Abstract
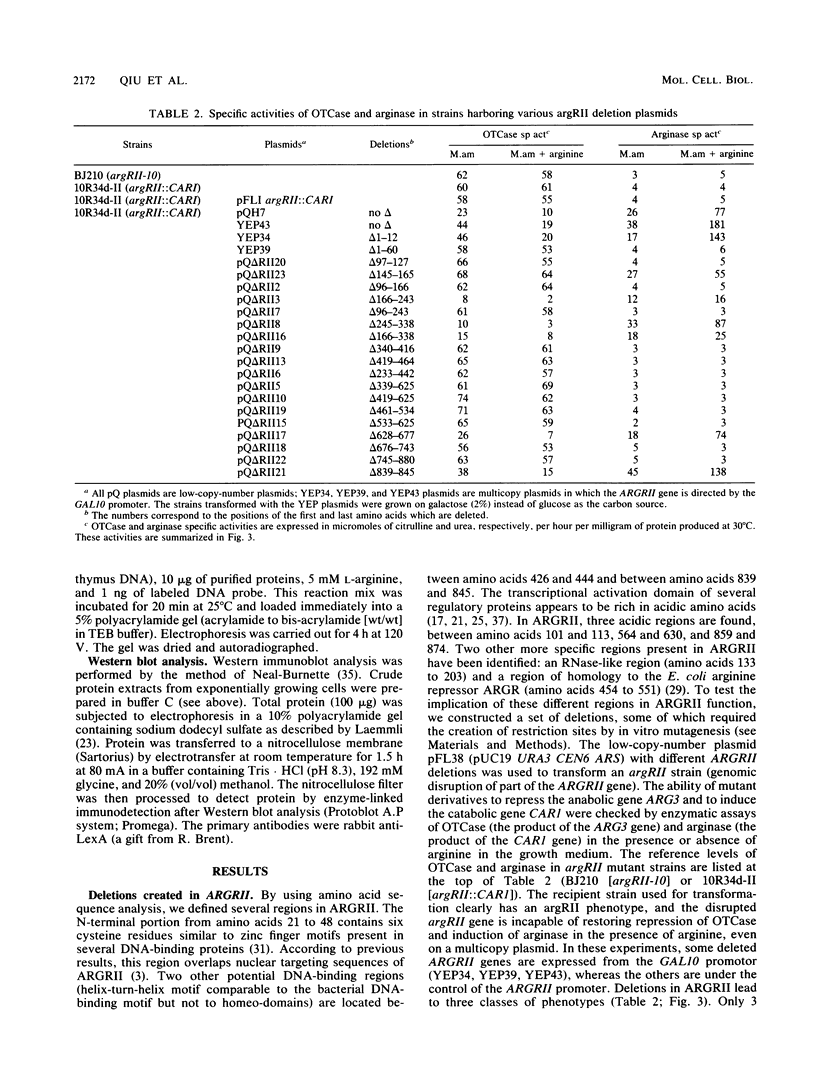
ARGRII is a regulatory protein which regulates the arginine anabolic and catabolic pathways in combination with ARGRI and ARGRIII. We have investigated, by deletion analysis and fusion to LexA protein, the different domains of ARGRII protein. In contrast to other yeast regulatory proteins, 92% of ARGRII is necessary for its anabolic repression function and 80% is necessary for its catabolic activator function. We can define three domains in this protein: a putative DNA-binding domain containing a zinc finger motif, a region more involved in the repression activity located around the RNase-like sequence, and a large activation domain.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ammerer G. Expression of genes in yeast using the ADCI promoter. Methods Enzymol. 1983;101:192–201. doi: 10.1016/0076-6879(83)01014-9. [DOI] [PubMed] [Google Scholar]
- Bechet J., Greenson M., Wiame J. M. Mutations affecting the repressibility of arginine biosynthetic enzymes in Saccharomyces cerevisiae. Eur J Biochem. 1970 Jan;12(1):31–39. doi: 10.1111/j.1432-1033.1970.tb00817.x. [DOI] [PubMed] [Google Scholar]
- Bercy J., Dubois E., Messenguy F. Regulation of arginine metabolism in Saccharomyces cerevisiae: expression of the three ARGR regulatory genes and cellular localization of their products. Gene. 1987;55(2-3):277–285. doi: 10.1016/0378-1119(87)90287-3. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brent R., Ptashne M. A bacterial repressor protein or a yeast transcriptional terminator can block upstream activation of a yeast gene. Nature. 1984 Dec 13;312(5995):612–615. doi: 10.1038/312612a0. [DOI] [PubMed] [Google Scholar]
- Brent R., Ptashne M. A eukaryotic transcriptional activator bearing the DNA specificity of a prokaryotic repressor. Cell. 1985 Dec;43(3 Pt 2):729–736. doi: 10.1016/0092-8674(85)90246-6. [DOI] [PubMed] [Google Scholar]
- Burnette W. N. "Western blotting": electrophoretic transfer of proteins from sodium dodecyl sulfate--polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal Biochem. 1981 Apr;112(2):195–203. doi: 10.1016/0003-2697(81)90281-5. [DOI] [PubMed] [Google Scholar]
- Chen E. Y., Seeburg P. H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985 Apr;4(2):165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Crabeel M., Huygen R., Verschueren K., Messenguy F., Tinel K., Cunin R., Glansdorff N. General amino acid control and specific arginine repression in Saccharomyces cerevisiae: physical study of the bifunctional regulatory region of the ARG3 gene. Mol Cell Biol. 1985 Nov;5(11):3139–3148. doi: 10.1128/mcb.5.11.3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabeel M., Lavalle R., Glansdorff N. Arginine-specific repression in Saccharomyces cerevisiae: kinetic data on ARG1 and ARG3 mRNA transcription and stability support a transcriptional control mechanism. Mol Cell Biol. 1990 Mar;10(3):1226–1233. doi: 10.1128/mcb.10.3.1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabeel M., Seneca S., Devos K., Glansdorff N. Arginine repression of the Saccharomyces cerevisiae ARG1 gene. Comparison of the ARG1 and ARG3 control regions. Curr Genet. 1988 Feb;13(2):113–124. doi: 10.1007/BF00365645. [DOI] [PubMed] [Google Scholar]
