Abstract
The exocyst is a hetero-octameric complex proposed to serve as the tethering complex for exocytosis, although it remains poorly understood at the molecular level. Here, we purified endogenous exocyst from Saccharomyces cerevisiae, and show that the purified complexes are stable and consist of all eight subunits with equal stoichiometry. Using a combination of biochemical and auxin-induced degradation experiments in yeast, we mapped the subunit connectivity, identified two stable four-subunit modules within the octamer, and demonstrated that several known exocyst binding partners are not necessary for exocyst assembly and stability. Furthermore, we visualized the structure of the yeast complex using negative stain electron microscopy; our results indicate that exocyst exists predominantly as a stable, octameric complex with an elongated architecture that suggests the subunits are contiguous helical bundles packed together into a bundle of long rods.
INTRODUCTION
Exocytosis is the evolutionarily conserved pathway by which protein and lipid cargos are trafficked from intracellular compartments to the plasma membrane in membrane-bound vesicles. This pathway is essential for cellular growth and division, as well as specialized processes such as cell migration, ciliogenesis, and autophagy1. To maintain the fidelity of the secretory pathway, numerous conserved protein families regulate every step of the process2. Tethering factors, including the multi-subunit tethering complexes (MTCs), serve as the first, long-range, reversible connection between a vesicle and its target membrane3,4. However, in many cases experimental evidence demonstrating tethering by these factors is lacking5. Tethers are proposed to provide specificity for vesicle targeting, but may also play a more active role in regulating SNARE-mediated membrane fusion3,6–8.
The exocyst complex is the MTC for secretory vesicles at the plasma membrane, and contains eight subunits including Sec3, Sec5, Sec6, Sec8, Sec10, Sec15, Exo70, and Exo84, which have orthologs in eukaryotes ranging from yeasts to humans9–14. Yeast exocyst mutants display severe growth and secretion defects and accumulate post-Golgi secretory vesicles in the cytoplasm15,16. Similarly, null mutants in mice and flies lead to embryonic and larval lethality, respectively17,18. While previous studies have revealed requirements for the exocyst in many critical cellular processes involving polarized vesicle trafficking, the structure and mechanisms of tethering by the exocyst remain unresolved1.
Similar to other tethering factors, the exocyst is a peripheral membrane protein complex that interacts with numerous GTPases, SNAREs, phospholipids, and the vesicle transport motor Myosin V1,3,19,20. The exocyst is proposed to interact with vesicles through Sec15 binding to the Rab GTPase Sec4 and Myosin V, as well as Sec6 binding the v-SNARE Snc16,19,21. On the target membrane side, both Sec3 and Exo70 interact with Rho GTPases and PI(4,5)P2 22–26, and Sec6 may interact with an as yet unidentified “anchor” factor at the plasma membrane27. It is through this myriad of connections that the exocyst is predicted to selectively capture secretory vesicles and tether them to the plasma membrane. A current model for exocyst function proposes that a subcomplex of exocyst subunits in S. cerevisiae is carried on vesicles to another subcomplex at the plasma membrane, and that assembly of these together drives vesicle tethering28, although this model has not yet been validated biochemically, nor have the putative subcomplexes been identified. Whether regulated assembly of the exocyst is required for tethering and SNARE complex regulation in yeast or other organisms, and if these mechanisms differ between different species, are important unanswered questions.
Mechanistic models for exocyst function must be informed by the structural arrangement of its subunits. Crystal structures of several exocyst subunits reveal a strikingly similar motif of contiguous helical bundles that pack together into long rods, classifying it in the evolutionarily conserved Complexes Associated with Tethering Containing Helical Rods (CATCHR) family3,20. Numerous pairwise subunit interactions were identified via yeast-2-hybrid assays, immunoprecipitations, and in vitro binding experiments using recombinant and in vitro translated proteins20,29. To examine the architecture and regulation of assembly of the exocyst, we developed a new robust exocyst purification method to reproducibly isolate stable exocyst complexes from S. cerevisiae. Using an auxin-inducible degradation system to deplete single subunits, we mapped the connectivity of the eight subunits and determined that most of the subunits are required for the association of two assembly modules within the exocyst. In contrast, depletion of known binding partners had no effect on the assembly status of the exocyst. Here we present the first structure of a fully assembled CATCHR MTC—we determined the structure of the fully assembled exocyst using negative stain electron microscopy (EM) and 2-dimensional averaging. Furthermore, we demonstrate that exocyst complexes are stoichiometric, with no detectable subcomplexes; therefore, we propose that the yeast exocyst functions predominantly as a fully assembled complex.
RESULTS
Purification of intact yeast exocyst complexes
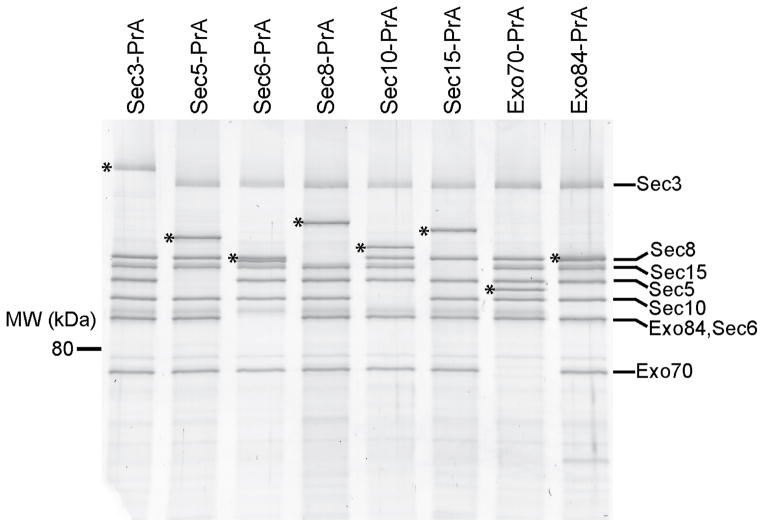
Biochemical and structural studies of the intact exocyst complex were previously limited by preparations with poor yield, stability and purity9,10,30–32 (Munson lab unpublished data). In order to answer critical questions regarding the architecture of the yeast exocyst complex and its putative assembly dynamics, we developed an improved protocol for isolating the entire native complex from yeast extract33,34. In order to maintain endogenous expression levels and function, we fused C-terminal Protein-A (PrA) affinity tags onto each exocyst subunit individually by integrating DNA encoding PrA at each genomic locus, creating eight different tagged haploid Saccharomyces cerevisiae strains (Supplementary Table 1). The C-terminal PrA tags did not confer growth defects (Supplementary Fig. 1a), thus demonstrating that each of the tagged subunits was functional. Yeast strains were grown, harvested in log phase as frozen noodles, and lysed using a planetary ball mill grinder (see Methods). The lysate powder was resuspended in a physiological buffer, bound to rabbit IgG-conjugated magnetic beads, and eluted from the beads either by proteolytic digestion, or by denaturation using SDS loading buffer (Fig. 1). Exocyst subunit identities were confirmed by the molecular weight shift of the PrA tag on SDS-PAGE (Fig. 1), MALDI-MS, and western blot analyses (data not shown).
Figure 1.
Purification of intact yeast exocyst complexes. Purified complexes were separated by SDS-PAGE and visualized by Krypton staining (Thermo Scientific). The asterisk corresponds to the PrA-tagged exocyst subunit used as purification handle (shifts the protein molecular weight by 25 kDa). Both the Sec3 and Exo84 protein bands often migrate as multiple species due to phosphorylation, which appear as slightly smeared bands on SDS-PAGE. The resuspension buffer used was 50 mM Hepes pH 7.4, 300 mM NaCl, plus protease inhibitors. Full-size images for this and most gels in Figures 2–5 are shown in Supplementary Data Set 1.
We isolated intact exocyst complexes from yeast extracts using each of the eight subunits as the PrA-tagged purification handle. The eight exocyst subunits co-purify with equal stoichiometry by both Coomassie-stained SDS-PAGE and densitometry using Krypton fluorescent protein stain (Fig. 1), consistent with earlier reports9,31. We next asked if the complexes purified by this method undergo disassembly and reassembly during the purification. When Sec10-GFP lysate was mixed with either Sec3-PrA or Exo70-PrA lysates, and the exocyst complexes were subsequently purified, no Sec10-GFP was detected in either pull-down, indicating that no exchange or assembly of subunits occurred during the incubation (1h at 4 °C) (Supplementary Fig. 1b), consistent with our previous studies27. Therefore, the purified complexes represent the state of the endogenous complex at the time of cell lysis.
