LNB medium) (1).
We recently reported that gsf2+, encoding a flocculin that binds to galactose residues located on cell surface glycoconjugates, is essential not only for nonsexual flocculation but also for filamentous invasive growth in S. pombe (7, 20). Cell surface glycoproteins play a key role in flocculation and filamentous invasive growth in yeasts. The budding yeast S. cerevisiae extends the core oligosaccharide with an α1,6-linked mannose backbone, which is further modified by the addition of α1,2- and α1,3-linked mannose side chains (3). Flo1, a lectin-like cell surface protein that aggregates cells into “flocs,” binds to mannose sugar chains on the surface of other cells (16, 38, 39). In contrast, the glycoproteins of fission yeast S. pombe contain large amounts of α-linked galactose residues in addition to α-linked mannose residues (4). The galactosylation of glycoproteins is a unique feature of fission yeast, and Gsf2 binds to galactose-containing sugar chains on the surfaces of other cells. N-linked galactomannans of S. pombe have pyruvylated galactose (PvGal) caps on a portion of the galactose residues in their outer chains (12). PvGal biosynthesis has been investigated by ethyl methanesulfonate mutagenesis in S. pombe, followed by the isolation of cells devoid of negatively charged N-glycans by Q-Sepharose exclusion and by the failure of such cells to bind the human serum amyloid P component, which acts as a lectin for terminal PvGal residues (2). Andreishcheva et al. isolated mutants that lack PvGal in cell surface glycoconjugates (pvg mutants) (2). The restoration of a negative charge to the cell surface by complementation with an S. pombe genomic library led to the identification of five genes involved in PvGal biosynthesis, designated pvg1+ to pvg5+. We previously reported that pyruvylation negatively regulates nonsexual flocculation by capping the galactose residues in N-linked galactomannan (20). However, it was not clear whether PvGal is involved in filamentous invasive growth.
In this study, we report that the deletion of the MADS box gene mbx2+/pvg4+ prevents invasive growth and flocculation and is associated with a decrease in gsf2+ mRNA levels. Mbx2/Pvg4 was found to induce nonsexual flocculation in both fission and budding yeast by upregulating flocculins.
MATERIALS AND METHODS
Strains and media.
The strains used in this study are listed in Table 1. Wild-type S. pombe strains ARC039 (h− ura4-C190T leu1-32) and ARC001 (h− leu1-32) were obtained from the Yeast Genetic Resource Center of Japan, which is supported by the National BioResource Project (NBRP). Rich YES medium (3% glucose, 0.5% yeast extract with minimal medium [MM] supplements), and synthetic MM for S. pombe were used as described previously (23). For filamentous growth, LNB medium was used (0.067 g/liter yeast nitrogen base without amino acids [Bacto], 20 g/liter glucose, and salts and vitamins as in MM). A previously described modified lithium acetate method was used for plasmid transformation and S. pombe gene disruption (24). The Escherichia coli strain used for all cloning procedures was XL1-Blue (Stratagene, CA).
Table 1.
S. pombe and S. cerevisiae strains used in this study
| Strain | Genotype | Reference or source |
|---|---|---|
| Schizosaccharomyces pombe | ||
| ARC039 | h−leu1-32 ura4-C190T | NBRPa |
| ARC001 | h−leu1-32 | NBRPa |
| gsf2Δ | h−leu1-32 ura4-C190T gsf2::ura4 | 22 |
| pvg1Δ | h−leu1-32 ura4-C190T pvg1::ura4 | 22 |
| pvg2Δ | h−leu1-32 ura4-C190T pvg2::ura4 | This study |
| pvg3Δ | h−leu1-32 ura4-C190T pvg3::ura4 | This study |
| mbx2Δ/pvg4Δ | h−leu1-32 ura4-C190T mbx2/pvg4::ura4 | This study |
| pvg5Δ | h−leu1-32 ura4-C190T pvg5::ura4 | This study |
| lkh1Δ | h−leu1-32 ura4-C190T lkh1::ura4 | 22 |
| lkh1Δ mbx2Δ | h−leu1-32 ura4-C190T lkh1 mbx2/pvg4::ura4 | This study |
| gsf1 | h−leu1-32 ura4-C190T | 41 |
| gsf1 mbx2Δ | gsf1 mbx2/pvg4::ura4 | This study |
| Saccharomyces cerevisiae | ||
| BY4742 | MATaleu2D0 ura3D0 his3D1 met15D0 | NBRPa |
| flo1Δ | MATaleu2D0 ura3D0 his3D1 met15D0 flo1::kanMX4 | Open Biosystems |
| flo10Δ | MATaleu2D0 ura3D0 his3D1 met15D0 flo10::kanMX4 | Open Biosystems |
| flo11Δ | MATaleu2D0 ura3D0 his3D1 met15D0 flo11::kanMX4 | Open Biosystems |
These strains were obtained from the Yeast Genetic Resource Center of Japan, which is supported by the National BioResource Project (NBRP).
