The transcription factor TCP15 represses anthocyanin biosynthesis in a redox-dependent manner during high-light intensity conditions.
Abstract
TCP proteins integrate a family of transcription factors involved in the regulation of developmental processes and hormone responses. It has been shown that most members of class I, one of the two classes in which the TCP family is divided, contain a conserved Cys that leads to inhibition of DNA binding when oxidized. In this work, we describe that the class-I TCP protein TCP15 inhibits anthocyanin accumulation during exposure of plants to high light intensity by modulating the expression of transcription factors involved in the induction of anthocyanin biosynthesis genes, as suggested by the study of plants that express TCP15 from the 35SCaMV promoter and mutants in TCP15 and the related gene TCP14. In addition, the effect of TCP15 on anthocyanin accumulation is lost after prolonged incubation under high light intensity conditions. We provide evidence that this is due to inactivation of TCP15 by oxidation of Cys-20 of the TCP domain. Thus, redox modulation of TCP15 activity in vivo by high light intensity may serve to adjust anthocyanin accumulation to the duration of exposure to high irradiation conditions.
TCP proteins constitute a family of plant-specific transcription factors involved in the regulation of cell proliferation and growth-related processes. TCP transcription factors contain a conserved DNA binding and dimerization domain, the TCP domain, that bears a resemblance to the bHLH domain found in many eukaryotic transcription factors (Cubas et al., 1999). Arabidopsis contains 24 TCP proteins that can be ascribed to classes I or II according to sequence conservation within the TCP domain (Cubas et al., 1999; Martín-Trillo and Cubas, 2010). Class-I proteins participate in the regulation of cell proliferation, leaf and flower development, stem elongation, and responses to hormones. They modulate jasmonic acid biosynthesis and cytokinin, auxin, and gibberellin responses (Martín-Trillo and Cubas, 2010; Manassero et al., 2013). Mutations in genes encoding two closely related class-I proteins from Arabidopsis, TCP14 (At3g47620) and TCP15 (At1g69690), affect several aspects of plant development, among them seed germination (Resentini et al., 2015), leaf shape (Kieffer et al., 2011), inflorescence stem growth (Davière et al., 2014), and gynoecium development (Lucero et al., 2015). Recently, it was also shown that mutations in these genes affect effector-triggered immunity (Kim et al., 2014), thus suggesting a role of class-I TCP proteins in responses to stress.
As part of the sequence conservation observed within the TCP domain of class-I proteins, we have previously identified a conserved Cys located at position 20 of the TCP domain. We have shown that this Cys undergoes redox interconversions that render the proteins unable to bind DNA when oxidized (Viola et al., 2013). According to this, we have postulated that class-I TCP proteins may participate in responses to internal or external conditions that cause changes in cell redox parameters. Changes in the production or amounts of redox compounds, mainly due to the generation of reactive oxygen species (ROS), are observed under a variety of stress conditions (Noctor and Foyer, 1998; Grant and Loake, 2000; Foyer and Noctor, 2005; Potters et al., 2009; Frederickson Matika and Loake, 2014). However, little is known about the participation of class-I TCP proteins in responses to stress.
A stress condition that causes changes in redox parameters is the growth of plants under high light intensities. Excess light causes changes in the redox state of photosynthetic electron transport chain components and thiol compounds and increases ROS production (Karpinski et al., 1997; Huner et al., 1998; Foyer and Allen, 2003; Li et al., 2009). These changes originate protective responses oriented to prevent cell damage under the new situation. A typical response to high light intensities is the accumulation of anthocyanins, a group of aromatic pigments that have protecting functions against excess light and ROS (Holton and Cornish, 1995; Grotewold, 2006; Albert et al., 2009; Agati et al., 2012; Page et al., 2012). Anthocyanins act as protecting pigments since they absorb light and thus reduce the amount of energy that reaches the photosynthetic apparatus (Hughes et al., 2005; Albert et al., 2009). It has been postulated that anthocyanins also act as antioxidant compounds in plants since they are ROS scavengers (Neill et al., 2002; Nakabayashi et al., 2014). Anthocyanin synthesis is modulated at the transcriptional level through the regulation of anthocyanin biosynthesis genes (i.e. genes that encode enzymes involved in anthocyanin biosynthesis) by transcription factors from the Myb, bHLH, and WD40 families (Winkel, 2006; Guo et al., 2014). These factors form a transcriptional complex, called MBW, which binds to anthocyanin biosynthesis gene promoters and induces gene expression (Quattrochio et al., 2006). Some members of the MBW complex are in turn also regulated by light conditions at the transcriptional and posttranslational levels (Lee et al., 2007; Shin et al., 2007; Dubos et al., 2008; Matsui et al., 2008; Maier et al., 2013; Albert et al., 2014). It was recently shown that the class-II TCP protein TCP3 induces anthocyanin biosynthesis by integrating into the MBW complex (Li and Zachgo, 2013).
In this work, we report that the class-I TCP protein TCP15 from Arabidopsis acts as a repressor of anthocyanin accumulation acting on the expression of anthocyanin biosynthesis genes and upstream transcriptional regulators. TCP15 prevents accumulation of anthocyanin under high light intensity after relatively short irradiation times but not after prolonged irradiation. We provide evidence indicating that loss of TCP15 action after prolonged irradiation is due to inactivation of TCP15 by oxidation of Cys-20 of the TCP domain. Redox interconversions of TCP15 and other class-I proteins would then help to modulate the response of plants to high light intensities depending on the duration of these external conditions.
RESULTS
Expression of a Repressor Form of TCP15 Causes an Increase in Anthocyanin Levels
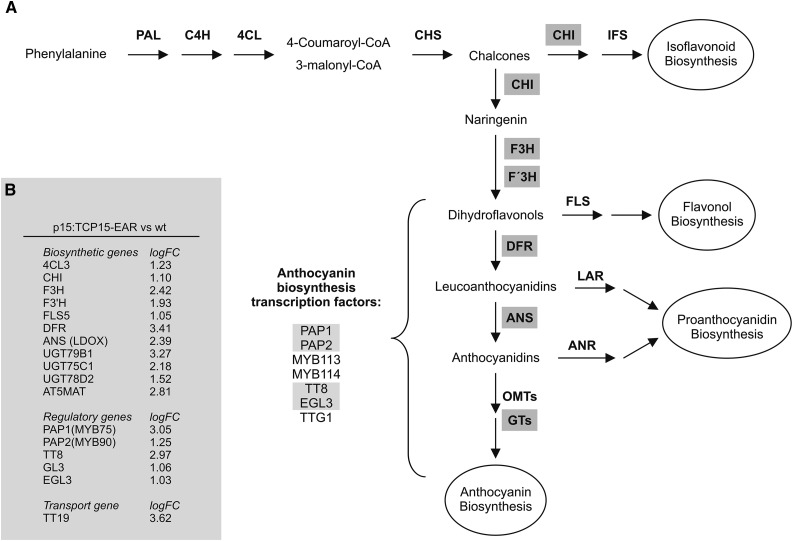
Previously, we described plants that express a fusion of the Arabidopsis TCP transcription factor TCP15 to the EAR repression domain under the control of the TCP15 promoter (p15:TCP15-EAR plants) (Uberti-Manassero et al., 2012). These plants express TCP15-EAR from a 1514-bp fragment containing sequences located upstream of the TCP15 translation start codon. This fragment promotes expression in developing leaves, petioles, and inflorescences (Uberti-Manassero et al., 2012), which is in agreement with expression data from microarray experiments (http://bar.utoronto.ca/). Upon growing these plants under standard conditions, as described in “Materials and Methods”, we observed typical signs of anthocyanin accumulation. Analysis of global gene expression changes in rosettes of these plants revealed the induction of 740 genes. Among them, several genes involved in anthocyanin biosynthesis were observed (Fig. 1; Supplemental Table S1). In fact, 42% (15 out of 36) of the anthocyanin biosynthesis or regulatory genes described by Guo et al. (2014) are induced in p15:TCP15-EAR plants, which is significantly higher than expected by chance (P < 0.001). Four of these genes encode transcription factors involved in the positive regulation of the expression of anthocyanin biosynthesis genes (PAP1, PAP2, TT8, and EGL3) and 11 encode enzymes involved in different steps of anthocyanin biosynthesis from chalcones (Fig. 1). In addition, TT19, a gene encoding a transporter involved in the accumulation of anthocyanins in the vacuole (Kitamura et al., 2004; Sun et al., 2012), and AT5MAT, which encodes an anthocyanin 5-O-glucoside-6″-O-malonyltransferase involved in the malonylation of anthocyanin (D’Auria et al., 2007), are also induced in p15:TCP15-EAR plants (Fig. 1). Notably, genes encoding enzymes involved in the synthesis of isoflavonoids, flavonols, and proanthocyanidins, which share metabolic precursors with anthocyanins, did not show changes in expression or were repressed, as in the case of the flavonol biosynthetic genes FLS2 (Owens et al., 2008; Guo et al., 2014), UGT78D1 (Jones et al., 2003), and UGT71D1 (Lim et al., 2004) (Supplemental Table S1).
