Acetylation of the SUN protein Mps3 by Eco1 regulates nuclear organization, including chromosome cohesion, telomere tethering, and gene silencing, without affecting Mps3 distribution in the nuclear membrane. It is one of the regulatory pathways known to affect chromosome positioning within the nucleus.
Abstract
The Saccharomyces cerevisiae SUN-domain protein Mps3 is required for duplication of the yeast centrosome-equivalent organelle, the spindle pole body (SPB), and it is involved in multiple aspects of nuclear organization, including telomere tethering and gene silencing at the nuclear membrane, establishment of sister chromatid cohesion, and repair of certain types of persistent DNA double-stranded breaks. How these diverse SUN protein functions are regulated is unknown. Here we show that the Mps3 N-terminus is a substrate for the acetyltransferase Eco1/Ctf7 in vitro and in vivo and map the sites of acetylation to three lysine residues adjacent to the Mps3 transmembrane domain. Mutation of these residues shows that acetylation is not essential for growth, SPB duplication, or distribution in the nuclear membrane. However, analysis of nonacetylatable mps3 mutants shows that this modification is required for accurate sister chromatid cohesion and for chromosome recruitment to the nuclear membrane. Acetylation of Mps3 by Eco1 is one of the few regulatory mechanisms known to control nuclear organization.
INTRODUCTION
Accurate transmission of genetic and epigenetic information requires the nonrandom positioning of chromosomes within the nucleus. Binding of chromosomes to inner nuclear membrane (INM) proteins is one mechanism to localize specific regions of the genome to the nuclear periphery. Tethering of chromosomes at the nuclear membrane serves a variety of functions, including the regulation of transcription, control of recombination, and repair of double-stranded DNA breaks (DSBs; reviewed in Mekhail and Moazed, 2010; Taddei et al., 2010). The evolutionarily conserved SUN-domain family of INM proteins is involved in multiple aspects of nuclear organization during vegetative growth and meiotic prophase (reviewed in Fridkin et al., 2009; Razafsky and Hodzic, 2009; Starr and Fridolfsson, 2010).
During meiosis, SUN proteins are required for the formation of a specialized chromosome arrangement known as the meiotic bouquet that facilitates homologue pairing and meiotic recombination (Hiraoka and Dernburg, 2009). SUN proteins also function in clustering of telomeres or centromeres at the nuclear envelope in mitotic cells and play a role in recruitment of certain types of DNA lesions to the nuclear periphery for repair after damage (Graf et al., 2004; Bupp et al., 2007; King et al., 2008; Kalocsay et al., 2009; Oza et al., 2009; Schober et al., 2009). In many organisms, SUN proteins are involved in attachment of the centrosome or spindle pole body (SPB) to the nuclear envelope (Malone et al., 2003; Jaspersen et al., 2006; Xiong et al., 2008; Zhang et al., 2009). Disruption of this interaction results in aberrant spindle formation and chromosome segregation defects, indicating that SUN proteins play a direct role in the maintenance of genomic stability.
The role of SUN proteins in nuclear organization has been extensively characterized in Saccharomyces cerevisiae, which contains a single SUN protein, Mps3 (Jaspersen et al., 2006). Mps3 is a structural component of the budding yeast SPB and is essential for SPB duplication once per cell cycle (Jaspersen et al., 2002; Nishikawa et al., 2003). Mps3 also localizes to the peripheral nuclear envelope, where it plays a role in chromosome positioning during both vegetative growth and sporulation (Bupp et al., 2007; Conrad et al., 2007, 2008; Rao et al., 2011). During S phase of the mitotic cell cycle, Mps3 links telomeric chromatin at the nuclear periphery through interactions with the silent information regulator Sir4 and possibly through Lrs4, a subunit of the nucleolar cohibin complex (Bupp et al., 2007; Chan et al., 2011). Telomere ends are also positioned at the nuclear periphery in both G1 and S phase by the yKu70/yKu80 heterodimer in a pathway that requires the telomerase complex subunits Est1 and Est2 (Taddei et al., 2004; Schober et al., 2009). We will refer to this pathway as the yKu/Est pathway of telomere tethering. Of interest, yKu70/yKu80, telomerase, and Mps3 all play a role in recruitment of nonrepairable DSBs to the nuclear periphery, presumably due to the formation of telomere-like ends at the break site (Oza et al., 2009; Oza and Peterson, 2010; Schober et al., 2009).
To achieve the multiple functions just mentioned, Mps3 must have multiple binding partners, which raises an important question: how do cells regulate the interaction between proteins to achieve the correct spatial and temporal organization of chromosomes within the nucleus? One possibility is that the binding protein is not synthesized until a particular time in the yeast life cycle. An example of this type of regulation occurs for Ndj1, which is needed only in meiosis and is not made in mitotically dividing cells (Conrad et al., 2007). A second possibility is that binding is regulated by posttranslational modification of either Mps3 or the partner. Mps3 is known to be glycosylated, but this modification occurs in the central region of the protein and would presumably not affect binding to nuclear proteins (Nishikawa et al., 2003). Other modifications of Mps3 have not been reported, although the Caenorhabditis elegans orthologue of Mps3, SUN-1, is phosphorylated on several sites during meiotic bouquet formation. Phosphorylation of SUN-1 requires the activity of the CHK-2 and PLK-2 kinases (Penkner et al., 2007; Harper et al., 2011; Labella et al., 2011). Posttranslational modification could directly regulate binding to nuclear proteins, or it could have an indirect effect through a change in subcellular localization of either Mps3 or its partner so that the two proteins are spatially separated.
The conserved Eco1/Ctf7 acetyltransferase (hereafter called Eco1) is an excellent candidate to regulate Mps3 functions in the nucleus. Eco1 is essential for the establishment of sister chromatid cohesion during S phase of the cell cycle due to its role in acetylation of two conserved lysine residues in Smc3, a structural component of the multisubunit cohesin complex (Skibbens et al., 1999; Toth et al., 1999; Rolef Ben-Shahar et al., 2008; Unal et al., 2008; Zhang et al., 2008; Rowland et al., 2009). Eco1 is also involved in maintaining cohesion after DNA DSBs, control of recombination, regulation of gene expression, condensation of chromosomes, and tethering of telomeres at the nuclear periphery (Strom et al., 2004, 2007; Unal et al., 2004, 2007; Gard et al., 2009; Lu et al., 2010). A number of additional Eco1 targets have been proposed based largely on acetylation by Eco1 in vitro; however, the biological importance of each is poorly understood (reviewed in Skibbens, 2009; Xiong and Gerton, 2010). Although Mps3 is not among these targets, Mps3 was previously identified as an Eco1-binding protein in a genome-wide yeast two-hybrid screen (Uetz et al., 2000). Eco1 and Mps3 can also interact in vitro and in vivo, at least when Eco1 is overproduced, further supporting the idea that Eco1 may regulate Mps3 function (Antoniacci et al., 2004). Analysis of an mps3 mutant unable to associate with Eco1 showed that Mps3 plays a nonessential role in the establishment of sister chromatid cohesion (Antoniacci et al., 2004; Antoniacci and Skibbens, 2006). However, because this mutant is inviable at high temperatures most likely due to a SPB duplication defect, it is possible that the failure in chromosome cohesion is an indirect result of aberrant spindle formation.
Intrigued by the possibility that Eco1 might regulate Mps3 function in nuclear organization, we further investigated the Mps3–Eco1 interaction. We found that Mps3 is an Eco1 substrate in vitro and in vivo. Analysis of an mps3 mutant that lacks the Eco1 acetylation sites revealed that acetylation is not essential for Mps3 function in SPB duplication, DNA damage repair, or Mps3 distribution in the nuclear membrane. However, the acetylation-site mutant shows defects in sister chromatid cohesion, as well as decreased telomere tethering and gene silencing at the nuclear periphery, indicating that Eco1 acetylation of Mps3 regulates its function in chromosome organization, possibly by affecting its ability to interact with certain telomere-associated proteins.
RESULTS
Mps3 is acetylated by Eco1 in vivo
One possible explanation for Mps3 binding to Eco1 in the two-hybrid system and its role in establishment of sister chromatid cohesion is that Mps3 is a substrate of Eco1. To test whether Eco1 acetylates Mps3 in vivo, we tagged the endogenous copy of MPS3 at its C-terminus with three copies of the FLAG epitope in wild-type and eco1-1 mutants. The eco1-1 mutant contains a glycine–to–aspartic acid substitution at position 211 in the catalytic domain and displays virtually no acetyltransferase activity when assayed in vitro (Ivanov et al., 2002; Zhang et al., 2008; Lu et al., 2010). Yeast cells harboring this allele arrest growth at high temperatures due to a defect in the establishment of sister chromatid cohesion, presumably because proteins involved in cohesion such as Smc3 are not acetylated (Toth et al., 1999; Rolef Ben-Shahar et al., 2008; Unal et al., 2008; Zhang et al., 2008; Rowland et al., 2009).
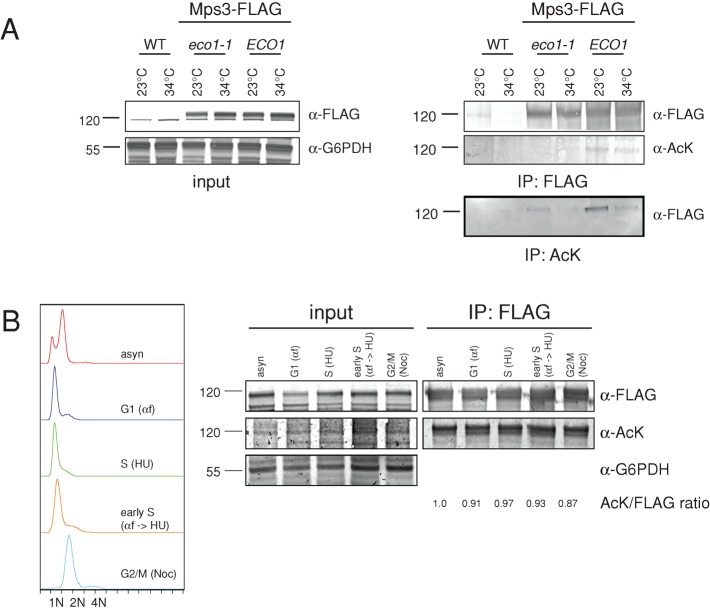
Proteins were immunoprecipitated with anti-FLAG antibodies from untagged, eco1-1 MPS3-3xFLAG, and MPS3-3xFLAG strains and were analyzed by immunoblotting with α-FLAG and anti–pan-acetyl lysine (α-Ack) antibodies to detect the presence of total Mps3-3xFLAG and acetylated Mps3-3xFLAG, respectively. Mps3-3xFLAG was readily detected in all samples. However, a dramatic reduction in the acetylated Mps3 levels was observed in the eco1-1 mutant at both the permissive and nonpermissive temperatures compared with wild-type cells using the α-Ack antibody (Figure 1A), demonstrating that Mps3 is acetylated in vivo and that a significant fraction of this acetylation requires the activity of Eco1. Acetylated proteins were also immunoprecipitated with α-Ack antibodies, and we found that Mps3-3xFLAG was present in these samples in wild-type cells and in eco1-1 mutants grown at the permissive temperature (Figure 1A). It is important to note that the level of acetylated Mps3-3xFLAG present in eco1-1 mutants at 23°C was dramatically reduced compared with wild-type cells, suggesting that the eco1-1 mutant is partially defective in acetylation even at the permissive temperature. This is consistent with previous reports showing that eco1-1 mutants display cohesion defects and lack acetyltransferase activity at 23°C (Toth et al., 1999; Ivanov et al., 2002; Zhang et al., 2008).
