Astrocytic tumors are highly invasive, but the mechanisms are unclear. PICK1 has reduced expression in astrocytic tumor cell lines and clinical cases. Exogenous PICK1 expression reduces invasion and anchorage-independent growth of a tumor cell line. The data suggest that PICK1 may be a target for therapeutic intervention.
Abstract
Astrocytic tumors are the most common form of primary brain tumor. Astrocytic tumor cells infiltrate the surrounding CNS tissue, allowing them to evade removal upon surgical resection of the primary tumor. Dynamic changes to the actin cytoskeleton are crucial to cancer cell invasion, but the specific mechanisms that underlie the particularly invasive phenotype of astrocytic tumor cells are unclear. Protein interacting with C kinase 1 (PICK1) is a PDZ and BAR domain–containing protein that inhibits actin-related protein 2/3 (Arp2/3)-dependent actin polymerization and is involved in regulating the trafficking of a number of cell-surface receptors. Here we report that, in contrast to other cancers, PICK1 expression is down-regulated in grade IV astrocytic tumor cell lines and also in clinical cases of the disease in which grade IV tumors have progressed from lower-grade tumors. Exogenous expression of PICK1 in the grade IV astrocytic cell line U251 reduces their capacity for anchorage-independent growth, two-dimensional migration, and invasion through a three-dimensional matrix, strongly suggesting that low PICK1 expression plays an important role in astrocytic tumorigenesis. We propose that PICK1 negatively regulates neoplastic infiltration of astrocytic tumors and that manipulation of PICK1 is an attractive possibility for therapeutic intervention.
INTRODUCTION
Astrocytic tumors are the most common form of primary brain tumor in humans (Furnari et al., 2007; Louis et al., 2007). Ninety-five percent of grade IV astrocytic tumors (glioblastomas) are “primary,” occurring de novo. The remaining 5% develop slowly from grade II tumors to form “secondary” grade IV tumors (Furnari et al., 2007; Louis et al., 2007; Sturm et al., 2014). Patients with grade IV tumors, both primary and secondary, have a life expectancy of approximately 1 yr. Two major characteristics of astrocytic tumors cause them to be refractory to current treatments: tumor heterogeneity and the highly invasive nature of the neoplastic cells (Giese et al., 2003; Furnari et al., 2007; Aum et al., 2014). Astrocytic tumor cells infiltrate deep into CNS tissue surrounding the primary tumor, allowing them to evade removal upon surgical resection of the primary tumor, and a common occurrence postsurgery is for the locally infiltrating cells to form tumors adjacent to the edge of the resection cavity. Astrocytic tumor cells can spread long distances from the initial tumor site to form tumors throughout the CNS (Giese et al., 2003). The mechanisms that underlie the particularly invasive phenotype of these cells are unclear.
Dynamic alterations to the actin cytoskeleton are crucial to all forms of cancer cell invasion (Nurnberg et al., 2011). Invasion of brain tissue by neoplastic astrocytes is initiated by the formation of membrane protrusions—lamellipodia and invadopodia (Maestro et al., 2001), which are driven by the polymerization of actin filaments that physically push the plasma membrane outward. Focal adhesions are formed at the protrusive leading edge to generate traction, and retraction of the rear of the cell then occurs through actin:myosin contractile forces (Olson et al., 2009). The actin-related protein (Arp) 2/3 complex is the major nucleator for the polymerization of branched actin networks necessary to drive membrane protrusions and hence cell motility (Welch et al., 1997; Mullins et al., 1998). A frequent observation in many forms of cancer, including astrocytic tumors, is deregulation of Arp2/3-dependent actin dynamics (Kurisu et al., 2010; Liu et al., 2013). Protein interacting with C-kinase 1 (PICK1) is a negative regulator of Arp2/3-dependent actin polymerization (Rocca et al., 2008), and PICK1 expression is up-regulated in a number of different cancers (Zhang et al., 2010). Of importance, however, PICK1 has not previously been studied in astrocytic tumors.
PICK1 is a cytosolic protein composed of a BAR and a PDZ domain, which mediate lipid membrane and protein interactions, respectively (Staudinger et al., 1995; Jin et al., 2006). PICK1 has a well-documented role in neuronal synaptic plasticity, in which it functions as a trafficking protein, particularly in regulating the endosomal trafficking and surface expression of α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid (AMPA) receptors, which are the major excitatory neurotransmitter receptors in the CNS (Xia et al., 1999; Hanley, 2008). The BAR and PDZ domains, as well as the Arp2/3-binding region of PICK1 in the C-terminal region, are critical for AMPA receptor trafficking (Xia et al., 1999; Jin et al., 2006; Rocca et al., 2008; Nakamura et al., 2011). PICK1 also interacts with the cytosolic domains of plasma membrane proteins involved in promoting tumorigenesis, such as Eph receptor tyrosine kinases, ephrins, Coxsackie-adenovirus receptor (CAR), and ErbB2/Her2 (Jaulin-Bastard et al., 2001; Excoffon et al., 2004), suggesting that PICK1 may play a role in cell motility or growth via such interactions. In addition, we recently demonstrated that endogenous PICK1 modulates the morphological plasticity of nontransformed astrocytes (Murk et al., 2013).
Here we report that PICK1 expression is reduced in astrocytic tumor cell lines and also in tumor tissue from patients with astrocytic tumors, particularly those that are high grade. We demonstrate that exogenous expression of PICK1 reduces the ability of astrocytic tumor cells to grow in an anchorage-independent manner, to migrate, and to invade in a three-dimensional (3D) environment. We therefore propose that PICK1 acts as a negative regulator of neoplastic infiltration by astrocytic tumors.
RESULTS
PICK1 expression is reduced in human astrocytic tumor cell lines
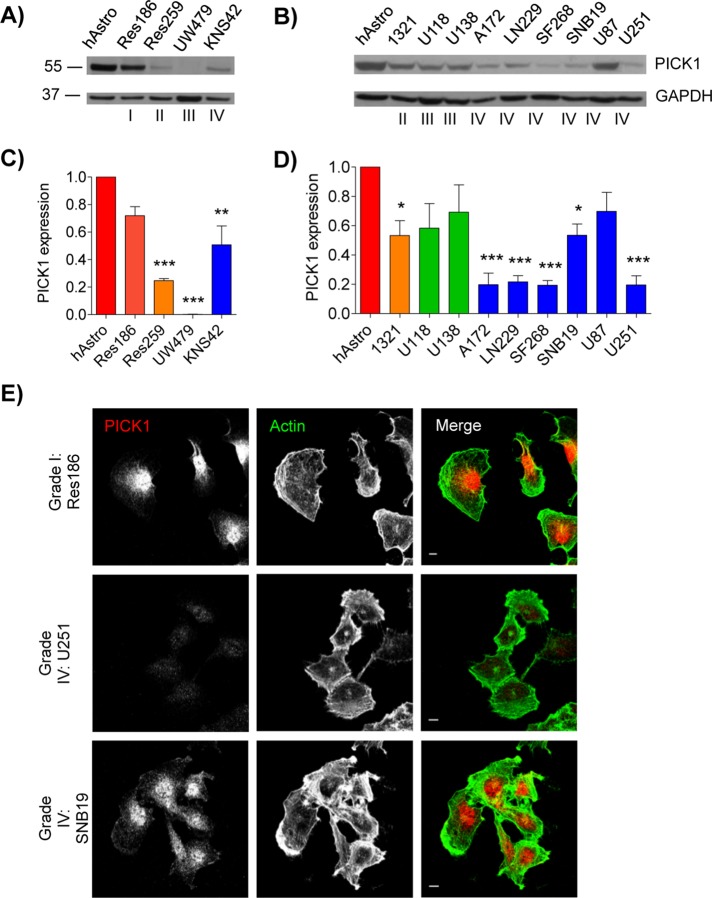
Although deregulation in PICK1 expression has been shown in a number of human cancers, it has not previously been investigated in astrocytic tumors (Zhang et al., 2010). Therefore we evaluated PICK1 expression in a number of astrocytic tumor cell lines originating from adult (Figure 1, B and D) and pediatric (Figure 1, A and C) tumors of grades I–IV. Most of the astrocytic tumor cell lines showed a reduction in PICK1 expression compared with that in the nontumorigenic human astrocyte control (Figure 1, A–D). In six of the seven grade IV cell lines, the reduction in PICK1 expression was statistically significant (Figure 1, C and D), suggesting that an aberration in PICK1 expression is a frequent occurrence in astrocytic tumors. The variability in PICK1 expression across the cell lines is consistent with the highly heterogeneous nature of astrocytic tumors (Furnari et al., 2007; Aum et al., 2014). The more dramatic reduction in PICK1 expression in cells derived from grade IV than with grade I tumors was confirmed by immunocytochemistry, which showed stronger labeling of PICK1 in the grade I Res186 tumor cells than in grade IV U251 (Figure 1E). An alternative grade IV line, SNB19, was also analyzed, which showed an intermediate level of PICK1 expression (Figure 1, D and E). Although PICK1 expression in U251 cells was at the limit of detection, we observed no differences in subcellular distribution of PICK1 among the three astrocytic tumor cell lines analyzed.