- Degols G. Functional analysis of the regulatory region adjacent to the cargB gene of Saccharomyces cerevisiae. Nucleotide sequence, gene fusion experiments and cis-dominant regulatory mutation analysis. Eur J Biochem. 1987 Nov 16;169(1):193–200. doi: 10.1111/j.1432-1033.1987.tb13597.x. [DOI] [PubMed] [Google Scholar]
- Deschamps J., Dubois E., Wiame J. M. L-Ornithine transaminase synthesis in Saccharomyces cerevisiae: regulation by inducer exclusion. Mol Gen Genet. 1979 Jul 24;174(3):225–232. doi: 10.1007/BF00267794. [DOI] [PubMed] [Google Scholar]
- Dubois E., Hiernaux D., Grennon M., Wiame J. M. Specific induction of catabolism and its relation to repression of biosynthesis in arginine metabolism of Saccharomyces cerevisiae. J Mol Biol. 1978 Jul 15;122(4):383–406. doi: 10.1016/0022-2836(78)90417-5. [DOI] [PubMed] [Google Scholar]
- Dubois E., Messenguy F. In vitro studies of the binding of the ARGR proteins to the ARG5,6 promoter. Mol Cell Biol. 1991 Apr;11(4):2162–2168. doi: 10.1128/mcb.11.4.2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois E., Messenguy F. Isolation and characterization of the yeast ARGRII gene involved in regulating both anabolism and catabolism of arginine. Mol Gen Genet. 1985;198(2):283–289. doi: 10.1007/BF00383008. [DOI] [PubMed] [Google Scholar]
- Gill G., Ptashne M. Mutants of GAL4 protein altered in an activation function. Cell. 1987 Oct 9;51(1):121–126. doi: 10.1016/0092-8674(87)90016-x. [DOI] [PubMed] [Google Scholar]
- Hinnen A., Hicks J. B., Fink G. R. Transformation of yeast. Proc Natl Acad Sci U S A. 1978 Apr;75(4):1929–1933. doi: 10.1073/pnas.75.4.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes D. S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981 Jun;114(1):193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- Hope I. A., Mahadevan S., Struhl K. Structural and functional characterization of the short acidic transcriptional activation region of yeast GCN4 protein. Nature. 1988 Jun 16;333(6174):635–640. doi: 10.1038/333635a0. [DOI] [PubMed] [Google Scholar]
- Hope I. A., Struhl K. Functional dissection of a eukaryotic transcriptional activator protein, GCN4 of yeast. Cell. 1986 Sep 12;46(6):885–894. doi: 10.1016/0092-8674(86)90070-x. [DOI] [PubMed] [Google Scholar]
- Johnston M., Dover J. Mutational analysis of the GAL4-encoded transcriptional activator protein of Saccharomyces cerevisiae. Genetics. 1988 Sep;120(1):63–74. doi: 10.1093/genetics/120.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lech K., Anderson K., Brent R. DNA-bound Fos proteins activate transcription in yeast. Cell. 1988 Jan 29;52(2):179–184. doi: 10.1016/0092-8674(88)90506-5. [DOI] [PubMed] [Google Scholar]