The improved yield and purity of our exocyst preparations are due to reduced proteolysis from cryogenic lysis (Supplementary Fig. 1c) and the use of rabbit IgG-conjugated magnetic beads, which has a tight affinity for PrA35,36. Additionally, protease cleavage allowed for increased purity and native elution of untagged complexes for structural studies (Supplementary Fig. 1d). Substoichiometric levels of co-purifying proteins were detected by mass spectrometry and krypton fluorescent protein staining, but they appear to primarily be highly expressed, non-specific contaminants or previously detected binding partners, including Rtn131.
We next tested the functionality of our exocyst preparations by western blotting for known exocyst interacting partners (Supplementary Fig. 2). The improved yield and rapid, gentle purification procedure allowed detection of binding of Sec1, Myo2, and Snc1/2 (redundant paralogues) to the exocyst. Previous studies revealed an interaction of the exocyst subunit Sec6 with both Sec1 and Snc2 7,21 and Sec15 with Myo2 19. Here, we show that these proteins can be pulled down with tagged exocyst subunits that are not their direct binding partners, suggesting that these interactions occur within the context of the assembled complex.
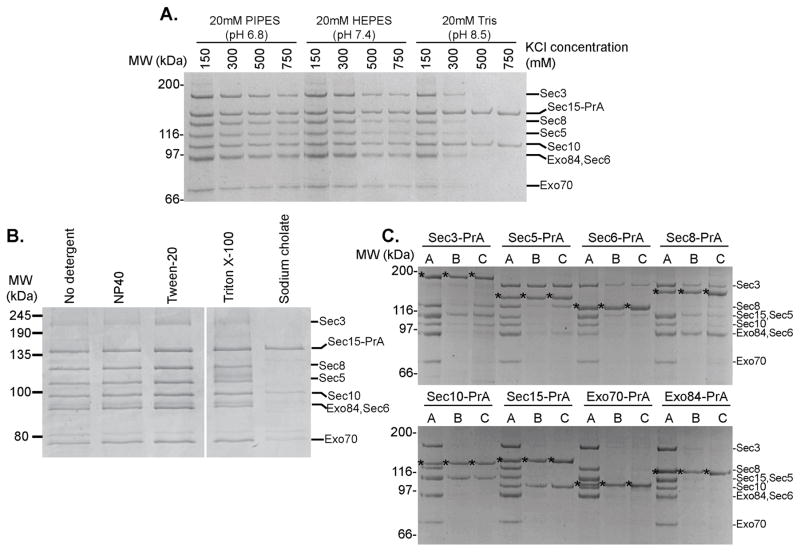
Using Sec15-PrA as the purification handle, we monitored exocyst integrity under a variety of pH and salt conditions (Fig. 2a). The presence of reducing agents had no effect on complex recovery, and the complex was stable across a range of pH solutions, in contrast to previous studies30. Increasing the pH above 8.5 rendered purified exocyst complexes sensitive to salt concentrations ≥300 mM. Using Tris, pH 8.5 and ≥500 mM salt, only Sec15 and Sec10 remained bound together, indicating a strong physical interaction between these two subunits, consistent with earlier studies16.
Figure 2.
Purified exocyst complexes are stable over a wide range of conditions and are comprised of discrete pairwise interactions. (a) Sec15-PrA exocyst complexes were purified using buffers of different pH and KCl concentration as indicated and visualized using Coomassie-stained SDS-PAGE (b) Sec15-PrA exocyst complexes were purified using 50 mM Hepes pH 7.4, 300 mM NaCl buffer and various commonly used detergents at the following concentrations: 0.1% NP-40, 0.1% Tween-20, 1% Triton X-100, 20 mM Sodium cholate. (c) Destabilizing buffer conditions were used with each exocyst subunit as PrA purification handle in order to isolate subcomplexes and stable subunit pairs. A=20 mM PIPES pH 6.8, 300 mM KCl. B=20 mM Tris pH 8.0, 500 mM KCl. C=20 mM Tris pH 8.0, 300 mM KCl, 500 mM Urea. Asterisks correspond to the PrA-tagged subunit.
The exocyst complex peripherally associates with vesicles and the plasma membrane37. We therefore tested the effect of detergents, particularly whether the stoichiometry changes due to the solubilization of membrane-bound subcomplexes or disruption of intersubunit interactions. We tested several non-ionic detergents including NP-40 (Igepal), Tween-20, and Triton X-100, and none affected the overall yield of assembled exocyst or the relative stoichiometry of the subunits (Fig. 2b). In contrast, the exocyst was severely disrupted by sodium cholate, a strong anionic detergent. Taken together, these results indicate that varying the ionic strength of the resuspension buffer has a pronounced effect on exocyst integrity, suggesting that ionic interactions may be a major stabilizing force for intersubunit connections.
We used our pull-down assay to identify stable intracomplex interactions within the endogenous exocyst complex using partially destabilizing buffer conditions with each of the eight PrA-tagged exocyst subunits (Fig. 2c). Several stable subunit pairs emerged: Sec3-Sec5, Sec8-Sec6, and Sec10-Sec15. Neither Exo70 nor Exo84 bound tightly to any of the other subunits under these destabilizing conditions. Although several of these pairwise interactions had been previously identified16,29,38,39, the relative stabilities of the subunit pairs compared to other intersubunit interactions were unknown.
Subunit connections and intra-complex assembly determinants
We applied a more targeted approach to answer additional architectural questions: How are these pairs of subunits assembled into the overall connectivity map of the assembled exocyst? Which of these intersubunit interactions are functionally important for maintaining exocyst integrity? Are some subunits more important for interactions with binding partners on the plasma membrane and vesicle? We decided to selectively eliminate individual exocyst subunits to define their role in maintaining overall complex assembly. All exocyst subunits except Sec3 are encoded by essential genes and, therefore, cannot be deleted from the yeast genome39,40. We tested the temperature-sensitive (ts) mutants sec3-2, sec5-24, sec6-4, sec8-6, and sec10-2 using Sec15-PrA as the purification handle and only sec8-6 had a major effect on exocyst integrity at the restrictive temperature (data not shown). These results were difficult to interpret, however, as the ts alleles vary in severity and amount of destabilization or degradation of the mutant protein. Previous studies using a similar panel of exocyst ts mutants showed greater disassembly for several of the mutants than we observed, even at the permissive temperature10. These differences are likely due to proteolysis of exocyst subunits during spheroplasting lysis, which destabilizes the complex (see Supplementary Fig. 1c). To overcome these challenges, we employed an auxin-inducible degradation (AID) system to specifically remove each individual exocyst subunit.
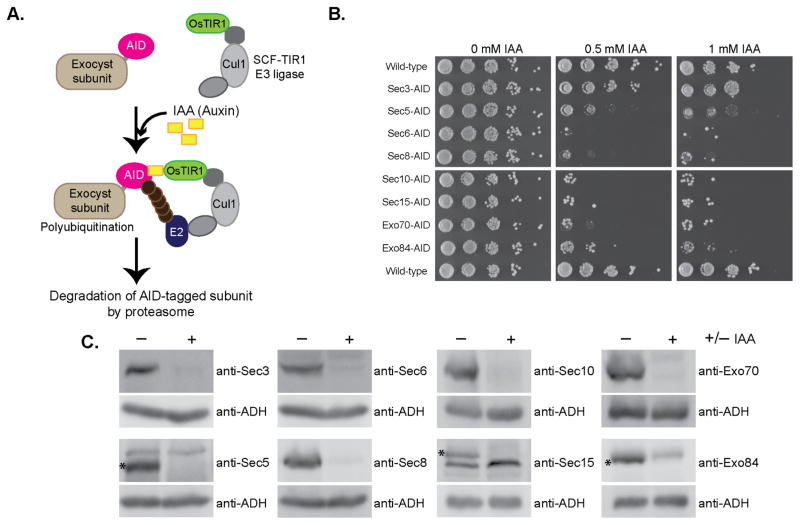
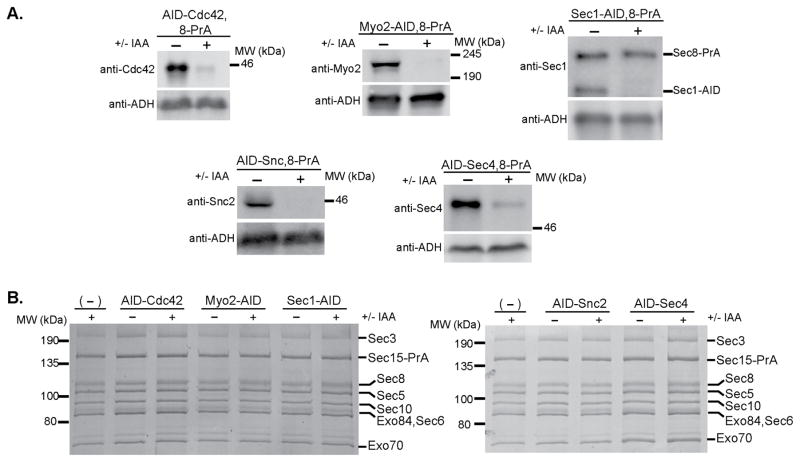
This degron system uses the IAA17 AID sequence from Arabidopsis thaliana, which is fused to each exocyst subunit. When co-expressed with OsTIR1, exposure to the plant hormone auxin leads to rapid proteosomal degradation of the tagged subunit41,42 (Fig. 3a). Addition of these tags to the C-terminal ends of exocyst subunits conferred no growth defects on their own, but when grown on plates containing auxin (Indole 3-acetic acid, IAA), all exocyst-AID strains were inviable except for Sec3-AID (Fig. 3b). We confirmed rapid and specific IAA-induced degradation of individual exocyst subunits in liquid culture by western blot analyses of yeast lysates. Each exocyst subunit was degraded to <12% of the starting level within 60 minutes of IAA treatment (Fig. 3c), whereas the protein levels of the remaining subunits were mostly unchanged (Supplementary Fig. 3).
