Significance
Specific and functional interactions between membrane lipids and proteins are increasingly evident across biology. The outer membrane (OM) of gram-negative bacteria such as Escherichia coli is a selective barrier formed by complex lipids (lipopolysaccharides; LPSs) and outer-membrane proteins. The high stability and low permeability of the OM are critical to bacterial growth and pathogenesis. Here, using biochemical and structural techniques, we reveal specific LPS binding sites on OM porin proteins that allow them to stabilize, rather than disrupt, the ordered network of LPS molecules. Furthermore, we demonstrate that one such site is essential for porin assembly in the OM.
Keywords: gram-negative bacteria, lipopolysaccharide, X-ray crystal structure, porin, outer membrane
Abstract
The outer membrane (OM) of gram-negative bacteria is an unusual asymmetric bilayer with an external monolayer of lipopolysaccharide (LPS) and an inner layer of phospholipids. The LPS layer is rigid and stabilized by divalent cation cross-links between phosphate groups on the core oligosaccharide regions. This means that the OM is robust and highly impermeable to toxins and antibiotics. During their biogenesis, OM proteins (OMPs), which function as transporters and receptors, must integrate into this ordered monolayer while preserving its impermeability. Here we reveal the specific interactions between the trimeric porins of Enterobacteriaceae and LPS. Isolated porins form complexes with variable numbers of LPS molecules, which are stabilized by calcium ions. In earlier studies, two high-affinity sites were predicted to contain groups of positively charged side chains. Mutation of these residues led to the loss of LPS binding and, in one site, also prevented trimerization of the porin, explaining the previously observed effect of LPS mutants on porin folding. The high-resolution X-ray crystal structure of a trimeric porin–LPS complex not only helps to explain the mutagenesis results but also reveals more complex, subtle porin–LPS interactions and a bridging calcium ion.
Specific membrane lipid–protein interactions are increasingly significant in cell biology but, due to their weak or transient nature, are often difficult to define (1). The gram-negative outer membrane (OM) is a highly asymmetric lipid bilayer with lipopolysaccharide (LPS) on the outer leaflet and phospholipids, mainly phosphatidylethanolamine, on the inner leaflet (2). Both the structure of LPS and the bilayer’s extreme asymmetry are unusual for biological membranes, and their combined effect upon the integral membrane proteins embedded in the OM has not been clearly investigated. The role of the OM is to create a robust and tight barrier toward the external environment such that transport across it is highly regulated by the outer-membrane proteins (OMPs) it contains. Uniquely, this also applies to hydrophobic molecules, for which the polar part of the LPS poses a significant permeation barrier. Thus, a tight interaction between the LPS barrier and the inserted OMPs is likely to be essential to maintain OM impermeability. With the rise of antimicrobial resistance it has become increasingly clear that the OM affords gram-negative bacteria an additional and effective means to withstand antibiotic therapy (3).
Several lines of evidence have indicated that there is a close and specific interaction between OMPs and LPS. In early studies of matrix porin (OmpF), LPS copurified with the protein and a ratio of 3:1 LPS:OmpF was determined (4, 5). It was also proposed that LPS was required for full ion-channel activity (6, 7). LPS binding to OmpF was later analyzed by free-flow electrophoresis (FFE), which separated four isoforms clearly visible as ladders on SDS/PAGE (8). All forms contained ∼1 mol of tightly bound LPS per trimer with three additional isoforms containing respectively, 1, 2, and 8 mol per trimer of loosely bound LPS, so that the highest stoichiometry observed was nine per trimer or a 3:1 LPS:OmpF molar ratio. Interestingly, analytical centrifugation of samples, which produced ladders on SDS/PAGE, showed a single species in solution, and addition of 3 mM EDTA partially removed the loosely bound LPS (8).