- Ma J., Ptashne M. Deletion analysis of GAL4 defines two transcriptional activating segments. Cell. 1987 Mar 13;48(5):847–853. doi: 10.1016/0092-8674(87)90081-x. [DOI] [PubMed] [Google Scholar]
- Messenguy F., Dubois E., Descamps F. Nucleotide sequence of the ARGRII regulatory gene and amino acid sequence homologies between ARGRII PPRI and GAL4 regulatory proteins. Eur J Biochem. 1986 May 15;157(1):77–81. doi: 10.1111/j.1432-1033.1986.tb09640.x. [DOI] [PubMed] [Google Scholar]
- Messenguy F., Dubois E. Participation of transcriptional and post-transcriptional regulatory mechanisms in the control of arginine metabolism in yeast. Mol Gen Genet. 1983;189(1):148–156. doi: 10.1007/BF00326068. [DOI] [PubMed] [Google Scholar]
- Messenguy F., Dubois E. The yeast ARGRII regulatory protein has homology with various RNases and DNA binding proteins. Mol Gen Genet. 1988 Jan;211(1):102–105. doi: 10.1007/BF00338399. [DOI] [PubMed] [Google Scholar]
- Messenguy F., Penninckx M., Wiame J. M. Interaction between arginase and ornithine carbamoyltransferase in Saccharomyces cerevisiae. The regulatory site for ornithine. Eur J Biochem. 1971 Sep 24;22(2):277–286. doi: 10.1111/j.1432-1033.1971.tb01542.x. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Pfeifer K., Arcangioli B., Guarente L. Yeast HAP1 activator competes with the factor RC2 for binding to the upstream activation site UAS1 of the CYC1 gene. Cell. 1987 Apr 10;49(1):9–18. doi: 10.1016/0092-8674(87)90750-1. [DOI] [PubMed] [Google Scholar]
- Pfeifer K., Kim K. S., Kogan S., Guarente L. Functional dissection and sequence of yeast HAP1 activator. Cell. 1989 Jan 27;56(2):291–301. doi: 10.1016/0092-8674(89)90903-3. [DOI] [PubMed] [Google Scholar]
- Pinkham J. L., Olesen J. T., Guarente L. P. Sequence and nuclear localization of the Saccharomyces cerevisiae HAP2 protein, a transcriptional activator. Mol Cell Biol. 1987 Feb;7(2):578–585. doi: 10.1128/mcb.7.2.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu H. F., Dubois E., Broën P., Messenguy F. Functional analysis of ARGRI and ARGRIII regulatory proteins involved in the regulation of arginine metabolism in Saccharomyces cerevisiae. Mol Gen Genet. 1990 Jul;222(2-3):192–200. doi: 10.1007/BF00633817. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumrada R. A., Cooper T. G. Isolation of the CAR1 gene from Saccharomyces cerevisiae and analysis of its expression. Mol Cell Biol. 1982 Dec;2(12):1514–1523. doi: 10.1128/mcb.2.12.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thukral S. K., Tavianini M. A., Blumberg H., Young E. T. Localization of a minimal binding domain and activation regions in yeast regulatory protein ADR1. Mol Cell Biol. 1989 Jun;9(6):2360–2369. doi: 10.1128/mcb.9.6.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]