FIGURE 1:
Mps3 is acetylated by Eco1 in vivo. (A) A wild-type untagged strain (WT; SLJ001), as well as wild-type (ECO1; SLJ2860) and eco1-1 (SLJ2853) mutant cells in which the endogenous copy of MPS3 was tagged at its C-terminus with three copies of the FLAG epitope, were grown at 23°C or shifted to 34°C for 3 h, and then lysates were prepared using liquid nitrogen grinding. The levels of Mps3-3xFLAG were examined by immunoblotting whole-cell extracts with anti-FLAG antibodies. Anti-G6PDH serves as a loading control. Proteins were immunoprecipitated with anti-FLAG M2 and analyzed by immunoblotting with anti-FLAG and α-Ack antibodies to detect the presence of total Mps3 and acetylated Mps3, respectively. In addition, acetylated proteins were immunoprecipitated from lysates using α-Ack antibodies and analyzed for the presence of Mps3-3xFLAG by immunoblotting with α-FLAG antibodies. (B) The strain containing Mps3-3xFLAG (SLJ2860) was grown asynchronously (asyn), or was arrested for 3 h in G1 phase using 1 μg/ml α-factor, in S-phase using 10 mM HU, or in G2/M using 10 μg/ml nocodazole. To arrest cells in early S phase, cells were arrested in G1 using 1 μg/ml α-factor and then released into media containing 10 mM HU. The cell cycle arrest was confirmed by flow cytometric analysis of DNA content, and levels of Mps3-3xFLAG and its acetylation were examined as in A. The level of acetylated Mps3 detected in each sample is shown. Positions of molecular mass markers (kDa) are indicated next to each blot.
The eco1-1 mutants exhibit a prolonged mitotic delay (Toth et al., 1999), so one explanation for the decrease in Mps3 acetylation in the mutant is that Mps3 is not acetylated in mitosis. To exclude this possibility, we examined Mps3 acetylation during cell cycle arrests using α-factor, hydroxyurea (HU), and nocodazole. Mps3-3xFLAG acetylation in cells arrested in G1 phase, S phase, or mitosis was equal to the asynchronous condition (Figure 1B), indicating that the decreased acetylation we observed in eco1-1 mutants is due to loss of acetyltransferase activity rather than a cell cycle arrest. Taken together, these data suggest that Mps3 is acetylated in vivo in an Eco1-dependent manner.
The Mps3 N-terminus is acetylated by Eco1 in vitro
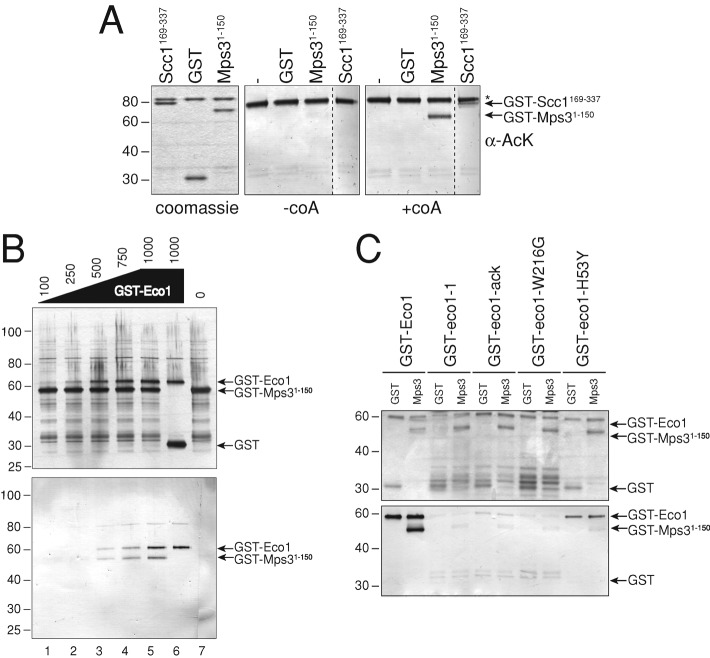
To determine whether Mps3 is a direct target of Eco1, we expressed and purified a fusion between glutathione S-transferase (GST) and the N-terminal 150 amino acids of Mps3, which are present in the nucleoplasm (GST-Mps31-150; Nishikawa et al., 2003); a fusion between GST and amino acids 169–337 of Scc1 (GST-Scc1169-337), which was shown to be acetylated by Eco1 in vitro (Ivanov et al., 2002; Lu et al., 2010); and GST alone. In addition, we purified enzymatically active GST-Eco1 from bacteria. We mixed GST-Eco1 with GST, GST-Scc1169-337, or GST-Mps31-150 in the presence and absence of acetyl-CoA to test whether Eco1 acetylates Mps3. Three lines of evidence demonstrate that the Mps3 N-terminus is acetylated by Eco1 in vitro. First, only when acetyl-CoA was present were we able to detect acetylation of GST-Scc1169-337 and GST-Mps31-150 (Figure 2A). Second, we observed a dose-dependent increase in the amount of GST-Mps31-150 that was acetylated when we added increasing amounts of GST-Eco1 (Figure 2B). Finally, we observed efficient acetylation of the Mps3 N-terminus only when the GST-Mps31-150 was incubated with wild-type GST-Eco1 but not with any of the four GST-eco1 mutant proteins with significantly reduced catalytic activity (Figure 2C; Ivanov et al., 2002; Zhang et al., 2008; Lu et al., 2010). Based on comparison to the bona fide Eco1 substrate, the GST-Scc1169-337 fragment, Mps3 appears to be acetylated at least as well, if not slightly more efficiently, under our reaction conditions in vitro (Figure 2A). The slowly migrating band in these reactions is due to autoacetylation of Eco1, which occurs in Escherichia coli and when activity is assayed in vitro (Ivanov et al., 2002; Lu et al., 2010).
FIGURE 2:
Mps3 is acetylated by Eco1 in vitro. (A) Recombinant GST, GST-Mps31-150, and GST-Scc1169-337 were treated with GST-Eco1 in the presence and absence of CoA in an acetylation reaction in vitro. Reaction products were analyzed by SDS–PAGE, followed by Coomassie blue staining (left) or by immunoblotting with α-Ack antibodies (right). The migration of GST-Mps31-150 and GST-Scc1169-337 is indicated by arrows. An asterisk marks the position of GST-Eco1, which autoacetylates in vitro even in the absence of exogenous CoA, as previously shown (Ivanov et al., 2002). (B) GST-Mps31-150 was incubated with increasing amounts of GST-Eco1 (nanograms; lanes 1–5), with GST alone (lane 6; 1000 ng), or in the absence of any acetyltransferase (lane 7) in the presence of 3H-CoA. Total protein was detected by Coomassie blue staining (top), and acetylated proteins were visualized by immunoblotting with α-Ack antibodies (bottom). (C) GST-Mps31-150 or GST alone was incubated in an acetylation assay with wild-type GST-Eco1 or with one of four different GST-eco1 mutant proteins that have differing catalytic activity (Ivanov et al., 2002; Lu et al., 2010). α-GST (top) and α-Ack (bottom) Western blots show total protein levels and acetylated proteins, respectively, in each reaction. Positions of molecular mass markers (kDa) are indicated next to each blot.
Eco1 acetylates lysines 147, 148, and 150 in Mps3 in vitro
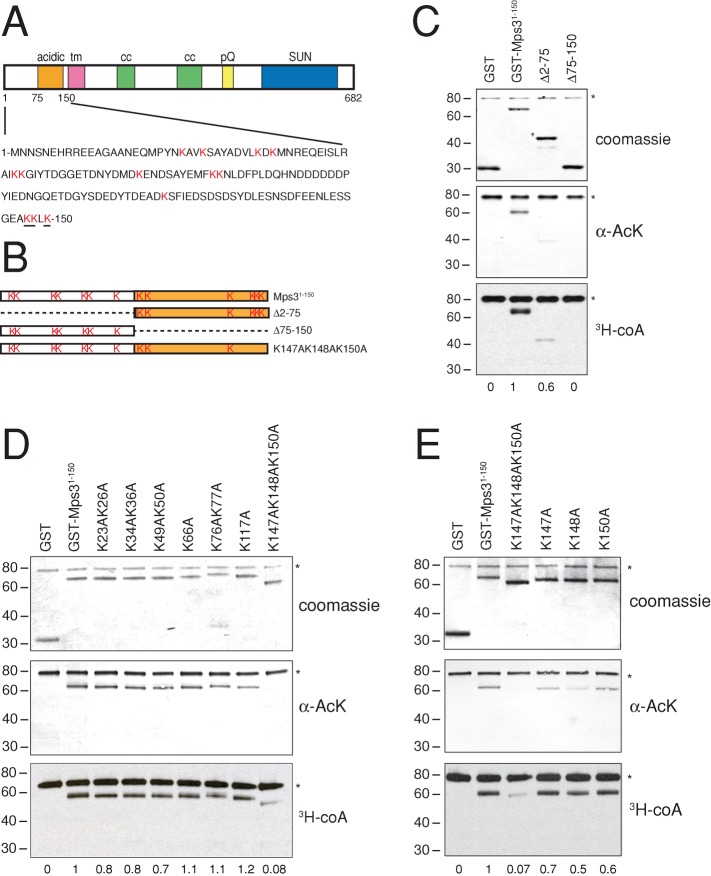
The Mps3 N-terminus has 13 lysine residues that are possible sites of acetylation (Figure 3A). Deletion of amino acids 2–75 resulted in a 40% decrease in acetylation, whereas removal of amino acids 75–150 led to a version of the N-terminus that was no longer acetylated, as determined by Western blotting with anti-AcK antibodies and 3H-CoA incorporation (Figure 3, B and C). This suggests that the primary acetylation site(s) is located between residues 75 and 150. To map the specific lysine residue(s), we created alanine substitution mutants in each lysine in the N-terminus and examined acetylation by Eco1 in vitro. Mutation of residues 147, 148, and 150 to alanine virtually abolished GST-Eco1 acetylation (Figure 3, D and E). Each of these sites contributes to overall acetylation by Eco1, as single and double mutants in these residues were still efficiently acetylated (Figure 3E; unpublished data). Although a small level of acetylation could be detected in GST-Mps31-150K147AK148AK150A, quantitation of 3H-CoA incorporation showed a reproducible reduction in acetylation by >90% compared with the wild-type GST-Mps31-150 in our in vitro reactions (Figure 3, D and E). On the basis of these data, we conclude that Eco1 acetylates Mps3 on three lysine residues in the N-terminus: K147, K148, and K150.
FIGURE 3:
Eco1 acetylates lysines 147, 148, and 150 on Mps3. (A) Schematic of Mps3 showing different domains, including the N-terminal acidic domain, the transmembrane domain (tm), coiled-coil domains (cc), a polyglutamine region (pQ), and the SUN domain. Amino acids located between 1 and 150 are shown, and the 13 lysine (K) residues in this region that could be possible acetylation sites are highlighted in red. The three lysines that appear to be acetylated by Eco1 are underlined. (B) Schematic of fusions of the Mps3 N-terminus fused to GST. GST-Mps31-150 contains amino acids 1–150 and includes all 13 lysine residues. The deletion constructs ∆2-75 and ∆75-150 remove amino acids 2–75 and 75–150, respectively, and the K147A K148A K150A mutant has three key lysine residues mutated to alanine. (C) GST, GST-Mps31-150, and the indicated deletion mutants were incubated with 3H-CoA in an acetylation assay. Reaction products were analyzed by Coomassie blue staining (top), by immunoblotting with α-Ack antibodies (center), and by autoradiography (bottom). (D, E). Similarly, specific lysine residues were mutated to the nonacetylatable residue alanine, and the ability of each to serve as an Eco1 substrate was tested in an acetylation reaction in vitro as in C, Acetylation level was quantitated based on 3H-CoA incorporation into the GST-Mps3 fusion protein. The levels of incorporation on GST only and GST-Mps31-150 were set to 0 and 1, respectively. Asterisks mark the position of GST-Eco1, which autoacetylates in vitro, as previously shown (Ivanov et al., 2002; Lu et al., 2010). Positions of molecular mass markers (kDa) are indicated next to each blot.