Figure 3.
Use of the auxin-inducible degron (AID) system to selectively degrade essential exocyst proteins from yeast. (a) Schematic of the AID system. The auxin-inducible degron (AID) tag from Arabidopsis thaliana was fused to the C-terminus of exocyst subunits at their genomic locus in yeast strains constitutively expressing OsTIR1 (F-box transport inhibitor response 1) protein. Upon treatment with the natural plant hormone auxin (IAA=Indole 3-acetic acid), the SCF-OsTIR1 E3 Ubiquitin ligase complex is activated, which then recruits E2 Ubiquitin ligases for polyubiquitination of the AID-tagged protein. The AID-tagged protein is then rapidly degraded by the proteasome41,42. (b) AID-tagged exocyst strains were tested for growth by serial dilution growth assay on YPD plates containing the indicated amount of IAA. Suppressor colonies can be seen in some dilutions. (c) Degradation of exocyst subunits in these strains was confirmed by western blotting lysates from NaOH/SDS lysis. (−) denotes untreated strains and (+) treated with IAA. All subunits were degraded to <10–12% of starting protein level. Asterisks indicate the AID-tagged exocyst subunit in blots where antibodies also bind non-exocyst subunits.
To assess the role of each individual subunit in maintaining the assembly of the endogenous exocyst complex, we combined this AID system with our PrA-tag purification approach. Genomic C-terminal PrA tags were added to Sec8, Sec15, or Sec6 in strains already expressing an AID-tagged exocyst subunit and OsTIR1. Two different PrA-tag handles were tested for each AID-tagged subunit in order to determine the fate of each of the exocyst subunits. Most of the dual-tagged exocyst strains grew normally, but were inviable on IAA plates, as expected (Supplementary Fig. 4a). Surprisingly, Sec10-AID, Sec15-PrA showed no growth defect on IAA plates and no loss of Sec10-AID in IAA-containing liquid culture; similarly, Sec15-AID was not degraded in combination with PrA-tagged exocyst subunits (data not shown). We speculate that the lack of degradation in these strains may be due to masking of the AID tag by the 25 kDa PrA tag on a neighboring exocyst subunit.
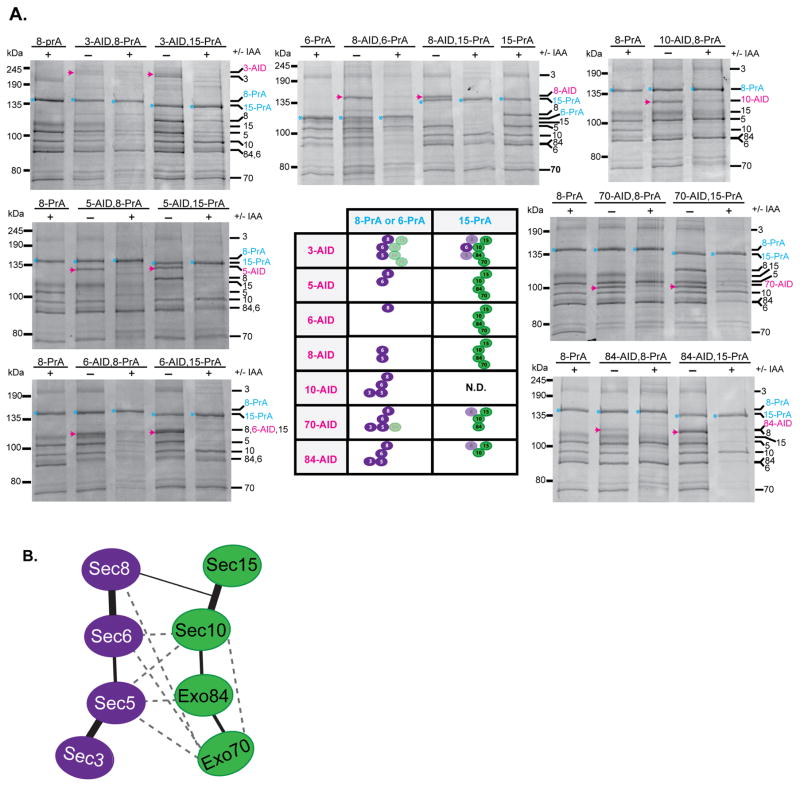
We purified the exocyst complex from both untreated and IAA-treated cultures for each exocyst-AID-PrA combination strain and visualized the complexes by Coomassie staining and western blots (Fig. 4a and Supplementary Figure 4b). Surprisingly, the loss of Sec5, Sec6, Sec8, Sec10, Exo70, or Exo84 resulted in the exocyst complex splitting into two distinct, stable modules: Sec3–Sec5–Sec6–Sec8 (3–5–6–8) and Sec10–Sec15–Exo70–Exo84 (10–15–70–84). The results from the different combinations of AID and PrA tags are summarized in the table in Fig. 4a, showing the division of the exocyst structure into two modules. Loss of Sec3 had the least destabilizing effect on exocyst complex assembly. Degradation of each of the other subunits had distinct effects on its own module, depending on the strength and connectivity of its interactions with its partners, but no effect on the integrity of the opposing module.
Figure 4.
Most exocyst subunits are critical for maintaining the assembly of two 4-subunit modules within the full octameric complex. (a) Exocyst complexes were purified using the indicated PrA purification handle (blue) from yeast strains where one AID-tagged subunit (magenta) is degraded. The resuspension buffer used was 50 mM Hepes pH 7.4, 150 mM NaCl. Purified complexes were run on SDS-PAGE and visualized with Coomassie staining. (−) denotes untreated and (+) treated with IAA. Exocyst subunits are denoted by their number (Sec3, Sec5, Sec6, Sec8, Sec10, Sec15, Exo70, Exo84 as 3,5,6,8,10,15,70,84). Degradation of 6 of the subunits tested led to the complete separation of exocyst into two 4-subunit modules: 3–5–6–8 and 10–15–70–84 with the connections depicted in the central table. Sec10-AID, Sec15-PrA was not determined (N.D.). Faded symbols represent subunits that showed partial loss from the complex. (b) Model depicting the subunit connectivity within and between each exocyst module (green and purple). Thick lines indicate the strong pairwise connections identified in Fig. 2, Fig. 4a, and Supplementary Figure 4b which are required for stability of the assembled exocyst. The thin line depicts a putative connection between Sec8 and Sec10–Sec15 identified in the AID studies, but Sec8’s direct binding partner within this pair is not known. Dashed lines represent interactions identified in previous in vitro studies using Y2H and recombinant proteins (summarized in 20); these are consistent with several additional, weaker pairwise interactions identified here.
