Abstract
Acyl carrier proteins play a central role in metabolism by transporting substrates in a wide variety of pathways including the biosynthesis of fatty acids and polyketides. However, despite their importance, there is a paucity of direct structural information concerning the interaction of ACPs with enzymes in these pathways. Here we report the structure of an acyl-ACP substrate bound to the E. coli fatty acid biosynthesis enoyl reductase (FabI), based on a combination of X-ray crystallography and molecular dynamics simulation. The structural data are in agreement with kinetic studies on wild-type and mutant FabIs, and reveal that the complex is primarily stabilized by interactions between acidic residues in the ACP helix α2 and a patch of basic residues adjacent to the FabI substrate-binding loop. Unexpectedly, the acyl-pantetheine thioester carbonyl is not hydrogen-bonded to Y156, a conserved component of the short chain alcohol dehydrogenase/reductase superfamily active site triad. FabI is a proven target for drug discovery, and the structure provides insight into the molecular determinants that regulate the interaction of ACPs with target proteins.
Acyl carrier proteins (ACPs) play an essential role in a diverse array of metabolic pathways including the biosynthesis of fatty acids (1,2), polyketides (3), membrane-derived oligosaccharides (4), lipopolysaccharides (5,6) and phospholipids (7). In each case the growing substrate is attached via a thioester to the ACP phosphopantetheine group. ACPs must therefore be able to recognize and interact, in an acyl group-dependent manner, with a wide variety of enzymes. In eukaryotic type I fatty acid synthesis (FASI) and in polyketide biosynthesis, the ACP occurs as part of a larger polypeptide that is also associated with other catalytic activities. In contrast, in bacterial type II fatty acid biosynthesis (FASII), each of the enzyme activities as well as the ACP are encoded by separate polypeptide chains (2). ACPs that function in FASII-mediated biosynthesis are small, highly soluble, acidic proteins that vary in molecular weight from 7.5 kDa (Escherichia coli) to 13 kDa (Mycobacterium tuberculosis) (1,8–11).
Despite the central role that ACPs play in metabolism, structural details of their interaction with target proteins are sparse. While the structures of ACPs from a variety of different species have been determined by X-ray crystallography (12) and NMR spectroscopy (see for example (13,14)), only one structure has been determined in complex with holo ACP synthase (AcpS) (15) and no structural information is available for the interaction between ACP and enzymes of the fatty acid biosynthesis pathway. AcpS attaches the phosphopantetheine to the ACP serine and thus, while valuable, the complex of AcpS and ACP differs fundamentally from other ACP-protein complexes and does not provide insight into the delivery of substrate by ACP.
The NMR studies reveal that ACPs are highly flexible, a structural feature that is thought to be important for transporting the growing acyl chain and for protein recognition. In addition, X-ray crystallographic analysis of butyryl-ACP resulted in the proposal that ACP adopts two major conformations, optimized for either delivering the acyl chain to the target enzyme or for transporting the growing acyl chain between enzymes (12). However, the flexibility of ACP, coupled with the relatively low affinity of ACP for target proteins has hindered crystallographic efforts to obtain direct structural data on the interaction of ACP with any of the target proteins (16,17).
Using docking and site directed mutagenesis, Rock and coworkers have rigorously analyzed the interaction of ACP with two FASII enzymes. In agreement with the X-ray structure of ACP bound to AcpS (15), these studies confirmed the importance of the ACP recognition helix (helix α2) in FASII protein binding and identified a patch of basic residues on the E. coli β-ketoacyl-ACP synthase (FabH) and β-ketoacyl-ACP reductase (FabG) enzymes responsible for interaction with the ACP helix α2 (16,17).
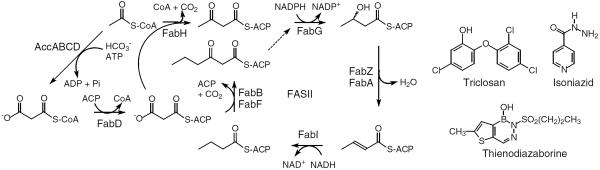
The efforts in our lab are focused on the FASII enoyl reductase enzyme FabI, a target for antibacterial diazaborine compounds (18) and triclosan (19–23), while the anti-TB drug isoniazid inhibits InhA, the FabI homolog from M. tuberculosis (24–26) (Scheme 1). Modeling studies, based on the information provided by the interaction of ACP with FabH and FabG (16,17), suggest that ACP should interact with a cluster of basic residues adjacent to the FabI substrate binding loop. This loop is disordered in binary FabI-cofactor complexes, but becomes ordered in the ternary FabI:NAD+:triclosan complex. The ordered loop provides two entries into the active site, termed the major and minor portals. Sacchettini and coworkers have determined the structure of a C16 substrate bound to InhA and have discussed the role of the major and minor portals in substrate recognition by the FabI enzymes (27). The location of the substrate and N-acetylcysteamine group in InhA suggest that substrates enter the FabI active site through the major portal.
Scheme 1.
The Type 2 Fatty Acid Biosynthesis Pathway and FabI Inhibitors.
In the current study, we have determined the structure of ACP bound to FabI from E. coli. X-ray crystallographic data obtained from a complex between FabI and dodecenoyl-ACP revealed most of the main chain electron density for both FabI and ACP. The observed relative orientation of ACP and FabI leaves the ACP S36 residue too far from the active site to deliver the substrate through the major portal.
Since some aspects of the structure were not resolved by the crystallographic data, we employed computational methods to model the missing details and importantly to ascertain whether the ACP could deliver substrate into the FabI active site in the observed complex. Through molecular dynamics simulations, we generated a model for a productive complex between ACP and FabI. The details of the interaction between FabI and ACP in the resulting model are supported by mutagenesis studies, and provide the first detailed description of ACP recognition by a FASII enzyme. Intriguingly, the structural data indicate that the substrate enters the FabI active site through the minor portal and furthermore suggest that the substrate thioester carbonyl group does not form a hydrogen bond with Y156, a conserved active site residue.
