SUMMARY
Porphyromonas gingivalis is the main causing agent of periodontitis. It deregulates the inflammatory and innate host immune responses through virulence factors, which include the immunodominant outer-membrane surface receptor antigens A (PgRagA) and B (PgRagB), co-transcribed from the rag pathogenicity island. The former is predicted to be a Ton-dependent porin-type translocator but the targets of this translocation and the molecular function of PgRagB are unknown. Phenomenologically, PgRagB has been linked with epithelial cell invasion and virulence according to murine models. It also acts as a Toll-like receptor agonist and promotes multiple mediators of inflammation. Hence, PgRagB is a candidate for the development of a periodontitis vaccine, which would be facilitated by the knowledge of its atomic structure. Here, we crystallized and solved the structure of 54-kDa PgRagB, which revealed a single domain centered on a curved helical scaffold. It consists of four tetratrico peptide repeats (TPR1-4), each arranged as two helices connected by a linker, plus two extra downstream capping helices. The concave surface bears four large intertwined irregular inserts (A–D), which contribute to an overall compact moiety. Overall, PgRagB shows substantial structural similarity with Bacteroides thetaiotaomicron SusD and Tannerella forsythia NanU, which are, respectively, engaged in binding and uptake of maltooligosaccharide/starch and sialic acid. This suggests a similar sugar-binding function for PgRagB for uptake by the cognate PgRagA translocator, and, consistently, three potential monosaccharide-binding sites were tentatively assigned on the molecular surface.
Keywords: periodontitis, sugar-binding proteins, X-ray crystal structure, SusD-like proteins, tetratricorepeat proteins
INTRODUCTION
Chronic periodontal disease (PD) is a inflammatory condition caused by bacteria, which affects the periodontium of 10%–15% of adults worldwide (Paster et al., 2006; Rocas et al., 2001; Socransky et al., 1998) and leads to tissue destruction and tooth loss in severe cases (Preshaw et al., 2012). Porphyromonas gingivalis, together with Tannerella forsythia and Treponema denticola, participates in the “red complex” of bacteria associated with PD (Socransky et al., 1998), and its importance as a periodontal pathogen has been highlighted by recent microbiome and dysbiosis studies (Hajishengallis, 2015; Park et al., 2015). P. gingivalis is found at the infection site of up to 85% of PD cases, and its presence is a marker of disease progression (van Winkelhoff et al., 2002; Yang et al., 2004). The pathogenic relationship between the human oral cavity and P. gingivalis is ancient, as revealed by its detection in the ~5,300-year old mummy of the Tyrolean Iceman “Ötzi” (Maixner et al., 2014).
During infection and colonization, the pathogen evades host defense mechanisms through several virulence factors that deregulate the inflammatory and innate immune responses (Hanley et al., 1999). Comparison between the serum of PD patients and healthy controls revealed two immunodominant outer-membrane proteins, the surface receptor antigens A (PgRagA; 115 kDa) and B (PgRagB; 55 kDa), which are expressed in vivo and interact with the immune systems of PD patients (Curtis et al., 1991; Curtis et al., 1999a; Hanley et al., 1999). They are contiguously encoded by the rag pathogenicity island and co-transcribed to a single ~4.7-kb mRNA (Hanley et al., 1999). The proteins interact (Nagano et al., 2007) to form hetero-oligomeric complexes of variable mass (Glew et al., 2014). The rag locus varies, which results in different alleles of RagA and RagB (Hall et al., 2005). Isolates that caused serious disease in mice were significantly more likely to carry the rag-1 allele—the equivalent in P. gingivalis strain W50 of rag from strain W83 (Hall et al., 2005)—than any other ones (Laine & van Winkelhoff, 1998). Expression of rag is temperature-dependent (Bonass et al., 2000) and up-regulated by cigarette smoke (Bagaitkar et al., 2009), and has been associated with severe PD (Su et al., 2010), as the locus is absent in avirulent P. gingivalis strains (Loos et al., 1990).
PgRagA is required for bacterial survival in Mariner-family transposon mutagenesis studies (Klein et al., 2012), and an ortholog from Bacteroides caccae is associated with inflammatory bowel disease (Wei et al., 2001). PgRagB, in turn, is an immunodominant antigen of P. gingivalis (Curtis et al., 1999a; Hutcherson et al., 2015; Imai et al., 2005; Nagano et al., 2007). It has been associated with epithelial cell invasion through comparative genomics (Dolgilevich et al., 2011), and virulence through murine soft-tissue infection models (Nagano et al., 2007). More recently, PgRagB has also been reported to be an unusual Toll-like receptor (TLR) agonist with several properties: it is recognized by both human monocytic TLR2 and TLR4; it induces signal transducer and activator of transcription, STAT4; it activates nuclear factor-κB; and it promotes multiple mediators of inflammation at the transcriptional and protein levels (Hutcherson et al., 2015). Hence, PgRagB is under investigation as a potential novel periodontitis vaccine (Hutcherson et al., 2015; Kong et al., 2015; Zheng et al., 2013). Current vaccine development includes structural vaccinology approaches, which depend on the three-dimensional (3D) structures of the antigenic proteins to identify the amino acids and epitopes that are crucial for immunogenicity (Dormitzer et al., 2012).
In contrast to pathogenic bacteria, commensal bacteria are beneficial for the host. Gut microbiota digest polysaccharides by fermenting and absorbing host-derived mucosal glycans (Sonnenburg et al., 2005). Bacteroidetes, the most prevalent phylum in the human gut (Eckburg et al., 2005), employs the starch-utilization system (Sus) for this (Tancula et al., 1992). The sus operon, first and best studied in Bacteroides thetaiotaomicron (Anderson & Salyers, 1989a; b; Cho & Salyers, 2001; Reeves et al., 1997), encodes outer-membrane proteins involved in maltooligosaccharide and starch binding (proteins SusC–F) and hydrolysis (SusA, SusB and SusG), as well as a maltose-activated transcriptional regulator, SusR, which modulates sus transcription. SusC–G may form a complex to bind, process, and import starch (Cho & Salyers, 2001), of which SusC and SusD would constitute the minimal functional unit (Martens et al., 2009). BtSusC, predicted to be a Ton-dependent β-barrel porin translocator (Reeves et al., 1996), requires direct interaction with BtSusD for starch import (Cho & Salyers, 2001). B. thetaiotaomicron has an additional 100+ gene pairs that are similar to susC-susD, each potentially targeting a different glycan (Koropatkin et al., 2008) and collectively making up 18% of the genome (Martens et al., 2008). Furthermore, several hundreds of such operons have been identified in both beneficial and pathogenic Bacteroidetes, consistently with their role in glycan uptake (Xu et al., 2007). One example is Tannerella forsythia, which requires the BtSusC/BtSusD-like neuraminate uptake system, TfNanO/TfNanU, to import sialic acid (Phansopa et al., 2014). This system is also found in B. thetaiotaomicron. PgRagA/PgRagB was also predicted to be similar to BtSusC/BtSusD and TfNanO/TfNanU (Hall et al., 2005; Hanley et al., 1999; Phansopa et al., 2014). Accordingly, PgRagA and PgRagB may contribute to a surface receptor complex functionally linked to TonB, and thus act as transporters on the cell surface (Hanley et al., 1999; Nagano et al., 2007).
To shed light on the PgRagA/PgRagB axis, we analyzed the 3D crystal structure of PgRagB, which provided hints for the molecular basis of its function and may assist in the future development of a vaccine against PD.