We found that the individual assembly of each module is predominantly based on the association of three stable subunit pairs (3–5, 6–8, and 10–15), instead of requiring the cooperative assembly of all four subunits together. If exocyst assembly was cooperative, we would expect to observe complete disassembly of all four subunits from each module upon loss of one subunit; instead, we generally find subcomplexes containing 2–3 subunits (e.g. Sec6 and Sec5 remain bound after Sec8 is degraded). This finding is consistent with our earlier biochemistry results demonstrating that these subunit pairs are stable enough to be co-purified (Fig. 2c). Therefore, the most robust interactions within the complex exist between pairs of subunits and the overall assembly appears to be mediated by a network of weaker interactions. Several additional rules for exocyst assembly can be drawn from these results (Fig. 4b). Sec8 requires Sec6 for assembly into the complex. Sec5 is required for Sec3’s assembly and for the stable interaction of Sec3 with Sec6 and Sec8. In the absence of Sec8, there was also loss of Sec3 from Sec5–Sec6, suggesting either a potential interaction between Sec3 and Sec8 or a potential conformational change that weakens Sec3’s association with Sec5–Sec6. In the case of the other module, Sec10 and Sec15 are a stable pair that require Exo84 for their association with Exo70. Although we were unable to test it, we predict that degradation of Sec15 would not disrupt Sec10’s connection with Exo84 and Exo70, as its only known stable exocyst partner is Sec10 16 (Fig. 2).
These studies only provide a few clues as to the interconnections between the modules. All subunits are required for the assembly of the two modules, including Exo70 and Exo84, which is perhaps surprising in light of our biochemical studies, which demonstrated that they were not tightly associated with any other subunits of the complex (Fig. 2). We propose that the interconnections between the modules are made up of a network of weaker subunit-subunit interactions, although we cannot rule out that the degradation of a subunit from one module may alter the structure of its respective subcomplex, making it incompatible for binding the opposing module. Other previously identified subunit interactions may contribute to this inter-module network but their relative contributions remain to be tested (Fig. 4b)20,29.
Exocyst binding partners have no effect on exocyst assembly
We wondered if any additional binding partners would be necessary to maintain this stable assembly. However, only substoichiometric amounts of known binding partners were detected in our exocyst preparations, suggesting that these partners do not need to remain bound to exocyst to maintain its integrity (Fig. 1 and Supplementary Fig. 2).
A major unresolved question is how the exocyst assembles in vivo and whether additional factors are required for regulating this assembly. Selective elimination of individual exocyst interacting partners along the late secretory pathway might identify subcomplexes, indicating a failure of the complex to fully assemble. To test this idea, we again employed our AID tag approach to deplete the master polarity regulator Cdc42 43, the type V myosin motor Myo2 19, the SNARE regulator Sec1 44, the v-SNARE Snc2 21, and the Rab GTPase Sec4 16 (Supplementary Fig. 5a). The functional consequences of each of these interactions are not known, and it is unclear at which stage in exocytosis these interactions occur7,23,39,45.
The AID-tagged partner strains were treated with IAA for 1 hour, which is sufficient time for numerous rounds of vesicle delivery and fusion in S. cerevisiae46. Degradation of Sec1 induced a severe vesicle accumulation phenotype, as expected47, while degradation of Myo2 and Cdc42 caused a more mild secretion defect consistent with previous reports (Supplementary Fig. 5b,c43,48). N-terminal AID-tagging Snc and Sec4 resulted in severe vesicle accumulation even before IAA treatment, suggesting that these N-terminal tags partially impair protein function (Supplementary Fig. 5b,c). Using a PrA tag on Sec8, we pulled out exocyst complexes after degradation of these partners (Fig. 5). For each of the proteins tested, we observed that the exocyst complexes were fully assembled, stoichiometric, and could be recovered with the same yield. This indicates that none of these components are required for driving or stabilizing the assembly of exocyst complexes. Together with the preceding observations that the exocyst subunits copurify in stoichiometric complexes, these data support a model where the exocyst functions predominantly in a fully assembled state in actively growing cells, even under conditions where vesicles are not being transported and the exocyst is not interacting with its partners.
Figure 5.
Depletion of known exocyst binding partners does not affect the assembly of exocyst complex. (a) Exocyst binding partners Cdc42, Myo2, Sec1, Snc2 (in snc1Δ strain background), and Sec4 were AID-tagged in strains with Sec8-PrA and constitutively expressing OsTIR1. (−) denotes untreated and (+) treated with IAA for 60 minutes. Western blots demonstrate degradation of these proteins from yeast lysate using antibodies specific to the AID-tagged protein of interest. In the Sec1 blot, the Sec1 antibody also reacts with the PrA tag on Sec8-PrA. (b) Exocyst complexes were purified using Sec8-PrA as the purification handle from untreated (−) versus IAA-treated (+) yeast lysates.
Visualization of exocyst structure by electron microscopy
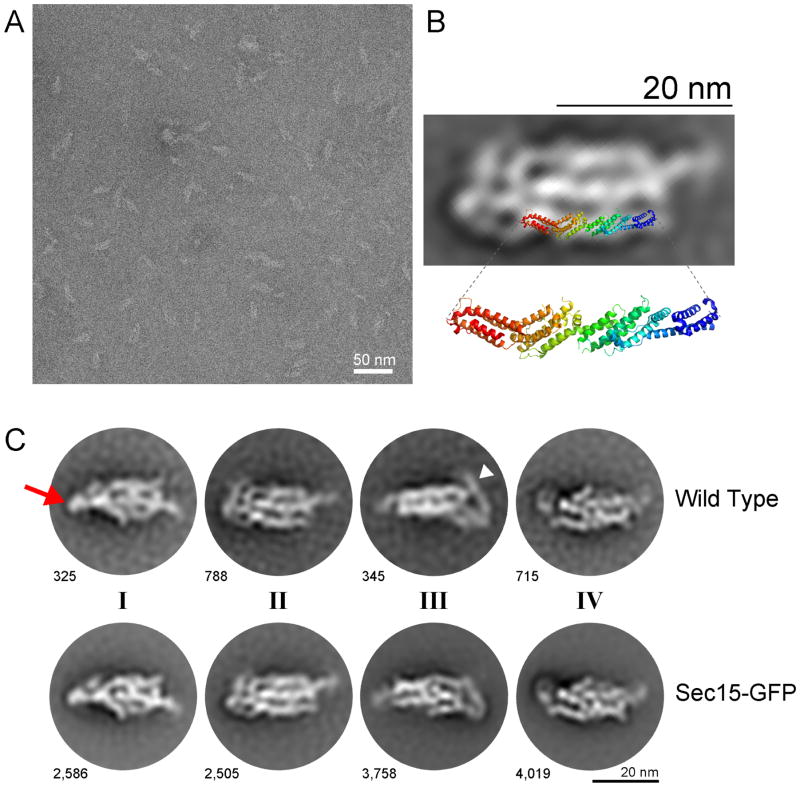
Our new purification method for the yeast exocyst complex allowed us to obtain pure complexes for structural studies. We purified both Sec15-GFP and wild type complexes and analyzed them using negative stain EM. Raw micrographs revealed distinct particles (Fig. 6A) with an ellipsoid structure, approximately 25 nm in length (Fig. 6A). Iterative rounds of unsupervised 2D classification and class averaging revealed multiple coherent views of the exocyst complex resolved between 17 – 25 Å resolution (Fig. 6b,c, Supplementary Fig. 6). However, this averaging failed to reveal a unique density attributable to GFP, precluding identification of Sec15’s location within the structure. At this resolution, the orientations and overall architecture of the exocyst were indistinguishable between these biologically and technically independent datasets (Fig. 6c and Supplementary Fig. 6). No apparent density or class averages were observed for smaller particles, such as subcomplexes.
Figure 6.
Negative stain electron microscopy of purified exocyst complexes. (a) A representative transmission electron micrograph of Sec15-GFP exocyst complexes after negative staining in uranyl acetate. Scale bar is 50 nm. (b) Representative 2D class average (Sec15-GFP) is shown, overlaid with a ribbon diagram of the structure of yeast Exo70 (residues 67–623), PDB ID 2B1E 51. The orientation and position of Exo70 were arbitrarily chosen to illustrate the similarities in the length and width of the “legs” of the complex and Exo70. (c) Highly populated 2D class averages generated by unsupervised classification for both wild type and Sec15-GFP image datasets, the number of particles per class is indicated next to each 2D average. Four apparent “faces” of the complex are labeled as I-IV. The red arrow points to the more “compact” end of the complex in class I, while the white arrowhead points to the more “open” flexible end in class III. Scale bar is 20 nm.