MATERIALS AND METHODS
Preparation of Substrates and Enzymes
Plasmids for wild-type FabI and the Y156F mutant were available from a previous study (23). The Y146F, R204E and K205E FabI mutations were introduced using the QuikChange mutagenesis kit (Stratagene). Wild-type and mutant FabI proteins were overexpressed and purified as described previously (23). Trans-2-dodecenoyl-CoA (DD-CoA) was synthesized from trans-2-dodecenoic acid using the mixed anhydride method (28). Apo-ACP was a gift from Dr. J. Shanklin and Dr. S. Booker. Trans-2-dodecenoyl-ACP (DD-ACP) was a gift from Dr. M. Schaeffer and was also synthesized from DD-CoA and apo-ACP using E. coli holo ACP synthase (AcpS) (29) which was overexpressed and purified from E. coli as described previously (30). Briefly, 0.9 mg of apo-ACP was incubated with 50 μM DD-CoA (1.4-fold excess) and 50 μg of AcpS in 1.4 mL of 50 mM Tris-HCl, 25 mM MgCl2, 1 mM DTT, pH 7.5 buffer for 1 h at 30°C, followed by quenching the reaction by placing it into a dry ice-ethanol bath for 5 min. Subsequently, an equal volume of isopropanol was added and the reaction mixture was incubated for 2 h at 10 °C. AcpS was removed by centrifugation at 6000 rpm for 15 min and the supernatant was applied to a 1-mL Q Sepharose column, equilibrated with 20 mM Bis-Tris, 1 mM DTT, pH 6.5 (Buffer B) containing 50% isopropanol. The column was washed three times with Buffer B containing 50% isopropanol, and then five times with Buffer B alone. DD-ACP was eluted with five column volumes of Buffer B containing 600 mM NaCl. The fractions containing DD-ACP were identified by SDS-PAGE, pooled, concentrated, dialyzed into 20 mM Tris·HCl, pH 7.0 and stored at −80°C. The concentration of protein was determined by a BCA assay.
Kinetic Assays
All kinetic assays were carried out on a Cary 100 Bio (Varian) spectrophotometer at 25°C in 30 mM PIPES, 150 mM NaCl, pH 8.0. Kinetic parameters were determined by following the oxidation of NADH to NAD+ at 340 nm (ε = 6.3 mM−1·cm−1). kcat and kcat/Km for DD-CoA and DD-ACP were determined at a fixed, saturating concentration of NADH (250 μM) and by varying the concentration of DD-CoA (5–85 μM) or DD-ACP (1–200 μM), respectively. Kinetic parameters were obtained by fitting the initial velocity data to the Michaelis-Menten equation using Grafit 3.09b (Erithacus Software Ltd.).
Crystallization, Data Collection and Refinement
The FabI:NAD+:DD-ACP complex was crystallized by the hanging drop vapor diffusion method. Crystals were grown at 22°C from a protein solution containing FabI, NAD+ and DD-ACP in a 1:10:1 ratio mixed with a precipitant solution containing 18–22% PEG 4000 and 100 mM HEPES, pH 7.0. Crystals were flash-frozen in liquid nitrogen with 30% (w/v) glycerol as cryoprotectant and data were collected at beam line X26C at the National Synchrotron Light Source at Brookhaven National Laboratory. Data were indexed, integrated and scaled using the HKL software package (31). Crystals belonged to the hexagonal spacegroup P6522, with a=b=127.7 Å, and c=206.7 Å and contained one FabI dimer and one ACP molecule in the asymmetric unit. The structure was solved by molecular replacement using the program MOLREP (32) and the FabI dimer as a search model (PDB entry 1C14 (33)). Two unambiguous solutions with R-factors of 0.418 and 0.426 and correlation coefficients 0.569 and 0.552, respectively, were obtained. Structure refinement was performed with REFMAC (34). In order to locate the ACP molecule, several cycles of rigid-body refinement followed by solvent flattening and NCS averaging were carried out using DM (35). Additional electron density was observed after DM, which to a large extent was a good fit for the main chain of butyryl-ACP (12). All residues of the ACP were changed to alanines, since no density for the side chains was observed and the loop region 195–199 in molecule B of FabI was disordered as well. The model was improved with restrained refinement using REFMAC (34) and then manually improved using the program O (36). The structure was refined to an Rfree of 0.263 and R of 0.226. An omit map calculated with CNS (37) for the final model showed no additional electron density. The stereochemistry of the model was good, with RMS deviations of 0.010 Å and 1.235° in bond lengths and bond angles, respectively. The structure was analyzed with SFCHECK (38) and PROCHECK (39) and 87.1% of all residues were in the core region, 11.7% allowed, 1.2% generously allowed and 0% in the disallowed regions of the Ramachandran diagram. Crystallographic statistics are given in Table 1. The coordinates and structure factors have been submitted to the RCSB (PDB ID 2FHS).
Table 1.
Data Collection and Refinement Statistics
| Data collection | |
| Space group | P6522 |
| Cell dimensions a, b, c (Å) | 127.7, 127.7, 206.7 |
| Resolution (Å) | 50.0–2.7 |
| Rsym (%) | 8.1(71.2) |
| Mean I/σ(I) | 25.4(2.7) |
| Completeness (%) | 97.1(98.4) |
| Multiplicity | 10.1(7.3) |
| Refinement | |
| Resolution range (Å) | 30.0–2.7 |
| Unique reflections | 25,906 |
| Ramachandran statistics | 87.1/ 11.7/1.2/0.0 |
| Rcryst | 0.226 |
| Rfree | 0.263 |
| Number of nonhydrogen atoms (Protein/solvent) | 3947/79 |
| R.m.s. deviations from ideal values in | |
| Bond length (Å) | 0.010 |
| Bond angle (°) | 1.235 |
Molecular Dynamics Simulations
Computational modeling and molecular dynamics simulations were performed with the Amber suite of programs (40). The missing ACP atoms were added using the butyryl-ACP crystal structure (pdb code 1L0I) (12) by overlapping the two structures, and replacing the partial coordinate set with the coordinates from 1LOI.. Similarly, the structure of triclosan bound to FabI in the presence of NAD+ (pdb code 1QSG) (21) was used to place the NADH cofactor. Missing FabI side chains and all hydrogens were added using Xleap The coordinates for the phosphopantetheine moiety were taken from the crystal structure of holo-acyl carrier protein-synthase in complex with holo-acyl carrier protein (1F80) (15). Maestro Molecular Modeling software was used to build the acyl chain for the phosphopantetheine moiety (PP). Force field parameters were the ff99 set for proteins (41) and published parameters (42,43) for NADH. The Amber antechamber module and GAFF force field (44) with am1bcc charges (45) were used to generate the parameters for PP and the attached acyl chain. In order to present the correct face of the crotonyl double bond to the NADH, the substrate must be bound in an s-trans conformation such that the si face of Cβ is oriented toward the NADH pro4(S) proton so that hydride transfer will generate the expected 3(S)-enoyl product (46). Since spontaneous isomerization of this double bond is unlikely to occur during relatively short MD simulations, we modeled the crotonyl in the s-trans conformation.
Initial minimization of the FabI-NADH-ACP ternary complex was performed in a stepwise fashion by restraining the backbone Cα atoms and allowing the side chains to move, with each step consisting of 1000 cycles. The restraints were gradually removed in each step (force constants from 10, 7, 4, 1 and 0 kcal mol−1 Å−2). Equilibration dynamics was performed on the minimized structure, with a constant temperature of 300K maintained by coupling to a thermostat using the Langevin algorithm with the collision frequency set to 1 ps−1. This reduced viscosity has been shown to facilitate rapid structural rearrangement (47). During dynamics, restraints on the backbone atoms were gradually released in a stepwise fashion in 25ps increments (force constants 10, 7, 4 to 1 kcal/mol/Å2). Further equilibration with no restraints was performed for 500ps at 300K. The time step was 1fs. During all simulations, all possible nonbonded interactions were evaluated at each time step (i.e. no cutoff was employed). Solvation effects were incorporated using the Generalized Born model as implemented in Amber (48–50). RMSD values were calculated using the initial model structure (prior to equilibration) as a reference.