METHODS
Protein production, purification, and characterization
PgRagB protein from strain W83 (UniProt access code F5H948) was obtained by recombinant heterologous overexpression in Escherichia coli as previously described (Hutcherson et al., 2015). Briefly, ragB was cloned into Escherichia coli by the pBAD202D TOPO expression plasmid (Invitrogen), which attaches a hexahistidine-tag and a thioredoxin fusion protein. RagB expression was induced by arabinose, cells were then lysed using CelLytic Buffer (Sigma-Aldrich), and protein was purified using a HiTrap Chelating HP column. The protein was then dialyzed using a Thermo Slide-A-Lyzer Cassettee 10000 MWCO in phosphate-buffered saline, and purity was verified by SDS-PAGE and mass spectrometry. Purified RagB was then digested with EKMax and washed in EK-Away resin (Invitrogen) to remove the thioredoxin tag. Selenomethione-derivatized protein was obtained in the same way. Finally, each protein pool was concentrated by ultrafiltration and subjected to size-exclusion chromatography (SEC) in a Superdex 200, 10/300 column (GE Healthcare Life Sciences) equilibrated with 20mM Tris-HCl, 150mM sodium chloride, pH7.5.
Protein identity and purity were assessed by 15% Tricine-SDS-PAGE (Schägger, 2006), peptide-mass fingerprinting of tryptic protein digests, N-terminal sequencing through Edman degradation and mass spectrometry. The latter three were carried out at the Protein Chemistry and Proteomics facilities of Centro de Investigaciones Biológicas (Madrid, Spain). Ultrafiltration steps were performed with Vivaspin 15 and Vivaspin 500 filter devices with a 10-kDa cut-off (Sartorius Stedim Biotech). Protein concentrations were calculated by measuring A280 in a spectrophotometer (NanoDrop) and applying the theoretical extinction coefficients. Concentrations were also measured by the BCA Protein Assay Kit (Thermo Scientific) with bovine serum albumin as a standard. The native molecular mass of the purified protein was determined by size exclusion chromatography on a Superdex 200 PG 16/60 column (GE Healthcare Ltd, UK). The molecular mass markers used for calibration of the column were thyroglobulin (669kDa), apoferritin (440kDa), aldolase (156kDa), conalbumin (75kDa), ovalbumin (44kDa), carbonic anhydrase (29kDa) and cytochrome (13.7kDa). Elution was performed with 50mM sodium phosphate, 150mM sodium chloride, pH 7.2 at a flow rate of 1.5 mL/min.
Crystallization and diffraction data collection
Crystallization assays were performed by the sitting-drop vapor diffusion method. Reservoir solutions were prepared by a Tecan robot and 100nL crystallization drops were dispensed on 96×2-well MRC plates (Innovadyne) by a Phoenix nanodrop robot (Art Robbins) or a Cartesian Microsys 4000 XL robot (Genomic Solutions) at the joint IBMB/IRB Automated Crystallography Platform of Barcelona Science Park. Plates were stored in Bruker steady-temperature crystal farms at 4°C or 20°C. Successful conditions were scaled up to the microliter range in 24-well Cryschem crystallization dishes (Hampton Research). The best crystals were obtained at 4°C in drops containing 1μL protein solution (at 20mg/mL in 20mM Tris-HCl pH7.4, 50mM sodium chloride) and 1μL reservoir solution (100mM 2-(N-morpholino)ethanesulfonic acid, pH6.0, 200mM calcium acetate, 20% [w/v] polyethylene glycol 10,000). Selenomethionine-containing crystals were obtained under the same conditions. Crystals were cryo-protected by rapid passage through drops containing increasing amounts of glycerol (up to 20% [v/v]). Complete diffraction datasets were collected at 100K from liquid-N2 flash cryo-cooled crystals (Oxford Cryosystems 700 series cryostream) on a Pilatus 6M pixel detector (from Dectris) at beam line XALOC of ALBA synchrotron (Cerdanyola, Barcelona; (Juanhuix et al., 2014)). Crystals were primitive orthorhombic, contained fragment D30–I501 as determined by Edman degradation and MALDI-TOF analysis, and comprised two protein molecules per asymmetric unit (solvent content 65%; VM=3.5Å3/Da, (Matthews, 1968); calculated for the best selenomethionine-containing crystal, see Table 1). Diffraction data were processed with programs XDS (Kabsch, 2010b) and XSCALE (Kabsch, 2010a), and transformed with XDSCONV to formats suitable for SHELX (Sheldrick, 2010; Sheldrick, 2011) and the CCP4 suite of programs (Winn et al., 2011)(see Table 1 for data processing statistics).
Table 1.
Crystallographic data.
| Dataset | PgRagB (Se absorption peak | PgRagB (Se absorption peak; merged Friedel mates) |
|---|---|---|
| Space group | P21212 | P21212 |
| Cell constants (a, b, c, in Å) | 56.6, 184.7, 144.3 | 56.6, 184.7, 144.3 |
| Wavelength (Å) | 0.9793 | 0.9793 |
| No. of measurements/unique reflections | 864,963/129,496 | 790,705/60,274 |
| Resolution range (Å) (outermost shell)a | 48.3 – 2.30 (2.44 – 2.30)f | 48.3 – 2.40 (2.46 – 2.40) |
| Completeness (%) | 99.6 (97.7) | 100 (100) |
| Rmergeb | 0.155 (1.288) | 0.155 (1.118) |
| Rr.i.m. [= Rmeas] c/CC(1/2)c | 0.168 (1.436)/0.996 (0.641) | 0.161 (1.169)/0.998 (0.890) |
| Average intensityd | 11.2 (1.4) | 15.4 (2.7) |
| B-Factor (Wilson) (Å2)/Aver. Multiplicity | 44.9/6.7 (5.1) | 44.5/13.1 (11.7) |
| Number of heavy-atom sites for phasing | 12 | |
| Resolution range used for refinement (Å) | 35.1 – 2.40 | |
| No. of reflections used (test set) | 60,258 (881) | |
| Crystallographic Rfactor (free Rfactor)b | 0.178 (0.208) | |
| No. of protein atoms/solvent molecules/neutral ligands/ionic ligands | 7,528/509/3 monosaccharides, 3 glycerols 1 acetate | |
| Rmsd from target values e | ||
| bonds (Å)/angles (°) | 0.010/1.03 | |
| Average B-factors (Å2) | 56.0 | |
| All-atom contacts and geometry analysise | ||
| Residues | ||
| in favored regions/outliers/all residues | 912 (97.0%)/0/940 | |
| with poor rotamers/bad bonds/bad angles | 15 (1.9%)/0/0 | |
| with Cβ deviations >0.25Å/clashscore | 0/3.88 (99th percentile) | |
| MolProbity score | 1.56 (99th percentile) | |
Data processing values in parenthesis refer to the outermost resolution shell.
For definitions, see Table 1 in (García-Castellanos et al., 2003).
For definitions, see (Karplus & Diederichs, 2012; Weiss, 2001).
Average intensity is <I/σ (I)> of unique reflections after merging according to the XDS program (Kabsch, 2010b).
According to MOLPROBITY (Chen et al., 2010; Davis et al., 2007).
For phasing, which included the free-lunch algorithm based on data extrapolation to very high resolution as implemented in program SHELXE, the data were processed to slightly higher resolution than for crystallographic model refinement.