FIGURE 1:
PICK1 expression is reduced in human astrocytic tumor cell lines. (A–D) Western blot analysis of PICK1 protein expression in lysates of astrocytic tumor cell lines compared with control human astrocytes (hAstro). (A, C) Pediatric tumor cell lines. (B, D) Adult tumor cell lines. (C, D) Quantification of blots after calibration of PICK1 to GAPDH levels and normalization to control hAstro value on each blot. Values are mean ± SEM, n = 3. ANOVA p = 0.0001 for C and D. *p < 0.05, **p < 0.01, ***p < 0.001 (one-way ANOVA with Bonferroni’s post hoc test). (E) Res186 (grade I), U251 (grade IV), and SNB19 (grade IV) cell lines were stained for PICK1 (red) and F-actin (green). Far right, merged images. Scale bar, 10 μm.
To investigate the role of reduced PICK1 expression in astrocytic tumor biology, we generated lentiviral constructs to exogenously increase PICK1 expression in the U251 grade IV cell line. The viral vectors bicistronically encode mCherry and PICK1 via an internal ribosome entry site (IRES) or mCherry-IRES alone as a control. Virally transduced cells were sorted by fluorescence activated cell sorting (FACS) to generate homogeneous populations by analysis of the mCherry fluorescence signal. The FACS-sorted cells were gated with parameters to select for a relatively low level of mCherry fluorescence and therefore a low level of exogenous PICK1 to avoid excessive PICK1 expression (see Supplemental Figure S1 for characterization of exogenous PICK1 expression in virally transduced U251 cells). We tested these cells in a variety of assays to define the effect of altered PICK1 expression on the functional characteristics of grade IV tumor cells.
PICK1 reduces astrocytic tumor cell growth in an anchorage- independent setting
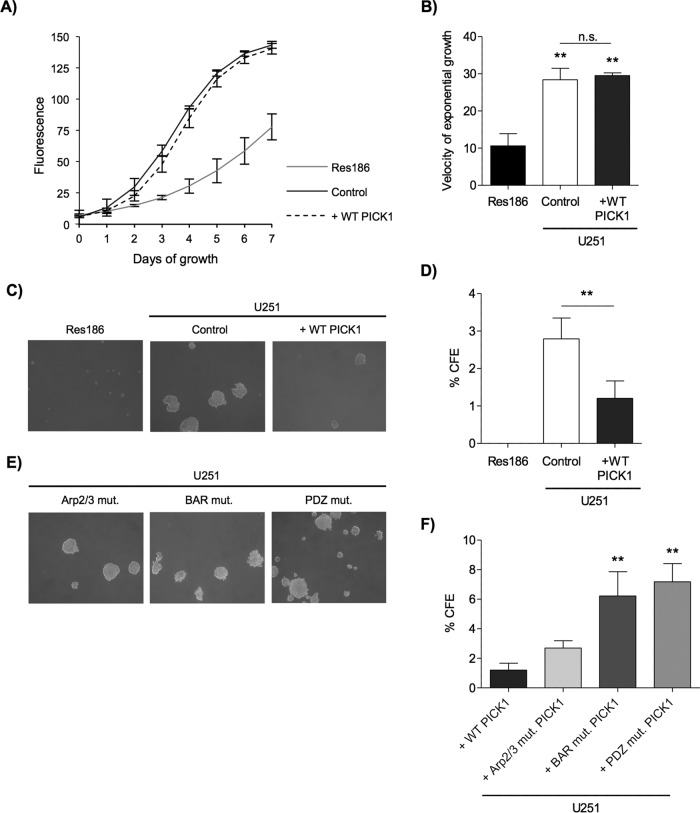
A defining characteristic of cancer is its limitless and uncontrolled proliferative capacity (Hanahan and Weinberg, 2000). Previous work suggested that PICK1 may play a role in cancer cell proliferation; for example, in cancers in which PICK1 is up-regulated, it is found to act as a proliferation-promoting factor (Zhang et al., 2010). We therefore investigated the effect of PICK1 on the proliferation of grade IV U251 cells. In the exponential phase (when grown in an adherent manner), the rate of control (mCherry alone) U251 cell proliferation did not differ from cells expressing exogenous wild-type (WT) PICK1 (Figure 2, A and B). A grade I cell line (Res186) was analyzed for comparison and, as expected, had much slower exponential growth than both populations of U251 cells (Figure 2, A and B). These results indicate that PICK1 expression has no effect on the proliferation of U251 cells when adhered to a solid substratum.
FIGURE 2:
Exogenous expression of PICK1 reduces the anchorage-independent growth but not anchorage-dependent proliferation in a grade IV astrocytic tumor cell line. (A) PICK1 expression had no effect on anchorage-dependent proliferation. Fluorescence is proportional to cell number for each well of cells plated in triplicate for each cell line, shown as sigmoidal logistic regression curves with four parameters. Values are mean ± SEM, n = 4. (B) Velocity of cell proliferation, calculated as slope coefficient in the linear exponential growth phase of each curve. ANOVA p = 0.0011. **p < 0.01 (one-way ANOVA with Bonferroni’s post hoc test, compared with Res186). (C) Exogenous PICK1 expression reduced anchorage-independent growth. Representative images after 1 wk of growth. Cells were seeded on soft agar at a density of 1 × 105 per 6-cm dish. (D) Quantification of experiments shown in C; values are mean percentage colony-forming efficiency (CFE) ± SEM, n = 6. Res186 cells never grew colonies larger than threshold size, and so Student’s t test was used to compare control and WT-PICK1–expressing cells, **p = 0.0044. (E) BAR and PDZ domain interactions were required for PICK1 to reduce anchorage- independent growth. Representative images after 1 wk of growth. Cells were seeded on soft agar at a density of 1 × 105 per 6-cm dish. (F) Quantification of experiments shown in E. ANOVA p = 0.0016, **p < 0.01 (repeat-measure ANOVA with Bonferroni post hoc test).
An important feature of cell transformation in high-grade malignant cancers is an ability to sustain anchorage-independent growth (Mori et al., 2009). To investigate whether the expression of PICK1 affects anchorage-independent growth, we monitored the formation of colonies generated from U251 cells grown in soft agar. Of interest, exogenous WT PICK1 expression caused a marked reduction in colony-forming efficiency (Figure 2, C and D), indicating reduced capacity to grow in an anchorage-independent manner. However, the colony-forming efficiency of the U251 cells expressing exogenous WT PICK1 was greater than that of the grade I cell line (Res186), which did not produce any colonies greater than the threshold size, indicating that the growth of the grade I cell line is exclusively anchorage dependent. This experiment demonstrates that low PICK1 expression plays a role in the capacity for anchorage- independent growth in astrocytic tumors.