The production phase consisted of 1 ns simulations at 300K with parameters as described above. Simulations were fully unrestrained with the exception of a distance restraint between the C3 atom of the substrate and C4 of the NADH nicotinamide ring, with a force constant of 10 kcal mol−1 Å−2. The initial separation was ~18 Å, and the final distance was 4.6 Å. We modified Amber to apply the restraint force only to the substrate C3 in order to avoid pulling NADH out of the binding pocket. After the substrate was drawn into the active site, a 100ps simulation was performed to equilibrate the system with the substrate in the active site. Finally, this restraint was released and an additional 100ps of fully unrestrained simulation was performed.
The sensitivity of the results to the force field parameters was studied by repeating the procedure using the ff99SB protein force field (51) and a newer variant of the GB solvation model (52,53). We recently showed that this combination of force field and solvent model was able to accurately reproduce experimental data for large conformational changes in HIV-1 protease that occur upon addition or removal of an inhibitor (54,55). After the substrate was drawn into the FabI active site, the system was simulated for an additional 2500 ps.
RESULTS
X-ray Crystallography
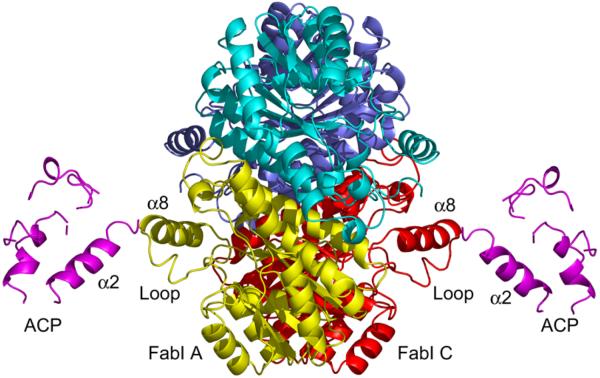
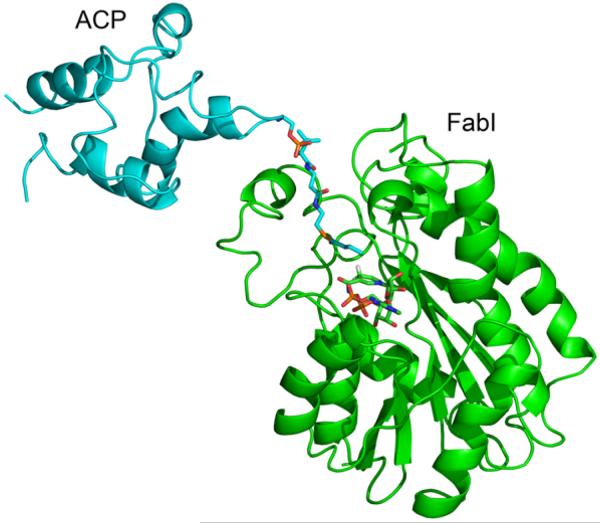
FabI forms a 110.9 kDa tetramer with 222 symmetry in which each monomer is formed by a central β-sheet that contains seven β-strands sandwiched by eight helices. Two FabI monomers (A and C) within the tetramer form a complex with ACP, resulting in a stoichiometry of 2:1 for the FabI:ACP interaction (Fig. 1). The ACP-bound FabI monomers and the uncomplexed FabI monomers are oriented in such a way that they form a nearly continuous β-sheet across the dimer interface.
Figure 1. X-ray Crystallographic Structure of the FabI-ACP Complex.
X-ray structure of the FabI-ACP complex. Two ACP molecules (pink) are bound to the FabI tetramer and interact with FabI monomers labeled A (yellow) and C (red). The principal interaction interface is between ACP helix α2 and FabI helix α8. The FabI helix α8 is adjacent to the FabI substrate binding loop (loop). The figure was made with pymol (62).
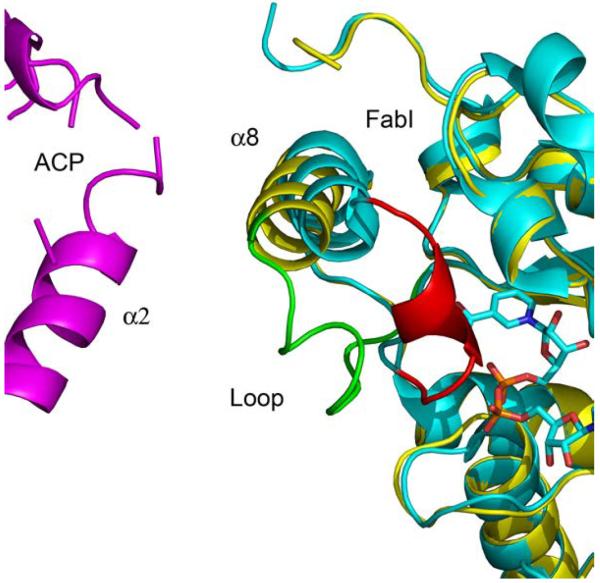
Superposition of the ACP-bound FabI monomers with FabI from the triclosan-bound structure (1QSG.pdb) (21) results in an RMS deviation of 0.58 Å for 252 Cα atoms for residues 2–194 and 200–259. Thus, the overall structure of FabI does not change upon binding to ACP. However, the substrate binding loop (residues 191–200) undergoes a major conformational change upon complexation with ACP while FabI helix α8 (residues 201–213) is shifted towards helix α2 of ACP (21) (Fig. 2). Although the density is missing for most of the side chains in ACP, the distance between the main chain atoms of these two helices indicates that helix α8 of FabI interacts with helix α2 of ACP with a main chain separation of about 8.5 Å, over a helix-helix interface length of roughly 11 Å. The substrate binding loop in the triclosan-bound structure, which is connected to helix α8, forms a lid on top of the triclosan moiety and the nicotinamide ring, thereby shielding them from the solvent. In contrast, this loop adopts an open lid conformation in the FabI-ACP structure, likely due to interaction with ACP, although the details of this interaction cannot be elucidated from the crystallographic data due to missing side chain density. Finally, the observation that only two ACP molecules are observed in the complex with the FabI tetramer could be due to crystal packing. We attempted to dock ACP molecules to the corresponding positions on the other monomers and obtained steric conflicts between the existing ACP molecules, suggesting that the lack of 1:1 stoichiometry is biologically relevant.
Figure 2. The Substrate Binding Loop in the FabI-ACP and FabI-Triclosan Complexes.