Gene disruptions.
pvg2+, pvg3+, mbx2+, and pvg5+ were disrupted using ura4+ as a selectable marker. pvg2+ was amplified by PCR using the primers 5′-GCAGCGTAGTTGTAATCACAAAGACGCGG-3′ and 5′-GAGAGAGAGAACAAACGGATGCTTAGGAG-3′ and was cloned into the pGEM T vector (Promega, Madison, WI). The ura4+ marker was inserted at the BamHI site. The wild-type strain ARC039 was transformed with the PCR products amplified from this construct. Gene disruption was confirmed by PCR using appropriate primers: 5′-GCCCTGCTCATTTCCTAGGAAAACCATGAC-3′ and 5′-GCTTCAGCTGCATCAAGACCATGATAGGGC-3′.
pvg3+ was amplified by PCR using the primers 5′-GTTACCAACGCTATGAGGGCATTAAGGG-3′ and 5′-GCTTTTAAGCTTATGGACGGCAATGGCGTC-3′ and was cloned into the pGEM T vector. The ura4+ marker was inserted at the SnaBI and BglII sites. The wild-type strain ARC039 was transformed with the PCR products amplified from this construct. Gene disruption was confirmed by PCR using appropriate primers: 5′-CTCTGAGATAAAGAAATGTAGCAACCCGG-3′ and 5′-CCAATTGCTTACCGTCGACAAGAGAGGG-3′.
mbx2+ was amplified by PCR using the primers 5′-CACGCACTTATAAACGCACACAGACAAC-3′ and 5′-GATAGTAGTAAGGAGCGAAGTGCGTTTGCC-3′ and cloned into the pGEM T vector. The ura4+ marker was inserted at the HpaI and NheI sites. The wild-type strain ARC039 was transformed with the PCR products amplified from this construct. Gene disruption was confirmed by PCR using appropriate primers: 5′-GTTGCGGTTTCCTAGTTATTGCTATCCCTG-3′ and 5′-GACTGGTCTTGTTGCAGTACATGAGTACCC-3′.
The pvg5+ upstream region and downstream region were cloned into the pBluescript ura4+ vector. pBluescript ura4+ contains the ura4+ marker at the ClaI site. The pvg5+ upstream region was amplified by PCR using the following primers, including the indicated restriction sites (underlined): 5′-GTTTTGGTACCGTGATTTCGTGAGACCGAAG-3′ (KpnI) and 5′-GTTTCTCGAGCCGATACCATTACCAGC-3′ (XhoI). Similarly, the pvg5+ downstream region was amplified using the primers 5′-GTTTTGGATCCTTTGCCCCTGATGCGTC-3′ (BamHI) and 5′-GTTTGAGCTCATAACGGTGTAAGAGG-3′ (SacI). The pvg5+ upstream region was digested with KpnI and XhoI, the pvg5+ downstream region was digested with BamHI and SacI, and the fragments were cloned into the corresponding sites of pBluescript ura4+. The wild-type strain ARC039 was transformed with the PCR products amplified from this construct. Gene disruption was confirmed by PCR using appropriate primers: 5′-CATCTTCCAAGTATAAATGCTCTGAGCCCC-3′ and 5′-CGACAGCATAAGAGGAGTAGAAATTGCCGC-3′.
lkh1 mutant (20) cells were plated on FOA-supplemented YNB medium (6.7 g/liter yeast nitrogen base without amino acids, 20 g/liter glucose with MM supplements) to select for the loss of ura4+ activity. The ura4− cells then were used as parent cells to generate an lkh1 mbx2 double mutant.
Plasmids.
To create the pTN197-mbx2+ plasmid, mbx2+ was amplified by PCR using the following primers, including the indicated restriction sites (underlined): 5′-GTTTTCTCGAGAATGGGGCGCAAAAAAATCTCCAT-3′ (XhoI) and 5′-GTTTTGCGGCCGCCGTATTTGACATCACTGAGTGA-3′ (NotI). The fragment was digested with XhoI and NotI and cloned into the corresponding sites of pTN197 (25).
To create the pREP1-mbx2+ plasmid, mbx2+ was amplified by PCR using the following primers, including the indicated restriction sites (underlined): 5′-GTTTTGTCGACATGGGGCGCAAAAAAATCTCC-3′ (SalI) and 5′-GTTTTGGATCCTTAGTATTTGACATCACTGAG-3′ (BamHI). The fragment was digested with SalI and BamHI and cloned into the corresponding sites of pREP1 (25). pTN197-mbx2+ plasmid was used for the expression of Mbx2-GFP, and the pREP1-mbx2+ plasmid was used for the overexpression of mbx2+ in S. pombe. These vectors include LEU2 as a nutritional marker for selection.
To create pBP73G-mbx2+ and pBP73G-FLO8, mbx2+ and FLO8 were amplified by PCR using the following primers, including the indicated restriction sites (underlined): for mbx2+, 5′-GTTTTTCTAGAATGGGGCGCAAAAAAATCTCCA-3′ (XbaI) and 5′-GTTTTGGATCCTTAGTATTTGACATCACTGAGTG-3′ (BamHI); for FLO8, 5′-GTTTTTCTAGAATGAGTTATAAAGTGAATAGTTCG-3′ (XbaI) and 5′-GTTTTGGATCCTCAGCCTTCCCAATTAATAAAATTG-3′ (BamHI). The amplified fragments then were digested and cloned into the corresponding sites of pBP73G (6). The pBP73G-mbx2+ and pBP73G-FLO8 plasmids were used for the overexpression of mbx2+ and FLO8, respectively, in S. cerevisiae. These vectors include URA3 as a nutritional marker for selection.
Alcian blue staining.
Standard methods for the alcian blue staining of yeast cells were used (10, 28). Cells were cultivated at 30°C, harvested while in logarithmic growth, and washed with 0.02 N HCl. Washed cells were suspended in 1 ml of 50 μg/ml alcian blue solution (Nacalai Tesque, Inc., Japan). The cells were allowed to settle for 10 min and then were collected by centrifugation at 25°C for 5 min at 15,000 rpm. The optical density at 600 nm (OD600) of the supernatant (OD600 sup) was measured, and the difference between the OD600 of the original solution (OD600 ori) and that of the supernatant was calculated using the following formula: T = 61.3 × (OD600 ori − OD600 sup), where T is the total amount (in μg) of absorbed alcian blue. Alcian blue binding (in μg/OD600 unit) is represented by the following equation: alcian blue binding = T × d/OD600 c.c., where d is a dilution ratio to measure the diluted cell density (OD600 c.c.).