Structure solution and refinement
The structure of PgRagB was solved by single-wavelength anomalous diffraction with data collected at the selenium absorption peak wavelength of a selenomethionine-derivatized crystal as any attempt to solve the structure by maximum-likelihood-scored molecular replacement or Patterson search failed. Data processed with separate Friedel mates served to identify all 12 selenium sites present based on anomalous differences with program SHELXD (Sheldrick, 2010). Subsequent phasing with SHELXE (Sheldrick, 2010; Sheldrick, 2011), which included application of the free-lunch algorithm, enabled distinction between the two possible hands based on the pseudo-free correlation coefficient, and yielded phases with an overall figure-of-merit of 66%. Subsequent density modification and automatic model building with the ARP/warp suite (Langer et al., 2008), which included calculations performed with program REFMAC5 (Murshudov et al., 2011), yielded a strongly improved map and a partial model. The latter was completed through subsequent manual model building with the COOT program (Emsley et al., 2010), which alternated with crystallographic refinement against the Friedel-merged selenomethionine-derivative diffraction data with programs PHENIX (Afonine et al., 2012) and BUSTER/TNT (Smart et al., 2012) under inclusion of TLS refinement, until the final refined model of PgRagB was obtained. This consisted of residues D30–I501 of either molecule A and B. In addition, three regions on the surface evinced clear extra density, which—after a number of different trials—were tentatively interpreted as three monosaccharide ligands based on best fitting to the experimental density: 3′-deoxy-D-ribofuranose-5′-phosphate (residue identifier RP3), 3′-deoxy-β-D-glucopyranose (3DO), and D-tagatose-6′-phosphate in its open keto form (TG6). These molecules, together with three glycerols, one acetate, and 509 solvent molecules, completed the structure. See Table 1 for final refinement and model quality statistics.
Miscellaneous
Ideal coordinates and parameters for crystallographic refinement of nonstandard ligands were obtained from the PRODRG server (Schüttelkopf & van Aalten, 2004). Structural similarity searches were performed with DALI (Holm & Rosenström, 2010), and figures were prepared with programs COOT and CHIMERA (Pettersen et al., 2004). The sequence of PgRagB was analyzed with the TPRpred program (http://toolkit.tuebingen.mpg.de/tprpred; (Karpenahalli et al., 2007)) for the presence of tetratrico peptide repeats. The final structure was validated with MOLPROBITY (Chen et al., 2010). The final coordinates of PgRagB are available from the Protein Data Bank (PDB) at www.pdb.org (access code 5CX8).
RESULTS AND DISCUSSION
Molecular structure of PgRagB
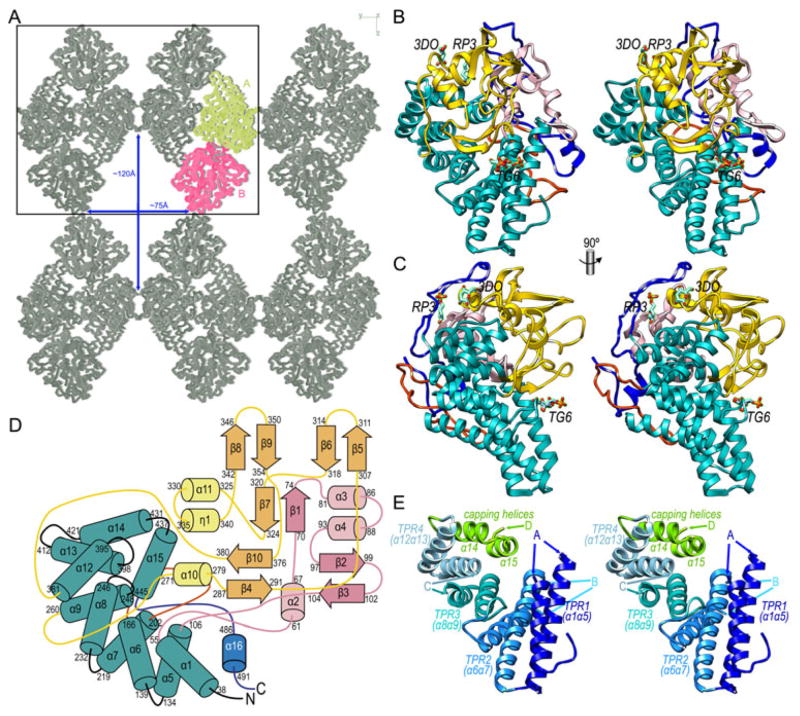
Full-length PgRagB includes a signal peptide for export to the outer membrane and a cysteine (C20) that is lipidated and anchors the protein to the cell membrane. Accordingly, to obtain a soluble variant of the protein, recombinant production in E. coli just included segment E21–I501 plus an extra threonine at the N-terminus replacing the cysteine owing to the cloning strategy (Hutcherson et al., 2015). The X-ray crystal structure of this 54-kDa PgRagB variant was solved by single-wavelength anomalous diffraction and refined with data from selenomethionine-derivatized protein, as crystals of the latter diffracted to higher resolution (the best to 2.4 Å; Table 1) than native crystals. The final 3D structure, which showed good geometry and conformational parameters, had two monomers (A and B) in the asymmetric unit, each spanning residues D30–I501, i.e. the first ten residues were missing in both molecules as confirmed by N-terminal Edman degradation. In addition, the crystal packing revealed long solvent channels along cell axis a, with the channel section spanning ~120Å in one direction and ~75Å in the perpendicular one, thus accounting for large solvent content of the crystals (65%; (Fig. 1A).
Figure 1. Molecular structure of P. gingivalis RagB.
(A) Crystal packing of PgRagB in Cα traces viewed along cell axis a. The two molecules in the asymmetric unit, A and B, are shown in yellow and magenta, respectively, symmetry equivalents are in grey. The box delineates the bc plane of the unit cell. Large solvent channels—of maximal dimensions ~75Å and ~120Å traverse the crystal along cell axis a. (B) Ribbon-type plot of PgRagB in cross-eye stereo, showing the TPRs in turquoise, insert A in light pink, insert B in red, insert C in gold, and the C-terminal insert D in dark blue. The three tentative sugar moieties—labeled RP,3 3DO and TG6—are depicted as stick models with carbons in turquoise. (C) Orthogonal view of (B). (D) Topology scheme of PgRagB depicting helices as rods (labeled α1-α16 and η1) and strands as arrows (β1-β10), the numbers of the delimiting residues are shown in each case. Color code similar to (B). (E) Cartoon in stereo depicting only the four tetratrico repeats (TPR1-4) of PgRagB and the capping helices (α14+α15), each shown in one color and labeled. The points of insertion of inserts A, B, C, and D are shown by arrows. Orientation as in (B).
PgRagB is a single-domain compact molecule of maximal approximate dimensions 70×55×50Å (Fig. 1B, C). It comprises a scaffold consisting of ten large α-helices (α1, α5-α9, α12-α15; Fig. 1D, E), the first eight giving rise to four helix-turn-helix repeats or α-hairpins (α1α5, α6α7, α8α9, and α12α13), sequentially arranged as a right-handed solenoid of helices (Fig. 1E). This superhelix is curved and succeeded by a two-helical tandem (α14+α15), which is folded toward the concave surface of the solenoid and nestles on the inner surface of hairpin α12α13, so that α14 is antiparallel to α12. This scaffold cradles large insertions in its concave surface, which include short helices and strands participating in β-ribbons or β-hairpins but not in higher-order β-sheets (Fig. 1D). The turn connecting the helices of the first α-hairpin (“insert A” in Fig. 1B–D) spans 50 residues (E56–G105) and includes three helices (α2-α4), a β-hairpin (β2β3) and one of the two strands of a β-ribbon (β1). The turn of the second α-hairpin is replaced by a 35-residue irregular double loop (“insert B”; F167–T201), which shapes the right back surface of the molecule and interacts with the N-terminus and helix α5, mainly through residue Y191. The segment connecting the third and fourth α-hairpins (“insert C”), in turn, spans 120 residues (D261–S380) and includes two α-helices (α10 and α11), a 310-helix (η1), a β-ribbon (β4β10), two β-hairpins (β5β6 and β8β9), and the second strand of the aforementioned β-ribbon involving β1, viz. β7 (Fig. 1D). Finally, downstream of the last helix of the helical scaffold (α15), the last 56 residues (“insert D”; S446–I501) are mainly irregular except for a short C-terminal helix, α16. Of these residues, the first ~30 are arranged in an extended loop that forms the top of the molecule and covers inserts A and C, and the final ~25 residues shape another loop that nestles on the concave surface of insert A.