To investigate the mechanism that underlies PICK1 suppression of U251 anchorage-independent growth, we generated PICK1 lentiviral vectors to express a number of PICK1 mutations that disrupt specific intermolecular interactions. These included the C-terminal PICK1 interaction with the Arp2/3 complex (W413A), the BAR domain interaction with membrane phospholipids and F-actin (KK251, 252EE), and PDZ domain interactions with various protein binding partners (KD27, 28AA; Terashima et al., 2004; Jin et al., 2006; Xu and Xia, 2006; Rocca et al., 2008, 2013). These constructs, hereafter referred to as Arp2/3 mutant, BAR mutant, and PDZ mutant, respectively, were expressed at comparable levels in U251 cells (Supplemental Figure S1, A and B), and immunocytochemical analysis suggests that these mutations have no detectable effect on the localization of FlagPICK1 in U251 cells (Supplemental Figure S1B). Figure 2, C, E, and F, shows that exogenous expression of the Arp2/3 mutant but not the BAR and PDZ mutants suppressed U251 anchorage-independent growth to a level similar to that of WT PICK1. These results indicate that both BAR and PDZ domain interactions of PICK1, but not its interaction with Arp2/3, are crucial for suppression of U251 anchorage-independent growth.
PICK1 impedes astrocytic tumor cell migration and invasion
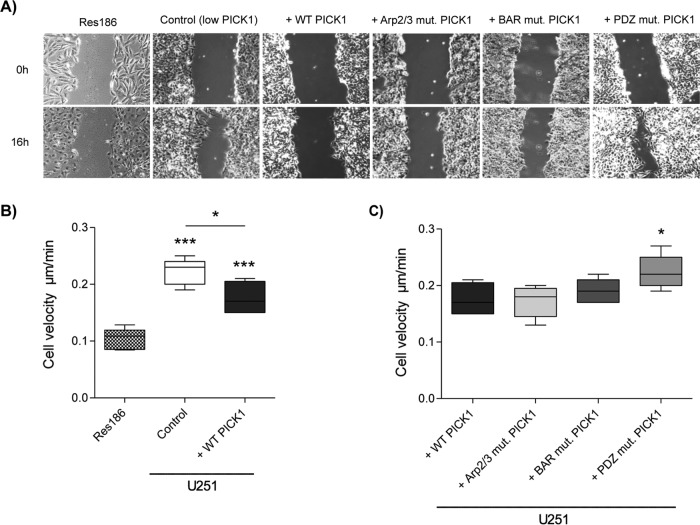
Migration and invasion through tissue is another key characteristic of cancer cells (Hanahan and Weinberg, 2000), and the invasion of astrocytic tumors through the CNS is known to be particularly aggressive (Furnari et al., 2007). To investigate the effect of PICK1 on U251 cell migration, we used a scratch-wound assay to assess the velocity of cell movement from a confluent cell monolayer into the wound area. As expected, U251 cells migrated faster than Res186 cells (Figure 3, A and B, and Supplemental Movies S1A and S1F). Exogenous expression of WT PICK1 caused a significant reduction in the migration rate of U251 cells (Figure 3, A and B, and Supplemental Movies S1A and S1B). Surprisingly, cells expressing the Arp2/3 mutant migrated at a similar rate to cells expressing exogenous WT PICK1, suggesting that the interaction with Arp2/3 is not required for the reduction in migration caused by PICK1 (Figure 3, A and C, and Supplemental Movie S1C). Cells expressing the BAR mutant also migrated at a similar rate, suggesting that BAR domain interactions with phospholipids and actin are also not involved (Figure 3, A and C, and Supplemental Movie S1D). In contrast, the suppression of cell migration by PICK1 was blocked by the PDZ mutation, indicating that PDZ domain protein–protein interactions of PICK1 are critically involved (Figure 3, A and C, and Supplemental Movie S1E). We also investigated the migration of another grade IV cell line, SNB19. Exogenous expression of WT-PICK1 by lentiviral transduction caused a small but significant reduction in migration rate of these cells (Supplemental Figure S2 and Supplemental Movies S2A and S2B; Sturm et al., 2014), providing further evidence that PICK1 reduces the migration of astrocytic tumor cells.
FIGURE 3:
Exogenous expression of PICK1 reduces the migration speed of U251 cells in a scratch-wound assay. (A) Representative phase contrast images of grade I Res186 (nontransduced) and grade IV U251 cells transduced with control (empty vector), WT-PICK1, or mutant PICK1 lentivirus, 0 and 16 h after scratching. (B, C) Cell velocity (micrometers/minute) was assessed by tracking individual cell trajectories every 20 min for 16 h. The box plots show the interquartile range, with mean and whiskers from the 10th to the 90th percentile, n = 5. (B) Res186 cells were compared with both control (empty vector) and WT-PICK1 U251 cells, ANOVA p < 0.0001 with Bonferroni post hoc tests also comparing control to WT PICK1, *p < 0.05, ***p < 0.001. (C) PICK1 mutants compared with WT PICK1, ANOVA p = 0.0293, *p < 0.05 (one-way ANOVA with Bonferroni post hoc tests).
To analyze actin dynamics in cells at the leading edge of the scratch wound, we generated similar lentiviral vectors that express Lifeact–green fluorescent protein (GFP) instead of mCherry and used live imaging to analyze the dynamics of filamentous (F)-actin structures in real time (Supplemental Movie 2, A and B; Louis et al., 2007). The movies demonstrate that exogenous WT PICK1 expression did not suppress dynamic alterations in the actin cytoskeleton.
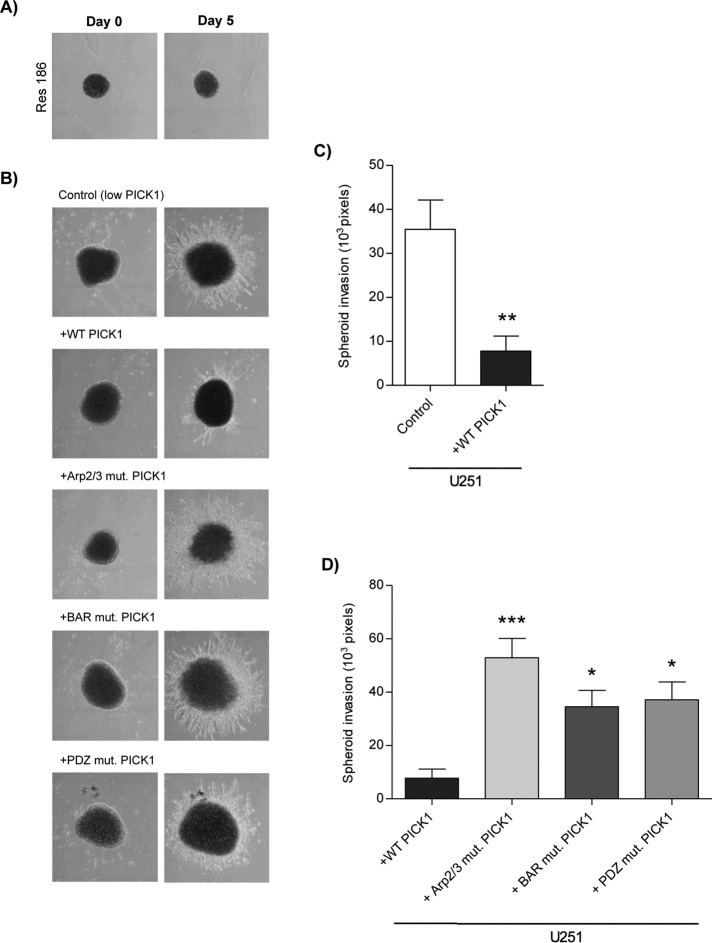
Cancer cells invade through a 3D environment, and although the process of invasion uses similar actin-based mechanisms to migration, there are key differences—in particular, the involvement of specialized invasive structures (invadopodia; Yamaguchi and Condeelis, 2007). To investigate 3D invasion, we used a cell spheroid assay to measure invasion of lentivirus-transduced U251 cells through Matrigel. As expected, Res186 cell spheroids were not able to invade into the Matrigel (Figure 4A). Exogenous expression of WT PICK1 dramatically reduced the invasion of cells away from the main “tumor-like” cell spheroid (Figure 4, B and C). All three PICK1 mutations blocked this effect, revealing that Arp2/3 binding, PDZ domain interactions, and BAR domain interactions are required for the suppressive activity of PICK1 on invasion (Figure 4, B and D). These experiments demonstrate that reduced PICK1 expression in U251 astrocytic tumor cells is a critical part of the mechanism that underlies the high level of invasiveness of these cells in a 3D environment.