Figure 1.
Genes involved in anthocyanin production are induced in p15:TCP15-EAR plants. A, Scheme of the anthocyanin biosynthesis pathway in Arabidopsis. The abbreviated names of the genes encoding enzymes involved in anthocyanin biosynthesis are shown, together with the names of transcription factors involved in the induction of anthocyanin biosynthesis genes. Names boxed in gray indicate genes whose expression is increased in p15:TCP15-EAR plants. B, List of genes involved in anthocyanin accumulation whose expression is induced in p15:TCP15-EAR plants grown under standard light conditions. logFC is the log2 of the fold-change in expression in p15:TCP15-EAR plants relative to wild-type (wt) plants. Details of gene names and AGI codes are given in Supplemental Table S1.
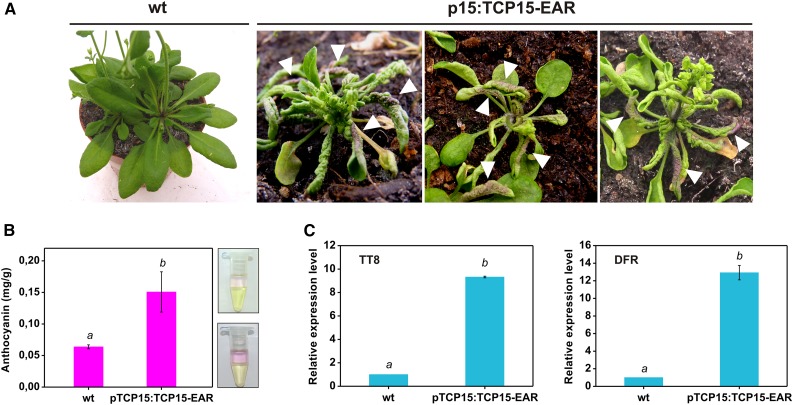
Accumulation of anthocyanins in p15:TCP15-EAR plants was evident after visual inspection (Fig. 2 A, arrowheads). Measurement of anthocyanin levels indicated that, indeed, p15:TCP15-EAR plants accumulate higher anthocyanin levels than wild-type plants (Fig. 2 B). We also measured transcript levels of TT8 and DFR, anthocyanin regulatory and biosynthesis genes, respectively (Fig. 1), by RT-qPCR. The results confirmed the induction of these genes in p15:TCP15-EAR plants (Fig. 2 C) and suggest that TCP15 may be involved in the modulation of anthocyanin levels through the regulation of the expression of anthocyanin biosynthesis and regulatory genes.
Figure 2.
p15:TCP15-EAR plants contain higher anthocyanin levels. A, A photograph of wild-type (wt) and three different lines of p15:TCP15-EAR plants showing accumulation of anthocyanin in leaves (arrowheads). B, Anthocyanin levels in extracts from rosettes of wt and p15:TCP15-EAR plants. Note that the higher anthocyanin content is readily evident by visual inspection of the extracts after chlorophyll extraction, as indicated by the photographs to the right (top: wt; bottom: p15:TCP15-EAR). The bars indicate the mean ± sd of three independent measurements. Columns with different letters are significantly different at P < 0.05 (Student’s t test). C, Relative transcript levels of TT8 and DFR in wt and p15:TCP15-EAR plants measured by RT-qPCR. The value in wild-type plants was set to 1. The bars indicate the mean ± sd of three technical replicates. Columns with different letters are significantly different at P < 0.05 (Student’s t test). The experiment was repeated three times with similar results.
TCP15 Is a Negative Regulator of Anthocyanin Accumulation under High Light Conditions
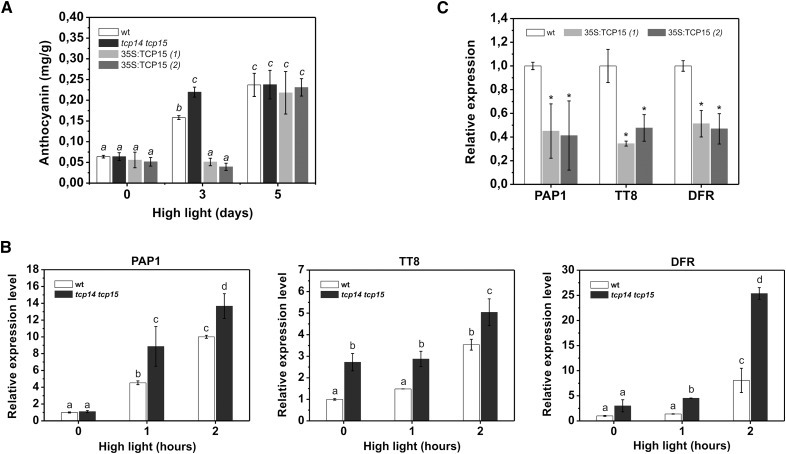
To further evaluate the role of TCP15 in anthocyanin accumulation, we analyzed anthocyanin levels in plants that express TCP15 from the 35SCaMV promoter (35S:TCP15 plants) and in double knock-out mutants in TCP15 and the related TCP protein TCP14. We used the double knock-out mutant with TCP14 for our studies, since it has been shown that there is a high degree of redundancy in the functions of TCP15 and TCP14 (Kieffer et al., 2011; Davière et al., 2014). No differences in anthocyanin levels were observed among mutant, wild-type, and overexpressing plants under standard conditions (Fig. 3 A). We then decided to apply a treatment that induces anthocyanin accumulation to analyze the response of plants in which the function of TCP15 was altered. For this purpose, we treated plants with high light intensity during the light period (see “Materials and Methods” for details) and analyzed anthocyanin levels at different times after the beginning of the treatment. At d 3, wild-type plants showed a 2.5-fold increase in anthocyanin levels (Fig. 3 A). These levels were similar to those observed in p15:TCP15-EAR plants either before or after the high light intensity treatment (Supplemental Figure S1). In two different lines of 35S:TCP15 plants, however, anthocyanin levels were significantly lower than in wild-type plants at d 3 of treatment, when tcp14 tcp15 mutants contained higher anthocyanin levels than wild-type plants (Fig. 3 A). These results suggest that TCP15 is a negative regulator of anthocyanin accumulation. Notably, anthocyanin levels were similar in all plants when analyzed at d 5 (Fig. 3 A).
Figure 3.
TCP15 inhibits anthocyanin accumulation during exposure of plants to high light intensity. A, Anthocyanin levels in wild-type plants (wt), tcp14 tcp15 loss-of-function mutants, and 35S:TCP15 plants (two independent lines) at different times after exposure to high light irradiation. Irradiation was started at the middle of the light period of d 1. Samples were collected at the end of the light period of d 3 and d 5. Control samples (d 0) were collected before the treatment was started. No increase in anthocyanin accumulation was observed in plants kept under normal illumination conditions during the treatment. The bars indicate the mean ± sd of three independent measurements. Columns with different letters are significantly different at P < 0.05 (ANOVA; Tukey test). B, Relative transcript levels of PAP1, TT8, and DFR in wild-type (wt) plants and tcp14 tcp15 mutants after exposure to high light irradiation. The bars indicate the mean ± sd of three biological replicates. Columns with different letters are significantly different at P < 0.05 (ANOVA; Tukey test). C, Relative transcript levels of PAP1, TT8, and DFR in wild-type (wt) and 35S:TCP15 plants (two independent lines) after 3 h of exposure to high light irradiation. The bars indicate the mean ± sd of three biological replicates. Asterisks indicate significant differences compared to wt (P < 0.05; ANOVA).
We then measured the response of genes involved in the regulation of anthocyanin biosynthesis (PAP1, TT8) and the anthocyanin biosynthesis gene DFR to high light treatment. As observed in Figure 3 B, tcp14 tcp15 plants showed higher transcript levels of the three genes after 1 h and 2 h of treatment, indicating that mutant plants respond more rapidly to high light conditions. For TT8 and DFR, transcript levels were already higher in tcp14 tcp15 plants before the treatment was started (Fig. 3 B). The increased response of anthocyanin regulatory and biosynthesis genes observed in tcp14 tcp15 plants may explain why these plants accumulate higher anthocyanin levels than wild-type. In the case of 35S:TCP15 plants, the opposite behavior was observed since transcript levels for PAP1, TT8, and DFR were significantly lower than those of wild-type plants after 3 h of incubation under high light intensity (Fig. 3 C).