Superposition of FabI bound to triclosan (cyan) and bound to ACP (yellow). The FabI substrate binding loop is colored red in the FabI-triclosan structure and green in the FabI-ACP structure. For clarity NAD+ is shown but triclosan is not. The figure was made with pymol (62).
Model Building and Molecular Dynamics
Since the side chains for both molecules in the region of the FabI-ACP interface were not resolved by the crystallographic data, we began our computational study by building a model that included these coordinates (see Methods for details), followed by a series of energy minimization and molecular dynamics (MD) simulations to relax the resulting structure of the complex. The proteins were stable, with the RMSD of FabI and ACP individually remaining below 3 Å. The relatively small contact interface between the proteins permitted a somewhat larger degree of relative motion of the two proteins, with the overall RMSD of the system reaching a plateau of ~3.5 Å.
Structural analysis of the resulting model indicates that the complex is predominantly stabilized through hydrogen bonding interactions between basic residues of FabI in helix α8 and acidic residues of ACP in helix α2, consistent with the structural and modeling studies on the interaction of ACP with AcpS, FabH and FabG (15–17). However, while the X-ray crystallographic data and computational modeling studies provide information on the nature of the interaction between FabI and ACP, the absence of electron density for the ACP pantetheine hindered our ability to predict how the ACP delivers the substrate into the FabI active site. Consequently, we set out to build a model of a productive FabI-ACP complex using the computationally refined crystallographic model as a starting point and attaching a crotonyl-phosphopantetheine group to the side chain of S36 in ACP. Importantly, these simulations also help determine whether the relative positions of FabI and ACP observed in the crystal structure permit delivery of substrate to the active site.
During MD simulations of this structure, the substrate remained outside the FabI active site. This is expected since diffusion of the substrate into the protein is likely an inaccessible event during standard MD simulations on the nanosecond timescale. We thus obtained a model for the productive complex by placing a distance restraint between the crotonyl C3 carbon and the NADH C4 carbon, reducing the target value during a 1 ns MD simulation to draw the substrate into the active site. No other restraints were employed to enforce any particular binding mode. Subsequently, the restraint between the substrate and FabI active site was released and a further completely unrestrained simulation was performed for 100 ps.
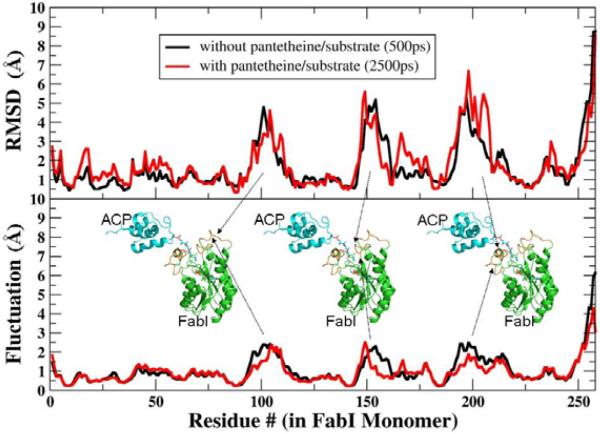
Similar to the behavior without substrate, the individual proteins remained relatively stable during these simulations, with backbone RMSD less than or equal to 3.0 Å as compared to the initial model. We observed a greater movement of the ACP molecule with respect to FabI, which allowed S36 to be oriented towards the active site cavity. In order to gain more specific insight into the structural changes that occurred during substrate entry, we performed a superposition of the initial and final structures of the entire protein and then separately calculated the RMSD values for each residue without refitting (Fig. 3). These show that the RMSD for the majority of FabI was < 2.0 Å, whereas the active site loop residues (95–115 and 190–210) showed a much larger deviation of > 5 Å. The larger fluctuations for these same loop regions even in the absence of substrate suggests that their flexibility may accommodate entry of substrate into the active site. The RMSD values for ACP are distributed more evenly across the sequence (3.5 Å); this is not unexpected since the initial model was based on the crystal structure of ACP in the absence of FabI with the butyryl moiety accommodated in an internal cavity of the ACP.
Figure 3. RMSD Values and Fluctuations for Each FabI Residue During MD Simulations of the Complex.
Average RMSD values and positional fluctuations for each residue in FabI during simulations of the complex without substrate (black lines) and after drawing the substrate into the active site (red lines). Most regions of the protein have deviations of only 1–2Å from the crystal structure, while several loop regions (indicated by the arrows) show larger RMSD values that arise from increased, as indicated by higher fluctuations. These loops show similar behavior in the presence and absence of the substrate.
Interactions in the Productive FabI-ACP Complex
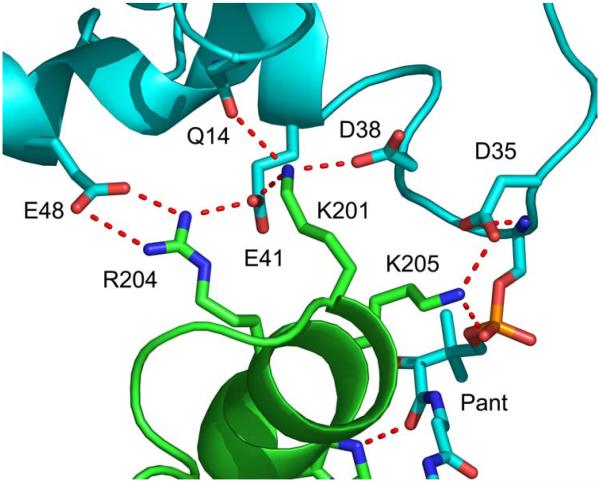
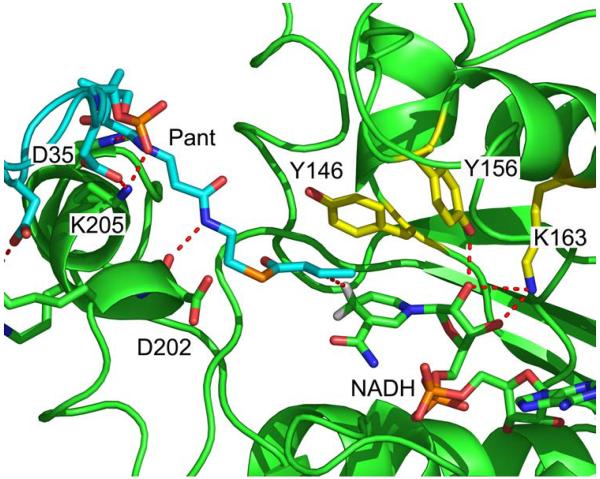
The structure of the final FabI-ACP structure is shown in Fig. 4. ACP interacts with FabI helix α8 and delivers the substrate to the active site between helix α8 and a loop comprised of FabI residues 152-156. Analysis of this structure reveals several important interactions at the FabI-ACP interface (Fig. 5) and also between the phosphopantetheine and the FabI protein. Residues K201, R204, and K205 from helix α8 of FabI contact residues D35, D38, E41, and E48 of ACP helix α2, while FabI K201 also interacts with Q14 in ACP. The side chain amino group of FabI K205, which interacts with D35 in ACP helix α2, is hydrogen bonded to the phosphopantetheine phosphate (O7). In addition, the backbone carbonyl of FabI K205 forms a hydrogen bond to the pantetheine hydroxyl group (O10) while Hε2 of H209 is hydrogen bonded to the pantetheine 4” amide carbonyl oxygen. Finally, the pantetheine 2” amide nitrogen forms a hydrogen bond to the backbone carbonyl of D202 (Fig. 6). These interactions between the ACP pantetheine and FabI undoubtedly play an important role in stabilizing the FabI-ACP complex and in positioning the substrate within the active site. In the final structure, the distance between the crotonyl C3 and the NADH pro4(S) proton is 3 Å. Analysis of the structure also reveals that the crotonyl thioester carbonyl is located 4 Å from the Y146 hydroxyl group, suggesting that Y146 may form a hydrogen bond to the thioester during substrate reduction (Fig. 6).