Q-Sepharose binding assay.
The Q-Sepharose binding assay has been described already (2, 11). Q-Sepharose fast flow (GE Healthcare, Japan) beads were sterilized by suspension in 95% ethanol. The beads then were washed five times with double-distilled water (DDW) to remove the ethanol. Cells were collected by centrifugation, washed three times with DDW, and resuspended in DDW. The sterile Q-Sepharose beads were added to the cell suspension, and the mixture was allowed to settle at room temperature for 5 min. Q-Sepharose and cells were observed by microscopy.
Fluorescence microscopy.
Cells were collected by centrifugation, resuspended with 5 μl of culture, and then placed on a slide glass and visualized. Fluorescent images of living cells were taken with a cooled charge-coupled device camera and stored digitally using MetaMorph software (Universal Imaging, Downingtown, PA). For fixed samples, the cultured cells were suspended in 70% ethanol, washed with phosphate-buffered saline, and suspended in 5 μl of phosphate-buffered saline containing Hoechst 33342 dye (0.1 mg/ml).
Northern blot analysis.
Total RNA from cultures with an OD600 of 0.8 to 1.0 was extracted by the glass bead method as described previously (31). Total RNA (30 μg) was separated on a 1% (wt/vol) agarose gel containing 20% formaldehyde. Subsequently, RNA was blotted onto Hybond-N membranes in 10× SSC buffer, pH 7 (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), and hybridized with gene-specific probes.
RT-PCR.
Total RNA from cultures with an OD600 of 0.8 to 1.0 was extracted by the glass bead method using the RNeasy kit (Qiagen, CA) with DNase treatment. A ReverTra Ace quantitative RT kit (Toyobo, Japan) was used for cDNA synthesis. Reverse transcription-PCR (RT-PCR) was performed using the following FLO- and ACT1-specific primer sets: FL01-specific primer set, 5′-CTACTTCTACCGAATTGACCACAGTCACTGGC-3′ and 5′-GCCAGCAATAAGGACGCAATGAAGACACTTAAAC-3′; FLO10-specific primer set, 5′-GCCTGTGGCTGCTCGATATATATTTTTGACCG-3′ and 5′-GACCCCTTTATGTCGGTAGGTGCATCTGCG-3′; FLO11-specific primer set, 5′-GTCACGACGGCTATTCCAACCACAGTTATTACC-3′ and 5′-GAATACAACTGGAAGAGCGAGTAGCAACCAC-3′; and ACT1-specific primer set, 5′-GACTCCTACGTTGGTGATGAAGCTCAA-3′ and 5′-GGAGGAGCAATGATCTTGACCTTCATGGA-3′.
RESULTS
Disruption of mbx2+/pvg4+ causes a defect in invasive growth.
Andreishcheva et al. isolated pvg mutants and identified five genes (pvg1+ to pvg5+) involved in PvGal biosynthesis (2). Pvg1 was predicted to be a pyruvyltransferase, because it shares an ∼300 to 350-amino-acid stretch of about 30% identity and about 45% similarity with nearly 20 identified or predicted pyruvyltransferases, including PssK exopolysaccharide polymerization proteins of Rhizobium leguminosarum bv. trifolii and R. leguminosarum bv. niciae (30). Pvg3 was predicted to be a galactosyltransferase (2). This corresponds with the finding that galactomannans purified from the pvg3 mutant lack both pyruvate and β1,3-linked galactose. Pvg2 and Pvg5 have no apparent orthologs, and Pvg4 (also called Mbx2) has an MEF2-type MADS box in its N-terminal region (2). The MADS box genes encode a eukaryotic family of transcriptional regulators involved in diverse and important biological functions. This class of proteins has been identified in yeasts, plants, insects, nematodes, lower vertebrates, and mammals (22). These proteins contain a conserved DNA binding and dimerization domain, which was named the MADS box after the five founding members of the family: Mcm1 (yeast) (29), Arg80 (yeast) (9), Agamous (plant) (41), Deficiens (plant) (34), and SRF (human) (26).
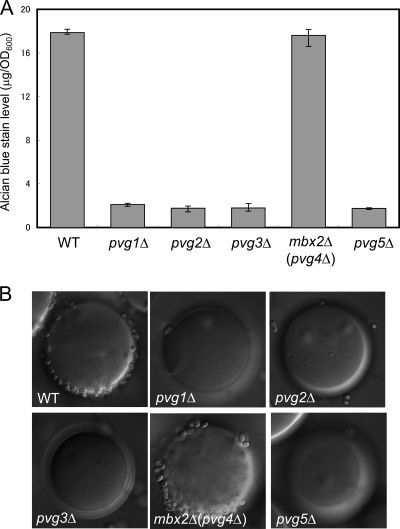
We examined whether pvg1Δ to pvg5Δ strains exhibited invasive growth on LNB plates and on a low-nitrogen and high-glucose plate relative to levels for the wild type. We constructed pvg1, pvg2, pvg3, mbx2/pvg4, and pvg5 deletion mutants and spotted them together with the wild type and a gsf2Δ mutant on LNB plates incubated at 30°C for 14 days. All strains grew equally well. The wild-type, pvg1Δ, pvg2Δ, pvg3Δ, and pvg5Δ cells remained on the agar surface after the plates were washed, while gsf2Δ and mbx2Δ/pvg4Δ cells were removed by washing (Fig. 1). This result indicates that pyruvylated galactose is not necessary for invasive growth, whereas Mbx2/Pvg4 is essential.