SPB duplication and DNA damage repair are independent of Mps3 acetylation
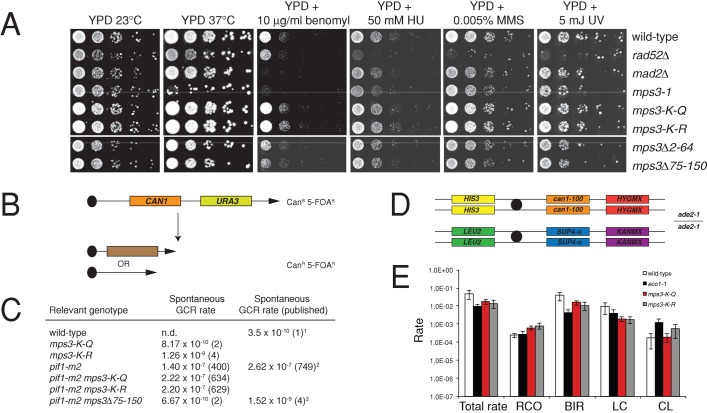
We created alleles of MPS3 in which lysines 147, 148, and 150 were changed to the nonacetylatable residue arginine (mps3-K-R) or to glutamine—which is believed to mimic acetylation (mps3-K-Q)—to determine the role of Eco1 acetylation on Mps3 function in vivo. Cells containing mps3-K-R or mps3-K-Q as the sole copy of the MPS3 gene are viable and do not show obvious growth defects at any temperature (Figure 4A; unpublished data). In addition, growth of both mutants is identical to that of wild type on media containing the microtubule inhibitor benomyl, the ribonucleotide reductase inhibitor HU, or the DNA-damaging agent methyl methanesulfonate (MMS) or after exposure to UV light (Figure 4A). Thus modification of lysines 147, 148, and 150 does not appear to be required for mitotic growth. Because the essential function of Mps3 is at the SPB, the fact that these mutants are viable and exhibit no apparent sensitivity to benomyl, increase in ploidy, or mitotic delay (unpublished data) suggests that acetylation is not required to regulate Mps3 function during SPB duplication.
FIGURE 4:
Loss of Eco1 acetylation does not affect chromosome rearrangements or mitotic recombination. (A) Wild-type (SLJ001), rad52∆ (SLJ2811), mad2∆ (SLJ5063), mps3-1 (SLJ4361), mps3-K-R (SLJ4754), mps3-K-Q (SLJ4753), mps3∆2-64 (SLJ2061), and mps3∆75-150 (SLJ2832) cells were serially diluted 10-fold and were stamped onto YPD plates containing nothing, 10 μg/ml benomyl, 50 mM HU, or 0.005% MMS. One set of YPD plates was exposed to 5 J of UV light. Plates were incubated for 3 d at 23°C. One YPD plate was incubated at 37°C. (B) Schematic of assay used to detect GCRs. Chromosome V contains CAN1 and URA3 at HXT13 between the centromere and the telomere, which renders cells sensitive to both canavanine and 5-FOA (Chen and Kolodner, 1999), allowing for rare rearrangements to be scored by determining the number of colonies that are resistant to canavanine and 5-FOA. Two types of rearrangements are shown, although many others are possible, as previously shown (Chen and Kolodner, 1999; Myung et al., 2001a). (C) The rate of spontaneous GCRs was determined in the indicated strains in three independent experiments. The numbers in parentheses refer to the fold increase in GCR rates over wild type. Also shown are previously described rates of GCRs from Chen and Kolodner (1999; note 1) and Oza et al. (2009; note 2). (D) The genotype of the starting diploid for detecting the products of mitotic recombination (Barbera and Petes, 2006). Phenotypic classes of unsectored canavanine-resistant colonies can be scored as outlined in Supplemental Figure S1 to determine the types of mitotic recombination events that have occurred. (E) This was done in diploids homozygous for MPS3 (SLJ4793 or SLJ5045), mps3-K-R (SLJ5020 or SLJ5021), mps3-K-Q (SLJ5017 or SLJ5019), and eco1-1 (SLJ5241 or SLJ5242). The total rate of recombination and the rate at which each type of recombination event was observed are shown for at least four independent colonies of each genotype in a minimum of four independent experiments. Error bars, SE of the mean. Although some values may appear to be slightly elevated or decreased, such as CL and BIR, respectively, in eco1-1 compared to wild type, the values obtained are not considered to be statistically significant (p < 0.01).
The ability of both mps3-K-R and mps3-K-Q to grow under conditions of low levels of DNA damage suggests that acetylation also is not required for Mps3's function in DNA damage repair. To further demonstrate that acetylation does not affect Mps3's response to damage, we used an assay that monitors gross chromosomal rearrangements (GCRs) caused by mutation of the Pif1 helicase (pif1-m2), which negatively regulates telomerase (Figure 4B; Schulz and Zakian, 1994; Myung et al., 2001a). GCRs such as translocations, inversions, deletion of a chromosome arm, or interstitial deletions are believed to occur between DNA sequences that have limited homology but may be located in the same region of the nucleus. The formation of GCRs results in aneuploidy, and their formation is correlated with tumorigenesis. Previously, deletion of the acidic region of the Mps3 N-terminus was shown to eliminate the increased level of GCRs in the pif1-m2 mutant, possibly by eliminating recruitment of broken chromosome ends to the nuclear periphery (Oza et al., 2009). In contrast, neither mps3-K-R nor mps3-K-Q had any significant effect on the rate of spontaneous GCR compared with wild type, nor did they decrease the rate of GCR in pif1-m2 mutants (Figure 4C). These mutants also did not affect the GCR rate in slx5∆ or mre11∆ mutants (unpublished data), confirming that acetylation of Mps3 is not required for DNA damage repair by this mechanism and that acetylation likely does not affect recruitment of broken chromosome arms to the nuclear periphery.
We also analyzed the mitotic recombination products in a diploid strain containing heteroallelic selectable markers along a 120-kb region of chromosome V (Barbera and Petes, 2006), since previous work showed that certain eco1 mutants are defective in reciprocal mitotic recombination, which is important for the maintenance of heterozygosity (Lu et al., 2010). Here one copy of chromosome V contains HIS3, can1-100, and HYGMX, and the second copy contains LEU2, SUP4-o, and KANMX at the same position (Figure 4D). In addition, the strain is homozygous for the ade2-1 allele, which is suppressible by the ochre suppressor SUP4-o: cells containing SUP4-o are white, whereas cells lacking SUP4-o are red due to the red pigment formed in ade2-1 mutants in the presence of limiting quantities of adenine. Using this strain, one can distinguish the products of break-induced replication (BIR), local gene conversion (LC), reciprocal crossovers (RCOs), and chromosome loss (CL; Supplemental Figure S1; Barbera and Petes, 2006).
We grew cells on 5 μg/ml bleomycin to induce a low level of DNA damage and elevate the rate of recombination. Growth on these plates did not inhibit cell proliferation in wild-type or mutant strains (unpublished data). Individual colonies were then resuspended and plated to selective medium such that only cells that had undergone a recombination event were able to grow. Colonies from the initial selection plate were further scored to assess the type of recombination event that occurred (Supplemental Figure S1; see also Materials and Methods). We found that a similar recombination rate and distribution of recombination products was observed in wild type and mps3-K-R, mps3-K-Q, and eco1-1 mutants in four independent analyses (Figure 4E). Although this result was somewhat surprising, given the previously described effects of eco1-W216G on recombination outcomes (Lu et al., 2010), the lack of an effect in eco1-1 mutants is consistent with our finding that acetylation of Mps3 does not significantly affect mitotic recombination. eco1-1, eco1-W216G, and eco1-ack contain mutations in conserved residues in the acetyltransferase catalytic cleft. Differences between these mutants have been previously reported in a number of biological assays (Strom et al., 2007; Unal et al., 2007, 2008; Gard et al., 2009; Lu et al., 2010). For reasons that are not entirely clear, the phenotypes observed in vivo do not always directly correlate with levels of acetyltransferase activity detected in the mutant eco1 proteins in vitro, suggesting that there are additional aspects to Eco1 regulation beyond its catalytic activity. Despite this issue, three lines of evidence show that acetylation of Mps3 is not required for its function in DSB repair: growth on media containing DNA-damaging agents and wild-type levels of mitotic recombination and chromosomal rearrangements.
Mps3 acetylation is required for establishment of sister chromatid cohesion
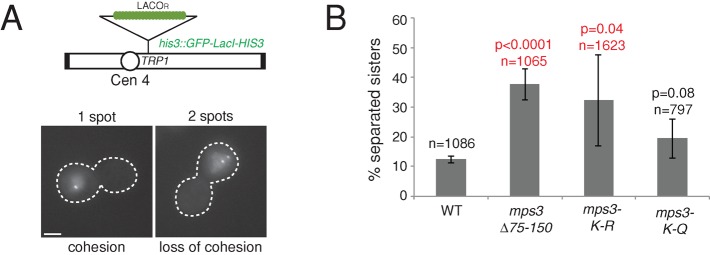
On the basis of the fact that Eco1 is essential for the establishment of sister chromatid cohesion and that Mps3 was previously shown to play a nonessential role in this process (Skibbens et al., 1999; Toth et al., 1999; Antoniacci et al., 2004; Antoniacci and Skibbens, 2006), we were interested in testing the possibility that acetylation is important for Mps3's function in establishment of cohesion. Wild-type, mps3∆75-150, mps3-K-R, and mps3-K-Q cells containing integrated arrays of the lactose operator (LacOR) near the centromere of chromosome IV and expressing the DNA-binding region of the lactose repressor fused to green fluorescent protein (GFP; LacI-GFP) were arrested with replicated DNA in G2/M using nocodazole (Figure 5A). If cohesion is correctly established, a single focus of GFP will be visible under these conditions due to the close proximity of the sister chromosomes. However, if there is a defect in cohesion, a significant fraction of cells will contain two GFP foci (Figure 5A). Eighty-seven percent of wild-type and 78% of mps3-K-Q cells correctly established cohesion (Figure 5B). In contrast, only 63 and 69% of mps3∆75-150 and mps3-K-R cells established cohesion, respectively; in the remaining 37 and 31% of cells, cohesion was lost (Figure 5B). This small but statistically significant decrease indicates that acetylation of Mps3 by Eco1 positively regulates the function of Mps3 in sister chromatid cohesion.
FIGURE 5:
Mps3 acetylation is required for its role in sister chromatid cohesion. (A) Sister chromatid cohesion was tested by arresting cells containing GFP-LacI and an integrated copy of ∼256 LacOR on the arm of chromosome IV in mitosis using nocodazole. Under these conditions, a single GFP focus is indicative of cohesion, whereas the appearance of two foci indicates that cohesion has been lost. The cell outline is based on the differential interference contrast (DIC) image. Bar, 5 μm. (B) Percentage of large-budded wild-type (SLJ1982), mps3∆75-150 (SLJ3130), mps3-K-R (SLJ3133), and mps3-K-Q (SLJ3134) cells that contained two distinct GFP foci. The number of cells analyzed in at least three independent experiments is indicated (n) along with the p value compared with wild type in Student's t test. Error bars, SD from the mean.