Figure 4. The Structure of ACP Bound to FabI Following MD Simulations.
Final structure of the FabI-ACP complex. FabI is colored green and ACP is colored cyan. The figure was made with pymol (62).
Figure 5. Interactions Between FabI and ACP.
Interactions between ACP (cyan) and FabI (green) at the helix α2 (ACP) - helix α8 (FabI) interface. The figure was made with pymol (62).
Figure 6. Interactions in the Active Site of the FabI-ACP Complex.
Interactions between crotonyl-pantetheine and FabI. The pantetheine (cyan) is hydrogen bonded to residues in FabI helix α8 (green). FabI residues in the conserved active site triad (Y146, Y156 and K163) are colored yellow. The crotonyl group of the substrate (cyan) is bound in the s-trans conformation and the crotonyl carbonyl group is oriented toward Y146 (yellow). The C3 carbon of the crotonyl group is 3 Å from the NADH pro4(S) proton (white). In addition, the NADH ribose (cyan) is hydrogen bonded to Y156 and K163. is The figure was made with pymol (62).
In order to investigate the influence of the computational protocol on the results, we repeated the process using a different protein force field and solvent model (see Methods). We recently employed this combination to successfully simulate ligand-induced conformational changes in HIV-1 protease (54,55). Importantly, we also demonstrated that this particular implicit solvent model accurately reproduces the stability of salt bridges between protein side chains as compared to simulations in explicit solvent (56). Simulations with these parameters gave very similar results to those described above. The crotonyl thioester carbonyl is located even closer (2.7 Å) to the Y146 hydroxyl group, providing further evidence that Y146 may form a hydrogen bond to the thioester during substrate reduction.
Kinetic Analysis of Wild-type and Mutant FabI Enzymes
The structural studies described above identify several key interactions in the FabI-ACP complex that have been evaluated using site-directed mutagenesis by replacing the basic FabI residues with acidic groups. Table 2 contains the kcat and Km values for the R204E and K205E FabI enzymes. Mutation of these two basic FabI residues at the FabI-ACP interface has no significant impact on the kinetic parameters for reduction of DD-CoA. In contrast, the kcat values for both mutants are reduced ca. 4-fold while the Km (DD-ACP) values are increased ca. 40-fold, compared to the values for the wild-type enzyme, when DD-CoA is replaced with DD-ACP. Thus, each mutation reduces the kcat/Km value for reduction of DD-ACP by more than 100-fold.
Table 2.
Kinetic Parameters for Wild-type and Mutant FabIs
| DD-CoAa | DD-ACPb | |||||
|---|---|---|---|---|---|---|
|
| ||||||
| Enzyme | kcat (min−1) | Km (μM) | kcat/Km (μM−1 min−1) | kcat (min−1) | Km (μM) | kcat/Km (μM−1 min−1) |
| Wild type | 248 ± 8 | 13 ± 1 | 19 ± 2 | 238 ± 3 | 3 ± 1 | 79 ± 27 |
| K205E | 281 ± 2 | 16 ± 3 | 18 ± 4 | 59 ± 9 | 112 ± 29 | 0.5 ± 0.2 |
| R204E | 295 ± 2 | 7 ± 2 | 42 ± 12 | 66 ± 1 | 112 ± 38 | 0.6 ± 0.2 |
| Y156F | 94 ± 2 | 22 ± 4 | 4 ± 1 | 52 ± 7 | 5 ± 2 | 10 ± 5 |
| Y146F | 23 ± 1 | 34 ± 3 | 0.7 ± 0.1 | NDc | NDc | NDc |
DD-CoA, trans-2-dodecenoyl-CoA.
DD-ACP, trans-2-dodecenoyl-ACP.
ND, not determined.
We have also examined the importance of Y156 and Y146, the two active site tyrosine residues, in substrate reduction. Replacement of Y156 with Phe has only a very small effect on substrate reduction. The kcat/Km values for reduction of DD-CoA and DD-ACP by Y156F FabI are reduced 5–8 fold compared to the wild-type enzyme. These data are in agreement with previous studies on both FabI (23) and InhA (22,28), which questioned the importance of Y156 (Y158 in InhA) in catalysis (57). In addition, mutagenesis of Y146 has a slightly larger effect on catalysis, with kcat and kcat/Km decreasing by 11 and 30-fold, respectively, compared to wild-type FabI.
DISCUSSION
ACPs are small, acidic proteins that fulfill an essential role in metabolism through their interactions with a diverse array of target enzymes. However, despite their central role, detailed structural information on the acyl-group specific recognition of ACPs by their target proteins has remained elusive, presumably partly as a result of the conformational flexibility of the ACP molecule. Here we report the first structural data for the direct interaction of an acyl-ACP substrate with a target enzyme, the FASII FabI enzyme, based on a combination of X-ray crystallography and computational modeling. FabI is the enoyl reductase in the bacterial FASII pathway and a target for antibacterial drug discovery. Thus, not only does the structure of the ACP:FabI complex provide general insight into how target proteins recognize and bind to acyl-ACPs, the present structure also provides a foundation for the development of novel FabI inhibitors that antagonize the interaction of FabI with its natural substrate.