Fig 1.
mbx2+/pvg4+ is essential for invasive growth. Wild-type, gsf2Δ, pvg1Δ, pvg2Δ, pvg3Δ, mbx2Δ/pvg4Δ, and pvg5Δ cells were spotted onto LNB medium, which was incubated at 30°C for 14 days (Before washing). Spots of growth were washed with tap water to score for invasive growth (After washing). leu1-32 and ura4-C190T strains were complemented with leu1+ and ura4+, respectively, to allow growth on LNB medium.
Andreishcheva et al. isolated pvg mutants and obtained strains pvg1+ to pvg5+ by complementation. However, they did not construct deletion strains for each pvg gene. We therefore constructed pvg1Δ, pvg2Δ, pvg3Δ, mbx2Δ/pvg4Δ, and pvg5Δ mutants and examined whether their respective N-glycans were negatively charged by alcian blue staining and Q-Sepharose binding. Because no phosphate or sulfate has previously been detected in S. pombe glycans, pyruvate appears to be the only negatively charged functional group on the cell surface (11). Alcian blue can bind to negatively charged glycans, such as pyruvylated galactose of S. pombe and mannosylphosphate of S. cerevisiae (28). Q-Sepharose beads are anion-exchange resins that bind to negatively charged cells to form a fluffy precipitate that quickly settles.
Whereas wild-type cells bound alcian blue at a ratio of about 19 μg/OD600 cells, pvg1Δ, pvg2Δ, pvg3Δ, and pvg5Δ cells did not bind alcian blue (Fig. 2A). In contrast, mbx2Δ/pvg4Δ cells bound alcian blue as well as wild-type cells did (Fig. 2A). In addition, wild-type and mbx2Δ/pvg4Δ cells adhered to Q-Sepharose beads, but pvg1Δ, pvg2Δ, pvg3Δ, and pvg5Δ cells did not (Fig. 2B). These results indirectly suggest that the biosynthesis of pyruvylated galactose does not require Mbx2/Pvg4, therefore we designated the gene Mbx2.
Fig 2.
Alcian blue and Q-Sepharose binding assays of pvgΔ cells. (A) Alcian blue binding assay of pvgΔ cells. Alcian blue binding (in μg/OD unit) was scored based on the amount of dye binding to cells. Values are means ± SEM (n = 3). (B) Q-Sepharose binding assay of pvgΔ cells. Negatively charged cells adhere tenaciously to positively charged beads, whereas uncharged cells do not bind the beads.
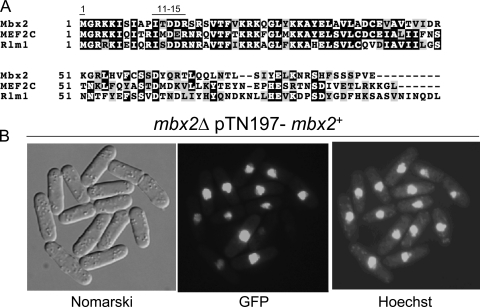
Mbx2 is a nuclear-localized MEF2-type MADS box transcription factor.
Mbx2 is a member of the MADS box protein family and is highly similar to Rlm1 in budding yeast. Here, we found that Mbx2-GFP localized to the nucleus (Fig. 3B). Rlm1 has been reported to be a key transcription factor that acts downstream of Slt2/Mpk1 mitogen-activated protein kinase (MAPK) to regulate cell wall integrity signaling (40). We compared the sequence of the putative DNA binding domain of Mbx2 to those of human MEF2C and S. cerevisiae Rlm1 (Fig. 3A). These putative DNA binding domains show significant similarity to that of Mbx2, and most importantly, MADS box residues 1 and 11 to 15, which together determine DNA binding specificity (27, 33), share significant conservation among Mbx2, Rlm1, and MEF2C. Although Mbx2 exhibits significant similarity to Rlm1, Takada et al. reported that Mbx2 seems to play only a minor role in cell wall integrity signaling (36). This indicates that MADS box domain sequence similarity is not necessarily linked to functional similarity. Interestingly, Dodou and Treisman reported that the microscopic examination of S. cerevisiae rlm1Δ cells grown in liquid medium limited for nitrogen revealed an absence of the cell aggregates observed for wild-type cells, suggesting that rlm1Δ cells are defective in flocculation (8).
Fig 3.
Mbx2 is an MEF2-like MADS box transcription factor. (A) Comparison of MADS box domains of Mbx2, Rlm1, and MEF2C. MADS box residues 1 and 11 to 15 are shown. (B) Localization of Mbx2-GFP. Nomarski differential interference contrast (Nomarski) microscopy, GFP fluorescence (GFP), and Hoechst 33324 staining (Hoechst) are shown.
Mbx2 is essential for adhesion and flocculation.
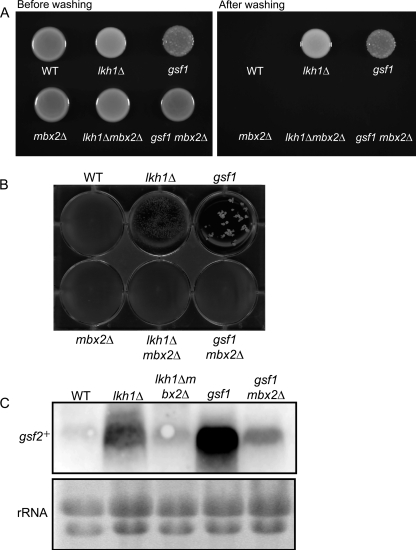
The fission yeast LAMMER kinase homolog, Lkh1, regulates Tup transcriptional repressors through phosphorylation, and an lkh1 null mutant flocculates upon reaching stationary phase in liquid media and adheres to agar surfaces (15). In addition, we previously isolated a fission yeast gsf1 mutant that flocculates constitutively during growth in liquid media and also adheres to agar (20, 37). The ability of cells to adhere to various substrates, such as host extracellular matrix, is considered a critical factor for many unicellular pathogenic organisms to establish an infection. A number of adhesive proteins have been identified in several fungal pathogens, including Candida albicans and Aspergillus fumigatus, and their expression has been shown to be an important virulence trait (21). The adhesion and nonsexual flocculation phenotypes of gsf1 and lkh1Δ mutants are completely abolished by the deletion of gsf2+, indicating their dependence on gsf2+ in S. pombe (20).