Acetylation of Mps3 is involved in peripheral localization of telomeres
In addition to its role in cohesion, Mps3 is required for telomere tethering at the nuclear periphery (Bupp et al., 2007; Schober et al., 2009; Chan et al., 2011). Despite considerable effort, we and others have been unable to detect an interaction between recombinant Mps3 and several proteins involved in recruitment of chromosome ends to the nuclear periphery (Antoniacci et al., 2007; Bupp et al., 2007; Schober et al., 2009). One possible explanation is that posttranslational modification of Mps3, including acetylation, is required for its binding to these proteins. To test this idea, we examined telomere position in mps3-K-R and mps3-K-Q mutants using LacOR arrays integrated into the chromosome adjacent to a telomere, which was visualized using GFP-LacI (Figure 6A). The nuclear surface was visualized by coexpression of the nucleoporin Nup49 fused to GFP (NUP49-GFP). Each telomeric focus was assigned a position within the nucleus based on its distance from the nuclear periphery, defined by Nup49-GFP, in the z-section where the spot was in focus. These distances were manually measured; however, a custom-written ImageJ plug-in ensured that the correct focal plane and pixel was chosen. To compare nuclei of different sizes, we derived a distance ratio by dividing the spot–periphery distance by the nuclear radius. If the distance ratio is close to 0, the telomere is likely to be tethered at the nuclear envelope, whereas a value close to 1 indicates that the focus is located near the center of the nucleus. Plots of the distance ratios for each telomere are shown in Figure 6B. If we consider that the nucleus is a sphere with two zones of equal volume, we can also organize foci into these zones using their distance ratio: foci in the outer zone are likely to have contacts with the nuclear periphery due to association of the telomere with proteins at the nuclear envelope, whereas foci in the inner zone are more likely not to contact the nuclear membrane and are considered to be untethered. This data are presented in Figure 6, B and C.
FIGURE 6:
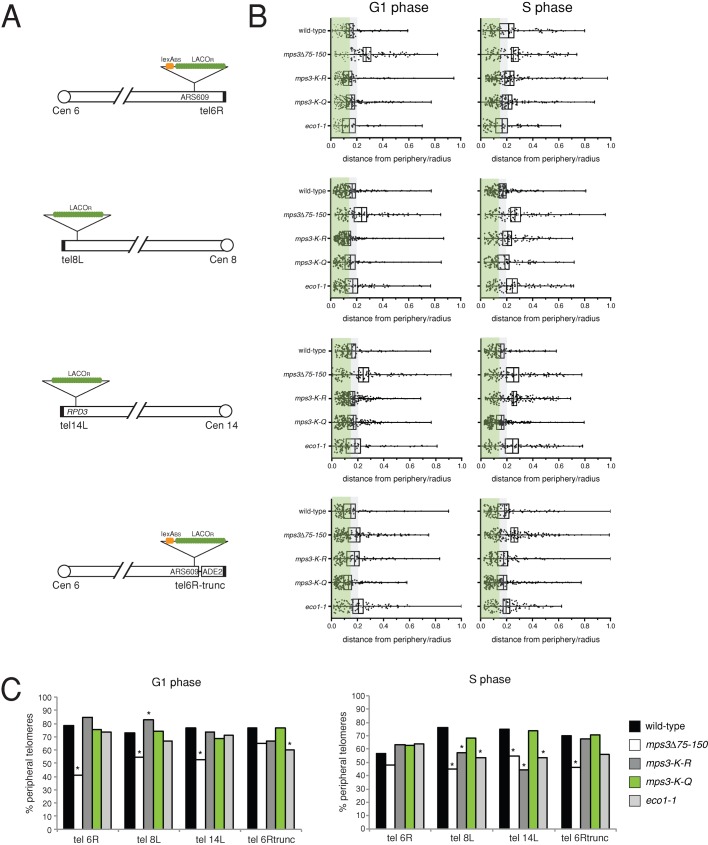
Mps3 acetylation is important for telomere tethering. (A) The subnuclear position of the indicated telomeres containing ∼256 copies of the LacOR was analyzed in cells expressing GFP-LacI and Nup49-GFP. Some telomeres also have four copies of the lexABS inserted adjacent to the LacOR. A truncated version of telomere 6R was constructed by inserting an ADE2 reporter and copies of telomeric repeats, as previously described (Hediger et al., 2002). (B) The distribution of telomeres was determined in asynchronously growing wild-type MPS3 (SLJ5697), mps3∆75-150 (SLJ5701), mps3-K-R (SLJ5705), mps3-K-Q (SLJ5709), and eco1-1 (SLJ5883) cells at 23°C. The ratio of the distance of the telomeric foci to the nuclear envelope compared with the nuclear radius was plotted for all foci examined. The small white box represents the mean, whereas the black box centered on the mean represents the SE. The third black line represents the median. A distance ratio of 0 is due to tethering, and a value of 1 represents a focus located in the center of the nucleus. Cells were grouped into G1 or S phase based on the bud morphology in the DIC image. G1 cells were defined as cells that had no visible bud, and S-phase cells were defined as cells that had a bud that was <50% of the size of the mother cell. To gauge the likelihood that these foci may interact with proteins at the nuclear periphery, one can divide the nucleus into two or three zones of equal volume: foci in the outer zone are likely to have contacts with the nuclear periphery due to association of the telomere with proteins at the nuclear envelope (Hediger et al., 2002; Meister et al., 2010). The gray and green shading correspond to the outermost zone using a two- or three-zone method of nuclear segmentation. (C) The percentage of cells from B in which telomeres were tethered at the nuclear periphery based on the two-zone method is shown (see Supplemental Table S1 for n and for p value from Fisher's exact probability test between wild-type and mutant distributions). *p ≤ 0.05.
We analyzed the position of telomere 6R, telomere 8L, telomere 14L, and a truncated version of telomere 6R that contains the ADE2 gene in addition to telomeric repeats (tel 6Rtrunc) in asynchronously growing mps3∆75-150, mps3-K-R, mps3-K-Q, and eco1-1 mutants and compared the distance ratio measurement to results in wild-type cells. Because the size of the bud can be used to approximate cell cycle position, the position of telomeres in G1- and S-phase nuclei could be scored in asynchronously growing cells. In nuclei from both G1- and S-phase cells, all four telomeres were strongly shifted to the peripheral zone in wild-type cells (Figure 6, B and C, and Supplemental Table S1). The one possible exception is telomere 6R, which shows 57% of foci in the peripheral zone in S phase compared with its 79% tethering in G1 phase. In all other wild-type samples, between 70 and 80% of telomeric foci were tethered in G1 and S phase. This is consistent with previous data showing that these telomeres are tethered at the nuclear periphery in both cell cycle phases (Hediger et al., 2002). However, the extent of tethering is greater than previously reported, especially in G1 nuclei (Hediger et al., 2002; Taddei et al., 2004). The exact source of this discrepancy is unknown, although it is not merely attributable to our use of a two-zone rather than a three-zone system (Figure 6B). One likely possibility is that use of confocal imaging rather than wide-field microscopy allowed us to achieve a greater level of resolution in the z-axis, allowing for an increased number of foci to be included in the analysis.
Telomeres from mps3∆75-150 mutants assume what appears to be a random distribution, with ∼40–60% of telomeric foci in the peripheral zone (Figure 6, B and C, and Supplemental Table S1). If nuclei from G1- and S-phase cells are compared, it appears that the mps3∆75-150 mutant has a more severe effect on telomere tethering in S phase than in G1. For example, tethering of telomere 8L shows a 26% reduction in G1 phase but a 41% reduction in S phase in mps3∆75-150 mutants compared with wild type. Telomere 6Rtrunc is also reduced by 15% in G1 phase but by 34% in S phase in mps3∆75-150 versus wild type. These findings are consistent with the idea that Mps3 is required for S-phase telomere tethering (Bupp et al., 2007; Schober et al., 2009). However, it is not possible to determine whether Mps3 is acting through Sir4 or yKu/Est based on these data. Tethering of telomere 14L and 6Rtrunc are believed to occur largely through Sir4 (Hediger et al., 2002; Taddei et al., 2004), and anchoring of both telomeres is reduced in S-phase nuclei in mps3∆75-150 mutants, but telomere 14L is also mislocalized in G1 phase (Figure 6C and Supplemental Table S1). Tethering of telomere 6R strongly depends on yKu80, and it shows a statistically significant decrease in G1 phase but not in S phase (Figure 6C and Supplemental Table S1). mps3∆75-150 may indirectly affect telomere tethering, perhaps through defects in membrane organization and/or protein synthesis (Witkin et al., 2010; Friederichs et al., 2011; Horigome et al., 2011).
Telomeres were localized in the peripheral zone near the nuclear envelope in mps3-K-Q mutants during both G1 and S phase, similar to what was observed in wild-type cells (Figure 6, B and C, and Supplemental Table S1). However, the enrichment of certain telomeres with the nuclear envelope was lost in mps3-K-R mutants (Figure 6, B and C, and Supplemental Table S1). A statistically significant decrease in the number of peripherally localized telomeric foci was observed in S-phase nuclei in mps3-K-R for telomere 8L and telomere 14L, whereas there was a 10% increase in the number of G1-phase cells with telomere 8L at the nuclear periphery compared with wild type (Figure 6C and Supplemental Table S1). The median distance of telomeres 6R, 8L, and 14L from the nuclear envelope, normalized based on nuclear size, also increased during S phase (Figure 6B and Supplemental Table S1): from a value of 0.10 to a value of 0.18, from 0.10 to 0.17, and from 0.13 to 0.24 for telomeres 6R, 8L, and 14L, respectively. The distance ratio for telomere 6Rtrunc was 0.12 in G1 phase and 0.13 in S phase, indicating that its tethering is not affected by Mps3 acetylation (Supplemental Table S1). From this we conclude that Mps3 acetylation is required for tethering of certain telomeres, particularly during S phase.
We also examined telomere position using LacOR arrays in cells containing the eco1-1 mutation grown at the permissive temperature of 23°C. In G1-phase nuclei, telomeres 6R, 8L, and 14L were enriched in the peripheral nuclear zone (Figure 6, B and C, and Supplemental Table S1). Only telomere 6Rtrunc was partially untethered. However, in S-phase nuclei, the eco1-1 mutant had a defect in peripheral localization of telomeres 8L and 14L but not telomere 6Rtrunc (Figure 6C and Supplemental Table S1). Therefore mutation of ECO1 leads to a defect in tethering of certain telomeres, particularly during S phase. When compared with mps3-K-R, the effect of eco1-1 on telomere positioning is similar but not completely identical. This could be due to other Eco1 targets or to defects associated with loss of chromosome integrity (reviewed in Skibbens, 2009; Xiong and Gerton, 2010). The mitotic arrest and nuclear morphology defects associated with eco1-1 prevented us from analyzing telomere position in cells grown at the nonpermissive temperature.
Telomere-silencing defects in mps3-K-R mutants
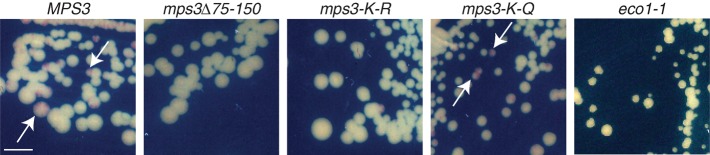
Strains containing the ADE2 marker at the truncated telomere 6R allowed us to determine whether mps3-K-R and eco1-1 mutants have defects in transcriptional regulation of a telomeric reporter gene. Wild-type cells appeared as sectored red and white colonies after growth on plates containing limiting amounts of adenine due to stochastic expression of ADE2 (Figure 7). As previously reported (Bupp et al., 2007), mps3∆75-150 mutants are partially defective in telomeric silencing and are unable to sector, appearing light pink to white in color due to increased expression of ADE2 at the truncated telomere (Figure 7). The mps3-K-R and eco1-1 mutants also did not show sectoring, and colonies were light pink to white (Figure 7). In contrast, red and white sectored colonies were observed in mps3-K-Q mutants (Figure 7). Thus Mps3 acetylation also affects telomeric gene expression. It is curious that telomere 6Rtrunc was not mislocalized in mps3-K-R mutants and its tethering was only partially affected in eco1-1 cells. Therefore the loss of silencing probably is due to an alteration in the distribution or stability of silencing proteins in the mutant cells rather than a direct consequence of loss of peripheral localization. Although the nuclear periphery is believed to contribute to inhibition of gene expression by providing a high local concentration of silencing factors, several studies have shown that telomeric silencing can occur at internal locations and is likely dependent on the levels and organization of silencing proteins (Mondoux et al., 2007; Ruault et al., 2011).