FIGURE 4:
Exogenous expression of PICK1 reduces the ability of U251 cells to invade in a 3D environment. (A, B) Representative images of spheroids generated from Res186 (nontransduced) and U251 cells transduced with control (empty vector) and PICK1 lentiviruses and placed in Matrigel. Images were taken on the day the spheroids were placed in Matrigel (day 0) and again on day 5. (C, D) Mean area of spheroid invasion ± SEM, n = 6. (C) WT-PICK1 compared with control, **p = 0.009 (t test). (D) PICK1 mutants compared with WT PICK1, ANOVA p = 0.0007, *p < 0.05, ***p < 0.001 (one-way ANOVA with Bonferroni post hoc tests).
PICK1 increases Rac1 activation in U251 cells
The small GTPase Rac1 plays a critical role in transducing external stimuli into intracellular signaling events in a range of cellular processes, including growth, migration, and invasion, and Rac1 activation is deregulated in astrocytic tumors (Jaffe and Hall, 2005; Khalil and El-Sibai, 2012). PICK1 interacts with Rac and also with the Rac GEF Kalirin7 (Penzes and Jones, 2008; Rocca and Hanley, 2015). PICK1 also regulates the trafficking of receptors known to be upstream of Rac signaling, for example, Eph receptors (Torres et al., 1998). To investigate possible mechanisms for PICK1 regulation of invasion, growth, and migration, we examined Rac1 activation using GST-PAK pulldowns. Exogenous expression of WT-PICK1 caused a marked increase in Rac1 activation compared with controls, which was blocked by PDZ, BAR, and Arp2/3 mutations (Figure 5). Because all three PICK1 mutants were ineffective in increasing Rac1 activation, these data are consistent with a role for PICK1 in regulating Rac1 signaling in astrocytic tumor cell invasion, which is similarly sensitive to all three mutations (Figure 4, B and D).
FIGURE 5:
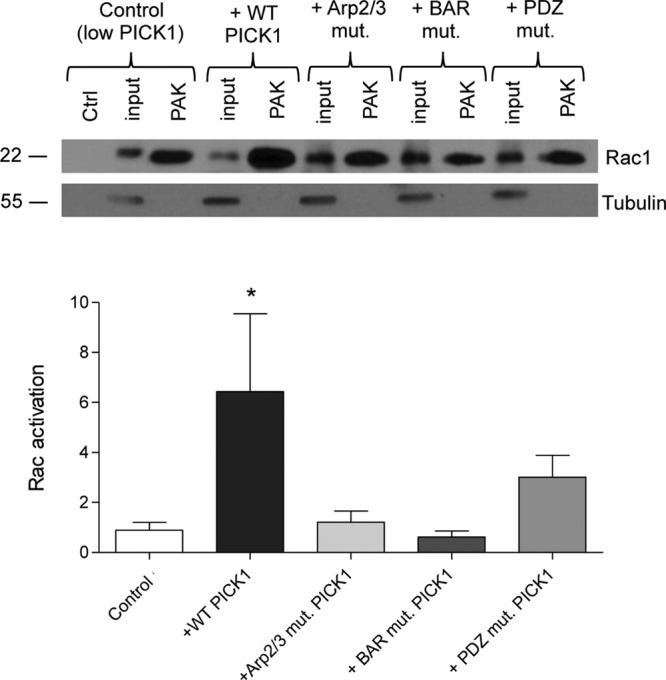
Exogenous expression of PICK1 WT in U251 cells increases levels of active Rac1. Lysates of U251 cells transduced with control (empty vector), and PICK1 lentiviruses were incubated with GST-PAK CRIB domain (PAK) immobilized on glutathione-agarose beads to isolate GTP-bound Rac1. Lysates from control cells were also incubated with GST alone as a negative control (Ctrl). Representative Western blots show the levels of active (PAK-bound) and total (input) Rac1. Tubulin was also analyzed as a loading control. Graph shows levels of active Rac1 normalized to the inputs and to tubulin; n = 5, ANOVA p = 0.0449, with Bonferroni post hoc tests, *p < 0.05.
Clinical astrocytic tumor tissue displays a reduction in PICK1 expression
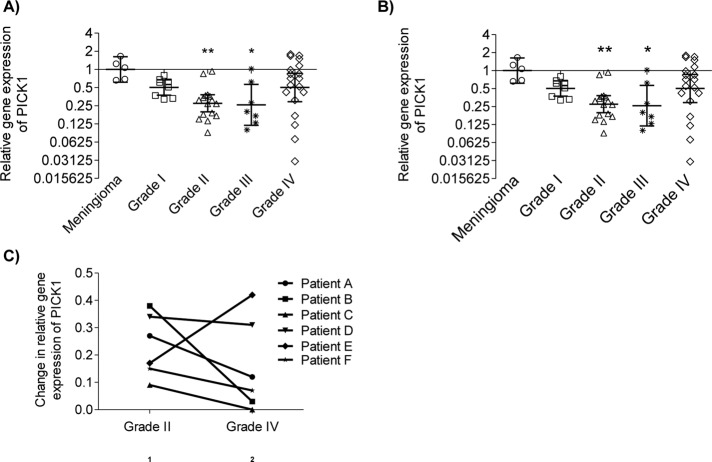
Our in vitro data demonstrate that PICK1 suppresses anchorage-independent growth, migration, and invasion in U251 cells, indicating that PICK1 acts as a suppressor of infiltration in astrocytic tumors. To evaluate how these in vitro observations relate to the human disease, we analyzed the expression of PICK1 in surgically resected tumor specimens by use of quantitative PCR (qPCR) to quantify PICK1 mRNA. In keeping with our in vitro findings, the tumors had an overall reduction in PICK1 mRNA compared with that in meningioma, which have a relatively noninvasive phenotype (Figure 6A; Louis et al., 2007; Alahmadi and Croul, 2011). However, post hoc testing showed the reduction in PICK1 mRNA expression to be significant only in the grade II and III astrocytic tumors, with the grade IV astrocytic tumors displaying a much wider variance in PICK1 expression that was not statistically significant from meningioma.
FIGURE 6:
PICK1 mRNA expression is reduced in clinical samples of human astrocytic tumors. (A, B) Vertical scatter plots of the relative levels of PICK1 mRNA in human astrocytic tumor tissue compared with the control group, a cohort of relatively noninvasive grade I meningiomas. The geometric mean and 95% confidence interval are shown for each tumor set (log 2 scale) ± SEM. (A) Entire cohort of excised astrocytic tumors, ANOVA p = 0.0058. (B) Only the progressive tumors from the cohort, ANOVA p = 0.0078. *p < 0.05, **p < 0.01 (Kruskal–Wallis test with Dunn’s post hoc tests). (C) Relative PICK1 mRNA expression of progressive tumors from six patients. Change in PICK1 expression accompanying the progression from grade II to grade IV in each patient.
Review of the clinical and neuropathological findings indicated that most of the grade IV tumors were primary glioblastomas (with a short clinical history and no mutation in the isocitrate dehydrogenase genes IDH1 or IDH2; Agnihotri et al., 2014) rather than secondary glioblastomas that had progressed from grade II and III neoplasms. The significantly lower PICK1 expression in the grade II and III progressive tumors and the wide variance in expression in the grade IV tumors led us to investigate whether reduced PICK1 expression is a specific characteristic of progressive rather than de novo astrocytic tumors. Therefore we separately analyzed the cases of progressive disease (i.e., all of the grade II and III tumors and a subpopulation of the grade IV tumors; Figure 6B). Indeed, PICK1 mRNA expression in all of the progressive tumors was significantly lower than that of meningiomas, suggesting that reduced PICK1 expression is particularly relevant in progressive forms of astrocytic neoplasia. Figure 6C highlights another important feature of PICK1 expression in progressive tumors: a reduction in PICK1 expression in five of the six patients who had repeat neurosurgical resections of tumor tissue as their disease progressed from grade II to grade IV neoplasia. The one patient who did not show a reduction in PICK1 with tumor progression was reviewed, and the histological findings indicated that this tumor differed from the other progressive tumors in being predominantly gemistocytic at the time of initial presentation (Supplemental Figure S3).