We found that adhesion and nonsexual flocculation by gsf1 and lkh1Δ mutants were completely abolished by the deletion of mbx2+ (Fig. 4). Wild-type, lkh1Δ, gsf1, mbx2Δ, lkh1Δ mbx2Δ, and gsf1 mbx2Δ cells were spotted on YES plates and incubated at 30°C for 5 days. Cells of the gsf1 and lkh1Δ mutants adhered to the agar surface after the plates were washed, but the disruption of mbx2+ in the lkh1Δ and gsf1 mutants completely abolished adherence (Fig. 4A). In addition, the nonsexual flocculation of the lkh1Δ and gsf1 mutants also disappeared after the deletion of mbx2+ (Fig. 4B). Based on Northern analysis, gsf2+ was highly expressed in lkh1Δ and gsf1 mutants grown in YES medium, whereas its expression decreased remarkably after the deletion of mbx2+ (Fig. 4C). mbx2Δ cells transformed with the nmt41-gsf2+ construct, which produces a high level of gsf2+ mRNA under the control of the thiamine-repressible nmt41 promoter (20), flocculated as well as wild-type cells transformed with the nmt41-gsf2+ construct (data not shown), indicating that the abolition of flocculation in the mbx2+ deletion mutants was linked to a reduction in gsf2+ mRNA levels.
Fig 4.
Mbx2 is required for gsf2+-dependent adhesion and nonsexual flocculation. (A) Wild-type, lkh1Δ, gsf1, mbx2Δ, lkh1Δ mbx2Δ, and gsf1 mbx2Δ cells were spotted onto YES plates and incubated at 30°C for 5 days, after which the spots were washed with a stream of water to score for adhesion. (B) Flocculation phenotypes of wild-type, lkh1Δ, gsf1, mbx2Δ, lkh1Δ mbx2Δ, and gsf1 mbx2Δ cells cultured on YES medium. (C) mRNA levels of gsf2+ in lkh1Δ, gsf1, mbx2Δ, lkh1Δ mbx2Δ, and gsf1 mbx2Δ cells. Cells were cultured in YES medium at 30°C to an OD600 of 1.0, at which time total RNA was extracted. Each RNA sample was separated on a 1% agarose gel in the presence of formaldehyde.
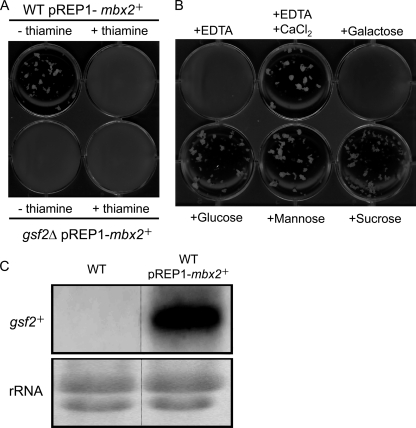
Overexpression of mbx2+ induces flocculation.
We next overexpressed mbx2+ in wild-type and gsf2Δ cells under the control of the nmt1 promoter. Wild-type and gsf2Δ cells transformed with pREP1-mbx2+ were cultured on MM-leucine medium at 30°C for 18 h with or without 20 μg/ml of thiamine. Flocculation was observed in the wild-type cells transformed with pREP1-mbx2+ but not in the gsf2Δ cells cultured without thiamine (Fig. 5A). The flocculation of the wild-type cells transformed with pREP1-mbx2+ was abolished by growth with thiamine, indicating that mbx2+ expression was required for flocculation. Flocculation in the wild-type strain overexpressing mbx2+ was inhibited by the addition of EDTA or galactose but was not inhibited by the addition of glucose, mannose, or sucrose (Fig. 5B). These flocculation phenotypes are consistent with the flocculation of gsf2+-overexpressing cells. Based on Northern analysis, no detectable expression of gsf2+ was observed in wild-type cells, whereas gsf2+ was highly expressed in mbx2+-overexpressing cells (Fig. 5C). These results indicate that Mbx2 mediates flocculation via the transcriptional activation of gsf2+ in fission yeast. Nonsexual flocculation induced by Mbx2 overexpression was abolished by deletion or point mutations of the MADS box domain of Mbx2, whereas the nuclear localization of Mbx2-GFP was unaffected by these alterations (data not shown), indicating that the MADS box domain is essential for function as a dominant flocculation gene but not for nuclear localization.
Fig 5.
mbx2+ is a dominant flocculation gene. (A) Wild-type and gsf2Δ cells transformed with pREP1-mbx2+ were cultured on MM-leucine medium at 30°C for 18 h with or without 20 μg/ml of thiamine, after which flocculation was assessed. (B) Effect of various sugars on flocculation of the mbx2+-overexpressing strain. EDTA (10 mM final concentration), CaCl2 (200 mM final concentration), and sugars (galactose, mannose, or sucrose; 200 mM final concentration) were added to mbx2+-overexpressing cells. (C) Northern analysis of the mbx2+-overexpressing strain. RNA was extracted from wild-type cells transformed with pREP1-mbx2+ and grown on MM-leucine medium at 30°C for 18 h.
Expression of S. pombe mbx2+ induces nonsexual flocculation of S. cerevisiae.