In agreement with previous predictions, several acidic residues in and close to the ACP helix α2 (D35, D38 and E41) form stable electrostatic interactions with three basic amino acids (K201, R204 and K205) located adjacent to the FabI substrate binding loop. Importantly, replacement of R204 or K205 by glutamic acid results in a large (40-fold) increase in Km and an overall 150-fold decrease in kcat/Km for reduction of DD-ACP, without affecting the kinetic parameters for reduction of the corresponding CoA substrate. Thus, these residues are involved in specific interactions with the protein portion of DD-ACP, as was shown in similar experiments with FabH and FabG (16,17). The activity of the K201E mutant is significant reduced even with DD-CoA substrate; thus it is not possible to reliably interpret the effect of the mutation on the binding of FabI and ACP. The FabI-ACP interactions position S36, the ACP residue that carries the phosphopantetheine, above an opening into the active site formed by the substrate binding loop, helix α8 comprised of residues 192-206 and a mobile loop comprised of residues 152-156. These two loops move apart to allow the phosphopantetheine to deliver the substrate to the active site through the minor portal (27). While the C3 carbon of the enoyl substrate is at a distance of approximately 3 Å from the pro4(S) NADH proton, the positioning of the substrate into the active site is unexpected given previous structural data on inhibitors bound to FabI and the hexadecenoyl-N-acetylcysteamine (C16-NAC) substrate (27) bound to InhA. Below we discuss the orientation of the substrate with respect to the catalytic triad in the FabI active site.
FabI is a member of the short chain alcohol dehydrogenase/reductase family. This superfamily is characterized by a conserved triad of active site residues. In FabI the triad is comprised of Y146, Y156 and K163, while in InhA, the M. tuberculosis enoyl reductase, the triad is F149, Y158 and K165. Mechanistic information on the role of these residues in catalysis has been provided by site-directed mutagenesis coupled with structural data primarily arising from enzyme-inhibitor rather than enzyme-substrate complexes. Inhibitors that have been structurally characterized in complex with these enzymes include compounds such as the diazaborines (18) and isoniazid (26) which modify the NAD(H) cofactor, and those, such as triclosan, that bind noncovalently to the enzyme-cofactor complex (20,21).
Triclosan, which binds with picomolar affinity to the E. coli FabI (23,58,59), has been proposed to bind to the enzyme as a substrate analog (60). This hypothesis has gained support from the structure of hexadecenoyl-N-acetylcysteamine (C16-NAC) bound in a stable ternary complex to InhA in the presence of NAD+ (27). Y156 of FabI (Y158 in InhA) forms a hydrogen bond to the phenol of triclosan and the carbonyl oxygen of the C16 substrate. Mutagenesis clearly supports the importance of Y156 (Y158) in triclosan binding, and replacement of this residue by Phe increases the Ki for triclosan inhibition by 400 (160)-fold (22,23). However, the impact of mutating Y156 (Y158) with respect to its role in catalysis is much less pronounced. In FabI, the Y156F variant has a kcat value that is only 5-fold reduced compared to wild-type FabI. The corresponding mutation in InhA (Y158F) has a slightly stronger effect on catalysis, reducing kcat 24-fold compared to wild-type. Intriguingly, however, the Y158S InhA mutant displays wild-type activity (28). Thus, Y156 does not play a major role in substrate reduction and, in agreement with these data, the substrate carbonyl group in the present FabI-ACP structure is pointing away from Y156 (Fig. 6).
The function of the second aromatic residue in the enoyl reductase triad (Y146/F149) is also not clear and Rozwarski et al. have argued that F149 in InhA is involved in directing the NADH to deliver a hydride to the correct position on the substrate and/or in modulating the interaction of the bound substrate with a channel of water molecules that leads away from the active site (27). In the simulation model, the FabI Y146 hydroxyl is located within hydrogen bonding distance from the substrate carbonyl, suggesting that Y146 rather than Y156 provides electrophilic assistance during substrate reduction. To probe the role of Y146 in catalysis, the Y146F mutant was characterized and shown to catalyze substrate reduction with a kcat/Km value 30-fold lower than wild-type FabI. Thus, if Y146 does hydrogen bond to the substrate carbonyl, then this interaction is not of major importance for catalysis. Indeed, Anderson and coworkers have suggested that the positively charged nicotinamide ring, formed following hydride transfer, could provide sufficient stabilization to the substrate enolate and that further stabilization of the enolate by the enzyme may not be required (57). We also note that the homologue of Y146 in InhA is a phenylalanine (F149), and so is unable to provide a hydrogen bond to the substrate. Indeed, Raman studies on deuterated NADD cofactor bound to InhA suggest that F149 most likely is involved in correctly positioning the cofactor for hydride transfer (data not shown).
Returning to previous structural studies, Sacchettini and coworkers have discussed two entry points for substrates into the active sites of enoyl reductases termed the major and minor portals (27). Based on the structures of inhibitors bound to FabI and InhA and that of the InhA:C16-NAC complex, the fatty acid substrate would have been expected to enter the FabI active site through the major portal. Instead, the present structure indicates that binding of ACP to FabI delivers the acyl-pantetheine to the active site between loops comprised of residues 192-206 and 152-156 such that the fatty acid enters the active site through, or adjacent to, the minor portal. This structure is consistent with our mutagenesis data and also the ACP-FabI binding interface proposed by Rock and coworkers (16,17). Indeed, interaction of ACP with the basic patch of residues on FabI leaves the ACP S36 residue too far from the active site to deliver the substrate through the major portal, thus predicating an alternative entry point for the substrate. Interestingly, in the recently determined structure of 5-octyl-2-phenoxyphenol bound to InhA, the analogous channel in InhA is occupied by the alkyl chain of the inhibitor which binds to the enzyme with a Ki value of 1 nM (2B37.pdb) (61). Taken together, these data indicate that substrate binding occurs in the opposite orientation to that expected from studies using inhibitors or the truncated C16-NAC substrate. In the case of InhA, we do not as yet have a structure of the relevant complex with ACP and it is possible that ACP delivers the substrate to this enzyme through the major portal, as suggested by the C16-NAC structure. Alternatively, without the ACP to locate it, the C16-NAC molecule could bind to InhA in a mode that more closely approximates an inhibitor rather than a substrate. The two resulting possibilities then are that the C16-NAC could be reduced by the enzyme in this alternative binding mode or that the C16-NAC is bound nonproductively to the enzyme in the complex used for the structural studies. While we are unable to differentiate between these possibilities at the present time, we note that the C16-NAC molecule is bound to the enzyme in the presence of NAD+ rather than NADH, as observed for the diphenyl ether inhibitors. Future studies on the interaction of ACP with InhA will shed light on these possibilities. In addition, we are also extending our efforts to probe the interaction of ACPs with other components of the FASII pathway and designing compounds to antagonize the interaction of the natural substrate with FabI.
SUMMARY
We have used a combination of X-ray crystallography and molecular dynamics to provide the first structural insight into the detailed interaction of ACP with a target enzyme. The structural data are substantiated by mutagenesis, and reveal that interactions between ACP and FabI are largely electrostatic in nature. Of key importance are interactions between acidic residues in ACP helix α2 and basic residues in FabI helix α8. The ACP phosphopantetheine delivers the enoyl substrate to the active site between helix α8 and a mobile loop comprised of residues 152-156. In agreement with site-directed mutagenesis, the conserved active site tyrosine Y156 is not directly involved in interactions with the substrate. While some of the specific interactions between FabI and ACP differ somewhat between simulations performed using different protein force fields and solvent models, they provide the consistent view that ACP is able to successfully deliver the substrate through the minor portal and into the FabI active site from the position observed in the crystal structure of the complex.