Prolonged High Light Treatment Abolishes the Effects of TCP15 Overexpression
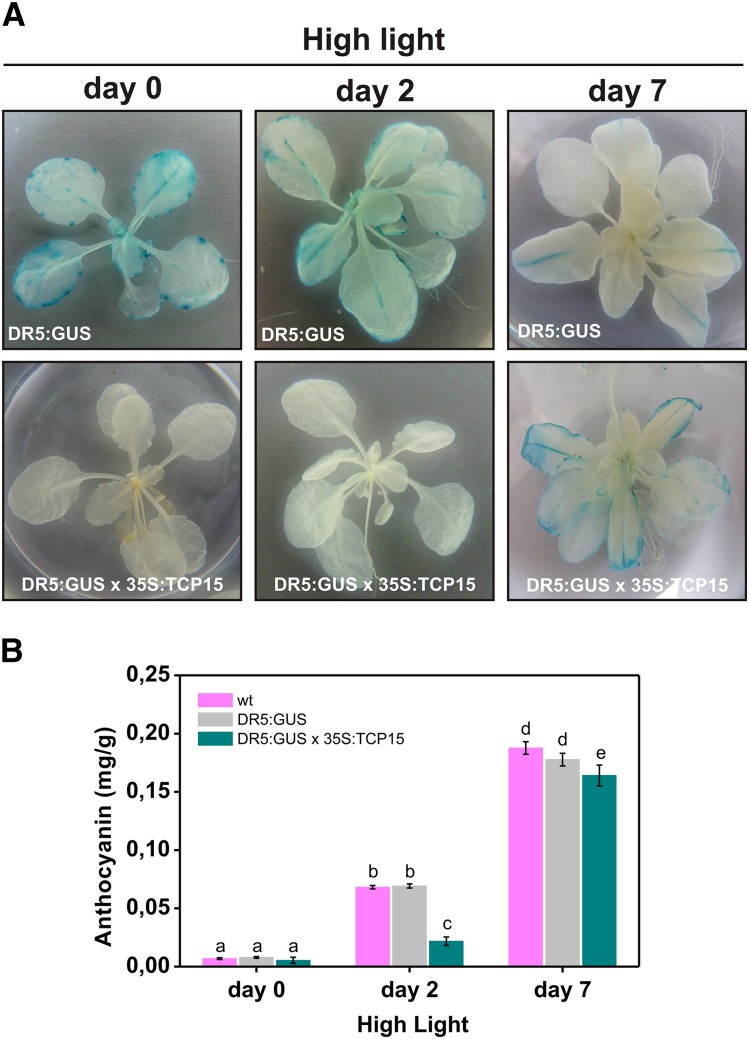
It is noteworthy that the effect of modifying TCP15 function on anthocyanin accumulation under high light was lost after five days of treatment (Fig. 3 A). One possibility is that TCP15 regulates the speed of the response to high light, but eventually anthocyanin levels reach a maximum that is similar regardless of the action of TCP15. Another possibility is that a TCP15-independent pathway becomes prevalent after prolonged high light treatment. Finally, a third possibility is that TCP15 action is inhibited by prolonged exposure to high light. To analyze this, we evaluated the effect of high light irradiation on the action of TCP15 in a supposedly different pathway, i.e. regulation of auxin homeostasis. In fact, it has been reported previously that TCP15 regulates auxin homeostasis and that expression of TCP15-EAR induces the expression of the auxin reporter DR5:GUS (Uberti-Manassero et al., 2012). We then crossed DR5:GUS plants with 35S:TCP15 plants and analyzed the effect of high light treatment on DR5:GUS expression. As expected from the results reported for TCP15-EAR, 35S:TCP15 plants showed significantly lower DR5:GUS expression levels than wild-type under normal illumination conditions (Fig. 4 A, left panels). When wild-type plants were kept under high light intensity, a progressive decay in DR5:GUS expression was evident (Fig. 4 A, top panels). This may be related to the fact that auxin responses are attenuated under stress conditions that generate ROS (Blomster et al., 2011; Tognetti et al., 2012; Peer et al., 2013). In turn, treatment of DR5:GUS x 35S:TCP15 plants caused a completely different response, since a significant increase in DR5:GUS expression was observed after prolonged high light treatment (Fig. 4 A, bottom panels). Repression of anthocyanin accumulation by 35S:TCP15 followed a similar pattern (Fig. 4 B), even if the treatment produced opposite behaviors of both processes in wild-type plants (i.e. increase in anthocyanin accumulation and decrease in DR5:GUS expression). This supports the idea that TCP15 is inactivated after prolonged high light irradiation, thus relieving the repression of DR5:GUS expression and anthocyanin accumulation.
Figure 4.
Repression of DR5:GUS by TCP15 is relieved after prolonged exposure to high light irradiation. A, Plants carrying the auxin reporter DR:GUS in a wild-type (DR5:GUS) or a 35S:TCP15 background (DR5:GUS x 35S:TCP15) were exposed to high light irradiation conditions. GUS activity was analyzed by histochemical staining at the beginning of the treatment (day 0) or at the end of the light period of days 2 and 7. B, Anthocyanin levels in the plants treated as described in (A). Wild-type plants (wt) were also included in the assay for comparison. The bars indicate the mean ± sd of three independent measurements. Columns with different letters are significantly different at P < 0.05 (ANOVA; Tukey test). The experiment was repeated three times with similar results.
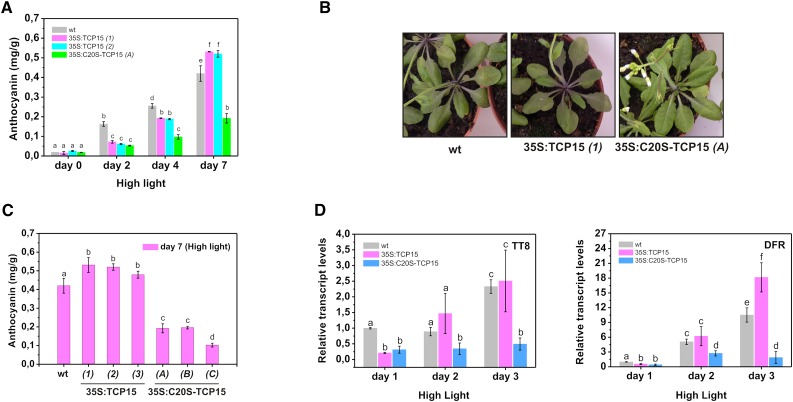
A C20S Mutant of TCP15 Causes Durable Repression of Anthocyanin Accumulation under High Light Intensity
One of the effects of high light intensity is the accumulation of ROS (Miller et al., 2009). ROS damage cellular structures, cause changes in redox conditions, and act as intermediates in signal transduction pathways involved in plant acclimation. We have previously reported that TCP15 and other class-I TCP proteins undergo redox interconversions that involve a conserved Cys (Cys-20 of the TCP domain) located at the beginning of the HLH motif. Oxidation of this Cys inhibits DNA binding and can be achieved in vivo by treatment of plants with H2O2 (Viola et al., 2013). We then asked whether oxidation of Cys-20 of the TCP domain may be the cause of the apparent inactivation of TCP15 after prolonged incubation under high light conditions. To analyze the role of Cys-20 in TCP15 action, we expressed in plants a constitutively active, redox-insensitive variant of TCP15 obtained by changing Cys-20 to Ser (C20S-TCP15). This replacement does not affect the DNA binding activity of TCP15 under reducing conditions (Viola et al., 2013). TCP15 transcript levels in lines that overexpress C20S-TCP15 were similar to those observed in plants that overexpress native TCP15 (Supplemental Figure S2). As shown in Figure 5 A, expression of C20S-TCP15 under the control of the 35SCaMV promoter caused a decrease in anthocyanin accumulation under high light conditions. At d 2 of treatment, the effect of C20S-TCP15 was similar to the one observed with native TCP15 (Fig. 5 A). However, after more prolonged incubation, repression of anthocyanin accumulation was maintained in 35S:C20S-TCP15 plants but was lost in 35S:TCP15 plants (Fig. 5, A and B). Similar results were obtained when different lines expressing either TCP15 or C20S-TCP15 were used (Fig. 5 C), indicating that this is not a line-specific effect. Repression of the expression of TT8 and DFR was also maintained after prolonged incubation under high light in 35S:C20S-TCP15 plants but not in 35S:TCP15 plants (Fig. 5 D), in agreement with the results of anthocyanin accumulation. The most likely explanation for this observation is that TCP15 becomes inactivated by oxidation of Cys-20 after prolonged exposure of plants to high light intensities.