FIGURE 7:
Silencing defect in mps3-K-R mutants. Expression of the telomeric ADE2 gene in wild-type MPS3 (SLJ2602), mps3∆75-150 (SLJ3081), mps3-K-Q (SLJ3077), mps3-K-R (SLJ3075), and eco1-1 (SLJ3191) was monitored by streaking cells to SD plates containing 10 mg/ml adenine. After growth for 3 d at 30°C, plates were incubated for 1 wk at 4°C to allow the red pigment to develop. In the case of eco1-1, cells were grown at 23°C. Expression of ADE2 results in white-colored cells and blocks the accumulation of the red pigment in this strain background (see arrows in MPS3 and mps3-K-Q); this occurs in cells that have lost telomeric silencing. Bar, 1 cm.
Acetylation of Mps3 does not affect its localization
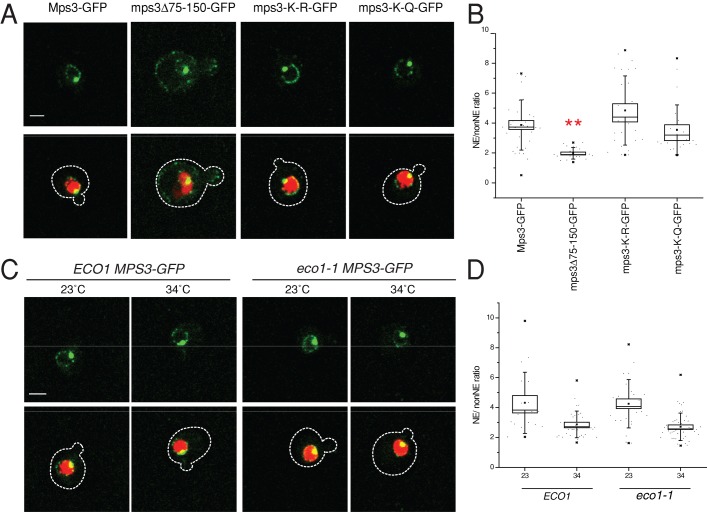
Our observation that mps3∆75-150-GFP does not localize to the peripheral nuclear membrane led to the hypothesis that INM distribution and Mps3's function in nuclear organization were interdependent (Bupp et al., 2007). Therefore we were interested in examining the localization of mps3-K-R-GFP and Mps3-GFP in eco1-1 to see whether acetylation controlled Mps3 distribution in the nuclear membrane. As we previously demonstrated, mps3∆75-150-GFP localizes to the SPB but not to foci on the nuclear envelope (Figure 8A). In contrast, localization of mps3-K-R-GFP and mps3-K-Q-GFP revealed a distribution that was virtually indistinguishable from that of wild-type Mps3-GFP (Figure 8A).
FIGURE 8:
Effect of Mps3 acetylation on INM localization. (A, C) Localization of Mps3-GFP (SLJ3330), mps3∆75-150-GFP (SLJ4025), mps3-K-R-GFP (SLJ5274), or mps3-K-Q-GFP (SLJ5273) (green) and H2B-mCherry (red) was examined by confocal imaging, and a representative single plane image of each is shown. The cell is outlined in white based on the DIC image. Mps3-GFP was also localized in wild-type (ECO1; SLJ5336) and eco1-1 (SLJ5338) mutants grown at 23°C or shifted to 34°C for 3 h. Bar, 2 μm. (B, D) Quantitation of NE/non-NE ratio of GFP in S-phase images from A and C was performed as described in Materials and Methods. Values for each data point (gray circles), the mean (black square), SE (box centered on mean), median (line), and SD (whiskers) are depicted. A double asterisk indicates mutants that displayed a statistically significant (U < 0.05) change in the mean NE/non-NE ratio compared with control using the Mann–Whitney test. Statistical values for all samples and quantitation of G1 cells from A and C are listed in Supplemental Table S2.
To quantify these images, we used the transmitted light image to determine the cell boundary and the histone 2B–mCherry signal to determine the nuclear boundary. We could then calculate the amount of Mps3-GFP at the nuclear envelope (NE; the signal defined by the outer boundary of the nuclear marker) and the levels of the protein present on nonnuclear membranes (non-NE; the signal inside the boundary of the transmitted light but outside that of the nucleus). A threshold cutoff allowed us to exclude Mps3-GFP at the bright SPB. We then determined the NE/non-NE ratio, which is a reflection of the degree to which Mps3-GFP localizes to the INM compared with other membranes throughout the cell (Gardner et al., 2011). Using this method of analysis, we found that there was no statistically significant change in the levels of mps3-K-R-GFP or mps3-K-Q-GFP at the INM compared with Mps3-GFP in either G1- or S-phase cells (Figure 8, A and B, and Supplemental Table S2). The level of protein at the SPB was also unchanged (Supplemental Figure S2A). However, the NE/non-NE ratio of mps3∆75-150-GFP was decreased in both G1- and S-phase cells, consistent with previous results (Gardner et al., 2011). In this experiment, statistical significance was analyzed using the Mann–Whitney test, which allows for comparison of two data sets that have unequal dispersion. A U value of <0.05 was considered to be statistically significant.
Inactivation of Eco1 by the temperature-sensitive eco1-1 allele also did not change Mps3-GFP localization in S-phase cells (Figure 8, C and D, Supplemental Figure S2B, and Supplemental Table S2). It is unclear why we see reduced levels of Mps3-GFP at the INM in eco1-1 mutants in G1 (NE/non-NE = 2.9 in eco1-1 compared with 3.98 in wild-type, U = 0.01); the defect in localization is likely not the result of changes in Mps3 acetylation, since INM distribution of both the nonacetylatable mps3-K-R-GFP and the constitutively acetylated mps3-K-Q-GFP are Eco1 independent (Supplemental Figure S3 and Supplemental Table S2). Thus, although the mps3-K-R allele is unable to tether certain telomeres, it still localizes to the INM, indicating the role of Mps3 in nuclear organization is controlled by at least two mechanisms: a pathway that regulates its distribution in the INM, and acetylation, which does not affect localization but instead controls interactions with proteins required for telomere tethering and sister chromatid cohesion.
DISCUSSION
In this article we showed that the INM protein Mps3 is a substrate for Eco1 acetyltransferase in vitro and in vivo. We mapped the sites of acetylation to three lysine residues adjacent to the Mps3 transmembrane domain and demonstrated that mutation of these residues leads to defects in multiple aspects of nuclear organization, including sister chromatid cohesion, telomere tethering, and gene silencing. This is a novel function for the highly conserved Eco1 acetyltransferase that is independent of its well-established role in sister chromatid cohesion.
We found that Mps3 was acetylated in vitro by Eco1, mapped the acetylation sites using site-directed mutagenesis, and found that the primary Eco1 targets are three lysine residues adjacent to the Mps3 transmembrane domain. The fact that Mps3 acetylation is virtually eliminated in eco1-1 mutants suggests that Mps3 is mainly acetylated by Eco1 in vivo, but it is also possible that it is modified at other sites by a different acetyltransferase. One leading candidate is the NuA4 acetyltransferase due to its role in telomeric silencing (Zhang et al., 2004) and its interaction with Mps2, a SPB component, and Mps3-binding protein (Le Masson et al., 2002; Jaspersen et al., 2006). Acetylation at additional sites may explain why mps3-K-R has a relatively mild phenotype compared with other mps3 alleles such as mps3∆75-150 mutants. During S phase, Smc3 is acetylated by Eco1 on two conserved lysine residues, K112 and K113 (Rolef Ben-Shahar et al., 2008; Unal et al., 2008; Zhang et al., 2008). Smc3 is believed to be the essential target of Eco1; however, several additional substrates have been proposed based largely on in vitro studies using recombinant proteins (Ivanov et al., 2002; Bellows et al., 2003; Williams et al., 2003; Unal et al., 2008). Included in this list is Scc1/Mcd1, which is acetylated in response to DNA damage on K84 and K210 (Heidinger-Pauli et al., 2009). Of interest, the Mps3 acetylation site that we mapped on Mps3 is reminiscent of that observed on Smc3, in that it contains a pair of adjacent lysine residues. However, the Mps3 site is not obviously conserved in other SUN proteins, and so although they might be substrates of Eco1, the modification probably occurs on distinct residues.
Examination of mps3-K-R mutants revealed that Eco1 acetylation of Mps3 is required for distinct cellular events. Although mps3-K-R has defects in sister chromatid cohesion and peripheral localization of some telomeres, the mutant is able to efficiently repair DSBs and duplicate the SPB. Our observation that mps3-K-R-GFP localizes to the SPB and the INM suggests that the defects in this mutant are not attributable merely to changes in localization but rather are due to loss of binding of critical nuclear factors to Mps3. Thus we would predict that acetylation would specifically affect the interaction between Mps3 and proteins involved in telomere tethering and sister chromatid cohesion. Candidates include the silencing proteins Sir2, Sir3, and Sir4, the telomere-binding proteins yKu70/yKu80, telomerase components such as Est1 and Est2, and the histone variant H2A.Z that is required for localization of Mps3 to the INM and is enriched in subtelomeric chromatin (Li et al., 2005; Zhang et al., 2005; Gardner et al., 2011). Coimmunoprecipitation and two-hybrid experiments have not revealed a change in binding between Sir4 or H2A.Z and Mps3 or mps3-K-R (unpublished data), suggesting that acetylation does not regulate Mps3 association with these proteins. We have been unable to observe a direct interaction between Mps3 and Est1, Est2, or yKu70/yKu80 by coimmunoprecipitation or by the two-hybrid test, indicating that additional proteins or modifications may regulate their interaction in vivo. One possibility is that Mps3 acetylation regulates its association with the cohibin complex. Originally identified based on its function in nucleolar organization, this tetrameric complex containing two copies of both Csm1 and Lrs4 also interacts with telomere factors and is required for Sir4 association with Mps3 (Smith et al., 1999; Mekhail et al., 2008; Corbett et al., 2010; Chan et al., 2011). The fact that the mps3-K-R mutant displays defects in nuclear organization but localizes normally to the INM suggests an additional level of complexity in Mps3 interaction with chromosomes that was not appreciated in previous studies (Antoniacci and Skibbens, 2006; Bupp et al., 2007; Oza et al., 2009; Schober et al., 2009).
Acetylation of Mps3 by Eco1 does not appear to fluctuate during the cell cycle, yet sister chromatid cohesion is established during S phase (Skibbens et al., 1999; Toth et al., 1999). In addition, we showed that Eco1-dependent telomere tethering is more affected in S phase than in G1 phase. This raises the interesting question of how Mps3 acetylation is regulated during the cell cycle and how this is translated into changes in its interaction with telomere-associated proteins or with proteins involved in cohesion. One possibility is that only a small fraction of Mps3 is controlled by acetylation/deacetylation, such as the population of Mps3 at the INM. Because the majority of Mps3 is at the SPB (Bupp et al., 2007), a change in modification of the smaller pool of INM Mps3 would be difficult to detect. Another possibility is that deacetylation occurs in a small time window of the cell cycle that we have not been able to observe in our arrested cells or in cells undergoing a synchronous cell cycle (unpublished data). Alternatively, Mps3 may be constitutively acetylated; only newly synthesized Mps3 may exist in the deacetylated form. Consistent with this latter hypothesis, we have been unable to deacetylate Mps3 with Hos1, the deacetylase that acts on Smc3 (unpublished data; Beckouet et al., 2010; Borges et al., 2010; Xiong et al., 2010). A final possibility is that Mps3 is constitutively acetylated, and it is cell cycle–dependent regulation of its binding partner that controls chromosome dynamics at the nuclear envelope.