The 2D class averages resolve into roughly four distinct views of the complex (Fig. 6c and Supplementary Fig. 6), which may represent four “faces” of the complex as it interacts with the EM grid. One end of the structure (left side of each of the 2D images; arrow) appears to be more tightly packed and ordered than the other end, which appears to be more flexible, often containing a long looping leg wrapping around the end (right side; arrowhead). Two of the faces of the complex (I and II) appear wider and contain three to four “legs” or columns of density packed together, whereas the two slightly narrower faces (III and IV) appear to have only two to three legs each. We speculate that the more tightly packed end of the long axis of the complex may be comprised of many of the N-terminal ends of exocyst subunits, as they generally have not been amenable to biochemical studies in isolation49. The C-terminal ends, therefore, would be present in the more flexible, “open” end of the structure; these regions contain many of the regions involved in binding GTPases and the plasma membrane23,24,50,51. The exception is Sec3, whose membrane-interaction domain is located at its N-terminal end26,52, and may therefore lie at the flexible open end of the exocyst (arrowhead), in an opposite orientation to the others.
Each of the individual legs observed in the 2D class averages of the exocyst complex are ~3 nm wide. Although the N- and C-terminal ends of the subunits cannot be unambiguously identified at this resolution, we can estimate the length of the legs in the range of ~15–35 nm, with the additional long leg at the flexible end ~25 nm longer than the others. The width and lengths of the legs are consistent with the crystal structure of nearly full-length yeast Exo70 (residues 67–623), which is ~16 nm long and ~3–3.5 nm wide51, as shown in Fig. 6b, in which the crystal structure of Exo70 is superimposed onto an arbitrarily chosen leg. Exo70 is the smallest exocyst subunit (71 kDa), the others range from 84 kDa to 155 kDa. The large size of Sec3 (155 kDa, estimated extended helical bundle length of ~38 nm) also suggests that it may be the subunit that wraps around the end of the complex (Fig. 6c, arrowhead). The other available crystal structures (Exo84CT, Sec6CT and Sec15CT) also revealed similar CATCHR family helical bundles that are ~3 nm wide; the other subunits are predicted to have similar folds49–51,53,54. The subunits of the complex appear to lie in a roughly parallel arrangement to each other, as suggested by previous interaction studies20,51,54. Our interpretation of the 2D averages suggests that this structure represents a fully assembled complex with an estimated volume of ~1800–2200 nm3. Using the volume and molecular weight of the structure of Exo70, and the assumption that all the subunits have roughly similar helical bundle structures, we calculate a comparable volume of ~1900 nm3 for the octameric complex. Therefore, we suggest that our structure contains all eight subunits, consistent with the biochemical and AID experiments. Furthermore, we speculate that the wider faces containing 3–4 legs represent the two distinct modules identified in our AID studies, with one module as the top face, and the other as the bottom face. However, we cannot rule out that the 2D averages could actually be showing the same face in alternative conformations; higher resolution data will therefore be necessary to resolve these models.
DISCUSSION
In this study we used biochemical, genetic, and structural methods to dissect the architecture of the yeast exocyst complex and examined mechanisms for its assembly and function. We purified endogenous, intact exocyst complexes from S. cerevisiae (Fig. 1) and our biochemical and structural characterization demonstrated an intrinsically stable, intact, octameric complex (Fig. 2 and Fig. 6). Our results using the AID system indicated that the presence of most of the exocyst subunits are critical to complex integrity and stability (Fig. 4). Degradation of 6 out of the 7 AID-tagged subunits tested, except Sec3, triggered complete separation of the exocyst into two modules (Fig. 4). Each of these modules (Sec3–5–6–8 and Sec10–15–Exo70–Exo84) is assembled by several critical pairwise interactions (3–5, 6–8, 10–15) with weaker contributions from 5–6, 70–84, 84–10, and 8–10 or 8–15 (Fig. 2 and Fig. 4); furthermore, the disassembly of one module does not affect the integrity of the other. Consistent with this, our negative stain EM 2D class averages demonstrate a stable, homogenous, octameric complex (Fig. 6). The assembly and stability of the exocyst structure is independent of the known binding partners Sec4, Snc1/2, Myo2, Sec1, and Cdc42 (Fig. 5). These components are not stable, stoichiometric partners of the exocyst complex, nor is their binding necessary to assemble or stabilize the exocyst complex during vesicle transport, tethering or fusion. We propose that the role of these interactions is to modulate the function, rather than the assembly, of the exocyst complex.
Our results do not support previous hypotheses that suggested a requirement for polarized vesicle transport in driving the assembly of a subcomplex of exocyst subunits (e.g. Sec15–10–6–8–Exo84) on vesicles with a subgroup (Sec3 and Exo70) serving as a “landmark” on the plasma membrane; assembly of these two subgroups would subsequently drive vesicle tethering28. Under physiological conditions, we do not detect any stable subcomplexes in our pulldowns. It is possible that we detect only stoichiometric complexes because uncomplexed subunits or unstable subcomplexes are degraded during the purification; however, our biochemical and AID experiments argue against this possibility, as we can easily purify individual subunits and subcomplexes from yeast lysate with equal yield to assembled complexes (Fig. 2 and Fig. 4). Furthermore, under conditions where we have disrupted vesicle transport, cell polarity, and exocyst binding to vesicles, no subcomplexes are detectable (Fig. 5). We cannot rule out the presence of either low levels of subcomplexes or free pools of exocyst subunits below our level of detection (<5–10%), however, the majority of the exocyst exists in the fully assembled state. On the other hand, subcomplexes appear to be present in mammalian cells: the components identified thus far (Exo84–Sec10 and Sec5–Sec6 in opposing groups) are consistent with the modules identified here55,56. Similarly, differences in subunit localization patterns in the growing hyphae of Neurospora crassa, Arabidopsis thaliana, and in different Drosophila melanogaster tissues suggest putative subgroups of exocyst subunits14,57,58. Regulated assembly and disassembly of the exocyst in different organisms may be an important mechanism by which the exocyst complex participates in a diverse array of processes in a variety of cell types.
Negative stain EM revealed, for the first time, the ellipsoid-shaped structure of the yeast exocyst complex, with its distinct helical bundle-shaped “legs” packed together (Fig. 6). Overall, the yeast exocyst structure is roughly similar to those of the mammalian exocyst complexes previously imaged using rotary shadowing EM32. However, unlike the individual Y-shape structures observed with glutaraldehyde-fixed mammalian exocyst particles, our yeast 2D averages do not appear to have the same short “arms.” The arms may be too flexible or heterogeneous to be observed in our 2D averages, they may represent mammalian specific domains (e.g. Ral binding domains in Sec5 and Exo84), or perhaps the mammalian exocyst was partially disassembled during processing. Future efforts will require the use of higher resolution data and other strategies to uniquely identify each exocyst subunit within the structure.
Members of the CATCHR family of MTCs, including exocyst, COG, GARP, and Dsl1, share functional similarity, as well as structural similarity at the individual subunit level. Thus, they might be expected to assemble into similar quaternary structures, although they contain different numbers of subunits3. Similar to the exocyst modules identified here, COG consists of two structurally and functionally distinct subassemblies with four subunits each59. However, in terms of their overall shapes, as determined by negative stain EM, the exocyst differs markedly from that of both COG and Dsl1. The COG and Dsl1 structures consist of ~3 nm wide legs emanating from a central flexible “joint”59,60, whereas the exocyst’s legs fold alongside each other to form a compact ellipsoid structure. It is possible that the COG and Dsl1 complexes might adopt more compact structures with all their subunits present, or perhaps they represent a different, biologically relevant conformation that is not captured in the exocyst EM particles. It will be interesting to determine whether there is a conserved distance for vesicle capture by MTCs at the target membrane and whether all MTCs undergo conformational changes to bring vesicles into closer proximity for SNARE assembly and vesicle fusion, as was previously suggested for the Dsl1 complex60.
We propose that, in contrast with models proposing that assembly of subcomplexes is required for exocyst function, the yeast exocyst complex functions as a stable, assembled octamer in the cell. The subunits pack together into an elongated structure. This structure could be a single conformation that functions through changing interactions with various partner proteins. Alternatively, the exocyst may undergo conformational changes in response to binding its protein or membrane partners. Defining the subunit positions and binding of partners at higher resolution is necessary for elucidating the mechanisms of vesicle tethering and SNARE complex regulation at the plasma membrane. This knowledge is also critical in determining whether the MTCs function by similar mechanisms, and how they are uniquely suited to specific trafficking pathways and cell types. Importantly, the ability to purify stable yeast exocyst complexes will now enable functional studies to obtain a detailed molecular understanding of its role in vesicle tethering and SNARE complex regulation.