These data demonstrate that PICK1 expression is significantly reduced in astrocytic tumors, particularly those that have progressed from low to high grade. This reflects our in vitro data and suggests that the increase in invasive behavior that accompanies malignant progression of astrocytic tumors may result, at least in part, from a reduction in PICK1 expression.
DISCUSSION
Our results demonstrate a role for PICK1 in regulating astrocytic tumor cell growth and invasion. PICK1 expression had not previously been examined in astrocytic tumors (Zhang et al., 2010), and here we show a reduction in PICK1 expression in both astrocytic tumor cell lines and human clinical samples. Using functional in vitro assays, we show that the exogenous expression of WT PICK1 in astrocytic tumor cells reduces anchorage-independent colony growth, two-dimensional (2D) migration, and invasion in a 3D environment.
A significant reduction in PICK1 expression compared with control astrocytes was shown for the majority but not all of the astrocytic tumor cell lines. A notable exception was the highly invasive grade IV U87 cell line. There is a precedent for distinct U87 expression profiles compared with other invasive cell lines. For example, high levels of ephrinB3 are found in U251 cells, and ephrin B3 knockdown reduces cell migration. In contrast, U87 cells have low ephrinB3 expression, and overexpression causes an increase in cell migration (Nakada et al., 2006). Therefore the experimental data from both cell lines support a role for ephrin B3 in increasing cell migration but suggest that the invasive phenotype in U87 involves an ephrinB3-independent mechanism. This observation, together with our data for PICK1, is consistent with the heterogeneous nature of astrocytic tumors.
The 2D migration assays demonstrate that exogenous expression of WT PICK1 reduces the migration rate of U251 and SNB19 cells into a scratch wound, indicating that the low PICK1 expression observed in these cells contributes to their migratory potential. By selective mutagenesis, we showed that the PICK1 PDZ domain is essential for this effect on 2D migration. PICK1 PDZ domain interacts with numerous cell surface proteins to regulate their trafficking through the endosomal system and also with cytosolic signaling proteins such as protein kinase C (PKC) and calcineurin (Xu and Xia, 2006). We propose that the highly motile and invasive phenotype of U251 cells is caused, at least in part, by a deficit in PICK1-mediated trafficking of or signaling through as-yet-unidentified proteins (Madsen et al., 2005). For example, PICK1 interacts with several members of the Eph receptor tyrosine kinase family, as well as with their ephrin ligands (Torres et al., 1998). Eph receptor–ephrin interactions are involved in sensing cell-to-cell contacts (Son et al., 2014), and initiate downstream signaling events that, via Rho GTPases, regulate actin dynamics in both the healthy state (Puschmann and Turnley, 2010; Batson et al., 2013) and numerous cancers, including astrocytic tumors (Nakada et al., 2006; Astin et al., 2010; Pasquale, 2010). Consistent with this hypothesis, we find that exogenous expression of PICK1 increases Rac1 activation. Although Rac1 is typically found to promote astrocytic tumor progression (Fortin Ensign et al., 2013), reduced levels of Rac1 have also been reported to correlate with an invasive phenotype, and increased activation of Rac1 can cause a reduction in astrocytic tumor invasion (Khalil and El-Sibai, 2012; Reyes et al., 2013). These latter studies are consistent with our results, since the high Rac1 activity found in the WT PICK1–expressing cells correlates with reduced tumorigenic characteristics.
In the scratch-wound experiments using Lifeact-GFP, dynamic F-actin was clearly visible in leading-edge filopodia and lamellipodia polarized toward the scratch wound. Although lamellipodia formation and dynamics are Arp2/3 dependent (Machesky et al., 1997; Mullins et al., 1998), increasing PICK1 expression appeared to have little effect on these structures. This is consistent with our observation that the interaction with Arp2/3 is not involved in PICK1-dependent inhibition of migration of U251 cells.
We also found that exogenous expression of PICK1 suppressed the invasion of U251 cells through a 3D matrix. This is further evidence that the low PICK1 expression level in these cells is at least partly responsible for their highly invasive phenotype. In contrast to 2D migration, however, in which only the PDZ mutation disrupted PICK1 function in the suppression of motility, all three mutations tested prevented exogenous PICK1 from suppressing the capacity of the U251 cells for 3D invasion. This demonstrates that interactions with PDZ domain ligands, the Arp2/3 complex, and phospholipid membranes are all necessary for this aspect of PICK1 function. These results indicating PICK1 manipulation of the actin cytoskeleton and trafficking via the BAR domain as necessary in 3D invasion but not 2D migration are consistent with the greater mechanistic complexity of 3D invasion than 2D migration (Olson and Sahai, 2009; Ridley, 2011). Moreover, previous evidence suggested that Arp2/3 inhibition is more effective in suppressing invasion than migration of U251 cells (Liu et al., 2013). This is consistent with our results, which suggest that a requirement for Arp2/3 inhibition by PICK1 is important for invasion but not for migration.
Although exogenous expression of WT PICK1 repressed the growth of anchorage-independent colonies, it did not alter the proliferation rate of U251 cells when adhered to a solid substratum. This indicates that the repressive effect of WT PICK1 on growth/proliferation depends on cell adhesion, suggesting that the mechanism by which WT PICK1 affects proliferation is indirect, through adhesive complex signaling. This hypothesis is supported by our results showing that the PDZ domain and the BAR domain, important for PICK1-mediated trafficking of cell surface receptors such as those important in adhesive interactions, are crucial for suppression of anchorage-independent colony growth by PICK1. Further support for this concept comes from the previous observation that the PICK1 PDZ domain interacts with plasma membrane adhesive proteins such as CAR, which plays an important role in astrocytic tumor growth (Kim et al., 2003; Excoffon et al., 2004).
Our results show that PICK1 PDZ domain interactions are essential for all aspects of U251 cell behavior tested. Further work will be necessary to identify PICK1-interacting proteins that show aberrant activity or localization in astrocytic tumor cells as a result of insufficient PICK1 to regulate relevant signaling pathways or trafficking events. The additional requirement for Arp2/3 binding in 3D invasion and BAR domain interactions in both invasion and anchorage-independent growth raises the possibility that these processes involve trafficking of distinct plasma membrane proteins with different requirements for PICK1 molecular interactions. Alternatively, additional PICK1-mediated mechanisms may be involved, such as regulation of actin-based structural dynamics.
The analysis of PICK1 expression in clinical samples of human astrocytic tumors showed that a reduction in PICK1 is a relevant phenotypic feature of the human disease. This was particularly evident in progressive tumors, with a reduction in PICK1 expression as the tumors progressed from grade II to IV. Our in vitro data suggest that this further reduction in PICK1 expression is likely to be biologically important in increasing invasiveness. Up-regulating PICK1 or stimulating its downstream effectors may offer a means of preventing this. Overall the primary grade IV tumors in our study did not show a significant reduction in PICK1; expression was low in some cases but high in others, reflecting the heterogeneity of this group. It seems likely that multiple molecular pathways may lead to the invasive behavior of tumor cells in primary glioblastomas, in which case, PICK1 manipulation would be a therapeutic option in only a subset of these tumors.
Our choice of U251 cells for the majority of our functional experiments was based on our initial observation that these cells have a consistently low level of PICK1 expression. They are therefore a good model system for investigating the importance of PICK1 in migration and invasion of astrocytic tumors. Our data from clinical samples indicate that this is most relevant to progressive rather than de novo tumors. Unfortunately, insufficient information is available to confirm such an origin of U251 cells.