PgRagB is a tetratrico peptide repeat protein
Tetratrico peptide repeat (TPR) proteins (TPRPs) were identified and named in 1990 in the absence of experimental 3D structures, based on a characteristic 34-residue motif, which was predicted to span two α-helices (Hirano et al., 1990; Sikorski et al., 1990). Initially, TPRs showed the consensus sequence Wn-Ln+4-Gn+5-Yn+7 in the first helix and Am-Fm+4-Am+7-Pm+12 in the second helix (D’Andrea & Regan, 2003). The first TPRP structure to be solved was that of protein phosphatase 5, which confirmed the bihelical architecture of TPRs (Das et al., 1998). Three such elements were found in the structure, each arranged in two antiparallel amphiphilic helices packing against each other at an angle of ~25° and linked by a turn, thus giving rise to an overall right-handed spiral of TPRs (D’Andrea & Regan, 2003). An extra helix was found C-terminally of the spiral and was dubbed “capping helix.” Subsequent TPRP structures indicated that additional secondary structure elements and loop segments may be inserted into such a basic scaffold of 3–4 concatenated TPRs (Cerveny et al., 2013; D’Andrea & Regan, 2003). In addition, the TPR sequence motifs became rather degenerate and variable, so positions were not generally conserved with the exception of Gn+5, Am and Am+7.
The arrangement of the four α-hairpins of PgRagB conforms to this TPR architecture and, thus, they constitute TPR1-to-4 (Fig. 1D, E). The angles at which the helices within a repeat cross over each other are canonical (~25°) in TPR2, 3 and 4 but close to zero within the first repeat, for here the helices are roughly antiparallel. The function of the capping helix in protein phosphatase 5 is exerted in PgRagB by helices α14+α15 (see previous section and Fig. 1E). Accordingly, PgRagB fulfills the structural requisites of TPRPs. In contrast, inspection of the PgRagB sequence revealed that even the minimal TPR consensus was only barely attained thanks to the presence of small residues at the points of helix intersection. This was attributable—at least partially—to the large inserts within and between TPRs (see above and Fig. 1B–E), which distort the minimal canonical structure. Consistently, a search within the PgRagB sequence using the TPRpred program revealed a probability of just 0.5% for PgRagB being a TPRP based on its sequence and solely identified TPR3 (P-value=3.6E-6). This program predicts pentatrico peptide repeats; 36–44 peptidic repeats as found in Caenorhabditis elegans suppressor-enhacer of lin gene 1; and TPRs in protein sequences (Karpenahalli et al., 2007).
Structural similarity points to potential function
To date, +20,000 TPRPs across all kingdoms of life have been identified or predicted. The evolutionary conservation of the TPR structure suggests relevance for living organisms, probably in mediating protein-protein interactions in a variety of cellular functions. Concatenated TPR backbones can be envisaged as a consensus scaffold, onto which specific ligand-binding residues are grafted thus furnishing distinct functions (Cerveny et al., 2013). Within prokaryotes, TPRPs have been directly related to sugar binding in commensal bacteria and to virulence of bacterial pathogens. Examples of the latter include molecules participating in pilus formation in Francisella tularensis, which is required for adhesion to the host (Chakraborty et al., 2008); in the inhibition of degradation of Mycobacterium tuberculosis in host phagolysosomes (Chao et al., 2010); and in class-II-chaperone-mediated translocation of virulence factors into host cells as shown for Pseudomonas aeruginosa (Bröms et al., 2006; Cerveny et al., 2013). As to P. gingivalis, TprA is apparently engaged in the transduction of stress signals from the environment, which regulates expression of proteins participating in secretion (Kondo et al., 2010).
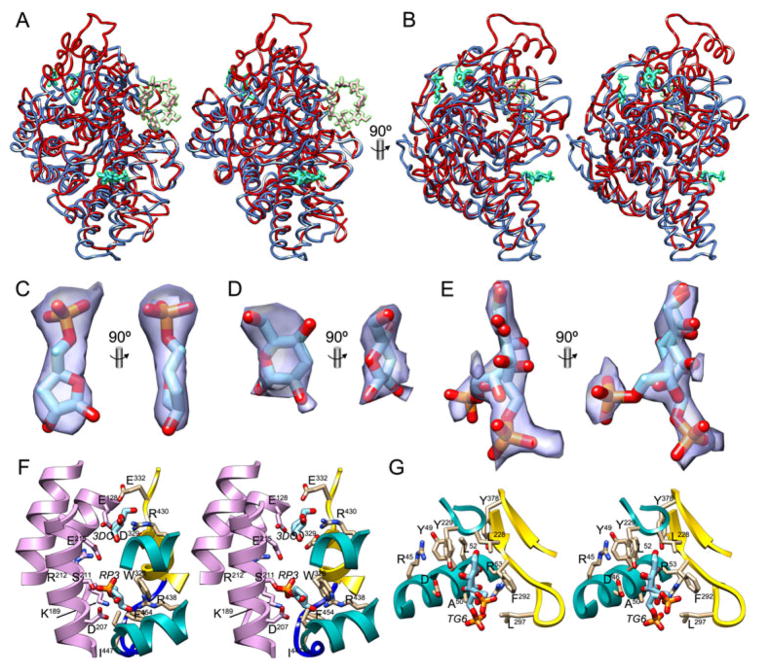
As could be anticipated from the similarity of the B. thetaiotaomicron susC-susD operons with the P. gingivalis ragA-ragB locus, 3D-structure similarity searches with PgRagB identified members of the SusD family of bacterial molecules as close relatives, with Z-score values spanning 37.1–17.2, rmsd values of 2.5–3.5Å, 418–332 aligned residues, but sequence identities of just 22–11%. Most of these structures have been deposited with the PDB but not published (PDB codes 3EJN, 3FDH, 3GZS, 3HDX, 3IHV, 3IH0, 3I4G, 3JQ0, 3JQ1, 3JYS, 3KEZ, 3L22, 3LEW, 3MCX, 3MX3, 3MYV, 3NQP, 3OTN, 3PLU, 3QNK, 3SGH, 3SNX, 4F53, 4F7A, 4G69, 4LER, and 4PUC). The only published structures are those of Bacteroides fragilis NanU (BfNanU; PDB 4L7T; (Phansopa et al., 2014)), and BtSusD (BtSusD; PDB 3CK7-3CK9, 3CKB, and 3CKC; (Koropatkin et al., 2008)) and SusD-like proteins BT3984 (3CGH; (Bakolitsa et al., 2010)) and BT1043 (PDB 3EHM, 3EHN; (Koropatkin et al., 2009)) from B. thetaiotaomicron. All these sugar-binding proteins are collectively polyhelical TPRPs, and thus significantly deviate from the predominantly β-type carbohydrate binding moieties of cellulases and other glycoside hydrolases and lectin proteins (Hashimoto, 2006). Superposition of PgRagB onto BtSusD (Fig. 2A, B), BfNanU, BT1043, and BT3984 reveals good general agreement of the helical scaffolds, both in length and position of the ten helices found in PgRagB, suggesting that this scaffold is conserved and required for domain integrity in these proteins. However, detailed inspection of the connecting loops and inserts shows substantial differences among these proteins, which explains why SusD-family proteins generally share only low sequence identity.