The purified isoforms were used to form OmpF 2D crystals observed by negative-stain electron microscopy. A protrusion on the central threefold axis of the trimer was linked to the single, tightly bound, molecule of LPS per trimer. Less distinct protrusions at the perimeter of each trimer were observed in samples with loosely bound LPS and thus ascribed to sites of partial LPS occupation (9). A similar central location of a single LPS molecule was also proposed for the structure of PhoE porin derived from cryoelectron microscopy (10). The samples with 1 mol of tightly bound LPS formed 3D crystals suitable for X-ray crystallography, but the structure has not been reported (8). Porin from Yersinia pseudotuberculosis was found to show two or three different binding sites depending upon the form of LPS acylation (11). The first trimeric porin structure solved to high resolution was from Rhodobacter capsulatus (12), and neither this, nor the later Escherichia coli OmpF and PhoE structures (13), showed any evidence of bound LPS. Furthermore, none of the structures of trimeric porins published since show identifiable LPS on the threefold axis or on the external surface of the barrel (14), and this is probably due to the stringent removal of LPS in many purification procedures (15). By in vitro folding in the absence of LPS, we were able to confirm earlier data (16) that LPS-free OmpF trimers show identical structural and electrophysiological properties to those that folded in vivo in the presence of LPS in the OM (17, 18).
The first high-resolution structure of an LPS–OMP complex was that of the E. coli siderophore transporter FhuA (19). It revealed for the first time the noncovalent interactions that could stabilize a tight association of LPS with protein. The lipid A acyl chains form van der Waals interactions with the membrane-exposed hydrophobic surface of the protein, whereas basic amino acids form salt bridges with the phosphate groups on LPS. The LPS–OMP structure enabled a bioinformatic search of the structural database that led to the definition of a likely LPS binding site, based upon a four-residue motif, in a variety of OM and innate immunity proteins (20). An obvious limitation of the bioinformatic approach is that the LPS binding-site definition was based on the structure of a single LPS–OMP complex, because FhuA is the only OMP for which structures with bound LPS have been reported. A crystal structure of the omptin protease Pla from Yersinia pestis showed density consistent with LPS acyl chains, but no density for the polar part of the glycolipid was observed (21, 22). Using the predictions from FhuA, we predicted likely LPS binding sites on the OmpF trimer and showed that these corresponded to changes in 2D crystal structure when the antibacterial toxin colicin N displaced tightly bound LPS from the OmpF trimer (18).
In addition to its role in creating the OM barrier, LPS is also the environment into which trimeric porins are inserted and folded. It is well-established that reduction in the amount or size of LPS molecules severely inhibits trimer formation (7, 23–25), whereas LPS and divalent cations are able to induce folding of porins in vitro (26). In recent years, the discovery and characterization of the relevant periplasmic chaperones and β‐barrel assembly machinery (BAM) complex have revealed in significant detail the protein components of the OMP biogenesis process (27). By contrast, how the LPS guides the folding and trimerization of porins is less clear.
Here we show by mutagenesis, small-angle X-ray and neutron scattering, and X-ray crystallography that there are at least two specific binding sites for LPS per porin monomer and that one of these is both calcium-dependent and essential for the assembly of trimers in the OM. The results reveal that trimeric OMPs form a specific, tight complex with LPS that maintains the permeability barrier of the OM.
Results
SDS/PAGE Can Resolve OmpF–LPS Complexes.
The addition of LPS (Fig. 1A) to LPS-free in vitro folded OmpF trimers (18) creates, on SDS/PAGE, a characteristic ladder of increasing molecular mass due to the slower mobility of LPS-bound OmpF (Fig. 1B) (8). Moreover, the results confirm that LPS–OmpF complexes form with a range of stoichiometries within each sample and are stable under SDS/PAGE conditions without boiling (18). Moreover, these ladders resemble those observed in preparations of native OmpF from E. coli OM fractions if LPS is not intentionally removed (18).