Figure 5.
Expression of a C20S-TCP15 mutant causes durable repression of anthocyanin accumulation under high irradiation conditions. A, Anthocyanin levels in wild-type (wt), 35S:TCP15 (two independent lines), and 35S:C20S-TCP15 plants at different times after exposure to high light irradiation. The bars indicate the mean ± sd of three independent measurements. B, Anthocyanin levels in wt and three independent lines of 35S:TCP15 and 35S:C20S-TCP15 plants at d 7 of exposure to high light irradiation. The bars indicate the mean ± sd of three independent measurements. C, Photograph of wt, 35S:TCP15, and 35S:C20S-TCP15 plants at d 7 of irradiation showing the decrease in anthocyanin accumulation in 35S:C20S-TCP15 plants. D, Relative transcript levels of TT8 and DFR in wild-type (wt), 35S:TCP15, and 35S:C20S-TCP15 plants at different times after exposure to high light irradiation. The bars indicate the mean ± sd of three biological replicates. Columns with different letters are significantly different at P < 0.05 (ANOVA; Tukey test).
TCP15 Is Inactivated after Exposure of Plants to High light Intensity
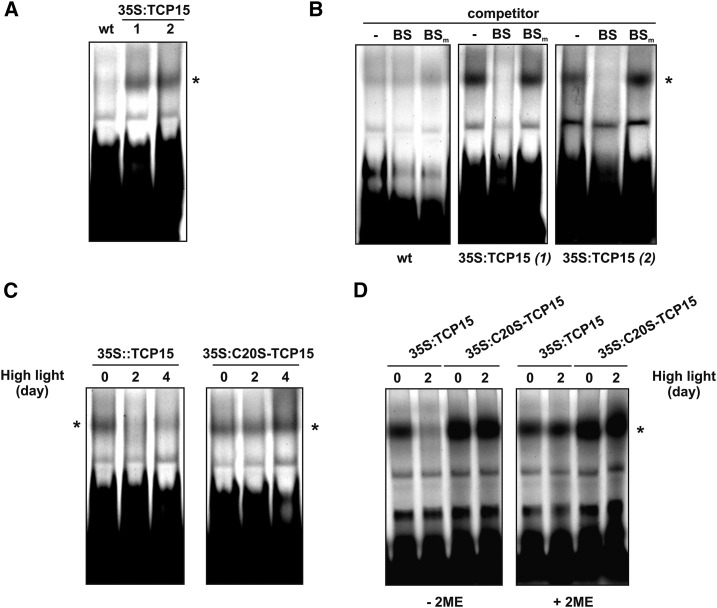
To effectively analyze whether TCP15 is inactivated after exposure of plants to high light, we analyzed the DNA binding activity of protein extracts prepared from 35S:TCP15 plants toward a double-stranded oligonucleotide that contains the sequence recognized by TCP15 using electrophoretic mobility shift assays (EMSAs) (Viola et al., 2011). In extracts from nontransformed plants, two to three faint retarded bands, probably arising from the binding of endogenous proteins, were observed (Fig. 6 A). The pattern is similar to the one observed earlier using seedling extracts and the same oligonucleotide (Viola et al., 2013). A significant increase in the intensity of one of the retarded bands was observed when extracts from 35S:TCP15 plants were used, suggesting that this represents the binding of TCP15 to the labeled oligonucleotide (Fig. 6 A, asterisk). The specificity of binding to the TCP15 target site was analyzed in a competition experiment (Fig. 6 B). Binding of 35S:TCP15 extracts was efficiently competed by a 30-fold molar excess of the same, unlabeled oligonucleotide, but not by a similar amount of an oligonucleotide carrying two point mutations in the TCP15 binding site (Fig. 6 B). This strongly suggests that this retarded band arises from TCP15 binding. In wild-type extracts, no significant competition was observed. This indicates that the faint retarded bands observed in wild-type extracts are most likely the consequence of unspecific protein-DNA interactions.
Figure 6.
High light intensity inhibits the DNA binding activity of TCP15 in a redox-dependent manner. A, EMSA showing the binding of proteins to DNA carrying a class-I TCP target site. Extracts from wild-type (wt) and two independent lines of 35S:TCP15 plants were used. The retarded band highlighted with an asterisk indicates binding of the expressed TCP15 to DNA. B, EMSA as in (A) showing the competition of binding by a 30-fold molar excess of unlabeled oligonucleotides carrying an unmodified (BS) or a mutated (BSm) target site. C, EMSA using extracts prepared from 35S:TCP15 and 35S:C20S-TCP15 plants exposed for different times to high light irradiation. D, EMSA using extracts from 35S:TCP15 and 35S:C20S-TCP15 plants exposed for 0 or 2 d to high light irradiation. Before loading in the EMSA, the samples were incubated for 10 min at 25°C either in the absence (−2ME) or presence (+2ME) of 10 mm 2-mercaptoethanol. The experiment was repeated twice with similar results.
We then performed EMSAs using extracts from plants that were kept under high light conditions for 2 or 4 days. As observed in Figure 6 C (left panel), binding of TCP15 was almost completely abolished under these conditions. However, binding was not affected by a high light treatment when extracts from 35S:C20S-TCP15 plants were used (Fig. 6 C, right panel). This may indicate that TCP15 is inactivated by oxidation after high light treatment of plants. To evaluate this, we treated the extracts with a reducing agent, since this treatment was shown previously to reactivate the oxidized protein (Viola et al., 2013). DTT, as used before in seedling extracts (Viola et al., 2013), was unsuccessful, since incubation in the presence of DTT caused a complete loss of binding in all samples. We speculate that DTT may activate a protease or another factor from the extract that inhibits binding of proteins to DNA. We then used 2-mercaptoethanol (2ME) as reducing agent and observed that the DNA binding activity in extracts from 35S:TCP15 plants subjected to high light intensity conditions was recovered after 2ME treatment (Fig. 6 D, right panel). The results indicate that TCP15 is present in an inactive form in extracts from 35S:TCP15 plants after the high light intensity treatment and that this form can be reactivated by a reducing agent.
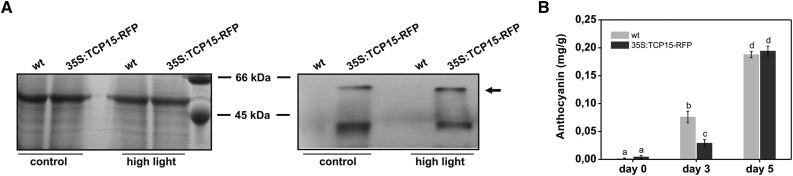
We also analyzed the effect of high light intensity on the stability of TCP15. For this purpose, we obtained plants that constitutively express a fusion of TCP15 to the red fluorescent protein (RFP) and analyzed protein levels using antibodies against RFP. Figure 7 A shows that these antibodies detected more than one band in 35S:TCP15-RFP plants but not in nontransformed plants. This indicates the presence of truncated products, probably arising from proteolytic processing since the RFP tag is located at the C-terminal end. The band of slowest migration (Fig. 7 A, arrow) corresponds to the expected Mr of the full-length protein. TCP15-RFP inhibited anthocyanin accumulation under high light at d 3 of treatment but not at d 5 (Fig. 7 B). This behavior is similar to the one observed with native TCP15. Analysis of protein levels showed that the amount of TCP15-RFP did not change significantly after five days of treatment (Fig. 7 A), indicating that lack of repression is not due to the disappearance of the protein.
Figure 7.
High light intensity does not affect the stability of TCP15. A, Western blot of extracts from wild-type plants (wt) and plants that express a fusion of TCP15 to RFP (35S:TCP15-RFP). The plants were either maintained under control or high light irradiation conditions for five days. Blots were revealed with an antibody against RFP. The left and right panels show images of a Coomassie Brilliant Blue stained gel and a Western blot, respectively, obtained using the same amounts of extracts. The rightmost lane in the left panel shows molecular-weight standards. The arrow in the right panel indicates the expected migration of full-length 35S:TCP15-RFP. B, Anthocyanin levels in wild-type (wt) and 35S:TCP15-RFP plants at different times after exposure to high light irradiation. The bars indicate the mean ± sd of three independent measurements. Columns with different letters are significantly different at P < 0.05 (ANOVA; Tukey test).