Knowledge of the structural determinants that control the interaction of ACP with target enzymes is of critical importance for the design of inhibitors against these enzymes and for fully understanding the multiple roles of ACP in metabolic pathways.
The abbreviations used are
- ACP
acyl carrier protein
- FabI
the enoyl reductase from Escherichia coli
- AcpS
holo ACP synthase
- DD-CoA
trans-2-dodecenoyl-CoA
- DD-ACP
trans-2-dodecenoyl-ACP
- C16-NAC
hexadecenoyl-N-acetylcysteamine
Footnotes
This research was supported by US National Institutes of Health grants AI44639 (to P.J.T.) and GM6167803 (to C.S.). Supercomputer time at NCSA (MCA02N028, C.S.) is gratefully acknowledged. Additional computer time was generously provided by the SGI Engineering group. P.J.T. is an Alfred P. Sloan Fellow. C.S. is a Cottrell Scholar of Research Corporation. C.K. is a PEW Scholar of the Biomedical Sciences.
REFERENCES
- 1.Rock CO, Cronan JE. Methods Enzymol. 1981;71(Pt C):341–351. doi: 10.1016/0076-6879(81)71043-7. [DOI] [PubMed] [Google Scholar]
- 2.Rock CO, Jackowski S. Biochem. Biophys. Res. Commun. 2002;292(5):1155–1166. doi: 10.1006/bbrc.2001.2022. [DOI] [PubMed] [Google Scholar]
- 3.Cane DE, Walsh CT. Chem. Biol. 1999;6(12):R319–325. doi: 10.1016/s1074-5521(00)80001-0. [DOI] [PubMed] [Google Scholar]
- 4.Tang L, Weissborn AC, Kennedy EP. J. Bacteriol. 1997;179(11):3697–3705. doi: 10.1128/jb.179.11.3697-3705.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brozek KA, Raetz CR. J. Biol. Chem. 1990;265(26):15410–15417. [PubMed] [Google Scholar]
- 6.Sweet CR, Williams AH, Karbarz MJ, Werts C, Kalb SR, Cotter RJ, Raetz CR. J. Biol. Chem. 2004;279(24):25411–25419. doi: 10.1074/jbc.M400597200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cronan JE. Annu. Rev. Microbiol. 2003;57:203–224. doi: 10.1146/annurev.micro.57.030502.090851. [DOI] [PubMed] [Google Scholar]
- 8.Vagelos PR, Majerus PW, Alberts AW, Larrabee AR, Ailhaud GP. Fed. Proc. 1966;25(5):1485–1494. [PubMed] [Google Scholar]
- 9.Majerus PW, Vagelos PR. Adv. Lipid Res. 1967;5:1–33. [PubMed] [Google Scholar]
- 10.Magnuson K, Jackowski S, Rock CO, Cronan JE., Jr. Microbiol. Rev. 1993;57(3):522–542. doi: 10.1128/mr.57.3.522-542.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heath RJ, White SW, Rock CO. Prog. Lipid Res. 2001;40(6):467–497. doi: 10.1016/s0163-7827(01)00012-1. [DOI] [PubMed] [Google Scholar]
- 12.Roujeinikova A, Baldock C, Simon WJ, Gilroy J, Baker PJ, Stuitje AR, Rice DW, Slabas AR, Rafferty JB. Structure. 2002;10(6):825–835. doi: 10.1016/s0969-2126(02)00775-x. [DOI] [PubMed] [Google Scholar]
- 13.Holak TA, Nilges M, Prestegard JH, Gronenborn AM, Clore GM. Eur. J. Biochem. 1988;175(1):9–15. doi: 10.1111/j.1432-1033.1988.tb14159.x. [DOI] [PubMed] [Google Scholar]
- 14.Wong HC, Liu G, Zhang YM, Rock CO, Zheng J. J. Biol. Chem. 2002;277(18):15874–15880. doi: 10.1074/jbc.M112300200. [DOI] [PubMed] [Google Scholar]
- 15.Parris KD, Lin L, Tam A, Mathew R, Hixon J, Stahl M, Fritz CC, Seehra J, Somers WS. Structure Fold. Des. 2000;8(8):883–895. doi: 10.1016/s0969-2126(00)00178-7. [DOI] [PubMed] [Google Scholar]
- 16.Zhang YM, Rao MS, Heath RJ, Price AC, Olson AJ, Rock CO, White SW. J. Biol. Chem. 2001;276(11):8231–8238. doi: 10.1074/jbc.M008042200. [DOI] [PubMed] [Google Scholar]
- 17.Zhang YM, Wu B, Zheng J, Rock CO. J. Biol. Chem. 2003;278(52):52935–52943. doi: 10.1074/jbc.M309874200. [DOI] [PubMed] [Google Scholar]
- 18.Baldock C, Rafferty JB, Sedelnikova SE, Baker PJ, Stuitje AR, Slabas AR, Hawkes TR, Rice DW. Science. 1996;274(5295):2107–2110. doi: 10.1126/science.274.5295.2107. [DOI] [PubMed] [Google Scholar]
- 19.McMurry LM, Oethinger M, Levy SB. Nature. 1998;394(6693):531–532. doi: 10.1038/28970. [DOI] [PubMed] [Google Scholar]
- 20.Heath RJ, Rubin JR, Holland DR, Zhang E, Snow ME, Rock CO. J. Biol. Chem. 1999;274(16):11110–11114. doi: 10.1074/jbc.274.16.11110. [DOI] [PubMed] [Google Scholar]
- 21.Stewart MJ, Parikh S, Xiao G, Tonge PJ, Kisker C. J. Mol. Biol. 1999;290:859–865. doi: 10.1006/jmbi.1999.2907. [DOI] [PubMed] [Google Scholar]
- 22.Parikh SL, Xiao G, Tonge PJ. Biochemistry. 2000;39(26):7645–7650. doi: 10.1021/bi0008940. [DOI] [PubMed] [Google Scholar]
- 23.Sivaraman S, Zwahlen J, Bell AF, Hedstrom L, Tonge PJ. Biochemistry. 2003;42(15):4406–4413. doi: 10.1021/bi0300229. [DOI] [PubMed] [Google Scholar]
- 24.Banerjee A, Dubnau E, Quemard A, Balasubramanian V, Um KS, Wilson T, Collins D, de Lisle G, Jacobs WR., J. r. Science. 1994;263(5144):227–230. doi: 10.1126/science.8284673. [DOI] [PubMed] [Google Scholar]
- 25.Quemard A, Sacchettini JC, Dessen A, Vilcheze C, Bittman R, Jacobs WR, Jr., Blanchard JS. Biochemistry. 1995;34(26):8235–8241. doi: 10.1021/bi00026a004. [DOI] [PubMed] [Google Scholar]
- 26.Rozwarski DA, Grant GA, Barton DHR, Jacobs WR, J. r., Sacchettini JC. Science. 1998;279(5347):98–102. doi: 10.1126/science.279.5347.98. [DOI] [PubMed] [Google Scholar]
- 27.Rozwarski DA, Vilcheze C, Sugantino M, Bittman R, Sacchettini JC. J. Biol. Chem. 1999;274:15582–15589. doi: 10.1074/jbc.274.22.15582. [DOI] [PubMed] [Google Scholar]