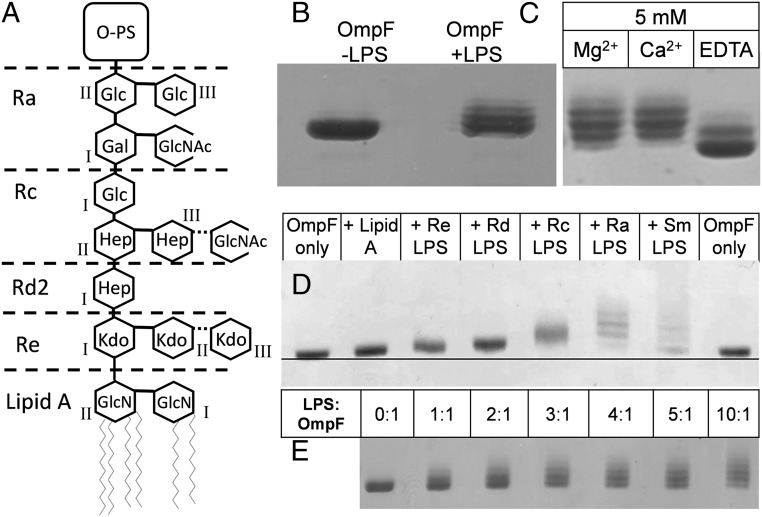
Fig. 1.
Binding of LPS to OmpF causes slower mobility of complexes on SDS/PAGE. (A) Structure of LPS from E. coli with the R3 core structure, including that of the Rc-LPS from E. coli J5 used in this study (63) (nonstoichiometric additions are shown with dotted lines). In lipid A, GlcNI and GlcNII are, respectively, the reducing and nonreducing glucosamine residues. The inner core usually comprises two or three Kdo and three Hep (l-glycero-d-manno-heptose) molecules. This region is phosphorylated at several sites. The variable trihexose backbone forms the outer core with varying side chains; shown here as present in the R3 form are glucose (Glc), N-acetylglucosamine (GlcNAc), and galactose (Gal). These link to the long O-antigen polysaccharide (O-PS) region found only in smooth strains. The depiction of the rough Ra-to-Re mutants that define the different chemotypes is based upon the original classification in S. minnesota (64). The Rd-LPS used here is from an Rd2 mutant, as shown. (B) Characteristic ladder, on 10% SDS/PAGE, of OmpF resulting from LPS binding. In vitro folded LPS-free OmpF (Left) and an identical sample mixed with a fivefold molar excess of Ra-LPS (Right). (C) Samples of in vitro folded OmpF + Ra-LPS as in B mixed with 5 mM MgCl2, CaCl2, or EDTA. (D) In vitro folded OmpF mixed with a fivefold molar ratio of LPS variants of increasing size. Sm, smooth LPS. (E) In vitro folded OmpF mixed with increasing molar ratios of Ra-LPS.
Divalent Cations Stabilize OmpF–LPS Complexes.
Because divalent cations are known, via interactions with phosphate groups, to stabilize LPS in the OM (28) and in protein complexes (8), we tested whether the complexes here were similarly stabilized. The addition of 5 mM CaCl2 or MgCl2 increased the resolution of bands displayed on SDS/PAGE by the complexes formed with a 5:1 ratio of Ra-LPS to OmpF. Addition of 5 mM EDTA reduced but did not abolish the ladder (Fig. 1C), implying the presence of some noncalcium-bridged LPS–OMP complexes.
OmpF Binds to All LPS Types Tested Including Minimal Lipid A.
In vitro folded LPS-free OmpF was mixed with LPS molecules of increasing length, starting with lipid A (Fig. 1D), at a molar ratio of five LPSs to one OmpF monomer (15 LPSs per trimer). The shift in mobility of the LPS-bound forms is proportional to the size of the LPS molecule added (29). Even though with lipid A a ladder cannot be resolved, a clear retardation indicates that even this minimal LPS molecule forms stable complexes with OmpF. Thus, the multiple bands that form the long LPS-induced ladder on SDS/PAGE are probably complexes containing different ratios of LPS:OmpF, stabilized by low ambient levels of divalent cations recruited from the LPS and buffer solutions. To further understand the ladder of complexes observed on SDS/PAGE, we attempted to saturate the LPS binding sites by adding higher molar ratios of LPS. We used the largest rough LPS molecule, Ra-LPS, to provide the clearest shift and added up to a tenfold molar ratio (30 LPSs per trimer). Increasing ratios of LPS led to increasingly higher ladders with no saturation nor clear population of a preferred complex (Fig. 1E).