ONLINE METHODS
Yeast methods
The strains used in this study are listed in Supplementary Table 1. Standard methods were used for yeast media and genetic manipulations. Cells were grown in YPD medium containing 1% Bacto-yeast extract (Fisher Scientific), 2% Bacto-peptone (Fisher Scientific), and 2% glucose (Sigma Aldrich). All protein-A (PrA) tags were integrated at the genomic loci in haploid yeast strains (BY4741 or BY4742) by integration of linear PCR products. PrA products were amplified from a plasmid (pProtAHIS5, Rout lab Rockefeller) encoding a PreScission Protease (PPX) site upstream of the PrA tag and a S. pombe HIS5 selection marker36. Approximately 60 bp of homology to the 3′ end of the coding sequence and 60 bp of homology to the 3′ flanking sequence were used for homologous recombination. All exocyst PrA tags were added at the C-terminal ends. AID tags (IAA17) and linker were amplified from BYP6740 (pMK43, Yeast Genome Resource Center (YGRC), Japan). For C-terminal AID tag strains, tags were added at the genomic locus of the strain BY25598 (YGRC), which expresses OsTIR1 under the ADH1 promoter (parent w303-1a), using linear PCR products and kanMX selection. N-terminal AID tags (SNC2, SEC4, and CDC42 only) were integrated at the genomic locus of BY4742 using the pRS306 integrating plasmid61. Inserts were amplified by overlap extension of PCR products to generate a product consisting of ~300bp of 5′ regulatory element, AID tag, linker, and homology to 5′ end of the gene of interest, and this was then inserted into pRS306 using NotI and XhoI restriction sites. The plasmids were linearized using restriction enzymes specific to the 5′ regulatory elements of each gene (SNC2: MluI, SEC4: BsrGI, CDC42: HpaI) prior to yeast transformation. For the AID-Snc2 strain, SNC1 was deleted by replacing the genomic locus with the kanMX cassette. Finally, for all N-terminal AID tag strains, the OsTIR1 gene was integrated at the MET15 locus using a URA3 marker and ADH1 promoter. The plasmid BYP6744 (pNHK53, YGRC) was used as template for generating the OsTIR1 PCR product and homology to the MET15 regulatory elements was added to the ends. For serial dilution growth assays, yeast were grown in YPD to OD 1.5 and serially diluted 10-fold across YPD plates or YPD plates containing indicated concentrations of Indole-3-acetic acid, IAA (VWR). Yeast plates were incubated at 30°C for 2 days before imaging on Fujifilm LAS3000 (GE).
Exocyst protein-A purification
2 liters of yeast cells were grown in YPD at 30°C to an OD of 1.3–1.5. Cells were washed with water, extruded through a syringe as frozen noodles into liquid nitrogen, and stored at −80°C until ready to be lysed36. Noodles were lysed in a 50 ml stainless steel Komfort jar with stainless steel ball bearings pre-chilled in liquid nitrogen using a PM100 machine (Retsch). The resulting yeast powder was stored at −80°C. 150 mg of yeast powder was added to 1.5 ml microfuge tubes prechilled in liquid nitrogen. 600 μl of resuspension buffer (50 mM Hepes pH 7.4, 150 mM NaCl unless noted otherwise in the text, with 1X cOmplete Mini EDTA-free protease inhibitor solution (Roche Life Science)) was added to the tube (buffer composition dependent upon experiment and noted in the relevant figure) then vortexed and pipetted briefly to resuspend completely. Spheroplasting and bead beating lysis were performed as previously described7 using 50 mM Hepes pH 7.4, 300 mM KCl lysis buffer. The use of NaCl versus KCl had no effect on exocyst preparations. Tubes were spun at 14,000xg for 10 minutes at 4°C and the supernatant is added to 5 μl home-made Rabbit IgG-magnetic bead slurry34,36. Binding was done for 45 minutes at 4°C on nutating platform. The beads were washed in resuspension buffer and eluted in either 1X SDS loading buffer or by 1 h treatment with PreScission Protease (GE Healthcare) at 4°C for a native elution. Samples were run on SDS-PAGE and stained with Coomassie Blue or Krypton fluorescent protein stain (Thermo Fisher Scientific). Western blot analyses were performed using rabbit polyclonal antibodies to Sec6, Sec8, Sec10, Exo70, and Exo84 7,27. Rabbit polyclonal antibodies to Sec3, Sec15, and Sec5 and mouse monoclonal antibodies to Cdc42 and Sec4 were gifts from P. Brennwald (University of North Carolina Chapel Hill). Rabbit polyclonal antibodies to Sec1 and Snc were gifts from C. Carr (Texas A&M University). Goat polyclonal antibody to Myo2 was a gift from L. Weisman (University of Michigan). Rabbit polyclonal antibody to ADH was purchased from Abcam (Catalog number ab20994). Mouse monoclonal antibody to GFP was purchased from Clontech (Catalog number 632380). Western blot analyses of exocyst protein levels in input versus unbound samples showed that ~60% of exocyst complexes are bound to the beads (varies slightly by bead preparation). The IgG beads are saturated in these experiments, however, as the exocyst complexes remaining in the lysates can be pulled down by sequential bead incubations. Krypton staining of the resulting gels showed no differences in stoichiometry in sequential pull-downs of either Sec5-PrA or Sec15-PrA. Coomassie-stained gels were imaged on a LAS 4000 (GE Healthcare Life Sciences) and Krypton gels were imaged on a Typhoon FLA9000 (GE Healthcare Life Sciences). Western blots were treated with ECL and imaged on a LAS 4000. Full size gels and western blots are available in Supplementary Data Set 1.
Auxin-induced degradation of exocyst subunits and exocyst regulators
2L of yeast cells were grown in YPD at 30°C to an OD of 1.0. Indole-3-acetic acid, or IAA, (VWR) dissolved in 100% ethanol at 500 mM was added to yeast cultures for a final concentration of 0.7 mM. The cells were allowed to grow in IAA for 45 min (with 15 minutes for post-processing) at 30°C until reaching an OD of about 1.5. The cells were then washed with water, harvested as frozen noodles, and lysed as described in purification method. NaOH/SDS lysis was used for visualizing IAA-induced degradation in yeast lysates for Fig. 3c and Supplementary Fig. 3. Briefly, 2.5 OD units of yeast were incubated in 100 mM NaOH for 5 minutes, centrifuged to remove the NaOH, resuspended in SDS loading buffer with DTT, and heated at 95°C before loading onto gel for SDS-PAGE and Western blot.
Bgl2 Secretion Assay
AID strains were grown at 30°C in YPD and treated for 1 hour with 0.7 mM IAA before harvesting. Bgl2 secretion assays were performed as described in Adamo et al.62 Internal Bgl2 levels were quantified by western blots and normalized to internal ADH levels. All strains were normalized relative to internal Bgl2 levels of the appropriate untreated, wild-type strain control.
Thin-section Electron Microscopy
EM on wild-type and AID-tagged yeast strains was performed as described63. Briefly, yeast were grown in YPD at 30 °C and treated with 0.7 mM IAA for 1 hr. 10 OD units were harvested, fixed for 1 h at room temperature with 3% gluteraldehyde, 2.5% sucrose, 5 mM CaCl2, 5 mM MgCl in 0.1 M sodium cacodylate, pH 7.4. Cells were spheroplasted using buffer containing 10% β-glucuronidase and 0.5 mg/ml zymolyase for 30 min at 30 °C, washed in 0.1 M cacodylate/1 M sorbitol, resuspended in 0.1 M sodium cacodylate, pH 6.8/1 M sorbitol, and embedded in 2% agarose. Agarose pieces were stained with 1% OsO4, 1% potassium ferrocyanide in 0.1 M sodium cacodylate, pH 6.8 for 30 min, then washed completely and stained in 1% thiocarbohydrazide for 5 min at rt. After washing completely, samples were treated for 5 min with 1% OsO4/1% potassium ferrocyanide and washed again. After ethanol dehydration and embedding in Epon resin (Electron Microscopy Science), thin sections were cut at 70 nm and added to uncoated copper grids. Grids were post-stained with uranyl acetate and lead citrate. Samples were viewed on a Philips CM10 at 80kV and recorded using a Gatan Erlangshen 785 CCD Digital Camera.