Figure 2. Structural similarities and possible ligands.
(A) Superposition in cross-eye stereo of PgRagB (blue ribbon, tentative sugars in turquoise) and BtSusD in its complex with maltoheptaose (red ribbon, sugar moieties in pink; PDB 3CK9; (Koropatkin et al., 2008)). Orientation as in Fig. 1B. (B) Orthogonal view of (A). (C) (2mFobs—DFcalc)-type Fourier omit map contoured at 1σ displayed as a purple semi-transparent surface in two orthogonal views around the final refined stick model of tentative ligand RP3 (carbons in light blue). (D) Same as (C) for tentative ligand 3DO. (E) Same as (C) for tentative ligand TG6, modeled as an open keto form with double conformation. (F) Close-up view in stereo of the binding sites of tentative ligands RP3 and 3DO, shown as stick models with carbons in light blue. Molecule A is shown in the colors of Fig. 1B, molecule B in light purple. Side chains participating in ligand binding are shown for their side chains (molecule A, carbons in tan; molecule B, carbons in light purple). (G) Same as (F) for the tentative TG6-binding site.
While in most TPRPs described ligand or protein binding occurs on the concave solenoid surface (Cerveny et al., 2013), in BtSusD and BT1043, the only SusD-family members that have been reported in complexes with glycan ligands (Koropatkin et al., 2008; 2009), it is the inserts that shape the glycan-binding sites. These are topologically equivalent in both proteins, located on the right-hand side surface of the molecules, and shaped by the respective inserts A, B and C. However, the inserts significantly deviate in sequence and trajectory, which contributes to BtSusD having a rather flexible binding site which facilitates binding to (predominantly cyclic) maltooligosaccharic molecules through recognition of the overall three-dimensional shape rather than the particular composite monosaccharides (Anderson & Salyers, 1989a; Koropatkin et al., 2008); in contrast, BT1403 has been crystallized with a disaccharide only (see Figs. 1 and 3 in (Koropatkin et al., 2009)).
Detailed comparison between PgRagB and BtSusD (Fig. 2A, B), which share an overall sequence identity of only 21%, reveals that insert A, while retaining the overall length of PgRagB, significantly deviates in BtSusD. It has an extra ~20-residue loop grafted onto the short linker found between α1 and α2 in RagB, and the region preceding α3 also deviates strongly. Immediately preceding helix α5, PgRagB shows β-ribbon β2β3, which is absent from BtSusD and partially occupies the space of the aforementioned ~20-residue loop of the latter. Next, insert B, while maintaining the general irregular shape in both proteins, is approximately half as long in BtSusD than in PgRagB (20 vs. 36 residues) and just comprises the second loop. Third, insert C, which contains many short regular structural elements in PgRagB (see above and Fig. 1D), displays completely different chain traces in the two proteins. In particular, BtSusD has a 44-residue insertion at the short linker connecting α11 and η1 in PgRagB. It features two extra helices and shapes the top of BtSusD, thus causing the latter moiety to be longer than the periodontal protein (Fig. 2A, B). Finally, insert D is similar in both proteins except for the central part of the first extended loop (see above), which is folded inward in BtSusD to accommodate the long extra part in insert C. Taken together, the strong general structural similarity of PgRagB with SusD-family proteins points to a similar function as a sugar-binding protein (see also next section). However, major differences in inserts A, B and C indicate that the BtSusD glycan-binding site is absent from PgRagB. This is reminiscent of BfNanU, for which likewise significant differences in the regions shaping the sugar-binding site in SusD entail that the sialic-acid binding site is currently unknown (Phansopa et al., 2014).
Potential sugar-binding pockets on the RagB surface
The Fourier-map of the PgRagB structure revealed that the protein had been serendipitously co-purified with three small-molecule ligands, possibly metabolites originating in the expression host (see Fig. 2C–E). By analogy with the other SusD-family members, PgRagB could function as a sugar-binding protein, and, hence, these densities were tentatively interpreted—after a number of different trials and based on best fitting to the Fourier omit maps—as three monosaccharides: 3′-deoxy-D-ribofuranose-5′-phosphate (residue identifier RP3), 3′-deoxy-β-D-glucopyranose (3DO), and D-tagatose-6′-phosphate in its open keto form (TG6). The latter was found in two conformations. Ligand RP3 is bound at the interface between the two molecules in the asymmetric unit (Fig. 2F). Participating residues include the side chains of D207, Y208, S211, R212 and E215 from helix α7 of molecule B; and D69 from loop Lα2β1, W326 from Lβ7α11, R438 from α15 and F454 from Lα15α16 of molecule A. Ligand 3DO is very close to RP3 (minimal distance 7.4Å) and bound by E128 from α5, K153 from α6, and E215 from α7 of molecule B; and the main-chain of segment D329–E332 plus the side chain of E332 from α11 and Lα11η1 of molecule A. Given the proximity of the two sites, it cannot be ruled out that they correspond to a single site for oligosaccharides occupied only for its two terminal sub-sites. Size-exclusion chromatography analysis calibrated with molecular mass standards indicated that the protein probably forms dimers, with an estimated molecular mass of 83kDa, which would result from somewhat slower migration than expectable (the theoretical molecular mass is 108.5kDa). This would be in agreement with the dimeric structure observed in the crystal, which is required to shape the binding sites for RP3 and 3DO. Finally, ligand TG6 was found only on the surface of molecule A, in the TPR region, bound by the main chain and side chains of segment D46–R53 from helix α1, L228–Y229 from Lα7α8, and R290–F292 plus L297 from β4 and Lβ4β5 (Fig. 2G). This site coincides with the presence of an ethylene glycol molecule in the structure of BtSusD in complex with maltoheptaose (PDB 3CK9; (Koropatkin et al., 2008)) and with a glycerol molecule in an uncharacterized SusD homolog from Bacteroides vulgatus (PDB 3JQ0). These PgRagB sites differ from those of BtSusD and BT1043 (Fig. 2A, B), in line with suggestions for BfNanU (Phansopa et al., 2014).
Concluding remarks and outlook
The present structural data document high structural similarity of PgRagB with BtSusD and indicate that P. gingivalis, a Gram-negative bacterium from the Bacteroidetes phylum, may have acquired a minimal Sus-unit—constituted by the SusC- and SusD-analogs PgRagA and PgRagB, respectively—from commensal Bacteroides spp by horizontal gene transfer. This acquisition had been suggested previously, on the basis of the lower G+C ratio found for the rag pathogenicity island compared to the rest of the genome, which is usually associated with mobility elements (Hall et al., 2005; Hanley et al., 1999); and on evolutionary studies (Su et al., 2010). This is reminiscent of the acquisition pathway postulated for NanO/NanU by red-complex-partner T. forsythia for sialic acid import (Phansopa et al., 2014).