The marked reduction in PICK1 expression in both the astrocytic tumor cell lines and human cases of the disease suggests that PICK1 acts as a tumor suppressor in this type of tumor. This is in contrast to previous reports of PICK1 up-regulation and purported oncogenic activity in other types of cancer (Zhang et al., 2010). This cancer type–specific deregulation of PICK1 is most likely due to the differences in the properties of the cells of origin and of the tumors themselves. Astrocytes have an inherent ability to move through CNS tissue (Barres, 2008), unlike epithelial cells. In contrast, even the highest-grade astrocytic tumors rarely enter the bloodstream and disseminate hematogenously, as many cancers do (Giese et al., 2003; Furnari et al., 2007). These distinctive characteristics of astrocytes and astrocytic tumor cells might explain why a protein that is involved in regulating invasion in one type of tumor might not play the same role in others. It might also be relevant that basal PICK1 expression is higher in the brain than in most other tissues (Staudinger et al., 1995), suggesting that PICK1 has functions that are specific to the CNS.
Further investigation is needed to elucidate the pathways responsible for down-regulation of PICK1 in astrocytic tumors and determine the molecular processes through which it influences the migration, invasion, and anchorage-independent growth of astrocytic tumors. The relevance of PICK1 suppression in at least a subset of the disease in humans makes manipulation of PICK1 an attractive possibility for therapeutic intervention in this notoriously aggressive and difficult-to-treat disease.
Materials and Methods
Cell culture
Human astrocytic tumor cell lines KNS42, LN229, Res186, Res259, SF268, SNB19, U87, U118, U138, U251, and UW479 (a generous gift from Chris Jones, Institute of Cancer Research, London, United Kingdom), A172 and U251 (kindly provided by Wilfried Roth, German Cancer Research Center, Heidelberg, Germany), 1321N1 (a generous gift from Amal Shervington, University of Lancashire, Preston, United Kingdom), and human astrocytes (normal human astrocytes, referred to here as hAstro; Lonza, Basel, Switzerland) were maintained in DMEM supplemented with 10% fetal bovine serum (FBS), glutamine, and penicillin/streptomycin (P/S) at 37°C, 4% CO2.
Antibodies and reagents
For immunoblotting, we used anti-PICK1 (Neuromab; University of California, Davis, Davis, CA), glyceraldehyde-3-phosphate dehydrogenase (GAPDH; Abcam, Cambridge, UK), Rac1 (BD Biosciences, San Jose, CA), tubulin (Sigma-Aldrich, Dorset, UK), and horseradish peroxidase–conjugated secondary antibodies (Sigma-Aldrich). Immunoreactive bands were detected by enhanced chemiluminescence. For immunofluorescence microscopy, we used anti-PICK1 (Abcam), anti-Flag (Sigma-Aldrich), and cy3-conjugated secondary antibody (Jackson ImmunoResearch Labs, West Grove, PA). F-actin was visualized with Alexa 647–conjugated phalloidin (Invitrogen, Paisley, UK).
Lentiviral constructs
mCherry IRES empty and mCherry IRES PICK1 cassettes were ligated into a pCDF vector (Systems Biosciences, Mountain View, CA). Mutagenesis was carried out by PCR to generate Arp2/3-binding (W413A), BAR (KK251, 252EE), and PDZ (KD27, 28AA) mutations (Terashima et al., 2004; Jin et al., 2006; Rocca et al., 2008, 2013).
Proliferation assay
The adherent cell growth assays were carried out using the CyQuant NF cell proliferation assay kit (Life Technologies, Paisley, UK) according to the manufacturer’s instructions (Liu et al., 2007). Fluorescence readout (proportional to cell number) was measured every day for 7 d on the Fluoroskan Ascent FL (ThermoScientific, Paisley, UK), exciting at 485 nm and detecting at 538 nm. Four-parameter sigmoidal logistic regression curves were fitted to the 7-d fluorescence readout from which the velocity of cell growth during the exponential linear phase of growth was calculated.
Anchorage-independent assay
This was carried out as previously described (Rutka et al., 1994; Chell et al., 2006). Cells were grown suspended in medium-enriched agar, and colonies that were of threshold size or above (threshold determined with reference to a central graticule in the viewing field) were counted 1 wk after seeding. The colony-forming efficiency (CFE) was then calculated for each cell line. The surface area of one field of view was 15.89 mm2, and the surface area of each dish was 2826 mm2. Therefore there were 177.84 possible fields of view that could be counted from. The percentage CFE was calculated in Excel (Microsoft) according to the equation
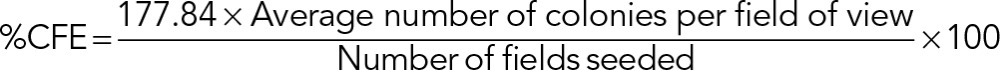 |
Migration assay
This was performed as previously described (Engelhard et al., 2001). Cells were grown until confluent on glass-bottomed microwell dishes (MatTek, Ashland, MA), after which medium was exchanged for CO2-independent medium (Leibovitz’s L-15 Life Technologies], 10% FBS, and P/S) containing 125 μM cytosine β-d-arabinofuranoside (Sigma-Aldrich) and then scratched with a p200 pipette tip. Images were acquired from multiple sites every 20 min for 16 h. Individual cells were traced, and the velocity of cell movement was calculated. The same migration assay was performed with cells expressing Lifeact-GFP imaged 2.5 h after scratching, with images taken every 30 s for a further 2.5 h.
Cell spheroid invasion assay
Spheroids were prepared by the hanging-drop method, adapted from a previous report (Del Duca et al., 2004), by suspending a small volume of cells on an inverted plate lid above wells containing 5 ml of DMEM. After 7 d, the spheroids were maneuvered onto glass-bottomed microwell dishes, and Matrigel (BD Biosciences) was prepared at 1:1 dilution with DMEM (without phenol red; Life Technologies) and added on top of the spheroids. Fluorescence and phase contrast images of the spheroids were acquired on days 0 and 5 by confocal microscopy. Spheroid invasion was calculated using ImageJ (National Institutes of Health, Bethesda, MD) as the area from the edge of the spheroid bulk to the most peripheral invasive extensions.
PAK Rac1 activation assay
Glutathione S-transferase (GST)–PAK or GST alone was immobilized on glutathione-agarose beads (Sigma-Aldrich). Beads were washed and incubated with 1.5–5 mg of lysate prepared from virally transduced U251 cells for 1 h at 4°C with rotation. Beads were washed four times, and then the bound fraction was eluted with SDS sample buffer. Proteins were separated by SDS–PAGE and detected by Western blotting with antibodies against Rac1 and tubulin. For each condition, the GST-PAK–bound signal was normalized to input and to tubulin loading controls. For each experiment, Rac1 activity of the PICK1 WT and mutant conditions was normalized to the control.
Clinical samples and histology
Frozen human tumor tissue was obtained from the diagnostic archive in the Department of Neuropathology, North Bristol NHS Trust (Bristol, United Kingdom) with approval by the National Research Ethics Service Committee South West (REC ref 11/H0107/2). This tissue had been removed during neurosurgical treatment of patients with astrocytic tumors. The tumors had all been subjected to detailed histological assessment and classified and graded according to World Health Organization (WHO) criteria (Louis et al., 2007). Samples of unfixed tumor tissue from the biopsies had been stored at −80°C. For this study, we used small samples (<1 mg) of the frozen tumor tissue. We analyzed 50 astrocytic and five meningioma tumor tissue samples.