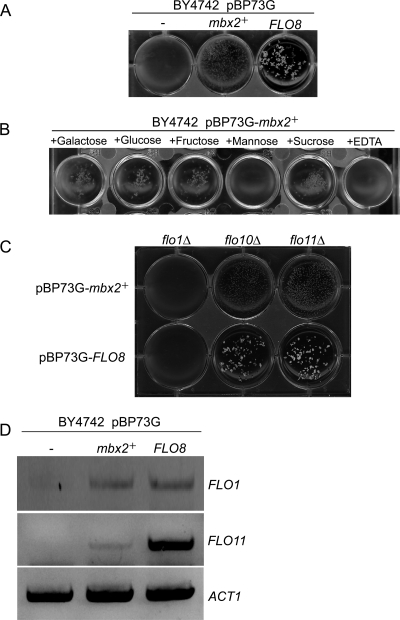
The budding yeast FLO8 gene, encoding a transcriptional activator of the dominant flocculation genes FLO1 and FLO11, induces nonsexual flocculation in S. cerevisiae (17). Interestingly, we found that nonsexual flocculation in S. cerevisiae haploid cells was induced by the expression of S. pombe mbx2+ under the control of the GPD promoter (Fig. 6A). The mbx2+-dependent flocculation of S. cerevisiae was inhibited by the addition of mannose or EDTA but was not inhibited by the addition of galactose, glucose, fructose, or sucrose (Fig. 6B). We also expressed mbx2+ and FLO8 in flo1Δ, flo10Δ, and flo11Δ cells under the control of the GPD promoter. Flocculation was observed in flo10Δ and flo11Δ cells expressing mbx2+ or FLO8 but not in flo1Δ cells (Fig. 6C). To clarify which FLO genes are induced by mbx2+ overexpression, the expression of the FLO genes was assayed by RT-PCR (Fig. 6D). Whereas FLO1 was induced in both mbx2+- and FLO8-expressing cells, FLO11 was induced highly in FLO8-expressing cells but only slightly by mbx2+. In addition, the adhesive growth of haploid cells and pseudohypha formation by diploid cells were observed in FLO8-expressing haploid and diploid cells, but these growth forms were not observed in mbx2+-expressing cells (data not shown). These results indicate that Mbx2 mediates flocculation in both S. pombe and S. cerevisiae via the transcriptional activation of gsf2+ and FLO1, respectively.
Fig 6.
S. pombe mbx2+ induces nonsexual flocculation in S. cerevisiae. (A) Nonsexual flocculation of S. cerevisiae cells expressing S. pombe mbx2+ and S. cerevisiae FLO8. pBP73G, containing the GPD promoter, was used as an overexpression vector. Wild-type (BY4742) cells transformed with pBP73G (−), pBP73G-mbx2+ (mbx2+), and pBP73G-FLO8 (FLO8) cells were cultured in SD-uracil medium at 30°C for 24 h, after which flocculation was assessed. (B) Effects of various sugars on flocculation of the mbx2+-overexpressing strain. EDTA (10 mM final concentration) and sugars (galactose, glucose, fructose, mannose, and sucrose; 200 mM final concentration) were added to the mbx2+-overexpressing cells. (C) Assessment of flocculation of flo1Δ, flo10Δ, and flo11Δ strains transformed with pBP73G-mbx2+ and pBP73G-FLO8. (D) mRNA levels of FLO1, FLO11, and ACT1 in cells transformed with pBP73G (−), pBP73G-mbx2+ (mbx2+), and pBP73G-FLO8 (FLO8). Total RNA was extracted from transformants, and gene expression was monitored by RT-PCR.
DISCUSSION
Previously, we characterized dominant genes involved in flocculation in fission yeast and cloned the gsf2+ gene that encodes a flocculin (20). In the present study, we cloned a new dominant flocculation gene, mbx2+. Mbx2 has a MADS box domain, induces nonsexual flocculation via the induction of gsf2+ (Fig. 5), and is essential for invasive growth (Fig. 1). Andreishcheva et al. gave Mbx2 the designation Pvg4, because the multicopy expression of mbx2+ complemented the defective pyruvylation phenotype of pvg4 mutants (2). However, we did not detect a difference between wild-type and mbx2Δ cells in alcian blue staining or Q-Sepharose binding assays (Fig. 2). Because Andreishcheva et al. did not carry out a genetic analysis and did not determine the mutation responsible for the pyruvylation defect in the pvg4 mutant (2), it is still unclear why the multicopy expression of mbx2+ complemented this mutation. We tested whether Mbx2 induced the expression of genes known to be involved in pyruvylation and glycosylation, including pvg1+, pvg2+, pvg3+, and pvg5+ and genes encoding glycosyltransferases, but we found no evidence for induction in mbx2+-overexpressing cells (data not shown).
In fungi, two distinct types of MADS box genes have been identified, the SRF-like class, including Mcm1 and Arg80, and the MEF2-like class, including Rlm1 and Smp1 (22). S. pombe has two MADS box-type transcription factors, Mbx1 and Mbx2. Mbx1, an S. cerevisiae Mcm1 homologue in fission yeast, belongs to the SRF-like class and is involved in regulating ecm33+, encoding a glycosyl-phosphatidylinositol (GPI)-anchored cell surface protein involved in calcium ion homeostasis as a transcriptional target of Pmk1 and Atf1 (35). Mbx2, a homologue of S. cerevisiae Rlm1 that is involved in the maintenance of cell integrity (13, 14), belongs to the MEF2-like class (Fig. 3A). Although Mbx2 shares sequence similarity with Rlm1, it has been reported that Mbx2 seems to play only a minor role in cell wall integrity signaling (36), suggesting that the S. pombe MEF2-like class transcription factor, Mbx2, is specialized in the induction of flocculation.