In the female gonad of C. elegans, SUN protein function is regulated by phosphorylation, which affects its ability to aggregate in the nuclear envelope (Penkner et al., 2009; Sato et al., 2009; Harper et al., 2011; Labella et al., 2011). Studies of Mps3 in meiosis also suggested that the formation of higher-order complexes mediate chromosome reorganization during prophase (Conrad et al., 2007, 2008; Rao et al., 2011). In mitosis, Mps3 was recently implicated in telomere clustering through association with the ribosome biogenesis factors Ebp2 and Rrs1 (Horigome et al., 2011), raising the possibility that acetylation affects Mps3 mobility in the membrane by neutralizing the negatively charged lysines adjacent to the transmembrane domain. Line-scanning fluorescence autocorrelation analysis of mps3-K-R-GFP and mps3-K-Q-GFP at the nuclear membrane did not reveal a statistically significant change in mobility or oligomerization compared with wild-type Mps3-GFP (unpublished data). Therefore we favor a simpler model in which Mps3 acetylation by Eco1 directly affects its ability to associate with telomere-associated proteins. Our observation that telomere tethering is most affected in S-phase cells when Eco1 levels are high is consistent with this model (Lyons and Morgan, 2011). We did not observe a strict requirement for Mps3 acetylation for tethering of certain classes of telomeres (i.e., mps3-K-R mutants failed to tether telomeres believed to be anchored primarily by yKu or Sir4 [ Hediger et al., 2002; Taddei et al., 2004]), perhaps suggesting that membrane receptors for telomeres do not exhibit a strict one-to-one correlation for telomeres. Instead, cooperativity and clustering may play a more important role. At least two other membrane-based receptors for telomeres exist during S-phase—Heh1 and Esc1 (Andrulis et al., 2002; Chan et al., 2011). Future studies aimed at a molecular analysis of functional silencing and tethering complexes on the membrane will help elucidate the role that acetylation and other posttranslational modifications, such as sumoylation of Sir4 and yKu80 (Ferreira et al., 2011), play in nuclear organization and cross-talk between telomere tethering pathways.
MATERIALS AND METHODS
Yeast strains and plasmids
All strains are derivatives of W303 (ade2-1, trp1-1, leu2-3112, ura3-1, his3-11,15, can1-100) unless indicated and are listed in Supplemental Table S3. Standard techniques were used for DNA and yeast manipulations. Construction of pSJ148 (pRS305-MPS3) and pSJ650 (pRS306-MPS3-GFP) has been previously described (Jaspersen et al., 2006; Bupp et al., 2007). Mutations were generated in these plasmids using the QuikChange Mutagenesis Kit (Stratagene, Santa Clara, CA). pSJ879 (pRS305-mps3-K147Q K148Q K150Q) and pSJ880 (pRS305-mps3-K147R K148R K150R) were digested with BstEII to integrate into the LEU2 locus, and pSJ1158 (pRS306-mps3-K147Q K148Q K150Q-GFP) and pSJ1160 (pRS306-mps3-K147R K148R K150R-GFP) were digested with ApaI to integrate into the URA3 locus. In some cases, mutants were integrated into the endogenous locus using the PCR strategy described in Tong and Boone (2006). Correct integration of mutants was confirmed by PCR and sequence analysis. MPS3 was fused to 3xFLAG and HTB2 to mCherry by PCR-mediated gene fusion (Sheff and Thorn, 2004).
Cytological techniques
To analyze nuclear envelope staining of Mps3-GFP, we grew cells to mid-log phase in synthetic complete media supplemented with 3× adenine and placed onto 25% gelatin pads as described previously (Bupp et al., 2007). Imaging was performed on a laser scanning microscope (LSM 510 META; Carl Zeiss, Jena, Germany) equipped with a ConfoCor3 module with avalanche photodiode detectors (Carl Zeiss) using a 100× α-Plan Fluar lens (numerical aperture [NA], 1.45) at room temperature. Excitation of GFP was performed with the 488-nm argon laser line and the appropriate filter sets. Data were acquired using AIM, version 4.2, software (Carl Zeiss). Images were collected with 8–10 image stacks with a 0.3-μm step size through the cells at room temperature. Localization of Mps3-GFP at the SPB and NE in mutant strains was quantitated as previously described (Gardner et al., 2011). Briefly, we created a mask based on the transmitted light image of cells using AxioVision software (Carl Zeiss). Next we selected all pixels above a predefined threshold of the H2B-mCherry signal to create an additional mask that describes the nucleus. These masks were used to quantify the amount of Mps3-GFP localized in the NE (INM signal) compared with the amount of protein that is mislocalized (non-NE/endoplasmic reticulum signal = non-NE) and to determine a ratio of mean intensity per unit area for each. The very bright SPB signal from Mps3-GFP is excluded from this calculation by using an upper intensity cutoff. At least 30 images of each genotype were quantified per cell cycle stage, and NE/non-NE ratio for each is shown in addition to a representative single-section image that contains the SPB(s) in-plane.
For telomere-tethering assays, mid-log-phase cells grown in synthetic complete media supplemented with 3× adenine and 5 μM CuSO4 were placed onto 1% agar pads as described (Meister et al., 2010). From 40 to 50 image stacks with a 0.15-μm step size through cells at room temperature were acquired with a 100×/1.4 NA oil objective on an inverted Zeiss 200m equipped with a Yokagawa CSU-10 spinning disk. We used 488-nm excitation for GFP and collected emission through a BP 500- to 550-nm filter onto a Hamamatsu electron-multiplying, charge-coupled device camera (C9000-13; Hamamatsu, Hamamatsu, Japan). Telomere tethering was quantitated by using a custom plug-in written in ImageJ (National Institutes of Health, Bethesda, MD). Three-dimensional images were binned by four (up to 0.6-μm steps) in the z-dimension to improve the accuracy of manual telomere selection. Next a line was manually drawn from the center of the telomere spot to the nearest edge (in the x, y-plane) of the nuclear envelope. By interpolating from the selected position to the maximum position within the 3 × 3 × 3 pixel neighborhood, we determined the most intense pixel, and the length of the line between this position and the nuclear envelope end of the drawn line gives the telomere-to-edge distance (dtxy). The x, y-projection of the nucleus was determined by performing a maximum projection in z followed by a Gaussian filter with a radius of 3 pixels. A threshold of 75% of the intensity at the telomere was applied to this projection. The area of the thresholded region was used to calculate the nuclear radius as  If dtxy was greater than the nuclear radius, foci were not counted. The degree of circularity was calculated using the formula 4π × area/perimeter2, and cells where this value was <0.5 were excluded from analysis (circularity is 1 for a perfect circle and 0 for a line). The distance of the interpolated telomere spot from the center of mass of the nuclear intensity in the z-direction was used to eliminate telomeres in the top or bottom 20% of the nucleus as described by Meister et al. (2010). The distance ratio was calculated for the remaining samples by dividing dxy by r. A scatter plot of distance ratio values for each strain was generated using Origin Pro (OriginLab, Northampton, MA). A completely tethered spot and a focus at the center of the nucleus would have values of 0 and 1, respectively. The mean, median, and SE were also calculated. Zones for tethering were chosen based on regions of equal volume and therefore equal occupation probability for random telomere positioning. For two zones, the nucleus is divided into two groups: an untethered group with a distance <0.79 times the radius and a tethered group with larger distances. Fisher's exact probability test was used to compare the distribution of mutant cells containing a particular telomere to wild type.
If dtxy was greater than the nuclear radius, foci were not counted. The degree of circularity was calculated using the formula 4π × area/perimeter2, and cells where this value was <0.5 were excluded from analysis (circularity is 1 for a perfect circle and 0 for a line). The distance of the interpolated telomere spot from the center of mass of the nuclear intensity in the z-direction was used to eliminate telomeres in the top or bottom 20% of the nucleus as described by Meister et al. (2010). The distance ratio was calculated for the remaining samples by dividing dxy by r. A scatter plot of distance ratio values for each strain was generated using Origin Pro (OriginLab, Northampton, MA). A completely tethered spot and a focus at the center of the nucleus would have values of 0 and 1, respectively. The mean, median, and SE were also calculated. Zones for tethering were chosen based on regions of equal volume and therefore equal occupation probability for random telomere positioning. For two zones, the nucleus is divided into two groups: an untethered group with a distance <0.79 times the radius and a tethered group with larger distances. Fisher's exact probability test was used to compare the distribution of mutant cells containing a particular telomere to wild type.
Sister chromatid cohesion was assayed in logarithmically growing cells that were arrested with 10 μg/ml nocodazole for 3 h. Cells were briefly fixed with 4% paraformaldehyde and stained with 4′,6-diamidino-2-phenylindole before cell counting. In addition, aliquots of cells were removed for flow cytometric analysis of DNA content to verify the mitotic arrest, as described (Jaspersen et al., 2002).
Immunoprecipitation and Western blotting
Liquid nitrogen–ground lysates were prepared from mid-log-phase cells as described previously (Bupp et al., 2007), except that a cocktail of deacetylase inhibitors (100 μM Trichostatin A, 50 mM nicotinamide, 50 mM sodium butyrate) was added during the immunoprecipitation process. A 100-μl amount of anti-FLAG M2 (Sigma-Aldrich, St. Louis, MO) resin was added to lysates to immunoprecipitate FLAG-tagged proteins. After 2 h of incubation at 4°C, beads were washed five times, and 1/10 of the bound protein was analyzed by SDS–PAGE, followed by Western blotting.
The following primary antibody dilutions were used: 1:1000 anti-FLAG M2 (Sigma-Aldrich), 1:1000 anti-GST (Covance, Berkeley, CA), 1:1000 anti–acetylated lysine (EMD Biosciences, San Diego, CA), and 1:2000 anti–glucose-6-phosphate dehydrogenase (G6PDH; Sigma-Aldrich). Alkaline phosphatase–conjugated secondary antibodies were used at 1:10000 (Promega, Madison, WI), and fluorescently conjugated secondary antibodies were used at 1:2500 for analysis using the Odyssey system (Li-Cor, Lincoln, NE).
Protein purification and acetylation
The N-terminal region of MPS3 from amino acids 1–150 was amplified by PCR and fused to GST in pGEX-3X (GE Healthcare, Piscataway, NJ). Similarly, amino acids 169–337 of Scc1 and the entire Eco1 open reading frame were cloned in pGEX-3X. Mutant versions of Eco1 have been previously described (Lu et al., 2010), and deletion and point mutations in the Mps3 N-terminus were made by site-directed mutagenesis or by PCR. Recombinant proteins were expressed in BL21(DE3) pLysS (GST, Mps3, and Scc1 constructs) or BL21(DE3) pLysY/I cells (Eco1 constructs) for 4 h at 23°C after addition of 0.3 mM isopropyl-β-d-thiogalactoside and purified using glutathione resin (GE Healthcare). Proteins were concentrated and exchanged into HAT buffer (50 mM Tris-HCl, pH 8.0, 0.1 mM EDTA, 50 mM KCl, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 5% glycerol) using YM-30 Centricon filters (Millipore, Billerica, MA). YM-10 filters were used for GST alone. The concentration of protein was analyzed using a Bradford assay (Bio-Rad, Hercules, Ca) and adjusted to 1 mg/ml for all proteins.
Acetylation reactions were carried out by mixing the indicated amounts of GST-Eco1 or GST with GST-Mps31-150, GST-Scc1169-337, or GST in HAT buffer supplemented with 0.1 mM acetyl-CoA for 30 min at 30°C. In some reactions, 0.25 μCi of 3H-CoA (GE Healthcare) was also added. Reactions were terminated by the addition of 2× SDS sample buffer, and the products were analyzed after SDS–PAGE by Western blotting or by Coomassie blue staining and autoradiography. Acetylation levels were quantitated based on the amount of 3H-CoA incorporation.