This structural similarity would suggest a role for PgRagB in the subgingival colonization of P. gingivalis through the binding of glycans. However, this binding would occur at pockets on the molecular surface that are distinct from those of BtSusD. These glycans would thereafter be internalized by PgRagA—following its similarity with BtSusC and TfNanO. Despite the classification of P. gingivalis as an asaccharolytic organism (van Steenbergen et al., 1993), sugar uptake is required for capsule generation and other non-ATP generating metabolic processes (Hutcherson et al., 2015). In addition, it was anticipated over 15 years ago that a possible PgRagA/PgRagB-mediated carbohydrate uptake would be linked to an anabolic process (Hanley et al., 1999). On the other hand, an equally plausible explanation for sugar uptake in P. gingivalis would be a source for its own glycosylation: a number of P. gingivalis surface proteins are glycosylated, such as the minor fimbria protein (Zeituni et al., 2010), the OmpA-like heterotrimeric complex formed by Pgm6 and Pgm7 (Murakami et al., 2014), protein Omp85 (Nakao et al., 2008), hemin-binding protein 35 (Shoji et al., 2011), peptidylargine deiminase (Glew et al., 2014; Goulas et al., 2015), and gingipains (Curtis et al., 1999b). They can contain up to 30% carbohydrates by weight (Gallagher et al., 2003). In this context, P. gingivalis encodes over 50 members of families of glycoside hydrolases, glycoside transferases, carbohydrate esterases and proteins containing carbohydrate-binding modules (see www.cazy.org/b161.html). Five mannosidases have been characterized and functionally validated (Rangarajan et al., 2013), as was an outer-membrane associated N-acetyl-β-D-glucosaminidase (Lovatt & Roberts, 1994). Compositional analyses of some of the aforementioned P. gingivalis glycoproteins have indicated an extensive repertoire of monosaccharides incorporated into glycan additions, and some of these might be derived exogenously ((Gallagher et al., 2003) and M.A. Curtis, personal communication). In this sense, red-complex-partner T. forsythia and further odontopathogen Prevotella intermedia can ferment carbohydrates (Hughes et al., 2003; Stubbs et al., 1996), and to possess surface proteins to bind and translocate sugar moieties may be a strategy of P. gingivalis to use resources generated by others.
Taken together, these results support the existance of an environmental sugar-uptake pathway in P. gingivalis but further experiments will be required to verify the sugar-binding capacity and translocation of the PgRagA/PgRagB axis.
Acknowledgments
We are grateful to Joan Pous and Xandra Kreplin from the joint IBMB/IRB Automated Crystallography Platform for assistance during crystallization experiments and to Graham Stafford, Mike Curtis, and Robin Rycroft for very helpful contributions to the manuscript. We further acknowledge the help provided by local contacts at the ESRF and ALBA synchrotrons. This study was funded in part by grants from US American, European, Polish, Spanish, and Catalan agencies (NIH NIDCR DE019826; UMO-2012/04/A/NZ1/00051, 137/7.PR/13/2014/2, NIH NIDCR DE09761; FP7-PEOPLE-2011-ITN-290246 “RAPID”; FP7-HEALTH-2012-306029-2 “TRIGGER”; BFU2012-32862; BIO2013-49320-EXP; MDM-2014-0435; and 2014SGR9). IGF acknowledges an FPU-fellowship (AP2010-3799) from the former Spanish Ministry for Education, Culture and Sport; and TG acknowledges a “Juan de la Cierva” research contract (JCI-2012-13573) from the Spanish Ministry for Economy and Competitiveness. The Department of Structural Biology of IBMB is a “María de Maeztu” Unit of Excellence from the Ministry of Economy and Competitiveness. Funding for diffraction data collection was provided in part by ESRF. The Faculty of Biochemistry, Biophysics and Biotechnology of the Jagiellonian University at Kraków (Poland) is a partner of the Leading National Research Center (KNOW) supported by the Polish Ministry of Science and Higher Education.
References
- Afonine PV, Grosse-Kunstleve RW, Echols N, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr sect D. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson KL, Salyers AA. Genetic evidence that outer membrane binding of starch is required for starch utilization by Bacteroides thetaiotaomicron. J Bacteriol. 1989a;171:3199–3204. doi: 10.1128/jb.171.6.3199-3204.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson KL, Salyers AA. Biochemical evidence that starch breakdown by Bacteroides thetaiotaomicron involves outer membrane starch-binding sites and periplasmic starch-degrading enzymes. J Bacteriol. 1989b;171:3192–3198. doi: 10.1128/jb.171.6.3192-3198.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagaitkar J, Williams LR, Renaud DE, et al. Tobacco-induced alterations to Porphyromonas gingivalis-host interactions. Environ Microbiol. 2009;11:1242–1253. doi: 10.1111/j.1462-2920.2008.01852.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakolitsa C, Xu Q, Rife CL, et al. Structure of BT_3984, a member of the SusD/RagB family of nutrient-binding molecules. Acta Crystallogr sect F. 2010;66:1274–1280. doi: 10.1107/S1744309110032999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonass WA, Marsh PD, Percival RS, et al. Identification of ragAB as a temperature-regulated operon of Porphyromonas gingivalis W50 using differential display of randomly primed RNA. Infect Immun. 2000;68:4012–4027. doi: 10.1128/iai.68.7.4012-4017.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bröms JE, Edqvist PJ, Forsberg A, Francis MS. Tetratricopeptide repeats are essential for PcrH chaperone function in Pseudomonas aeruginosa type III secretion. FEMS Microbiol Lett. 2006;256:57–66. doi: 10.1111/j.1574-6968.2005.00099.x. [DOI] [PubMed] [Google Scholar]
- Cerveny L, Straskova A, Dankova V, et al. Tetratricopeptide repeat motifs in the world of bacterial pathogens: role in virulence mechanisms. Infect Immun. 2013;81:629–635. doi: 10.1128/IAI.01035-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty S, Monfett M, Maier TM, Benach JL, Frank DW, Thanassi DG. Type IV pili in Francisella tularensis: roles of pilF and pilT in fiber assembly, host cell adherence, and virulence. Infect Immun. 2008;76:2852–2861. doi: 10.1128/IAI.01726-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao J, Wong D, Zheng X, et al. Protein kinase and phosphatase signaling in Mycobacterium tuberculosis physiology and pathogenesis. Biochim Biophys Acta. 2010;1804:620–627. doi: 10.1016/j.bbapap.2009.09.008. [DOI] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, 3rd, Headd JJ, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr sect D. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho KH, Salyers AA. Biochemical analysis of interactions between outer membrane proteins that contribute to starch utilization by Bacteroides thetaiotaomicron. J Bacteriol. 2001;183:7224–7230. doi: 10.1128/JB.183.24.7224-7230.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MA, Slaney JM, Carman RJ, Johnson NW. Identification of the major surface protein antigens of Porphyromonas gingivalis using IgG antibody reactivity of periodontal case-control serum. Oral Microbiol Immunol. 1991;6:321–326. doi: 10.1111/j.1399-302x.1991.tb00502.x. [DOI] [PubMed] [Google Scholar]
- Curtis MA, Hanley SA, Aduse-Opoku J. The rag locus of Porphyromonas gingivalis: a novel pathogenicity island. J Periodontal Res. 1999a;34:400–405. doi: 10.1111/j.1600-0765.1999.tb02273.x. [DOI] [PubMed] [Google Scholar]
- Curtis MA, Thickett A, Slaney JM, et al. Variable carbohydrate modifications to the catalytic chains of the RgpA and RgpB proteases of Porphyromonas gingivalis W50. Infect Immun. 1999b;67:3816–3823. doi: 10.1128/iai.67.8.3816-3823.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Andrea LD, Regan L. TPR proteins: the versatile helix. Trends Biochem Sci. 2003;28:655–662. doi: 10.1016/j.tibs.2003.10.007. [DOI] [PubMed] [Google Scholar]