We previously reported that S. cerevisiae FLO8, encoding a transcriptional activator of the dominant flocculation genes FLO1 and FLO11 (5, 17, 18), induces nonsexual flocculation in both S. cerevisiae and S. pombe (20). In the present study, we demonstrated that S. pombe mbx2+ also induced nonsexual flocculation in S. cerevisiae via the induction of FLO1, although Mbx2 does not share sequence similarity with Flo8. To investigate possible functional differences between these transcription factors in fission yeast, microarray analysis was performed in wild-type and mbx2+-expressing cells. We showed that only eight genes, gsf2+, SPCC1450.08c, SPAC977.14, spn3+, SPAC22A12.04c, SPBC211.03c, bms1+, and adh4+, were upregulated in FLO8-expressing cells (20). In contrast, at least 50 genes, including gsf2+, were induced by the overexpression of mbx2+ (data not shown). gsf2+, SPCC1450.08c, and SPAC977.14 were induced by both FLO8 and mbx2+, and five other genes were specific for FLO8. The analysis of the promoter regions of gsf2+, SPCC1450.08c, and SPAC977.14 of S. pombe and FLO1 of S. cerevisiae and the binding specificities of Mbx2 and Flo8 in S. pombe will be addressed in a future study.
The major environmental signal for S. pombe to differentiate into hyphae appears to be a lack of nitrogen, provided that cells are within a fairly narrow temperature range (∼30°C) and a preferred carbon source is present (1). Our ongoing efforts are focused on analyzing the regulatory machinery that controls Mbx2, including the identification of upstream regulators.
ACKNOWLEDGMENTS
We thank Naotaka Tanaka for valuable discussions and the Yeast Genetic Resource Center Japan, which is supported by the National BioResource Project (YGRC/NBRP; http://yeast.lab.nig.ac.jp/nig/), for strains.
T.M. was supported in part by the Japan Society for the Promotion of Science (JSPS).
Footnotes
Published ahead of print 16 December 2011
REFERENCES
- 1. Amoah-Buahin E, Bone N, Armstrong J. 2005. Hyphal growth in the fission yeast Schizosaccharomyces pombe. Eukaryot. Cell 4:1287–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Andreishcheva EN, Kunkel JP, Gemmill TR, Trimble RB. 2004. Five genes involved in biosynthesis of the pyruvylated Galbeta1,3-epitope in Schizosaccharomyces pombe N-linked glycans. J. Biol. Chem. 279:35644–35655 [DOI] [PubMed] [Google Scholar]
- 3. Ballou L, Hernandez LM, Alvarado E, Ballou CE. 1990. Revision of the oligosaccharide structures of yeast carboxypeptidase Y. Proc. Natl. Acad. Sci. U. S. A. 87:3368–3372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bush DA, Horisberger M, Horman I, Wursch P. 1974. The wall structure of Schizosaccharomyces pombe. J. Gen. Microbiol. 81:199–206 [DOI] [PubMed] [Google Scholar]
- 5. Cao F, et al. 2006. The Flo8 transcription factor is essential for hyphal development and virulence in Candida albicans. Mol. Biol. Cell 17:295–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P. 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene 110:119–122 [DOI] [PubMed] [Google Scholar]
- 7. Dodgson J, et al. 2009. Functional genomics of adhesion, invasion, and mycelial formation in Schizosaccharomyces pombe. Eukaryot. Cell 8:1298–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dodou E, Treisman R. 1997. The Saccharomyces cerevisiae MADS-Box transcription factor Rlm1 is a target for the Mpk1 mitogen-activated protein kinase pathway. Mol. Cell. Biol. 17:1848–1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dubois E, Bercy J, Messenguy F. 1987. Characterization of two genes, ARGRI and ARGRIII required for specific regulation of arginine metabolism in yeast. Mol. Gen. Genet. 207:142–148 [DOI] [PubMed] [Google Scholar]
- 10. Friis J, Ottolenghi P. 1970. The genetically determined binding of alcian blue by a minor fraction of yeast cell walls. C. R. Trav. Lab. Carlsberg 37:327–341 [PubMed] [Google Scholar]
- 11. Gemmill TR, Trimble RB. 1996. Schizosaccharomyces pombe produces novel pyruvate-containing N-linked oligosaccharides. J. Biol. Chem. 271:25945–25949 [DOI] [PubMed] [Google Scholar]
- 12. Gemmill TR, Trimble RB. 1998. All pyruvylated galactose in Schizosaccharomyces pombe N-glycans is present in the terminal disaccharide, 4,6-O-[(R)-(1-carboxyethylidine)]-Galbeta1,3Galalpha1. Glycobiology 8:1087–1095 [DOI] [PubMed] [Google Scholar]
- 13. Jung US, Levin DE. 1999. Genome-wide analysis of gene expression regulated by the yeast cell wall integrity signalling pathway. Mol. Microbiol. 34:1049–1057 [DOI] [PubMed] [Google Scholar]
- 14. Jung US, Sobering AK, Romeo MJ, Levin DE. 2002. Regulation of the yeast Rlm1 transcription factor by the Mpk1 cell wall integrity MAP kinase. Mol. Microbiol. 46:781–789 [DOI] [PubMed] [Google Scholar]
- 15. Kang WH, Park YH, Park HM. 2010. The LAMMER kinase homolog, Lkh1, regulates Tup transcriptional repressors through phosphorylation in Schizosaccharomyces pombe. J. Biol. Chem. 285:13797–13806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kobayashi O, Hayashi N, Kuroki R, Sone H. 1998. Region of FLO1 proteins responsible for sugar recognition. J. Bacteriol. 180:6503–6510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kobayashi O, Suda H, Ohtani T, Sone H. 1996. Molecular cloning and analysis of the dominant flocculation gene FLO8 from Saccharomyces cerevisiae. Mol. Gen. Genet. 251:707–715 [DOI] [PubMed] [Google Scholar]