Yeast chromosome assays
The assay to select and analyze reciprocal mitotic crossovers has been extensively described (Barbera and Petes, 2006). Briefly, individual colonies isolated from 5 μg/ml bleomycin plates were resuspended in 50 μl of H2O, diluted 10- to 100-fold, and plated on synthetic defined medium lacking Arg (SD-ARG ) plus canavanine or diluted 10,000-fold and plated on SD-ARG to calculate the rate of recombination. Colonies from SD-ARG plus canavanine were further analyzed to determine different subclasses of recombination events. Sectored colonies allow scoring of RCO events. For the determination of LC and CL, up to 48 red colonies were picked from each plate and tested for resistance to G418 and growth on SD-LEU. The remaining red colonies that are G418S and LEU+ are due to BIR.
The rate of gross chromosomal rearrangement was determined by fluctuation analysis using the method of the median (Lea and Coulson, 1948). Analysis was performed two or more times with multiple cultures for each clone, and the average value is reported for each as previously described (Myung et al., 2001b). Briefly, single colonies were used to inoculate overnight cultures in yeast extract/peptone/dextrose (YPD). Cells were then pelleted and washed in 1 ml of sterile water, and either 5 × 108 or 1 × 107 cells were plated to 2–12 SD-ARG plus canavanine plus 5-fluoroorotic acid (5-FOA) plates at 30°C for 4–6 d. The number of colonies on each plate was counted.
Supplementary Material
Acknowledgments
We thank Susan Gasser, Richard Kolodner, Craig Peterson, Marc Gartenberg, Jennifer Gerton, and Tom Petes for sharing strains and plasmids. We are grateful to Jennifer Gerton and Scott Hawley for helpful discussions throughout the course of this project and for comments on the manuscript. S.L.J. is supported by the Stowers Institute for Medical Research and the American Cancer Society (RSG-11-030-01-CSM).
Abbreviations used:
- BIR
break-induced recombination
- CL
chromosome loss
- DSB
double-stranded DNA break
- 5-FOA
5-fluoroorotic acid
- GCR
gross chromosomal rearrangement
- HU
hydroxyurea
- INM
inner nuclear membrane
- LC
local conversion
- MMS
methyl methanesulfonate
- NE
nuclear envelope
- RCO
reciprocal crossover
- SPB
spindle pole body
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-07-0600) on May 16, 2012.
REFERENCES
- Andrulis ED, Zappulla DC, Ansari A, Perrod S, Laiosa CV, Gartenberg MR, Sternglanz R. Esc1, a nuclear periphery protein required for Sir4-based plasmid anchoring and partitioning. Mol Cell Biol. 2002;22:8292–8301. doi: 10.1128/MCB.22.23.8292-8301.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniacci LM, Kenna MA, Skibbens RV. The nuclear envelope and spindle pole body-associated Mps3 protein bind telomere regulators and function in telomere clustering. Cell Cycle. 2007;6:75–79. doi: 10.4161/cc.6.1.3647. [DOI] [PubMed] [Google Scholar]
- Antoniacci LM, Kenna MA, Uetz P, Fields S, Skibbens RV. The spindle pole body assembly component Mps3p/Nep98p functions in sister chromatid cohesion. J Biol Chem. 2004;279:49542–49550. doi: 10.1074/jbc.M404324200. [DOI] [PubMed] [Google Scholar]
- Antoniacci LM, Skibbens RV. Sister-chromatid telomere cohesion is nonredundant and resists both spindle forces and telomere motility. Curr Biol. 2006;16:902–906. doi: 10.1016/j.cub.2006.03.060. [DOI] [PubMed] [Google Scholar]
- Barbera MA, Petes TD. Selection and analysis of spontaneous reciprocal mitotic cross-overs in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2006;103:12819–12824. doi: 10.1073/pnas.0605778103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckouet F, Hu B, Roig MB, Sutani T, Komata M, Uluocak P, Katis VL, Shirahige K, Nasmyth K. An Smc3 acetylation cycle is essential for establishment of sister chromatid cohesion. Mol Cell. 2010;39:689–699. doi: 10.1016/j.molcel.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellows AM, Kenna MA, Cassimeris L, Skibbens RV. Human EFO1p exhibits acetyltransferase activity and is a unique combination of linker histone and Ctf7p/Eco1p chromatid cohesion establishment domains. Nucleic Acids Res. 2003;31:6334–6343. doi: 10.1093/nar/gkg811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges V, Lehane C, Lopez-Serra L, Flynn H, Skehel M, Rolef Ben-Shahar T, Uhlmann F. Hos1 deacetylates Smc3 to close the cohesin acetylation cycle. Mol Cell. 2010;39:677–688. doi: 10.1016/j.molcel.2010.08.009. [DOI] [PubMed] [Google Scholar]
- Bupp JM, Martin AE, Stensrud ES, Jaspersen SL. Telomere anchoring at the nuclear periphery requires the budding yeast Sad1-UNC-84 domain protein Mps3. J Cell Biol. 2007;179:845–854. doi: 10.1083/jcb.200706040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JN, Poon BP, Salvi J, Olsen JB, Emili A, Mekhail K. Perinuclear cohibin complexes maintain replicative life span via roles at distinct silent chromatin domains. Dev Cell. 2011;20:867–879. doi: 10.1016/j.devcel.2011.05.014. [DOI] [PubMed] [Google Scholar]
- Chen C, Kolodner RD. Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants. Nature genetics. 1999;23:81–85. doi: 10.1038/12687. [DOI] [PubMed] [Google Scholar]
- Conrad MN, Lee CY, Chao G, Shinohara M, Kosaka H, Shinohara A, Conchello JA, Dresser ME. Rapid telomere movement in meiotic prophase is promoted by NDJ1, MPS3, and CSM4 and is modulated by recombination. Cell. 2008;133:1175–1187. doi: 10.1016/j.cell.2008.04.047. [DOI] [PubMed] [Google Scholar]
- Conrad MN, Lee CY, Wilkerson JL, Dresser ME. MPS3 mediates meiotic bouquet formation in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2007;104:8863–8868. doi: 10.1073/pnas.0606165104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett KD, Yip CK, Ee LS, Walz T, Amon A, Harrison SC. The monopolin complex crosslinks kinetochore components to regulate chromosome-microtubule attachments. Cell. 2010;142:556–567. doi: 10.1016/j.cell.2010.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira HC, Luke B, Schober H, Kalck V, Lingner J, Gasser SM. The PIAS homologue Siz2 regulates perinuclear telomere position and telomerase activity in budding yeast. Nat Cell Biol. 2011;13:867–874. doi: 10.1038/ncb2263. [DOI] [PubMed] [Google Scholar]
- Fridkin A, Penkner A, Jantsch V, Gruenbaum Y. SUN-domain and KASH-domain proteins during development, meiosis and disease. Cell Mol Life Sci. 2009;66:1518–1533. doi: 10.1007/s00018-008-8713-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friederichs JM, Ghosh S, Smoyer CJ, McCroskey S, Miller BD, Weaver KJ, Delventhal KM, Unruh J, Slaughter BD, Jaspersen SL. The SUN protein Mps3 is required for spindle pole body insertion into the nuclear membrane and nuclear envelope homeostasis. PLoS Genetics. 2011;7:e1002365. doi: 10.1371/journal.pgen.1002365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gard S, Light W, Xiong B, Bose T, McNairn AJ, Harris B, Fleharty B, Seidel C, Brickner JH, Gerton JL. Cohesinopathy mutations disrupt the subnuclear organization of chromatin. J Cell Biol. 2009;187:455–462. doi: 10.1083/jcb.200906075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner JM, Smoyer CJ, Stensrud ES, Alexander R, Gogol M, Wiegraebe W, Jaspersen SL. Targeting of the SUN protein Mps3 to the inner nuclear membrane by the histone variant H2A.Z. J Cell Biol. 2011;193:489–507. doi: 10.1083/jcb.201011017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf R, Daunderer C, Schulz I. Molecular and functional analysis of the Dictyostelium centrosome. Int Rev Cytol. 2004;241:155–202. doi: 10.1016/S0074-7696(04)41003-1. [DOI] [PubMed] [Google Scholar]
- Harper NC, Rillo R, Jover-Gil S, Assaf ZJ, Bhalla N, Dernburg AF. Pairing centers recruit a polo-like kinase to orchestrate meiotic chromosome dynamics in C. elegans. Dev Cell. 2011;21:934–947. doi: 10.1016/j.devcel.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hediger F, Neumann FR, Van Houwe G, Dubrana K, Gasser SM. Live imaging of telomeres: yKu and Sir proteins define redundant telomere-anchoring pathways in yeast. Curr Biol. 2002;12:2076–2089. doi: 10.1016/s0960-9822(02)01338-6. [DOI] [PubMed] [Google Scholar]
- Heidinger-Pauli JM, Unal E, Koshland D. Distinct targets of the Eco1 acetyltransferase modulate cohesion in S phase and in response to DNA damage. Mol Cell. 2009;34:311–321. doi: 10.1016/j.molcel.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraoka Y, Dernburg AF. The SUN rises on meiotic chromosome dynamics. Dev Cell. 2009;17:598–605. doi: 10.1016/j.devcel.2009.10.014. [DOI] [PubMed] [Google Scholar]
- Horigome C, Okada T, Shimazu K, Gasser SM, Mizuta K. Ribosome biogenesis factors bind a nuclear envelope SUN domain protein to cluster yeast telomeres. EMBO J. 2011;30:3799–3811. doi: 10.1038/emboj.2011.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov D, Schleiffer A, Eisenhaber F, Mechtler K, Haering CH, Nasmyth K. Eco1 is a novel acetyltransferase that can acetylate proteins involved in cohesion. Curr Biol. 2002;12:323–328. doi: 10.1016/s0960-9822(02)00681-4. [DOI] [PubMed] [Google Scholar]
- Jaspersen SL, Giddings TH, Jr, Winey M. Mps3p is a novel component of the yeast spindle pole body that interacts with the yeast centrin homologue Cdc31p. J Cell Biol. 2002;159:945–956. doi: 10.1083/jcb.200208169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen SL, Martin AE, Glazko G, Giddings TH, Jr, Morgan G, Mushegian A, Winey M. The Sad1-UNC-84 homology domain in Mps3 interacts with Mps2 to connect the spindle pole body with the nuclear envelope. J Cell Biol. 2006;174:665–675. doi: 10.1083/jcb.200601062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalocsay M, Hiller NJ, Jentsch S. Chromosome-wide Rad51 spreading and SUMO-H2A.Z-dependent chromosome fixation in response to a persistent DNA double-strand break. Mol Cell. 2009;33:335–343. doi: 10.1016/j.molcel.2009.01.016. [DOI] [PubMed] [Google Scholar]
- King MC, Drivas TG, Blobel G. A network of nuclear envelope membrane proteins linking centromeres to microtubules. Cell. 2008;134:427–438. doi: 10.1016/j.cell.2008.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labella S, Woglar A, Jantsch V, Zetka M. Polo kinases establish links between meiotic chromosomes and cytoskeletal forces essential for homolog pairing. Dev Cell. 2011;21:948–958. doi: 10.1016/j.devcel.2011.07.011. [DOI] [PubMed] [Google Scholar]
- Le Masson I, Saveanu C, Chevalier A, Namane A, Gobin R, Fromont-Racine M, Jacquier A, Mann C. Spc24 interacts with Mps2 and is required for chromosome segregation, but is not implicated in spindle pole body duplication. Mol Microbiol. 2002;43:1431–1443. doi: 10.1046/j.1365-2958.2002.02844.x. [DOI] [PubMed] [Google Scholar]
- Lea DE, Coulson CA. The distribution of the numbers of mutants in bacterial populations. J Genet. 1948;49:264–285. doi: 10.1007/BF02986080. [DOI] [PubMed] [Google Scholar]