DISCUSSION
TCP15 Is a Repressor of Anthocyanin Accumulation
Plants accumulate anthocyanins in response to several stresses and metabolic conditions (Kubasek et al., 1992; Batschauer et al., 1996; Winkel-Shirley, 2002; Lepiniec et al., 2006). One of the functions of anthocyanins is to act as protective pigments to prevent cell damage under conditions of high light intensity; they may also act as antioxidants under situations that cause an increase in ROS accumulation (Grotewold, 2006; Albert et al., 2009; Agati et al., 2012; Page et al., 2012). Expression of a repressor form of TCP15 (TCP15-EAR) in the TCP15 expression domain (i.e. under the TCP15 promoter) causes an increase in anthocyanin levels and in the expression of several anthocyanin biosynthesis genes, suggesting that TCP15 modulates anthocyanin accumulation. This is further supported by the fact that a tcp14 tcp15 mutant accumulates higher anthocyanin levels after exposure to high light intensity. This and the results obtained with plants that express TCP15 from the 35SCaMV promoter indicate that TCP15 is a repressor of anthocyanin accumulation. The fact that anthocyanin levels were similar to wild-type in tcp14 tcp15 mutants under normal illumination suggests that the role of TCP15 and the related protein TCP14 is especially important under conditions that promote anthocyanin accumulation, like the high light intensity treatment used here. Since p15:TCP15-EAR plants have higher anthocyanin levels already under normal conditions, this may indicate the existence of redundancy, probably with other class-I TCP proteins, in the repression of anthocyanin biosynthesis.
Anthocyanin biosynthesis is modulated at the transcriptional level by several transcription factors from the Myb, bHLH, and WD40 families (Quattrochio et al., 2006). These factors form the so-called MBW complex that induces the expression of genes encoding different enzymes of the anthocyanin biosynthesis pathway. The genes encoding several of these transcription factors are also induced under conditions that promote anthocyanin biosynthesis, suggesting the existence of upstream components in the signaling pathway. Since several of these genes, such as PAP1, PAP2, TT8, and EGL3, were induced in p15:TCP15-EAR plants, it can be postulated that TCP15 is one of these upstream components. In addition, the fact that a repressor form of TCP15 induces the expression of these genes further suggests that the regulation exerted by TCP15 is indirect and probably acts by inducing the expression of a repressor of PAP1 and other anthocyanin regulatory genes. Known repressors of anthocyanin biosynthesis include AtMYBL2 (Dubos et al., 2008; Matsui et al., 2008), CPC (Zhu et al., 2009), SPL9 (Gou et al., 2011), AtMYB4 (Jin et al., 2000; Fornalé et al., 2014), and AtMYB7 (Fornalé et al., 2014). However, the genes encoding these transcription factors did not show changes in expression in p15:TCP15-EAR plants according to the microarray experiment.
It was previously reported that TCP3, a class-II TCP protein, also regulates anthocyanin biosynthesis. In this case, TCP3 acts as an inducer of anthocyanin biosynthesis genes by incorporating into the MBW complex (Li and Zachgo, 2013). This fits into an earlier proposition that class-I and class-II TCP proteins have antagonistic functions (Li et al., 2005). Opposing functions of class-I and class-II TCP proteins have been shown for the regulation of senescence and leaf development associated with jasmonic acid biosynthesis and also suggested for the regulation of cell proliferation (Schommer et al., 2008; Sarvepalli and Nath, 2011; Danisman et al., 2012). The initial proposition of antagonistic functions was made based on the similarities of class-I and class-II TCP recognition sequences and assumed that they were the result of interactions with the same target sites in gene promoters (Kosugi and Ohashi, 2002). Although it is highly probable that this occurs, examples of interactions of class-I and class-II TCPs with the same target sites are scarce. In the case of jasmonic acid biosynthesis, class-I and class-II TCPs do interact with the same target gene (LOX2) but through different promoter elements (Danisman et al., 2012). In addition, it is not completely clear that these interactions always lead to antagonistic effects. In the case of anthocyanin biosynthesis, it has been described that TCP3 interacts with components of the MBW complex and strengthens the activation capacity of Myb proteins bound to this complex (Li and Zachgo, 2013). It would be interesting to analyze whether TCP15 is capable of interacting with members of the MBW complex producing the opposite effect, thus inhibiting anthocyanin biosynthesis. Even if this occurs, our results suggest that the mechanism involved in TCP15 action is different. (1) An effect on transcription through incorporation of TCP15 to the MBW complex would lead to repression by TCP15-EAR and activation by TCP15, which is opposite to a role of TCP15 in inhibiting anthocyanin accumulation, as observed here. (2) A role of TCP15 in interfering or destabilizing the MBW complex or the interaction of the complex with TCP3 through protein-protein interactions would lead to a decrease in anthocyanin levels independently of the TCP15 form, either native TCP15 or TCP15-EAR, that is used. The fact that opposing results are obtained when native or repressor forms are expressed strongly suggests that TCP15 acts by directly modulating the expression of one or more factors that negatively regulate anthocyanin accumulation. However, the primary TCP15 target genes that relate TCP15 action to anthocyanin biosynthesis remain unknown.
TCP15 Is Inactivated by Oxidation under High Light Conditions
It seemed intriguing that the action of TCP15 on anthocyanin accumulation was lost after prolonged exposure of plants to high light intensity. However, as shown under Results, a persistent inhibition of anthocyanin biosynthesis was observed when a redox-insensitive form of TCP15 was expressed in plants. This form, C20S-TCP15, carries a mutation in a conserved Cys present at the beginning of the HLH motif. It has been reported that oxidation of this Cys inhibits binding of TCP15 to DNA (Viola et al., 2013). Oxidation of Cys-20, located near the DNA binding domain, may disrupt DNA binding due to the formation of intermolecular disulfide bonds between two TCP15 monomers (Viola et al., 2013). This is suggested by the fact that covalent dimers were observed in nonreducing sodium dodecyl sulfate-polyacrylamide gel electrophoresis recombinant proteins treated with oxidizing agents. In addition, Cys-20 from both monomers are probably located at a short distance in dimers according to models of the TCP domain based on its resemblance with the bHLH domain (Viola et al., 2013). Formation of covalent dimers may impose spatial constraints that impair binding of both TCP15 monomers to DNA. It is also possible that oxidation of Cys-20 to sulphenic or sulphinic acid, which is also reversible, may impair DNA binding. Experiments using the two-hybrid system in yeast have shown that the interaction between TCP15 monomers is not disrupted after H2O2 treatment (our unpublished data). Mass spectrometry analysis of TCP15 after inactivation is required to ascertain the nature of the modification that occurs in vivo.
The results presented in this work indicate that oxidation of Cys-20 occurs after prolonged exposure of plants to high light intensity. In agreement with this, binding to DNA was lost in extracts from 35S:TCP15 plants exposed to high light and was recovered after treatment of the extracts with a reducing agent, suggesting that TCP15 is present in the extract in its oxidized, inactive form. Indeed, binding was not affected by the high light intensity treatment when C20S-TCP15 was expressed in plants. It has been shown that excess light causes changes in the pools of redox compounds and increase the production of ROS, such as H2O2 and singlet oxygen (Li et al., 2009). Treatment of leaves with H2O2 was shown previously to inactivate endogenous TCP15 activity (Viola et al., 2013). Progressive inactivation of TCP15 after exposure to high light may then be caused by changes in redox conditions of the cellular environment.
Even if excess light increases ROS production and promotes anthocyanin accumulation, current evidence does not support the existence of a direct role of ROS in the induction of anthocyanin biosynthesis genes. Contrary to that, Vanderauwera et al. (2005) reported that the induction of anthocyanin biosynthesis genes by high light is impaired in peroxisomal catalase-deficient plants, which accumulate high levels of H2O2. These results may imply that ROS produced during exposure to high light intensity repress, rather than induce, the expression of anthocyanin biosynthesis genes, while our model suggests that ROS act to enhance anthocyanin accumulation after prolonged exposure due to inactivation of TCP15 and related proteins. One possibility is that the effect of ROS depends on the location or the species involved. High light intensity causes redox changes mainly in the chloroplast (Li et al., 2009) and this may affect redox balance in other compartments in a different way than H2O2 from peroxisomes. In addition, catalase-deficient plants are constitutively stressed and this may affect a proper response to ROS produced during the high light intensity treatment.