- Das AK, Cohen PW, Barford D. The structure of the tetratricopeptide repeats of protein phosphatase 5: implications for TPR-mediated protein-protein interactions. EMBO J. 1998;17:1192–1199. doi: 10.1093/emboj/17.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis IW, Leaver-Fay A, Chen VB, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolgilevich S, Rafferty B, Luchinskaya D, Kozarov E. Genomic comparison of invasive and rare non-invasive strains reveals Porphyromonas gingivalis genetic polymorphisms. J Oral Microbiol. 2011;3:5764. doi: 10.3402/jom.v3i0.5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dormitzer PR, Grandi G, Rappuoli R. Structural vaccinology starts to deliver. Nat Rev Microbiol. 2012;10:807–813. doi: 10.1038/nrmicro2893. [DOI] [PubMed] [Google Scholar]
- Eckburg PB, Bik EM, Bernstein CN, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr sect D. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher A, Aduse-Opoku J, Rangarajan M, Slaney JM, Curtis MA. Glycosylation of the Arg-gingipains of Porphyromonas gingivalis and comparison with glycoconjugate structure and synthesis in other bacteria. Curr Protein Pept Sci. 2003;4:427–441. doi: 10.2174/1389203033486974. [DOI] [PubMed] [Google Scholar]
- García-Castellanos R, Marrero A, Mallorquí-Fernández G, Potempa J, Coll M, Gomis-Rüth FX. Three-dimensional structure of MecI : Molecular basis for transcriptional regulation of staphylococcal methicillin resistance. J Biol Chem. 2003;278:39897–39905. doi: 10.1074/jbc.M307199200. [DOI] [PubMed] [Google Scholar]
- Glew MD, Veith PD, Chen D, Seers CA, Chen Y, Reynolds EC. Blue native-PAGE analysis of membrane protein complexes in Porphyromonas gingivalis. J Proteomics. 2014;110:72–92. doi: 10.1016/j.jprot.2014.07.033. [DOI] [PubMed] [Google Scholar]
- Goulas T, Mizgalska D, Garcia-Ferrer I, et al. Structure and mechanism of a bacterial host-protein citrullinating virulence factor, Porphyromonas gingivalis peptidylarginine deiminase. Sci Rep. 2015;5:11969. doi: 10.1038/srep11969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G. Periodontitis: from microbial immune subversion to systemic inflammation. Nat Rev Immunol. 2015;15:30–44. doi: 10.1038/nri3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall LM, Fawell SC, Shi X, et al. Sequence diversity and antigenic variation at the rag locus of Porphyromonas gingivalis. Infect Immun. 2005;73:4253–4262. doi: 10.1128/IAI.73.7.4253-4262.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanley SA, Aduse-Opoku J, Curtis MA. A 55-kilodalton immunodominant antigen of Porphyromonas gingivalis W50 has arisen via horizontal gene transfer. Infect Immun. 1999;67:1157–1171. doi: 10.1128/iai.67.3.1157-1171.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H. Recent structural studies of carbohydrate-binding modules. Cell Mol Life Sci. 2006;63:2954–2967. doi: 10.1007/s00018-006-6195-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T, Kinoshita N, Morikawa K, Yanagida M. Snap helix with knob and hole: essential repeats in S. pombe nuclear protein nuc2+ Cell. 1990;60:319–328. doi: 10.1016/0092-8674(90)90746-2. [DOI] [PubMed] [Google Scholar]
- Holm L, Rosenström P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes CV, Malki G, Loo CY, Tanner AC, Ganeshkumar N. Cloning and expression of α-D-glucosidase and N-acetyl-β-glucosaminidase from the periodontal pathogen, Tannerella forsythensis (Bacteroides forsythus) Oral Microbiol Immunol. 2003;18:309–312. doi: 10.1034/j.1399-302x.2003.00091.x. [DOI] [PubMed] [Google Scholar]
- Hutcherson JA, Bagaitkar J, Nagano K, Yoshimura F, Wang H, Scott DA. Porphyromonas gingivalis RagB is a proinflammatory signal transducer and activator of transcription 4 agonist. Mol Oral Microbiol. 2015;30:242–252. doi: 10.1111/omi.12089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai M, Murakami Y, Nagano K, Nakamura H, Yoshimura F. Major outer membrane proteins from Porphyromonas gingivalis: strain variation, distribution, and clinical significance in periradicular lesions. Eur J Oral Sci. 2005;113:391–399. doi: 10.1111/j.1600-0722.2005.00235.x. [DOI] [PubMed] [Google Scholar]
- Juanhuix J, Gil-Ortiz F, Cuni G, et al. Developments in optics and performance at BL13-XALOC, the macromolecular crystallography beamline at the ALBA synchrotron. J Synchrotron Radiat. 2014;21:679–689. doi: 10.1107/S160057751400825X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr sect D. 2010a;66:133–144. doi: 10.1107/S0907444909047374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. XDS. Acta Crystallogr sect D. 2010b;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpenahalli MR, Lupas AN, Soding J. TPRpred: a tool for prediction of TPR-, PPR- and SEL1-like repeats from protein sequences. BMC Bioinformatics. 2007;8:2. doi: 10.1186/1471-2105-8-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karplus PA, Diederichs K. Linking crystallographic model and data quality. Science. 2012;336:1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein BA, Tenorio EL, Lazinski DW, Camilli A, Duncan MJ, Hu LT. Identification of essential genes of the periodontal pathogen Porphyromonas gingivalis. BMC Genomics. 2012;13:578. doi: 10.1186/1471-2164-13-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo Y, Ohara N, Sato K, et al. Tetratricopeptide repeat protein-associated proteins contribute to the virulence of Porphyromonas gingivalis. Infect Immun. 2010;78:2846–2856. doi: 10.1128/IAI.01448-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong F, Zheng D, She P, et al. Porphyromonas gingivalis B cell antigen epitope vaccine, pIRES-ragB′-mGITRL, promoted RagB-specific antibody production and Tfh cells expansion. Scand J Immunol. 2015;81:476–482. doi: 10.1111/sji.12281. [DOI] [PubMed] [Google Scholar]
- Koropatkin NM, Martens EC, Gordon JI, Smith TJ. Starch catabolism by a prominent human gut symbiont is directed by the recognition of amylose helices. Structure. 2008;16:1105–1115. doi: 10.1016/j.str.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koropatkin NM, Martens EC, Gordon JI, Smith TJ. Structure of a SusD homologue, BT1043, involved in mucin O-glycan utilization in a prominent human gut symbiont. Biochemistry. 2009;48:1532–1542. doi: 10.1021/bi801942a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laine ML, van Winkelhoff AJ. Virulence of six capsular serotypes of Porphyromonas gingivalis in a mouse model. Oral Microbiol Immunol. 1998;13:322–325. doi: 10.1111/j.1399-302x.1998.tb00714.x. [DOI] [PubMed] [Google Scholar]
- Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat Protoc. 2008;3:1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loos BG, Mayrand D, Genco RJ, Dickinson DP. Genetic heterogeneity of Porphyromonas (Bacteroides) gingivalis by genomic DNA fingerprinting. J Dent Res. 1990;69:1488–1493. doi: 10.1177/00220345900690080801. [DOI] [PubMed] [Google Scholar]
- Lovatt A, Roberts IS. Cloning and expression in Escherichia coli of the nahA gene from Porphyromonas gingivalis indicates that β-N-acetylhexosaminidase is an outer-membrane-associated lipoprotein. Microbiology. 1994;140(Pt 12):3399–3406. doi: 10.1099/13500872-140-12-3399. [DOI] [PubMed] [Google Scholar]