Clinical sample preparation
The tissue was homogenized and lysed in TRIzol reagent (Life Technologies) according to the manufacturer’s instructions. The Ribogreen RNA Assay kit (Molecular Probes Quan-iT; Life Technologies) was used to quantify RNA extract according to the manufacturer’s instructions, with fluorescence measured in a FLUOstar OPTIMA reader (BMG Labtech, Aylesbury, UK). The high-capacity cDNA reverse transcriptase kit (Applied Biosystems, Life Technologies) was used to reverse transcribe the RNA, and the resulting cDNA was quantified using the Picogreen cDNA assay kit.
qPCR
We performed qPCR using TaqMan Assay on Demand (AOD) Primer and Probe combination kits (Life Technologies) Hs00202661_m1 for PICK1 and Hs00427620_m1 for the calibrator, Tata box–binding protein (TBP). The qPCR measurements were performed according to manufacturer’s instructions on the Applied Biosystems ViiA 7 by use of the comparative cycle threshold (CT) method with a PCR cycling profile of 95°C for 20 s with 40 cycles of 95°C for 1 s and 60°C for 20 s. The CT was set at 0.2, and PICK1 astrocytic tumor transcript levels were calculated relative to TBP and PICK1 expression in the control group by means of the Livak 2−ΔΔCt method (Livak and Schmittgen, 2001). Because it was not possible to obtain normal, nonneoplastic astrocytic tissue from the human patients, we used tissue from five WHO grade I meningiomas as an arbitrary comparator, selected because low-grade meningiomas are relatively noninvasive tumors (Louis et al., 2007).
Statistics
Analyses were performed using two-tailed t tests or one-way analysis of variance (ANOVA) for parametric data and the Kruskal–Wallis test for nonparametric data, with the Bonferroni or Dunn test for post hoc pairwise comparisons.
Supplementary Material
Acknowledgments
We thank Chris Jones, Amal Shervington, and Wilfried Roth for the astrocytic tumor cell lines, Ann Williams for invaluable advice throughout the project, and Harry Mellor for critical reading of the manuscript. L.C. was funded by a Medical Research Council Studentship. The cell imaging in this study was carried out in the Wolfson Bioimaging Facility, University of Bristol, Bristol, United Kingdom.
Abbreviations used:
- Arp2/3
actin-related protein 2/3
- BAR
Bin-Amphiphysin-Rvs
- CAR
coxsackie and adenovirus receptor
- FACS
fluorescence-activated cell sorting
- IRES
internal ribosome entry site
- PAK
p21-activating kinase
- PDZ
PSD95, Dlg1, ZO-1
- PICK1
protein interacting with C kinase 1.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-05-0270) on October 14, 2015.
REFERENCES
- Agnihotri S, Aldape KD, Zadeh G. Isocitrate dehydrogenase status and molecular subclasses of glioma and glioblastoma. Neurosurg Focus. 2014;37:E13. doi: 10.3171/2014.9.FOCUS14505. [DOI] [PubMed] [Google Scholar]
- Alahmadi H, Croul SE. Pathology and genetics of meningiomas. Semin Diagn Pathol. 2011;28:314–324. doi: 10.1053/j.semdp.2011.01.002. [DOI] [PubMed] [Google Scholar]
- Astin JW, Batson J, Kadir S, Charlet J, Persad RA, Gillatt D, Oxley JD, Nobes CD. Competition amongst Eph receptors regulates contact inhibition of locomotion and invasiveness in prostate cancer cells. Nat Cell Biol. 2010;12:1194–1204. doi: 10.1038/ncb2122. [DOI] [PubMed] [Google Scholar]
- Aum DJ, Kim DH, Beaumont TL, Leuthardt EC, Dunn GP, Kim AH. Molecular and cellular heterogeneity: the hallmark of glioblastoma. Neurosurg Focus. 2014;37:E11. doi: 10.3171/2014.9.FOCUS14521. [DOI] [PubMed] [Google Scholar]
- Barres BA. The mystery and magic of glia: a perspective on their roles in health and disease. Neuron. 2008;60:430–440. doi: 10.1016/j.neuron.2008.10.013. [DOI] [PubMed] [Google Scholar]
- Batson J, Astin JW, Nobes CD. Regulation of contact inhibition of locomotion by Eph-ephrin signalling. J Microsc. 2013;251:232–241. doi: 10.1111/jmi.12024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chell SD, Witherden IR, Dobson RR, Moorghen M, Herman AA, Qualtrough D, Williams AC, Paraskeva C. Increased EP4 receptor expression in colorectal cancer progression promotes cell growth and anchorage independence. Cancer Res. 2006;66:3106–3113. doi: 10.1158/0008-5472.CAN-05-3702. [DOI] [PubMed] [Google Scholar]
- Del Duca D, Werbowetski T, Del Maestro RF. Spheroid preparation from hanging drops: characterization of a model of brain tumor invasion. J Neurooncol. 2004;67:295–303. doi: 10.1023/b:neon.0000024220.07063.70. [DOI] [PubMed] [Google Scholar]
- Engelhard HHD, Holly AK, Samuel CPS, Van Eldik L. Therapeutic effects of sodium butyrate on glioma cells in vitro and in the rat C6 glioma model. Neurosurgery 48. 2001:616–625. doi: 10.1097/00006123-200103000-00035. [DOI] [PubMed] [Google Scholar]
- Excoffon KJ, Hruska-Hageman A, Klotz M, Traver GL, Zabner J. A role for the PDZ-binding domain of the coxsackie B virus and adenovirus receptor (CAR) in cell adhesion and growth. J Cell Sci. 2004;117:4401–4409. doi: 10.1242/jcs.01300. [DOI] [PubMed] [Google Scholar]
- Fortin Ensign SP, Mathews IT, Symons MH, Berens ME, Tran NL. Implications of Rho GTPase signaling in glioma cell invasion and tumor progression. Front Oncol. 2013;3:241. doi: 10.3389/fonc.2013.00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- Giese A, Bjerkvig R, Berens ME, Westphal M. Cost of migration: invasion of malignant gliomas and implications for treatment. J Clin Oncol. 2003;21:1624–1636. doi: 10.1200/JCO.2003.05.063. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Hanley JG. PICK1: a multi-talented modulator of AMPA receptor trafficking. Pharmacol Ther. 2008;118:152–160. doi: 10.1016/j.pharmthera.2008.02.002. [DOI] [PubMed] [Google Scholar]
- Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- Jaulin-Bastard F, Saito H, Le Bivic A, Ollendorff V, Marchetto S, Birnbaum D, Borg JP. The ERBB2/HER2 receptor differentially interacts with ERBIN and PICK1 PSD-95/DLG/ZO-1 domain proteins. J Biol Chem. 2001;276:15256–15263. doi: 10.1074/jbc.M010032200. [DOI] [PubMed] [Google Scholar]
- Jin W, Ge WP, Xu J, Cao M, Peng L, Yung W, Liao D, Duan S, Zhang M, Xia J. Lipid binding regulates synaptic targeting of PICK1, AMPA receptor trafficking, and synaptic plasticity. J Neurosci. 2006;26:2380–2390. doi: 10.1523/JNEUROSCI.3503-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil BD, El-Sibai M. Rho GTPases in primary brain tumor malignancy and invasion. J Neurooncol. 2012;108:333–339. doi: 10.1007/s11060-012-0866-8. [DOI] [PubMed] [Google Scholar]
- Kim M, Sumerel LA, Belousova N, Lyons GR, Carey DE, Krasnykh V, Douglas JT. The coxsackievirus and adenovirus receptor acts as a tumour suppressor in malignant glioma cells. Br J Cancer. 2003;88:1411–1416. doi: 10.1038/sj.bjc.6600932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurisu S, Takenawa T. WASP and WAVE family proteins: friends or foes in cancer invasion? Cancer Sci. 2010;101:2093–2104. doi: 10.1111/j.1349-7006.2010.01654.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Hannafon B, Gill L, Kelly W, Benbrook D. Flex-Hets differentially induce apoptosis in cancer over normal cells by directly targeting mitochondria. Mol Cancer Ther. 2007;6:1814–1822. doi: 10.1158/1535-7163.MCT-06-0279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Yang X, Chen C, Liu B, Ren B, Wang L, Zhao K, Yu S, Ming H. Expression of the Arp2/3 complex in human gliomas and its role in the migration and invasion of glioma cells. Oncol Rep. 2013;30:2127–2136. doi: 10.3892/or.2013.2669. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky LM, Reeves E, Wientjes F, Mattheyse FJ, Grogan A, Totty NF, Burlingame AL, Hsuan JJ, Segal AW. Mammalian actin-related protein 2/3 complex localizes to regions of lamellipodial protrusion and is composed of evolutionarily conserved proteins. Biochem J. 1997;328:105–112. doi: 10.1042/bj3280105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen KL, Beuming T, Niv MY, Chang CW, Dev KK, Weinstein H, Gether U. Molecular determinants for the complex binding specificity of the PDZ domain in PICK1. J Biol Chem. 2005;280:20539–20548. doi: 10.1074/jbc.M500577200. [DOI] [PubMed] [Google Scholar]