Negative Stain Electron Microscopy and Image Analysis
Sec15-PrA and Sec15-GFP, Sec6-PrA complexes were purified in 20mM PIPES at pH 6.8 and 300mM KCl. The complexes were released from IgG beads after PPX cleavage to produce purified wild-type and Sec15-GFP complexes. Those complexes were absorbed to glow discharged carbon-coated copper grids and stained with 1% uranyl acetate. Micrographs of wild-type complex were collected on FEI Tecnai F20 electron microscope operated at 200kV and 20,400x nominal magnification. The defocus value ranged from 0.5 to 2.0 μm. Images were collected with a Gatan K2 summit direct detector with final pixel size 2.45 Angstroms. We semi-automatically picked 67,509 Sec15-GFP particles and 24,891 wild type particles, and gray-scale normalized with Relion-1.3 64. Micrographs of Sec15-GFP complex were collected on FEI Titan Krios electron microscope operated at 300kV and 29,000x nominal magnification. The defocus value ranged from 0.5–3.0 μm. Images were collected automatically using EPU (FEI) with final pixel size 2.87 Angstroms. Particles were selected manually and gray-scaled normalized with BOXER as implemented in EMAN2 65. For the Sec15-GFP dataset, there were: 2,568 unique micrographs; 67,509 particles picked; and 60,751 particles survived. For the untagged wild-type dataset, there were 298 unique micrographs; 24,891 particles picked; and 17,420 particles survived. Contrast Transfer Function (CTF) estimation was performed with CTFFIND3 66. CTF-correction, two-dimensional classification and averaging were performed via Maximum A Posteriori refinement as implemented in RELION64.
Supplementary Material
Acknowledgments
We thank P. Brennwald (University of North Carolina Chapel Hill), C. Carr (Texas A&M University), and L. Weisman (University of Michigan) for antibodies, P. Novick (University of California San Diego) for gifts of yeast strains, the Yeast Genome Resource Center in Japan for the AID system reagents, and the Wendland lab (Johns Hopkins University) for technical advice. Thanks to W. Holmes, M. Jacques, R. Kalia, L. Hassinger and members of the UMass Medical School Core EM Facility for technical assistance. Thanks to R. Gilmore, S. Ryder, P. Pryciak, C. Carr and members of the Munson lab for critical reading of this manuscript and advice. Work in our laboratories is supported by US National Institutes of Health Grants GM068803 (M.M. and A.F.), 1DP2GM110772 (A.F.), U54 GM103511 and P41 GM109824 (M.P.R.), and a Searle Scholars Award (A.F.).
Footnotes
AUTHOR CONTRIBUTIONS
M.R.H. and M.M. conceived the study, designed the biochemical and cell biological experiments, and wrote the manuscript; M.R.H. made yeast strains and performed most of the biochemistry and cell biology experiments with assistance from A.M.M., C.M.D., L.M.M., and A.C.W.; M.G. and A.F. designed, performed, and analyzed the EM experiments; early EM optimization work was done by N.F.; development of the purification method was done by Z.H., C.M.D., M.P.R., and M.C.F.; all authors contributed to discussion and approved the final manuscript.
References
- 1.Heider MR, Munson M. Exorcising the exocyst complex. Traffic. 2012;13:898–907. doi: 10.1111/j.1600-0854.2012.01353.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wickner W, Schekman R. Membrane fusion. Nat Struct Mol Biol. 2008;15:658–64. doi: 10.1038/nsmb.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu IM, Hughson FM. Tethering factors as organizers of intracellular vesicular traffic. Annu Rev Cell Dev Biol. 2010;26:137–56. doi: 10.1146/annurev.cellbio.042308.113327. [DOI] [PubMed] [Google Scholar]
- 4.Chia PZ, Gleeson PA. Membrane tethering. F1000prime reports. 2014;6:74. doi: 10.12703/P6-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brunet S, Sacher M. Are MTCs tethering complexes or trafficking complexes that may act as tethers? Traffic. 2014 doi: 10.1111/tra.12200. [DOI] [PubMed] [Google Scholar]
- 6.Sivaram MV, Saporita JA, Furgason MLM, Boettcher AJ, Munson M. Dimerization of the exocyst protein Sec6p and its interaction with the t-SNARE Sec9p. Biochemistry. 2005;44:6302–11. doi: 10.1021/bi048008z. [DOI] [PubMed] [Google Scholar]
- 7.Morgera F, et al. Regulation of exocytosis by the exocyst subunit Sec6 and the SM protein Sec1. Molecular biology of the cell. 2012;23:337–46. doi: 10.1091/mbc.E11-08-0670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laufman O, Hong W, Lev S. The COG complex interacts with multiple Golgi SNAREs and enhances fusogenic assembly of SNARE complexes. Journal of cell science. 2013;126:1506–16. doi: 10.1242/jcs.122101. [DOI] [PubMed] [Google Scholar]
- 9.TerBush DR, Maurice T, Roth D, Novick P. The Exocyst is a multiprotein complex required for exocytosis in Saccharomyces cerevisiae. EMBO J. 1996;15:6483–94. [PMC free article] [PubMed] [Google Scholar]
- 10.TerBush DR, Novick P. Sec6, Sec8, and Sec15 are components of a multisubunit complex which localizes to small bud tips in Saccharomyces cerevisiae. J Cell Biol. 1995;130:299–312. doi: 10.1083/jcb.130.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo W, Grant A, Novick P. Exo84p is an exocyst protein essential for secretion. J Biol Chem. 1999;274:23558–64. doi: 10.1074/jbc.274.33.23558. [DOI] [PubMed] [Google Scholar]
- 12.Hsu SC, et al. The mammalian brain rsec6/8 complex. Neuron. 1996;17:1209–19. doi: 10.1016/s0896-6273(00)80251-2. [DOI] [PubMed] [Google Scholar]
- 13.Koumandou VL, Dacks JB, Coulson RM, Field MC. Control systems for membrane fusion in the ancestral eukaryote; evolution of tethering complexes and SM proteins. BMC Evol Biol. 2007;7:29. doi: 10.1186/1471-2148-7-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Riquelme M, et al. The Neurospora crassa exocyst complex tethers Spitzenkorper vesicles to the apical plasma membrane during polarized growth. Molecular biology of the cell. 2014;25:1312–26. doi: 10.1091/mbc.E13-06-0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Novick P, Field C, Schekman R. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell. 1980;21:205–15. doi: 10.1016/0092-8674(80)90128-2. [DOI] [PubMed] [Google Scholar]
- 16.Guo W, Roth D, Walch-Solimena C, Novick P. The exocyst is an effector for Sec4p, targeting secretory vesicles to sites of exocytosis. EMBO J. 1999;18:1071–80. doi: 10.1093/emboj/18.4.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedrich GA, Hildebrand JD, Soriano P. The secretory protein Sec8 is required for paraxial mesoderm formation in the mouse. Dev Biol. 1997;192:364–74. doi: 10.1006/dbio.1997.8727. [DOI] [PubMed] [Google Scholar]
- 18.Murthy M, Garza D, Scheller RH, Schwarz TL. Mutations in the exocyst component Sec5 disrupt neuronal membrane traffic, but neurotransmitter release persists. Neuron. 2003;37:433–47. doi: 10.1016/s0896-6273(03)00031-x. [DOI] [PubMed] [Google Scholar]
- 19.Jin Y, et al. Myosin V transports secretory vesicles via a Rab GTPase cascade and interaction with the exocyst complex. Developmental Cell. 2011;21:1156–70. doi: 10.1016/j.devcel.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Munson M, Novick P. The exocyst defrocked, a framework of rods revealed. Nat Struct Mol Biol. 2006;13:577–81. doi: 10.1038/nsmb1097. [DOI] [PubMed] [Google Scholar]
- 21.Shen D, et al. The synaptobrevin homologue Snc2p recruits the exocyst to secretory vesicles by binding to Sec6p. The Journal of cell biology. 2013;202:509–26. doi: 10.1083/jcb.201211148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu H, Rossi G, Brennwald P. The ghost in the machine: small GTPases as spatial regulators of exocytosis. Trends Cell Biol. 2008;18:397–404. doi: 10.1016/j.tcb.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu H, Turner C, Gardner J, Temple B, Brennwald P. The Exo70 subunit of the exocyst is an effector for both Cdc42 and Rho3 function in polarized exocytosis. Mol Biol Cell. 2010;21:430–42. doi: 10.1091/mbc.E09-06-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He B, Xi F, Zhang X, Zhang J, Guo W. Exo70 interacts with phospholipids and mediates the targeting of the exocyst to the plasma membrane. EMBO J. 2007;26:4053–65. doi: 10.1038/sj.emboj.7601834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang X, et al. Membrane association and functional regulation of Sec3 by phospholipids and Cdc42. J Cell Biol. 2008;180:145–58. doi: 10.1083/jcb.200704128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baek K, et al. Structure-function study of the N-terminal domain of exocyst subunit Sec3. J Biol Chem. 2010;285:10424–33. doi: 10.1074/jbc.M109.096966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Songer JA, Munson M. Sec6p anchors the assembled exocyst complex at sites of secretion. Mol Biol Cell. 2009;20:973–82. doi: 10.1091/mbc.E08-09-0968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boyd C, Hughes T, Pypaert M, Novick P. Vesicles carry most exocyst subunits to exocytic sites marked by the remaining two subunits, Sec3p and Exo70p. J Cell Biol. 2004;167:889–901. doi: 10.1083/jcb.200408124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Katoh Y, Nozaki S, Hartanto D, Miyano R, Nakayama K. Architectures of multisubunit complexes revealed by a visible immunoprecipitation assay using fluorescent fusion proteins. Journal of cell science. 2015;128:2351–62. doi: 10.1242/jcs.168740. [DOI] [PubMed] [Google Scholar]