It is noteworthy that inactivation of TCP15 by high light was observed already at d 2, while differences in anthocyanin accumulation persisted until d 3. Gene expression changes, on the other hand, showed correlation with TCP15 activity, since differences with wild-type were lost at d 2. This indicates that, once TCP15 is inactivated, a certain time is required for plants to reach similar anthocyanin levels than wild-type. It should be kept in mind that the treatment applied consists of illumination under high light intensity only during the light period (8 h) and that some recovery may occur during the night. In any case, the results show that TCP15 inactivation precedes anthocyanin accumulation, which is consistent with a role of TCP15 redox changes in modulating anthocyanin levels under high light conditions. The fact that differences in anthocyanin accumulation in tcp14 tcp15 mutants were also evident at d 3 but not at d 5 is also consistent with a role of TCP15 and TCP14 only after relatively short exposure times. After prolonged treatment, inactivation of the proteins in wild-type plants would eliminate the differences with mutant plants.
According to our results, we propose that TCP15 and related class-I TCP proteins act to attenuate anthocyanin accumulation after short times of exposition to high light intensity. If high light intensity conditions persist, inhibition of anthocyanin biosynthesis by TCP15 is progressively relieved by oxidation of the protein. This would prevent excessive anthocyanin accumulation after short periods of high light intensity but allow a proper protective response under persistent high illumination. In addition, since TCP15 and other class-I TCP proteins are developmental regulators, high light-intensity-dependent inhibition of class-I TCP protein action may transduce changes in light intensity into morphological or developmental responses. Finally, the redox-dependent modulation of class-I TCP protein action described here for anthocyanin biosynthesis under high light intensity conditions may also operate to adjust the action of TCP proteins under other conditions that change the cellular redox state.
MATERIALS AND METHODS
Plant Materials
Arabidopsis Columbia-0 (Col-0) was used as the wild-type. Plants expressing TCP15 fused to the EAR repressor domain (Hiratsu et al., 2003) under the control of the TCP15 promoter (pTCP15:TCP15-EAR) were reported previously (Uberti-Manassero et al., 2012). The tcp14-4 tcp15-3 double mutant was kindly provided by Dr. Simona Masiero (Universitá degli Studi di Milano, Italy) and was described previously (Kieffer et al., 2011). The DR5:GUS reporter was obtained from the Arabidopsis Biological Resource Center (Ohio State University).
DNA Constructs and Plant Transformation
To generate 35S:TCP15 and 35S:C20S-TCP15 plants, the TCP15 coding sequence or a mutated version encoding Ser at position 20 of the TCP domain (Viola et al., 2013) were amplified using primers TCP15-F and TCP15-R (Supplemental Table S2). The amplified PCR products were digested with XbaI and XhoI and inserted into a modified binary plasmid pBi121 (which contains an XhoI site at the end of the GUS coding sequence) under the control of the 35SCaMV promoter. To obtain lines expressing TCP15 fused to RFP under the control of the 35SCaMV promoter, the TCP15 coding sequence was cloned into entry vector pENTR-3C (Life Technologies) and then transferred to destination vector pGWB554 (Nakagawa et al., 2007), using the Gateway cloning system (Life Technologies). This vector allows C-terminal fusions of proteins to monomeric red fluorescent protein (mRFP or mCherry). The primers used are listed in Supplemental Table S2. All constructs were checked by DNA sequencing and introduced into Agrobacterium tumefaciens strain LB4404. Arabidopsis plants were transformed by the floral dip procedure (Clough and Bent, 1998). Transformed plants were selected on the basis of kanamycin resistance and genotyping.
35S:TCP15 x DR5:GUS plants were obtained by crossing 35S:TCP15 (line 2) plants with DR5:GUS plants, followed by selfing and selection based on kanamycin resistance. The presence of DR5:GUS in the genome was confirmed by PCR analysis of genomic DNA with primers GUS-F and GUS-R (Supplemental Table S2).
Plant-Growth Conditions
Plants were grown in soil at 22 to 24°C under a long-day photoperiod (16 h of illumination by a mixture of cool-white and GroLux fluorescent lamps) at an intensity of 100 µE m−2 s−1. For high light intensity treatments, 2-week-old Arabidopsis plants were illuminated by light at 800 µE m−2 s−1, obtained by supplementing normal light with a metal-halide lamp, under a short-day (8 h of illumination) photoperiod. The high light intensity treatment was started at the middle of the illumination period of d 1. Unless otherwise stated, samples were collected at the end of the light period of each day. Experiments were performed in two different growth chambers and we observed that levels and rates of anthocyanin accumulation varied between them, probably because of differences in ambient conditions. The effects described here for the different plant lines under study, however, were consistently observed under both growth conditions.
Anthocyanin Measurement
Anthocyanin measurement was performed using a method adapted from Neff and Chory (1998). Seventy-five milligrams of tissue were collected from three plants in three replicates. Tissue was ground into a fine powder in liquid nitrogen, extracted with 1 mL of methanol acidified with 1% HCl, incubated overnight at 4°C, and centrifuged at 12,000 rpm for 5 min. Distilled water (0.25 mL) was added to 0.3 mL of supernatants and anthocyanins were separated from chlorophylls by extraction with 0.5 mL of chloroform. Total anthocyanin content in the aqueous phase was measured at 529 nm in a spectrophotometer and the amount of anthocyanin was calculated as mg/gram of fresh weight using a molar extinction coefficient of anthocyanin of 30,000 l/(mol × cm) and a Mr of 449.2.
Gene-Expression Analysis
Total RNA was extracted from entire rosettes with Trizol reagent (Invitrogen). One microgram of total RNA was used for cDNA synthesis with oligo(dT) primer and MMLV reverse transcriptase (Promega) under standard conditions. PCR was performed with an aliquot of the cDNA synthesis reaction with primers specific for the genes under analysis in 20 µL final volume containing 1 µL SYBR Green, 10 pmol of primers, 3 mm MgCl2, and 0.2 µL platinum Taq DNA polymerase (Invitrogen). The amplification was monitored in real time on an MJ Research Chromo4 apparatus. Based on primer efficiency, fold expression was calculated after normalization to actin (ACT2 and ACT8; Charrier et al., 2002) by a comparative Ct method. Triplicate PCR reactions of single lines were analyzed for each gene. Similar results were obtained with at least two additional lines in each case. The primers used for PCR are listed in Supplemental Table S2. GUS staining was performed as described in Uberti-Mansassero et al. (2012).
Microarray Results
The microarray experiment was performed in triplicate with total RNA isolated from rosettes of 25-day-old wild-type and p15:TCP15-EAR plants grown under standard conditions using Agilent Arabidopsis (V4) 4×44K arrays (two samples in two-color arrays with reciprocal labeling plus one sample in one-color arrays). The data are deposited in the GEO database under Accession nos. GSE57742, GSE57743, and GSE57744. For the analysis of differentially expressed genes, intensities were background-corrected employing the “normexp” method with an offset of 16, except for those probes with log2 background intensity above 10, which were excluded. After normalization using the loess procedure (for two-color arrays) or quantile normalization (for one-color arrays), a MA object (Minus/Average of log2 intensities) was generated. Differentially expressed genes were identified as those with absolute log2 fold change above 1 after applying a FDR-adjusted p-value of 0.05. Genes related with anthocyanin metabolism were extracted manually from the list of differentially expressed genes.
DNA-Binding Assays
For EMSAs, 10 µg of total protein extracts from transgenic plants prepared as described previously in Viola et al., 2013 were incubated with 50 ng of dsDNA generated by hybridization of the complementary oligonucleotides 5′-AATTCAGATCTGTGGGACCGGGAG-3′ (5′-end labeled with FAM) and 5′-GATCCTCCCGGTCCCACAGATCTG-3′ (TCP15 binding site underlined). Binding reactions were performed in 20 mm HEPES (pH 7.5), 50 mm KCl, 2 mm MgCl, 0.5 mm EDTA, 0.5% Triton X-100, 1 mg of poly(dI-dC), and 10% glycerol. After incubation on ice for 20 min, the reactions were supplemented with 2.5% Ficoll and immediately loaded onto a running gel (5% acrylamide, 0.08% bis-acrylamide in 0.5× TBE plus 2.5% glycerol; 1× TBE is 90 mm Tris-borate, pH 8.3, 2 mm EDTA). The gel was run in 0.5× TBE at 20 mA for 2 h and directly scanned for fluorescence using a Typhoon scanner (GE Healthcare Life Sciences). For competitions, complementary oligonucleotides with changes in positions 3 and 8 of the class-I TCP binding site (5′-GTGGGACC-3′), 5′-AATTCAGATCTGTAGGACTGGGAG-3′ and 5′-GATCCTCCCAGTCCTACAGATCTG-3′, were used.