- Maestro RD, Shivers R, McDonald W, Maestro AD. Dynamics of C6 astrocytoma invasion into three-dimensional collagen gels. J Neuro-Oncol. 2001;53:87–98. doi: 10.1023/a:1012236830230. [DOI] [PubMed] [Google Scholar]
- Mori S, Chang JT, Andrechek ER, Matsumura N, Baba T, Yao G, Kim JW, Gatza M, Murphy S, Nevins JR. Anchorage-independent cell growth signature identifies tumors with metastatic potential. Oncogene. 2009;28:2796–2805. doi: 10.1038/onc.2009.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins RD, Heuser JA, Pollard TD. The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc Natl Acad Sci USA. 1998;95:6181–6186. doi: 10.1073/pnas.95.11.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murk K, Blanco Suarez EM, Cockbill LM, Banks P, Hanley JG. The antagonistic modulation of Arp2/3 activity by N-WASP, WAVE2 and PICK1 defines dynamic changes in astrocyte morphology. J Cell Sci. 2013;126:3873–3883. doi: 10.1242/jcs.125146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada M, Drake KL, Nakada S, Niska JA, Berens ME. Ephrin-B3 ligand promotes glioma invasion through activation of Rac1. Cancer Res. 2006;66:8492–8500. doi: 10.1158/0008-5472.CAN-05-4211. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Wood CL, Patton AP, Jaafari N, Henley JM, Mellor JR, Hanley JG. PICK1 inhibition of the Arp2/3 complex controls dendritic spine size and synaptic plasticity. EMBO J. 2011;30:719–730. doi: 10.1038/emboj.2010.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurnberg A, Kitzing T, Grosse R. Nucleating actin for invasion. Nat Rev Cancer. 2011;11:177–187. doi: 10.1038/nrc3003. [DOI] [PubMed] [Google Scholar]
- Olson MF, Sahai E. The actin cytoskeleton in cancer cell motility. Clin Exp Metastasis. 2009;26:273–287. doi: 10.1007/s10585-008-9174-2. [DOI] [PubMed] [Google Scholar]
- Pasquale EB. Eph receptors and ephrins in cancer: bidirectional signalling and beyond. Nat Rev Cancer. 2010;10:165–180. doi: 10.1038/nrc2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzes P, Jones KA. Dendritic spine dynamics—a key role for kalirin-7. Trends Neurosci. 2008;31:419–427. doi: 10.1016/j.tins.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puschmann TB, Turnley AM. Eph receptor tyrosine kinases regulate astrocyte cytoskeletal rearrangement and focal adhesion formation. J Neurochem. 2010;113:881–894. doi: 10.1111/j.1471-4159.2010.06655.x. [DOI] [PubMed] [Google Scholar]
- Reyes SB, Narayanan AS, Lee HS, Tchaicha JH, Aldape KD, Lang FF, Tolias KF, McCarty JH. alphavbeta8 integrin interacts with RhoGDI1 to regulate Rac1 and Cdc42 activation and drive glioblastoma cell invasion. Mol Biol Cell. 2013;24:474–482. doi: 10.1091/mbc.E12-07-0521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley AJ. Life at the leading edge. Cell. 2011;145:1012–1022. doi: 10.1016/j.cell.2011.06.010. [DOI] [PubMed] [Google Scholar]
- Rocca DL, Amici M, Antoniou A, Blanco Suarez E, Halemani N, Murk K, McGarvey J, Jaafari N, Mellor JR, Collingridge GL, Hanley JG. The small GTPase Arf1 modulates Arp2/3-mediated actin polymerization via PICK1 to regulate synaptic plasticity. Neuron. 2013;79:293–307. doi: 10.1016/j.neuron.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocca DL, Hanley JG. PICK1 links AMPA receptor stimulation to Cdc42. Neurosci Lett. 2015;585:155–159. doi: 10.1016/j.neulet.2014.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocca DL, Martin S, Jenkins EL, Hanley JG. Inhibition of Arp2/3-mediated actin polymerization by PICK1 regulates neuronal morphology and AMPA receptor endocytosis. Nat Cell Biol. 2008;10:259–271. doi: 10.1038/ncb1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutka JT, Hubbard SL, Fukuyama K, Matsuzawa K, Dirks PB, Becker LE. Effects of antisense glial fibrillary acidic protein complementary DNA on the growth, invasion, and adhesion of human astrocytoma cells. Cancer Res. 1994;54:3267–3272. [PubMed] [Google Scholar]
- Son J, Park MS, Park I, Lee HK, Lee SH, Kang B, Min BH, Ryoo J, Lee S, Bae JS, et al. Pick1 modulates ephrinB1-induced junctional disassembly through an association with ephrinB1. Biochem Biophys Res Commun. 2014;450:659–665. doi: 10.1016/j.bbrc.2014.06.027. [DOI] [PubMed] [Google Scholar]
- Staudinger J, Zhou J, Burgess R, Elledge SJ, Olson EN. PICK1: a perinuclear binding protein and substrate for protein kinase C isolated by the yeast two-hybrid system. J Cell Biol. 1995;128:263–271. doi: 10.1083/jcb.128.3.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm D, Bender S, Jones DT, Lichter P, Grill J, Becher O, Hawkins C, Majewski J, Jones C, Costello JF, et al. Paediatric and adult glioblastoma: multiform (epi)genomic culprits emerge. Nat Rev Cancer. 2014;14:92–107. doi: 10.1038/nrc3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terashima A, Cotton L, Dev KK, Meyer G, Zaman S, Duprat F, Henley JM, Collingridge GL, Isaac JT. Regulation of synaptic strength and AMPA receptor subunit composition by PICK1. J Neurosci. 2004;24:5381–5390. doi: 10.1523/JNEUROSCI.4378-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres R, Firestein BL, Dong H, Staudinger J, Olson EN, Huganir RL, Bredt DS, Gale NW, Yancopoulos GD. PDZ proteins bind, cluster, and synaptically colocalize with Eph receptors and their ephrin ligands. Neuron. 1998;21:1453–1463. doi: 10.1016/s0896-6273(00)80663-7. [DOI] [PubMed] [Google Scholar]
- Welch MD, DePace AH, Verma S, Iwamatsu A, Mitchison TJ. The human Arp2/3 complex is composed of evolutionarily conserved subunits and is localized to cellular regions of dynamic actin filament assembly. J Cell Biol. 1997;138:375–384. doi: 10.1083/jcb.138.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, Zhang X, Staudinger J, Huganir RL. Clustering of AMPA receptors by the synaptic PDZ domain-containing protein PICK1. Neuron. 1999;22:179–187. doi: 10.1016/s0896-6273(00)80689-3. [DOI] [PubMed] [Google Scholar]
- Xu J, Xia J. Structure and function of PICK1. NeuroSignals. 2006;15:190–201. doi: 10.1159/000098482. [DOI] [PubMed] [Google Scholar]
- Yamaguchi H, Condeelis J. Regulation of the actin cytoskeleton in cancer cell migration and invasion. Biochim Biophys Acta. 2007;1773:642–652. doi: 10.1016/j.bbamcr.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Cao W, Zhang F, Zhang L, Niu R, Niu Y, Fu L, Hao X, Cao X. Protein interacting with C alpha kinase 1 (PICK1) is involved in promoting tumor growth and correlates with poor prognosis of human breast cancer. Cancer Sci. 2010;101:1536–1542. doi: 10.1111/j.1349-7006.2010.01566.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.