- Li B, Pattenden SG, Lee D, Gutierrez J, Chen J, Seidel C, Gerton J, Workman JL. Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling. Proc Natl Acad Sci USA. 2005;102:18385–18390. doi: 10.1073/pnas.0507975102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Goering M, Gard S, Xiong B, McNairn AJ, Jaspersen SL, Gerton JL. Eco1 is important for DNA damage repair in S. cerevisiae. Cell Cycle. 2010;9:3315–3327. doi: 10.4161/cc.9.16.12673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons NA, Morgan DO. Cdk1-dependent destruction of Eco1 prevents cohesion establishment after S phase. Mol Cell. 2011;42:378–389. doi: 10.1016/j.molcel.2011.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CJ, Misner L, Le Bot N, Tsai MC, Campbell JM, Ahringer J, White JG. The C. elegans hook protein, ZYG-12, mediates the essential attachment between the centrosome and nucleus. Cell. 2003;115:825–836. doi: 10.1016/s0092-8674(03)00985-1. [DOI] [PubMed] [Google Scholar]
- Meister P, Gehlen LR, Varela E, Kalck V, Gasser SM. Visualizing yeast chromosomes and nuclear architecture. Methods Enzymol. 2010;470:535–567. doi: 10.1016/S0076-6879(10)70021-5. [DOI] [PubMed] [Google Scholar]
- Mekhail K, Moazed D. The nuclear envelope in genome organization, expression and stability. Nat Rev Mol Cell Biol. 2010;11:317–328. doi: 10.1038/nrm2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekhail K, Seebacher J, Gygi SP, Moazed D. Role for perinuclear chromosome tethering in maintenance of genome stability. Nature. 2008;456:667–670. doi: 10.1038/nature07460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondoux MA, Scaife JG, Zakian VA. Differential nuclear localization does not determine the silencing status of Saccharomyces cerevisiae telomeres. Genetics. 2007;177:2019–2029. doi: 10.1534/genetics.107.079848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung K, Chen C, Kolodner RD. Multiple pathways cooperate in the suppression of genome instability in Saccharomyces cerevisiae. Nature. 2001a;411:1073–1076. doi: 10.1038/35082608. [DOI] [PubMed] [Google Scholar]
- Myung K, Datta A, Kolodner RD. Suppression of spontaneous chromosomal rearrangements by S phase checkpoint functions in Saccharomyces cerevisiae. Cell. 2001b;104:397–408. doi: 10.1016/s0092-8674(01)00227-6. [DOI] [PubMed] [Google Scholar]
- Nishikawa S, Terazawa Y, Nakayama T, Hirata A, Makio T, Endo T. Nep98p is a component of the yeast spindle pole body and essential for nuclear division and fusion. J Biol Chem. 2003;278:9938–9943. doi: 10.1074/jbc.M210934200. [DOI] [PubMed] [Google Scholar]
- Oza P, Jaspersen SL, Miele A, Dekker J, Peterson CL. Mechanisms that regulate localization of a DNA double-strand break to the nuclear periphery. Genes Dev. 2009;23:912–927. doi: 10.1101/gad.1782209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oza P, Peterson CL. Opening the DNA repair toolbox: localization of DNA double strand breaks to the nuclear periphery. Cell Cycle. 2010;9:43–49. doi: 10.4161/cc.9.1.10317. [DOI] [PubMed] [Google Scholar]
- Penkner A, Tang L, Novatchkova M, Ladurner M, Fridkin A, Gruenbaum Y, Schweizer D, Loidl J, Jantsch V. The nuclear envelope protein Matefin/SUN-1 is required for homologous pairing in C. elegans meiosis. Dev Cell. 2007;12:873–885. doi: 10.1016/j.devcel.2007.05.004. [DOI] [PubMed] [Google Scholar]
- Penkner AM, et al. Meiotic chromosome homology search involves modifications of the nuclear envelope protein Matefin/SUN-1. Cell. 2009;139:920–933. doi: 10.1016/j.cell.2009.10.045. [DOI] [PubMed] [Google Scholar]
- Rao HB, Shinohara M, Shinohara A. Mps3 SUN domain is important for chromosome motion and juxtaposition of homologous chromosomes during meiosis. Genes Cells. 2011;16:1081–1096. doi: 10.1111/j.1365-2443.2011.01554.x. [DOI] [PubMed] [Google Scholar]
- Razafsky D, Hodzic D. Bringing KASH under the SUN: the many faces of nucleo-cytoskeletal connections. J Cell Biol. 2009;186:461–472. doi: 10.1083/jcb.200906068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolef Ben-Shahar T, Heeger S, Lehane C, East P, Flynn H, Skehel M, Uhlmann F. Eco1-dependent cohesin acetylation during establishment of sister chromatid cohesion. Science. 2008;321:563–566. doi: 10.1126/science.1157774. [DOI] [PubMed] [Google Scholar]
- Rowland BD, et al. Building sister chromatid cohesion: smc3 acetylation counteracts an antiestablishment activity. Mol Cell. 2009;33:763–774. doi: 10.1016/j.molcel.2009.02.028. [DOI] [PubMed] [Google Scholar]
- Ruault M, De Meyer A, Loiodice I, Taddei A. Clustering heterochromatin: Sir3 promotes telomere clustering independently of silencing in yeast. J Cell Biol. 2011;192:417–431. doi: 10.1083/jcb.201008007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato A, Isaac B, Phillips CM, Rillo R, Carlton PM, Wynne DJ, Kasad RA, Dernburg AF. Cytoskeletal forces span the nuclear envelope to coordinate meiotic chromosome pairing and synapsis. Cell. 2009;139:907–919. doi: 10.1016/j.cell.2009.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schober H, Ferreira H, Kalck V, Gehlen LR, Gasser SM. Yeast telomerase and the SUN domain protein Mps3 anchor telomeres and repress subtelomeric recombination. Genes Dev. 2009;23:928–938. doi: 10.1101/gad.1787509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz VP, Zakian VA. The Saccharomyces PIF1 DNA helicase inhibits telomere elongation and de novo telomere formation. Cell. 1994;76:145–155. doi: 10.1016/0092-8674(94)90179-1. [DOI] [PubMed] [Google Scholar]
- Sheff MA, Thorn KS. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast. 2004;21:661–670. doi: 10.1002/yea.1130. [DOI] [PubMed] [Google Scholar]
- Skibbens RV. Establishment of sister chromatid cohesion. Curr Biol. 2009;19:R1126–R1132. doi: 10.1016/j.cub.2009.10.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbens RV, Corson LB, Koshland D, Hieter P. Ctf7p is essential for sister chromatid cohesion and links mitotic chromosome structure to the DNA replication machinery. Genes Dev. 1999;13:307–319. doi: 10.1101/gad.13.3.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JS, Caputo E, Boeke JD. A genetic screen for ribosomal DNA silencing defects identifies multiple DNA replication and chromatin-modulating factors. Mol Cell Biol. 1999;19:3184–3197. doi: 10.1128/mcb.19.4.3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr DA, Fridolfsson HN. Interactions between nuclei and the cytoskeleton are mediated by SUN-KASH nuclear-envelope bridges. Annu Rev Cell Dev Biol. 2010;26:421–444. doi: 10.1146/annurev-cellbio-100109-104037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom L, Karlsson C, Lindroos HB, Wedahl S, Katou Y, Shirahige K, Sjogren C. Postreplicative formation of cohesion is required for repair and induced by a single DNA break. Science. 2007;317:242–245. doi: 10.1126/science.1140649. [DOI] [PubMed] [Google Scholar]
- Strom L, Lindroos HB, Shirahige K, Sjogren C. Postreplicative recruitment of cohesin to double-strand breaks is required for DNA repair. Mol Cell. 2004;16:1003–1015. doi: 10.1016/j.molcel.2004.11.026. [DOI] [PubMed] [Google Scholar]
- Taddei A, Hediger F, Neumann FR, Bauer C, Gasser SM. Separation of silencing from perinuclear anchoring functions in yeast Ku80, Sir4 and Esc1 proteins. EMBO J. 2004;23:1301–1312. doi: 10.1038/sj.emboj.7600144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei A, Schober H, Gasser SM. The budding yeast nucleus. Cold Spring Harb Perspect Biol. 2010;2:a000612. doi: 10.1101/cshperspect.a000612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong AH, Boone C. Synthetic genetic array analysis in Saccharomyces cerevisiae. Methods Mol Biol. 2006;313:171–192. doi: 10.1385/1-59259-958-3:171. [DOI] [PubMed] [Google Scholar]
- Toth A, Ciosk R, Uhlmann F, Galova M, Schleiffer A, Nasmyth K. Yeast cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev. 1999;13:320–333. doi: 10.1101/gad.13.3.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- Unal E, Arbel-Eden A, Sattler U, Shroff R, Lichten M, Haber JE, Koshland D. DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol Cell. 2004;16:991–1002. doi: 10.1016/j.molcel.2004.11.027. [DOI] [PubMed] [Google Scholar]
- Unal E, Heidinger-Pauli JM, Kim W, Guacci V, Onn I, Gygi SP, Koshland DE. A molecular determinant for the establishment of sister chromatid cohesion. Science. 2008;321:566–569. doi: 10.1126/science.1157880. [DOI] [PubMed] [Google Scholar]
- Unal E, Heidinger-Pauli JM, Koshland D. DNA double-strand breaks trigger genome-wide sister-chromatid cohesion through Eco1 (Ctf7) Science. 2007;317:245–248. doi: 10.1126/science.1140637. [DOI] [PubMed] [Google Scholar]
- Williams BC, Garrett-Engele CM, Li Z, Williams EV, Rosenman ED, Goldberg ML. Two putative acetyltransferases, san and deco, are required for establishing sister chromatid cohesion in Drosophila. Curr Biol. 2003;13:2025–2036. doi: 10.1016/j.cub.2003.11.018. [DOI] [PubMed] [Google Scholar]
- Witkin KL, Friederichs JM, Cohen-Fix O, Jaspersen SL. Changes in the nuclear envelope environment affect spindle pole body duplication in Saccharomyces cerevisiae. Genetics. 2010;186:867–883. doi: 10.1534/genetics.110.119149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong B, Gerton JL. Regulators of the cohesin network. Annu Rev Biochem. 2010;79:131–153. doi: 10.1146/annurev-biochem-061708-092640. [DOI] [PubMed] [Google Scholar]
- Xiong B, Lu S, Gerton JL. Hos1 is a lysine deacetylase for the Smc3 subunit of cohesin. Curr Biol. 2010;20:1660–1665. doi: 10.1016/j.cub.2010.08.019. [DOI] [PubMed] [Google Scholar]
- Xiong H, Rivero F, Euteneuer U, Mondal S, Mana-Capelli S, Larochelle D, Vogel A, Gassen B, Noegel A. Dictyostelium Sun-1 connects the centrosome to chromatin and ensures genome stability. Traffic. 2008;9:708–724. doi: 10.1111/j.1600-0854.2008.00721.x. [DOI] [PubMed] [Google Scholar]
- Zhang H, Richardson DO, Roberts DN, Utley R, Erdjument-Bromage H, Tempst P, Cote J, Cairns BR. The Yaf9 component of the SWR1 and NuA4 complexes is required for proper gene expression, histone H4 acetylation, and Htz1 replacement near telomeres. Mol Cell Biol. 2004;24:9424–9436. doi: 10.1128/MCB.24.21.9424-9436.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Roberts DN, Cairns BR. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell. 2005;123:219–231. doi: 10.1016/j.cell.2005.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, et al. Acetylation of Smc3 by Eco1 is required for S phase sister chromatid cohesion in both human and yeast. Mol Cell. 2008;31:143–151. doi: 10.1016/j.molcel.2008.06.006. [DOI] [PubMed] [Google Scholar]
- Zhang X, Lei K, Yuan X, Wu X, Zhuang Y, Xu T, Xu R, Han M. SUN1/2 and Syne/Nesprin-1/2 complexes connect centrosome to the nucleus during neurogenesis and neuronal migration in mice. Neuron. 2009;64:173–187. doi: 10.1016/j.neuron.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.