- Maixner F, Thomma A, Cipollini G, Widder S, Rattei T, Zink A. Metagenomic analysis reveals presence of Treponema denticola in a tissue biopsy of the Iceman. PLoS One. 2014;9:e99994. doi: 10.1371/journal.pone.0099994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens EC, Chiang HC, Gordon J. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe. 2008;4:447–457. doi: 10.1016/j.chom.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens EC, Koropatkin NM, Smith TJ, Gordon J. Complex glycan catabolism by the human gut microbiota: the Bacteroidetes Sus-like paradigm. J Biol Chem. 2009;284:24673–24677. doi: 10.1074/jbc.R109.022848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews BW. Solvent content of protein crystals. J Mol Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- Murakami Y, Hasegawa Y, Nagano K, Yoshimura F. Characterization of wheat germ agglutinin lectin-reactive glycosylated OmpA-like proteins derived from Porphyromonas gingivalis. Infect Immun. 2014;82:4563–4571. doi: 10.1128/IAI.02069-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Skubak P, Lebedev AA, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr sect D. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano K, Murakami Y, Nishikawa K, Sakakibara J, Shimozato K, Yoshimura F. Characterization of RagA and RagB in Porphyromonas gingivalis: study using gene-deletion mutants. J Med Microbiol. 2007;56:1536–1548. doi: 10.1099/jmm.0.47289-0. [DOI] [PubMed] [Google Scholar]
- Nakao R, Tashiro Y, Nomura N, et al. Glycosylation of the OMP85 homolog of Porphyromonas gingivalis and its involvement in biofilm formation. Biochem Biophys Res Commun. 2008;365:784–789. doi: 10.1016/j.bbrc.2007.11.035. [DOI] [PubMed] [Google Scholar]
- Park OJ, Yi H, Jeon JH, et al. Pyrosequencing analysis of subgingival microbiota in distinct periodontal conditions. J Dent Res. 2015;94:921–927. doi: 10.1177/0022034515583531. [DOI] [PubMed] [Google Scholar]
- Paster BJ, Olsen I, Aas JA, Dewhirst FE. The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Periodontol 2000. 2006;42:80–87. doi: 10.1111/j.1600-0757.2006.00174.x. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, et al. UCSF Chimera - A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Phansopa C, Roy S, Rafferty JB, et al. Structural and functional characterization of NanU, a novel high-affinity sialic acid-inducible binding protein of oral and gut-dwelling Bacteroidetes species. Biochem J. 2014;458:499–511. doi: 10.1042/BJ20131415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preshaw PM, Alba AL, Herrera D, et al. Periodontitis and diabetes: a two-way relationship. Diabetologia. 2012;55:21–31. doi: 10.1007/s00125-011-2342-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangarajan M, Aduse-Opoku J, Hashim A, Paramonov N, Curtis MA. Characterization of the α- and β-mannosidases of Porphyromonas gingivalis. J Bacteriol. 2013;195:5297–5307. doi: 10.1128/JB.00898-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves AR, D’Elia JN, Frias J, Salyers AA. A Bacteroides thetaiotaomicron outer membrane protein that is essential for utilization of maltooligosaccharides and starch. J Bacteriol. 1996;178:823–830. doi: 10.1128/jb.178.3.823-830.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves AR, Wang GR, Salyers AA. Characterization of four outer membrane proteins that play a role in utilization of starch by Bacteroides thetaiotaomicron. J Bacteriol. 1997;179:643–649. doi: 10.1128/jb.179.3.643-649.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocas IN, Siqueira JF, Jr, Santos KR, Coelho AM. “Red complex” (Bacteroides forsythus, Porphyromonas gingivalis, and Treponema denticola) in endodontic infections: a molecular approach. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2001;91:468–471. doi: 10.1067/moe.2001.114379. [DOI] [PubMed] [Google Scholar]
- Schägger H. Tricine-SDS-PAGE. Nat Protoc. 2006;1:16–22. doi: 10.1038/nprot.2006.4. [DOI] [PubMed] [Google Scholar]
- Schüttelkopf AW, van Aalten DM. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr sect D. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- Sheldrick GM. Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Crystallogr sect D. 2010;66:479–485. doi: 10.1107/S0907444909038360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldrick GM. The SHELX approach to experimental phasing of macromolecules. Acta crystallogr sect A. 2011;67:C13. doi: 10.1107/S2059798317015121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji M, Sato K, Yukitake H, et al. Por secretion system-dependent secretion and glycosylation of Porphyromonas gingivalis hemin-binding protein 35. PLoS One. 2011;6:e21372. doi: 10.1371/journal.pone.0021372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Boguski MS, Goebl M, Hieter P. A repeating amino acid motif in CDC23 defines a family of proteins and a new relationship among genes required for mitosis and RNA synthesis. Cell. 1990;60:307–317. doi: 10.1016/0092-8674(90)90745-z. [DOI] [PubMed] [Google Scholar]
- Smart OS, Womack TO, Flensburg C, et al. Exploiting structure similarity in refinement: automated NCS and target-structure restraints in BUSTER. Acta Crystallogr sect D. 2012;68:368–380. doi: 10.1107/S0907444911056058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–144. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- Sonnenburg JL, Xu J, Leip DD, et al. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307:1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- Stubbs S, Lewis MO, Waddington RJ, Embery G. Hydrolytic and depolymerising enzyme activity of Prevotella intermedia and Prevotella nigrescens. Oral Dis. 1996;2:272–278. doi: 10.1111/j.1601-0825.1996.tb00237.x. [DOI] [PubMed] [Google Scholar]
- Su Z, Kong F, Wang S, et al. The rag locus of Porphyromonas gingivalis might arise from Bacteroides via horizontal gene transfer. Eur J Clin Microbiol Infect Dis. 2010;29:429–437. doi: 10.1007/s10096-010-0880-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tancula E, Feldhaus MJ, Bedzyk LA, Salyers AA. Location and characterization of genes involved in binding of starch to the surface of Bacteroides thetaiotaomicron. J Bacteriol. 1992;174:5609–5616. doi: 10.1128/jb.174.17.5609-5616.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steenbergen TJM, van Winkelhoff AJ, de Graaff J. Classification and typing methods of black-pigmented Gram-negative anaerobes. FEMS Immunol Med Microbiol. 1993;6:83–88. doi: 10.1111/j.1574-695X.1993.tb00307.x. [DOI] [PubMed] [Google Scholar]
- van Winkelhoff AJ, Loos BG, van der Reijden WA, van der Velden U. Porphyromonas gingivalis, Bacteroides forsythus and other putative periodontal pathogens in subjects with and without periodontal destruction. J Clin Periodontol. 2002;29:1023–1028. doi: 10.1034/j.1600-051x.2002.291107.x. [DOI] [PubMed] [Google Scholar]
- Wei B, Dalwadi H, Gordon LK, et al. Molecular cloning of a Bacteroides caccae TonB-linked outer membrane protein identified by an inflammatory bowel disease marker antibody. Infect Immun. 2001;69:6044–6054. doi: 10.1128/IAI.69.10.6044-6054.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss MS. Global indicators of X-ray quality. J Appl Cryst. 2001;34:130–135. [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr sect D. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Mahowald MA, Ley RE, et al. Evolution of symbiotic bacteria in the distal human intestine. PLoS Biol. 2007;5:e156. doi: 10.1371/journal.pbio.0050156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HW, Huang YF, Chou MY. Occurrence of Porphyromonas gingivalis and Tannerella forsythensis in periodontally diseased and healthy subjects. J Periodontol. 2004;75:1077–1083. doi: 10.1902/jop.2004.75.8.1077. [DOI] [PubMed] [Google Scholar]
- Zeituni AE, McCaig W, Scisci E, Thanassi DG, Cutler CW. The native 67-kilodalton minor fimbria of Porphyromonas gingivalis is a novel glycoprotein with DC-SIGN-targeting motifs. J Bacteriol. 2010;192:4103–4110. doi: 10.1128/JB.00275-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D, Sun Q, Su Z, et al. Enhancing specific-antibody production to the ragB vaccine with GITRL that expand Tfh, IFN-gamma(+) T cells and attenuates Porphyromonas gingivalis infection in mice. PLoS One. 2013;8:e59604. doi: 10.1371/journal.pone.0059604. [DOI] [PMC free article] [PubMed] [Google Scholar]