Western Blot Analysis
For Western blot analysis, 45 µg of protein extracts were separated through reducing sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then transferred to Hybond-ECL membranes (GE Healthcare Life Sciences). Membranes were subsequently probed with an antibody against RFP (Living colors DsRed Polyclonal Antibody, Clontech) at a dilution of 1:1,000, and developed with anti-rabbit immunoglobin conjugated with horseradish peroxidase using the SuperSignal West Pico Chemiluminescent Substrate (Pierce).
Accession Numbers
Microarray data from this article can be found in the GEO database under Accession nos. GSE57742, GSE57743, and GSE57744.
Supplemental Data
The following supplemental materials are available.
Supplemental Table S1. Genes involved in anthocyanin biosynthesis showing significantly altered expression in p15:TCP15-EAR plants.
Supplemental Table S2. Oligonucleotides used in this study.
Supplemental Figure S1. Anthocyanin levels in extracts from wild-type and p15:TCP15-EAR plants after a high light intensity treatment.
Supplemental Figure S2. TCP15 transcript levels in 35S:TCP15 and 35S:C20S-TCP15 plants.
Supplementary Material
Acknowledgments
We thank the Arabidopsis Biological Resource Center, Martin Kieffer, and Simona Masiero for seed stocks. We also thank Elina Welchen and Leandro Lucero, from our lab, for generating the 35S:TCP15-RFP line and Agustin Arce for help with the analysis of microarray data.
Glossary
- ROS
reactive oxygen species
- EMSA
electrophoretic mobility shift assay
- 2ME
2-mercaptoethanol
- RFP
red fluorescent protein
Footnotes
This work was supported by grants from the Agencia Nacional de Promoción Científica y Tecnológica, Argentina, the Consejo Nacional de Investigaciones Científicas y Técnicas, Argentina, and the Universidad Nacional del Litoral. D.H.G. and I.L.V. are members of CONICET. A.C. is a CONICET fellow.
This article is available without a subscription.
References
- Agati G, Azzarello E, Pollastri S, Tattini M (2012) Flavonoids as antioxidants in plants: location and functional significance. Plant Sci 196: 67–76 [DOI] [PubMed] [Google Scholar]
- Albert NW, Davies KM, Lewis DH, Zhang H, Montefiori M, Brendolise C, Boase MR, Ngo H, Jameson PE, Schwinn KE (2014) A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. Plant Cell 26: 962–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert NW, Lewis DH, Zhang H, Irving LJ, Jameson PE, Davies KM (2009) Light-induced vegetative anthocyanin pigmentation in Petunia. J Exp Bot 60: 2191–2202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batschauer A, Rocholl M, Kaiser T, Nagatani A, Furuya M, Schäfer E (1996) Blue and UV-A light-regulated CHS expression in Arabidopsis independent of phytochrome A and phytochrome B. Plant J 9: 63–69 [Google Scholar]
- Blomster T, Salojärvi J, Sipari N, Brosché M, Ahlfors R, Keinänen M, Overmyer K, Kangasjärvi J (2011) Apoplastic reactive oxygen species transiently decrease auxin signaling and cause stress-induced morphogenic response in Arabidopsis. Plant Physiol 157: 1866–1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrier B, Champion A, Henry Y, Kreis M (2002) Expression profiling of the whole Arabidopsis shaggy-like kinase multigene family by real-time reverse transcriptase-polymerase chain reaction. Plant Physiol 130: 577–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cubas P, Lauter N, Doebley J, Coen E (1999) The TCP domain: a motif found in proteins regulating plant growth and development. Plant J 18: 215–222 [DOI] [PubMed] [Google Scholar]
- Danisman S, van der Wal F, Dhondt S, Waites R, de Folter S, Bimbo A, van Dijk AD, Muino JM, Cutri L, Dornelas MC, Angenent GC, Immink RG (2012) Arabidopsis class I and class II TCP transcription factors regulate jasmonic acid metabolism and leaf development antagonistically. Plant Physiol 159: 1511–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Auria JC, Reichelt M, Luck K, Svatos A, Gershenzon J (2007) Identification and characterization of the BAHD acyltransferase malonyl CoA: anthocyanidin 5-O-glucoside-6′'-O-malonyltransferase (At5MAT) in Arabidopsis thaliana. FEBS Lett 581: 872–878 [DOI] [PubMed] [Google Scholar]
- Davière JM, Wild M, Regnault T, Baumberger N, Eisler H, Genschik P, Achard P (2014) Class I TCP-DELLA interactions in inflorescence shoot apex determine plant height. Curr Biol 24: 1923–1928 [DOI] [PubMed] [Google Scholar]
- Dubos C, Le Gourrierec J, Baudry A, Huep G, Lanet E, Debeaujon I, Routaboul JM, Alboresi A, Weisshaar B, Lepiniec L (2008) MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J 55: 940–953 [DOI] [PubMed] [Google Scholar]
- Fornalé S, Lopez E, Salazar-Henao JE, Fernández-Nohales P, Rigau J, Caparros-Ruiz D (2014) AtMYB7, a new player in the regulation of UV-sunscreens in Arabidopsis thaliana. Plant Cell Physiol 55: 507–516 [DOI] [PubMed] [Google Scholar]
- Foyer CH, Allen JF (2003) Lessons from redox signaling in plants. Antioxid Redox Signal 5: 3–5 [DOI] [PubMed] [Google Scholar]
- Foyer CH, Noctor G (2005) Redox homeostasis and antioxidant signaling: a metabolic interface between stress perception and physiological responses. Plant Cell 17: 1866–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederickson Matika DE, Loake GJ (2014) Redox regulation in plant immune function. Antioxid Redox Signal 21: 1373–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou JY, Felippes FF, Liu CJ, Weigel D, Wang JW (2011) Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell 23: 1512–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant JJ, Loake GJ (2000) Role of reactive oxygen intermediates and cognate redox signaling in disease resistance. Plant Physiol 124: 21–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotewold E. (2006) The genetics and biochemistry of floral pigments. Annu Rev Plant Biol 57: 761–780 [DOI] [PubMed] [Google Scholar]
- Guo N, Cheng F, Wu J, Liu B, Zheng S, Liang J, Wang X (2014) Anthocyanin biosynthetic genes in Brassica rapa. BMC Genomics 15: 426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsu K, Matsui K, Koyama T, Ohme-Takagi M (2003) Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J 34: 733–739 [DOI] [PubMed] [Google Scholar]
- Holton TA, Cornish EC (1995) Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 7: 1071–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes NM, Neufeld HS, Burkey KO (2005) Functional role of anthocyanins in high-light winter leaves of the evergreen herb Galax urceolata. New Phytol 168: 575–587 [DOI] [PubMed] [Google Scholar]
- Huner NPA, Öquist G, Sarhan F (1998) Energy balance and acclimation to light and cold. Trends Plant Sci 3: 224–230 [Google Scholar]
- Jin H, Cominelli E, Bailey P, Parr A, Mehrtens F, Jones J, Tonelli C, Weisshaar B, Martin C (2000) Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J 19: 6150–6161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P, Messner B, Nakajima J, Schäffner AR, Saito K (2003) UGT73C6 and UGT78D1, glycosyltransferases involved in flavonol glycoside biosynthesis in Arabidopsis thaliana. J Biol Chem 278: 43910–43918 [DOI] [PubMed] [Google Scholar]
- Karpinski S, Escobar C, Karpinska B, Creissen G, Mullineaux PM (1997) Photosynthetic electron transport regulates the expression of cytosolic ascorbate peroxidase genes in Arabidopsis during excess light stress. Plant Cell 9: 627–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer M, Master V, Waites R, Davies B (2011) TCP14 and TCP15 affect internode length and leaf shape in Arabidopsis. Plant J 68: 147–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Son GH, Bhattacharjee S, Kim HJ, Nam JC, Nguyen PD, Hong JC, Gassmann W (2014) The Arabidopsis immune adaptor SRFR1 interacts with TCP transcription factors that redundantly contribute to effector-triggered immunity. Plant J 78: 978–989 [DOI] [PubMed] [Google Scholar]