- 18. Kobayashi O, Yoshimoto H, Sone H. 1999. Analysis of the genes activated by the FLO8 gene in Saccharomyces cerevisiae. Curr. Genet. 36:256–261 [DOI] [PubMed] [Google Scholar]
- 19. Madhani HD, Fink GR. 1998. The control of filamentous differentiation and virulence in fungi. Trends Cell Biol. 8:348–353 [DOI] [PubMed] [Google Scholar]
- 20. Matsuzawa T, Morita T, Tanaka N, Tohda H, Takegawa K. 2011. Identification of a galactose-specific flocculin essential for non-sexual flocculation and filamentous growth in Schizosaccharomyces pombe. Mol. Microbiol. 82:1531–1544 [DOI] [PubMed] [Google Scholar]
- 21. Mendes-Giannini MJ, Soares CP, da Silva JL, Andreotti PF. 2005. Interaction of pathogenic fungi with host cells: molecular and cellular approaches. FEMS Immunol. Med. Microbiol. 45:383–394 [DOI] [PubMed] [Google Scholar]
- 22. Messenguy F, Dubois E. 2003. Role of MADS box proteins and their cofactors in combinatorial control of gene expression and cell development. Gene 316:1–21 [DOI] [PubMed] [Google Scholar]
- 23. Moreno S, Klar A, Nurse P. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194:795–823 [DOI] [PubMed] [Google Scholar]
- 24. Morita T, Takegawa K. 2004. A simple and efficient procedure for transformation of Schizosaccharomyces pombe. Yeast 21:613–617 [DOI] [PubMed] [Google Scholar]
- 25. Nakamura T, Kishida M, Shimoda C. 2000. The Schizosaccharomyces pombe spo6+ gene encoding a nuclear protein with sequence similarity to budding yeast Dbf4 is required for meiotic second division and sporulation. Genes Cells 5:463–479 [DOI] [PubMed] [Google Scholar]
- 26. Norman C, Runswick M, Pollock R, Treisman R. 1988. Isolation and properties of cDNA clones encoding SRF, a transcription factor that binds to the c-fos serum response element. Cell 55:989–1003 [DOI] [PubMed] [Google Scholar]
- 27. Nurrish SJ, Treisman R. 1995. DNA binding specificity determinants in MADS-box transcription factors. Mol. Cell. Biol. 15:4076–4085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Odani T, Shimma Y, Wang XH, Jigami Y. 1997. Mannosylphosphate transfer to cell wall mannan is regulated by the transcriptional level of the MNN4 gene in Saccharomyces cerevisiae. FEBS Lett. 420:186–190 [DOI] [PubMed] [Google Scholar]
- 29. Passmore S, Elble R, Tye BK. 1989. A protein involved in minichromosome maintenance in yeast binds a transcriptional enhancer conserved in eukaryotes. Genes Dev. 3:921–935 [DOI] [PubMed] [Google Scholar]
- 30. Sadykov MR, Ivashina TV, Kanapin AA, Shliapnikov MG, Ksenzenko VN. 1998. Structure-functional organization of exopolysaccharide biosynthetic genes in Rhizobium leguminosarum bv. viciae VF39. Mol. Biol. (Moscow) 32:797–804 [PubMed] [Google Scholar]
- 31. Sambrook J, Fritsch E, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 32. Sanchez-Martinez C, Perez-Martin J. 2001. Dimorphism in fungal pathogens: Candida albicans and Ustilago maydis–similar inputs, different outputs. Curr. Opin. Microbiol. 4:214–221 [DOI] [PubMed] [Google Scholar]
- 33. Sharrocks AD, von Hesler F, Shaw PE. 1993. The identification of elements determining the different DNA binding specificities of the MADS box proteins p67SRF and RSRFC4. Nucleic Acids Res. 21:215–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sommer H, et al. 1990. Deficiens, a homeotic gene involved in the control of flower morphogenesis in Antirrhinum majus: the protein shows homology to transcription factors. EMBO J. 9:605–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Takada H, et al. 2010. The cell surface protein gene ecm33+ is a target of the two transcription factors Atf1 and Mbx1 and negatively regulates Pmk1 MAPK cell integrity signaling in fission yeast. Mol. Biol. Cell 21:674–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Takada H, et al. 2007. Atf1 is a target of the mitogen-activated protein kinase Pmk1 and regulates cell integrity in fission yeast. Mol. Biol. Cell 18:4794–4802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tanaka N, et al. 1999. Cell surface galactosylation is essential for nonsexual flocculation in Schizosaccharomyces pombe. J. Bacteriol. 181:1356–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Teunissen AW, Steensma HY. 1995. Review: the dominant flocculation genes of Saccharomyces cerevisiae constitute a new subtelomeric gene family. Yeast 11:1001–1013 [DOI] [PubMed] [Google Scholar]
- 39. Verstrepen KJ, Derdelinckx G, Verachtert H, Delvaux FR. 2003. Yeast flocculation: what brewers should know. Appl. Microbiol. Biotechnol. 61:197–205 [DOI] [PubMed] [Google Scholar]
- 40. Watanabe Y, Irie K, Matsumoto K. 1995. Yeast RLM1 encodes a serum response factor-like protein that may function downstream of the Mpk1 (Slt2) mitogen-activated protein kinase pathway. Mol. Cell. Biol. 15:5740–5749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yanofsky MF, et al. 1990. The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 346:35–39 [DOI] [PubMed] [Google Scholar]