- 28.Parikh S, Moynihan DP, Xiao G, Tonge PJ. Biochemistry. 1999;38(41):13623–13634. doi: 10.1021/bi990529c. [DOI] [PubMed] [Google Scholar]
- 29.Schaeffer ML, Agnihotri G, Kallender H, Brennan PJ, Lonsdale JT. Biochim. Biophys. Acta. 2001;1532(1–2):67–78. doi: 10.1016/s1388-1981(01)00116-0. [DOI] [PubMed] [Google Scholar]
- 30.Lambalot RH, Walsh CT. J. Biol. Chem. 1995;270(42):24658–24661. doi: 10.1074/jbc.270.42.24658. [DOI] [PubMed] [Google Scholar]
- 31.Otwinowski Z, Minor W. Meth. Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 32.Vagin A, Teplyakov A. J. Appl. Crystallogra. 1997;30:1022–1025. [Google Scholar]
- 33.Qiu XY, Janson CA, Court RI, Smyth MG, Payne DJ, Abdel-Meguid SS. Protein Sci. 1999;8(11):2529–2532. doi: 10.1110/ps.8.11.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murshudov GN, Vagin AA, Dodson EJ. Acta Crystallogra. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 35.Cowtan K. Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography. 1994;31:34–38. [Google Scholar]
- 36.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Acta Crystallogra. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 37.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Acta Crystallogra. 1998;D54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 38.Vaguine AA, Richelle J, Wodak SJ. Acta Crystallogra. 1999;D55:191–205. doi: 10.1107/S0907444998006684. [DOI] [PubMed] [Google Scholar]
- 39.Laskowski RA, Macarthur MW, Moss DS, Thornton JM. J. Appl. Crystallogra. 1993;26:283–291. [Google Scholar]
- 40.Case DA, Cheatham TE, Darden T, Gohlke H, Luo R, Merz KM, Onufriev A, Simmerling C, Wang B, Woods RJ. J. Comput. Chem. 2005;26(16):1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang JM, Cieplak P, Kollman PA. Journal of Computational Chemistry. 2000;21(12):1049–1074. [Google Scholar]
- 42.Pavelites JJ, Gao JL, Bash PA, Mackerell AD. Journal of Computational Chemistry. 1997;18(2):221–239. [Google Scholar]
- 43.Walker RC, de Souza MM, Mercer IP, Gould IR, Klug DR. Journal of Physical Chemistry B. 2002;106(44):11658–11665. [Google Scholar]
- 44.Wang JM, Wolf RM, Caldwell JW, Kollman PA, Case DA. J. Comput. Chem. 2004;25(9):1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 45.Jakalian A, Bush BL, Jack DB, Bayly CI. J. Comput. Chem. 2000;21(2):132–146. [Google Scholar]
- 46.Fillgrove KL, Anderson VE. Biochemistry. 2000;39(23):7001–7011. doi: 10.1021/bi0000566. [DOI] [PubMed] [Google Scholar]
- 47.Zagrovic B, Pande V. Journal of Computational Chemistry. 2003;24(12):1432–1436. doi: 10.1002/jcc.10297. [DOI] [PubMed] [Google Scholar]
- 48.Still WC, Tempczyk A, Hawley RC, Hendrickson T. J. Am. Chem. Soc. 1990;112(16):6127–6129. [Google Scholar]
- 49.Hawkins GD, Cramer CJ, Truhlar DG. Chem. Phys. Lett. 1995;246(1–2):122–129. [Google Scholar]
- 50.Hawkins GD, Cramer CJ, Truhlar DG. J. Phys. Chem. 1996;100(51):19824–19839. [Google Scholar]
- 51.Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Proteins: Structure, Function and Genetics. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Feig M, Onufriev A, Lee MS, Im W, Case DA, Brooks CL. Journal of Computational Chemistry. 2004;25(2):265–284. doi: 10.1002/jcc.10378. [DOI] [PubMed] [Google Scholar]
- 53.Onufriev A, Bashford D, Case DA. Proteins-Structure Function and Bioinformatics. 2004;55(2):383–394. doi: 10.1002/prot.20033. [DOI] [PubMed] [Google Scholar]
- 54.Hornak V, Okur A, Rizzo RC, Simmerling C. Journal of the American Chemical Society. 2006;128(9):2812–2813. doi: 10.1021/ja058211x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hornak V, Okur A, Rizzo RC, Simmerling C. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(4):915–920. doi: 10.1073/pnas.0508452103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Geney R, Layten M, Gomperts R, Hornak V, Simmerling C. Journal of Chemical Theory and Computation. 2006;2(1):115–127. doi: 10.1021/ct050183l. [DOI] [PubMed] [Google Scholar]
- 57.Fillgrove KL, Anderson VE. Biochemistry. 2001;40(41):12412–12421. doi: 10.1021/bi0111606. [DOI] [PubMed] [Google Scholar]
- 58.Ward WHJ, Holdgate GA, Rowsell S, McLean EG, Pauptit RA, Clayton E, Nichols WW, Colls JG, Minshull CA, Jude DA, Mistry A, Timms D, Camble R, Hales NJ, Britton CJ, Taylor IWF. Biochemistry. 1999;38(38):12514–12525. doi: 10.1021/bi9907779. [DOI] [PubMed] [Google Scholar]
- 59.Sivaraman S, Sullivan TJ, Johnson F, Novichenok P, Cui G, Simmerling C, Tonge PJ. J. Med. Chem. 2004;47:509–518. doi: 10.1021/jm030182i. [DOI] [PubMed] [Google Scholar]
- 60.Levy CW, Roujeinikova A, Sedelnikova S, Baker PJ, Stuitje AR, Slabas AR, Rice DW, Rafferty JB. Nature. 1999;398(6726):383–384. doi: 10.1038/18803. [DOI] [PubMed] [Google Scholar]
- 61.Sullivan TJ, Truglio JJ, Boyne ME, Novichenok P, Zhang X, Stratton CF, Li H-J, Kaur T, Amin A, Johnson F, Slayden RA, Kisker C, Tonge PJ. ACS Chem. Biol. 2006;1(1):43–53. doi: 10.1021/cb0500042. [DOI] [PubMed] [Google Scholar]
- 62.Delano WL. 2002 http://www.pymol.org.