- Kitamura S, Shikazono N, Tanaka A (2004) TRANSPARENT TESTA 19 is involved in the accumulation of both anthocyanins and proanthocyanidins in Arabidopsis. Plant J 37: 104–114 [DOI] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y (2002) DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J 30: 337–348 [DOI] [PubMed] [Google Scholar]
- Kubasek WL, Shirley BW, McKillop A, Goodman HM, Briggs W, Ausubel FM (1992) Regulation of flavonoid biosynthetic genes in germinating Arabidopsis seedlings. Plant Cell 4: 1229–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, He K, Stolc V, Lee H, Figueroa P, Gao Y, Tongprasit W, Zhao H, Lee I, Deng XW (2007) Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19: 731–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepiniec L, Debeaujon I, Routaboul JM, Baudry A, Pourcel L, Nesi N, Caboche M (2006) Genetics and biochemistry of seed flavonoids. Annu Rev Plant Biol 57: 405–430 [DOI] [PubMed] [Google Scholar]
- Li C, Potuschak T, Colón-Carmona A, Gutiérrez RA, Doerner P (2005) Arabidopsis TCP20 links regulation of growth and cell division control pathways. Proc Natl Acad Sci USA 102: 12978–12983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Zachgo S (2013) TCP3 interacts with R2R3-MYB proteins, promotes flavonoid biosynthesis and negatively regulates the auxin response in Arabidopsis thaliana. Plant J 76: 901–913 [DOI] [PubMed] [Google Scholar]
- Li Z, Wakao S, Fischer BB, Niyogi KK (2009) Sensing and responding to excess light. Annu Rev Plant Biol 60: 239–260 [DOI] [PubMed] [Google Scholar]
- Lim EK, Ashford DA, Hou B, Jackson RG, Bowles DJ (2004) Arabidopsis glycosyltransferases as biocatalysts in fermentation for regioselective synthesis of diverse quercetin glucosides. Biotechnol Bioeng 87: 623–631 [DOI] [PubMed] [Google Scholar]
- Lucero LE, Uberti-Manassero NG, Arce AL, Colombatti F, Alemano SG, Gonzalez DH (2015) TCP15 modulates cytokinin and auxin responses during gynoecium development in Arabidopsis. Plant J 84: 267–282 [DOI] [PubMed] [Google Scholar]
- Maier A, Schrader A, Kokkelink L, Falke C, Welter B, Iniesto E, Rubio V, Uhrig JF, Hülskamp M, Hoecker U (2013) Light and the E3 ubiquitin ligase COP1/SPA control the protein stability of the MYB transcription factors PAP1 and PAP2 involved in anthocyanin accumulation in Arabidopsis. Plant J 74: 638–651 [DOI] [PubMed] [Google Scholar]
- Manassero NG, Viola IL, Welchen E, Gonzalez DH (2013) TCP transcription factors: architectures of plant form. Biomol Concepts 4: 111–127 [DOI] [PubMed] [Google Scholar]
- Martín-Trillo M, Cubas P (2010) TCP genes: a family snapshot ten years later. Trends Plant Sci 15: 31–39 [DOI] [PubMed] [Google Scholar]
- Matsui K, Umemura Y, Ohme-Takagi M (2008) AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J 55: 954–967 [DOI] [PubMed] [Google Scholar]
- Miller G, Schlauch K, Tam R, Cortes D, Torres MA, Shulaev V, Dangl JL, Mittler R (2009) The plant NADPH oxidase RBOHD mediates rapid systemic signaling in response to diverse stimuli. Sci Signal 2: ra45. [DOI] [PubMed] [Google Scholar]
- Nakabayashi R, Yonekura-Sakakibara K, Urano K, Suzuki M, Yamada Y, Nishizawa T, Matsuda F, Kojima M, Sakakibara H, Shinozaki K, Michael AJ, Tohge T, et al. (2014) Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant J 77: 367–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Suzuki T, Murata S, Nakamura S, Hino T, Maeo K, Tabata R, Kawai T, Tanaka K, Niwa Y, Watanabe Y, Nakamura K, et al. (2007) Improved Gateway binary vectors: high-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci Biotechnol Biochem 71: 2095–2100 [DOI] [PubMed] [Google Scholar]
- Neff MM, Chory J (1998) Genetic interactions between phytochrome A, phytochrome B, and cryptochrome 1 during Arabidopsis development. Plant Physiol 118: 27–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neill SO, Gould KS, Kilmartin PA, Mitchell KA, Markham KR (2002) Antioxidant activities of red versus green leaves in Elatostema rugosum. Plant Cell Environ 25: 539–547 [Google Scholar]
- Noctor G, Foyer CH (1998) Ascorbate and glutathione: keeping active oxygen under control. Annu Rev Plant Physiol Plant Mol Biol 49: 249–279 [DOI] [PubMed] [Google Scholar]
- Owens DK, Alerding AB, Crosby KC, Bandara AB, Westwood JH, Winkel BS (2008) Functional analysis of a predicted flavonol synthase gene family in Arabidopsis. Plant Physiol 147: 1046–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page M, Sultana N, Paszkiewicz K, Florance H, Smirnoff N (2012) The influence of ascorbate on anthocyanin accumulation during high light acclimation in Arabidopsis thaliana: further evidence for redox control of anthocyanin synthesis. Plant Cell Environ 35: 388–404 [DOI] [PubMed] [Google Scholar]
- Peer WA, Cheng Y, Murphy AS (2013) Evidence of oxidative attenuation of auxin signalling. J Exp Bot 64: 2629–2639 [DOI] [PubMed] [Google Scholar]
- Potters G, Pasternak TP, Guisez Y, Jansen MA (2009) Different stresses, similar morphogenic responses: integrating a plethora of pathways. Plant Cell Environ 32: 158–169 [DOI] [PubMed] [Google Scholar]
- Quattrochio F, Baudry A, Lepiniec L, Grotewold E (2006) The regulation of flavonoid biosynthesis. In E Grotewold, ed, The Science of Flavonoids. Springer, New York, pp 97–122 [Google Scholar]
- Resentini F, Felipo-Benavent A, Colombo L, Blázquez MA, Alabadí D, Masiero S (2015) TCP14 and TCP15 mediate the promotion of seed germination by gibberellins in Arabidopsis thaliana. Mol Plant 8: 482–485 [DOI] [PubMed] [Google Scholar]
- Sarvepalli K, Nath U (2011) Hyper-activation of the TCP4 transcription factor in Arabidopsis thaliana accelerates multiple aspects of plant maturation. Plant J 67: 595–607 [DOI] [PubMed] [Google Scholar]
- Schommer C, Palatnik JF, Aggarwal P, Chételat A, Cubas P, Farmer EE, Nath U, Weigel D (2008) Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol 6: e230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Park E, Choi G (2007) PIF3 regulates anthocyanin biosynthesis in an HY5-dependent manner with both factors directly binding anthocyanin biosynthetic gene promoters in Arabidopsis. Plant J 49: 981–994 [DOI] [PubMed] [Google Scholar]
- Sun Y, Li H, Huang J-R (2012) Arabidopsis TT19 functions as a carrier to transport anthocyanin from the cytosol to tonoplasts. Mol Plant 5: 387–400 [DOI] [PubMed] [Google Scholar]
- Tognetti VB, Mühlenbock P, Van Breusegem F (2012) Stress homeostasis—the redox and auxin perspective. Plant Cell Environ 35: 321–333 [DOI] [PubMed] [Google Scholar]
- Uberti-Manassero NG, Lucero LE, Viola IL, Vegetti AC, Gonzalez DH (2012) The class I protein AtTCP15 modulates plant development through a pathway that overlaps with the one affected by CIN-like TCP proteins. J Exp Bot 63: 809–823 [DOI] [PubMed] [Google Scholar]
- Vanderauwera S, Zimmermann P, Rombauts S, Vandenabeele S, Langebartels C, Gruissem W, Inzé D, Van Breusegem F (2005) Genome-wide analysis of hydrogen peroxide-regulated gene expression in Arabidopsis reveals a high light-induced transcriptional cluster involved in anthocyanin biosynthesis. Plant Physiol 139: 806–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viola IL, Güttlein LN, Gonzalez DH (2013) Redox modulation of plant developmental regulators from the class I TCP transcription factor family. Plant Physiol 162: 1434–1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viola IL, Uberti Manassero NG, Ripoll R, Gonzalez DH (2011) The Arabidopsis class I TCP transcription factor AtTCP11 is a developmental regulator with distinct DNA-binding properties due to the presence of a threonine residue at position 15 of the TCP domain. Biochem J 435: 143–155 [DOI] [PubMed] [Google Scholar]
- Winkel BSJ. (2006) The biosynthesis of flavonoids. In E Grotewold, ed, The Science of Flavonoids. Springer, New York, pp 71–95 [Google Scholar]
- Winkel-Shirley B. (2002) Biosynthesis of flavonoids and effects of stress. Curr Opin Plant Biol 5: 218–223 [DOI] [PubMed] [Google Scholar]
- Zhu H-F, Fitzsimmons K, Khandelwal A, Kranz RG (2009) CPC, a single-repeat R3 MYB, is a negative regulator of anthocyanin biosynthesis in Arabidopsis. Mol Plant 2: 790–802 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.