- 30.Terbush DR, Guo W, Dunkelbarger S, Novick P. Purification and characterization of yeast exocyst complex. Methods Enzymol. 2001;329:100–10. doi: 10.1016/s0076-6879(01)29071-5. [DOI] [PubMed] [Google Scholar]
- 31.De Craene JO, et al. Rtn1p is involved in structuring the cortical endoplasmic reticulum. Mol Biol Cell. 2006;17:3009–20. doi: 10.1091/mbc.E06-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsu SC, et al. Subunit composition, protein interactions, and structures of the mammalian brain sec6/8 complex and septin filaments. Neuron. 1998;20:1111–22. doi: 10.1016/s0896-6273(00)80493-6. [DOI] [PubMed] [Google Scholar]
- 33.Oeffinger M, et al. Rrp17p is a eukaryotic exonuclease required for 5′ end processing of Pre-60S ribosomal RNA. Molecular Cell. 2009;36:768–81. doi: 10.1016/j.molcel.2009.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hakhverdyan Z, et al. Rapid, optimized interactomic screening. Nature methods. 2015;12:553–60. doi: 10.1038/nmeth.3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Richman DD, Cleveland PH, Oxman MN, Johnson KM. The binding of staphylococcal protein A by the sera of different animal species. Journal of immunology. 1982;128:2300–5. [PubMed] [Google Scholar]
- 36.Oeffinger M, et al. Comprehensive analysis of diverse ribonucleoprotein complexes. Nature Methods. 2007;4:951–6. doi: 10.1038/nmeth1101. [DOI] [PubMed] [Google Scholar]
- 37.Bowser R, Muller H, Govindan B, Novick P. Sec8p and Sec15p are components of a plasma membrane-associated 19.5S particle that may function downstream of Sec4p to control exocytosis. J Cell Biol. 1992;118:1041–56. doi: 10.1083/jcb.118.5.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roth D, Guo W, Novick P. Dominant negative alleles of SEC10 reveal distinct domains involved in secretion and morphogenesis in yeast. Mol Biol Cell. 1998;9:1725–39. doi: 10.1091/mbc.9.7.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wiederkehr A, De Craene JO, Ferro-Novick S, Novick P. Functional specialization within a vesicle tethering complex: bypass of a subset of exocyst deletion mutants by Sec1p or Sec4p. J Cell Biol. 2004;167:875–87. doi: 10.1083/jcb.200408001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haarer BK, et al. SEC3 mutations are synthetically lethal with profilin mutations and cause defects in diploid-specific bud-site selection. Genetics. 1996;144:495–510. doi: 10.1093/genetics/144.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nature methods. 2009;6:917–22. doi: 10.1038/nmeth.1401. [DOI] [PubMed] [Google Scholar]
- 42.Nishimura K, Kanemaki MT. Rapid Depletion of Budding Yeast Proteins via the Fusion of an Auxin-Inducible Degron (AID) Current protocols in cell biology. 2014;64:20 9 1–20 9 16. doi: 10.1002/0471143030.cb2009s64. [DOI] [PubMed] [Google Scholar]
- 43.Adamo JE, et al. Yeast Cdc42 functions at a late step in exocytosis, specifically during polarized growth of the emerging bud. J Cell Biol. 2001;155:581–92. doi: 10.1083/jcb.200106065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hashizume K, Cheng YS, Hutton JL, Chiu CH, Carr CM. Yeast Sec1p functions before and after vesicle docking. Molecular biology of the cell. 2009;20:4673–85. doi: 10.1091/mbc.E09-02-0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang X, et al. Cdc42 interacts with the exocyst and regulates polarized secretion. J Biol Chem. 2001;276:46745–50. doi: 10.1074/jbc.M107464200. [DOI] [PubMed] [Google Scholar]
- 46.Donovan KW, Bretscher A. Myosin-V is activated by binding secretory cargo and released in coordination with Rab/exocyst function. Developmental cell. 2012;23:769–81. doi: 10.1016/j.devcel.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Novick P, Schekman R. Secretion and cell-surface growth are blocked in a temperature-sensitive mutant of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1979;76:1858–62. doi: 10.1073/pnas.76.4.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Govindan B, Bowser R, Novick P. The role of Myo2, a yeast class V myosin, in vesicular transport. J Cell Biol. 1995;128:1055–68. doi: 10.1083/jcb.128.6.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Croteau NJ, Furgason ML, Devos D, Munson M. Conservation of helical bundle structure between the exocyst subunits. PLoS ONE. 2009;4:e4443. doi: 10.1371/journal.pone.0004443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu S, Mehta SQ, Pichaud F, Bellen HJ, Quiocho FA. Sec15 interacts with Rab11 via a novel domain and affects Rab11 localization in vivo. Nat Struct Mol Biol. 2005;12:879–885. doi: 10.1038/nsmb987. [DOI] [PubMed] [Google Scholar]
- 51.Dong G, Hutagalung AH, Fu C, Novick P, Reinisch KM. The structures of exocyst subunit Exo70p and the Exo84p C-terminal domains reveal a common motif. Nat Struct Mol Biol. 2005;12:1094–1100. doi: 10.1038/nsmb1017. [DOI] [PubMed] [Google Scholar]
- 52.Yamashita M, et al. Structural basis for the Rho- and phosphoinositide-dependent localization of the exocyst subunit Sec3. Nat Struct Mol Biol. 2010;17:180–6. doi: 10.1038/nsmb.1722. [DOI] [PubMed] [Google Scholar]
- 53.Hamburger ZA, Hamburger AE, West AP, Weis WI. Crystal Structure of the S. cerevisiae Exocyst Component Exo70p. J Mol Biol. 2006;356:9–21. doi: 10.1016/j.jmb.2005.09.099. [DOI] [PubMed] [Google Scholar]
- 54.Sivaram MV, Furgason MLM, Brewer DN, Munson M. The structure of the exocyst subunit Sec6p defines a conserved architecture with diverse roles. Nat Struct Mol Biol. 2006;13:555–6. doi: 10.1038/nsmb1096. [DOI] [PubMed] [Google Scholar]
- 55.Moskalenko S, et al. Ral GTPases regulate exocyst assembly through dual subunit interactions. J Biol Chem. 2003;278:51743–8. doi: 10.1074/jbc.M308702200. [DOI] [PubMed] [Google Scholar]
- 56.Bodemann BO, et al. RalB and the exocyst mediate the cellular starvation response by direct activation of autophagosome assembly. Cell. 2011;144:253–67. doi: 10.1016/j.cell.2010.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fendrych M, et al. Visualization of the exocyst complex dynamics at the plasma membrane of Arabidopsis thaliana. Molecular biology of the cell. 2013;24:510–20. doi: 10.1091/mbc.E12-06-0492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Murthy M, et al. Sec6 mutations and the Drosophila exocyst complex. J Cell Sci. 2005;118:1139–50. doi: 10.1242/jcs.01644. [DOI] [PubMed] [Google Scholar]
- 59.Lees JA, Yip CK, Walz T, Hughson FM. Molecular organization of the COG vesicle tethering complex. Nature structural & molecular biology. 2010;17:1292–7. doi: 10.1038/nsmb.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ren Y, et al. A structure-based mechanism for vesicle capture by the multisubunit tethering complex Dsl1. Cell. 2009;139:1119–29. doi: 10.1016/j.cell.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Adamo JE, Rossi G, Brennwald P. The Rho GTPase Rho3 has a direct role in exocytosis that is distinct from its role in actin polarity. Mol Biol Cell. 1999;10:4121–33. doi: 10.1091/mbc.10.12.4121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Perkins EM, McCaffery JM. Mitochondria. Springer; 2007. Conventional and immunoelectron microscopy of mitochondria; pp. 467–483. [DOI] [PubMed] [Google Scholar]
- 64.Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. Journal of structural biology. 2012;180:519–30. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. Journal of structural biology. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 66.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. Journal of structural biology. 2003;142:334–47. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.