Mutagenesis Indicates Possible LPS Binding Sites of OmpF.
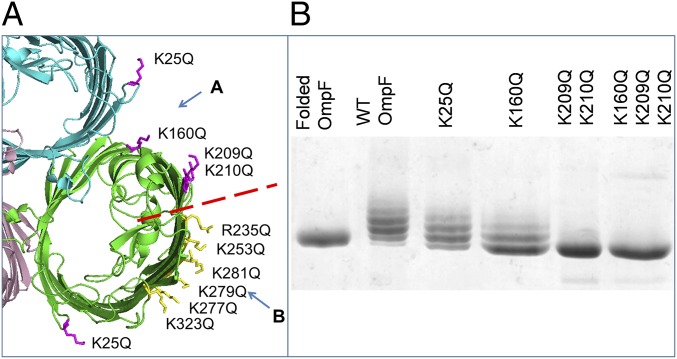
From the structure of FhuA (19) and subsequent bioinformatic analysis of the structural database (20) it was predicted that the LPS binding site(s) on OmpF should consist of about four basic amino acid residues facing the membrane near the OM extracellular surface, where they can interact with the phosphate groups on the lipid A core (18). Ten arginine and lysine side chains form a belt at the correct height on the OmpF surface for LPS binding, and these can be divided into two distinct groups we termed sites A and B (Fig. 2A). A series of site A mutations was designed, purified from the OM, and tested on SDS/PAGE for their ability to purify as a complex with the natural LPS from their host E. coli. We initially chose glutamine to replace the lysine and arginine chains to minimize the changes to the local structure. Single mutants showed marginally decreased ladders, but significant effects were only seen with double and triple mutations (Fig. 2B).
Fig. 2.
Removing positively charged residues decreases the amount of LPS bound to OmpF. (A) Localization of positively charged residues on the extracellular side of an OmpF trimer (PDB ID code 2OMF). We divided these residues into group A in the cleft (magenta) and group B at the perimeter (yellow) separated by the red dashed line. This and other structural images were created using PyMOL (65). (B) Native OmpF, WT, and site A mutant proteins, purified from the OM with bound LPS, analyzed on 10% SDS/PAGE stained with Coomassie blue. The double- and triple-glutamine mutations retain a tail of LPS-bound forms, which contrasts with the quadruple-glutamate mutations (Fig. 3F).
LPS Binding at Site B Is Essential for Stable Trimer Formation in Vivo.
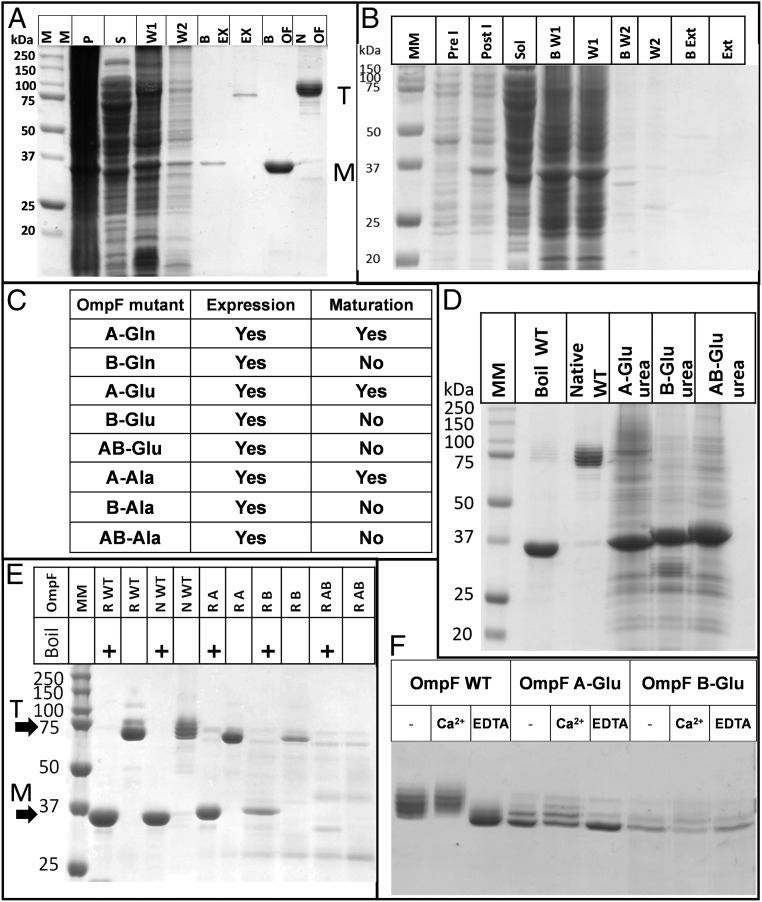
To comprehensively study the roles of the two sites, we performed complete mutagenesis of all four basic residues in the A site and all six in the B site (for details of mutated residues, see Fig. 2A) to polar uncharged glutamine (A-Gln and B-Gln), negatively charged glutamate (A-Glu, B-Glu, and both sites AB-Glu), and nonpolar alanine (A-Ala, B-Ala, and AB-Ala). Each mutant protein was purified from the E. coli OM, and trimer formation was assessed by SDS/PAGE. Only the site A mutants (site B intact) were able to mature and form trimers in the OM. In all of the site B mutants, the expressed OmpF remained as monomers (Fig. 3 A–C). To test whether site B mutants could still form trimers in vitro, we purified the most radical mutants (lysine/arginine-to-glutamate substitutions, which reverse the charge of the sites) as inclusion bodies by expressing them without signal sequences (17). The inclusion bodies were solubilized in urea and ran on SDS/PAGE as monomers with slightly altered mobilities (Fig. 3D). Following in vitro folding, both the individual A-Glu and B-Glu site mutants formed trimers, but the double-site mutant AB-Glu did not fold properly (Fig. 3E). This showed that OmpF lacking site B can still fold to form SDS-stable trimers in vitro. The in vitro folded mutant proteins A-Glu and B-Glu were then mixed with a 5:1 ratio of Ra-LPS:OmpF and analyzed on SDS/PAGE. Each mutant protein showed simpler ladders than WT and bound Ra-LPS in a calcium-dependent manner. The effect of EDTA on the OmpF–LPS ladders was more pronounced in the A-Glu compared with the B-Glu mutant (Fig. 3F).
Fig. 3.
LPS binding site B is essential for OmpF trimerization in vivo but not in vitro. The abbreviations describe the mutations applied at each site; for example, A-Gln, all basic residues in site A replaced by glutamine; AB-Glu, all basic residues in sites A and B replaced by glutamate. (A) Purification of A-Gln mutant protein. BEX and EX, boiled and native OmpF extraction samples showing monomers and trimers, respectively; BOF and NOF, boiled and native WT OmpF control samples from in vitro folded stock; M, position of the monomer band on the gel; MM, molecular weight markers; P, membrane pellet; S, supernatant; T, position of the trimer band on the gel; W1 and W2, supernatants after washes 1 and 2. (B) Purification of B-Gln mutant protein. B W1/2 and W1/2, boiled and native supernatants from wash 1/2; B Ext and Ext, as above, showing lack of trimeric OmpF extracted with SDS and NaCl; Pre I, whole-cell pellet before induction; Post I, whole-cell pellet after 1-h induction showing an OmpF band at 37 kDa; Sol, supernatant after cell breakage. (C) Summary table of results for mutants. “Expression” indicates a band observed at 37 kDa after induction; “maturation” indicates that intact trimers were purified from the OM. (D) Solubilization of A-Glu, B-Glu, and AB-Glu inclusion bodies in urea, showing the different migration of unfolded monomers on SDS/PAGE. (E) In vitro folding of trimeric porins. N WT, native WT purified from the OM; R A, R B, and R AB, in vitro folded A-Glu, B-Glu, and AB-Glu mutants with and without boiling; R WT, in vitro folded WT. A-Glu and B-Glu fold fully in vitro, but no heat-modifiable trimer is evident in AB-Glu samples. (F) LPS binding to in vitro folded mutants. WT, A-Glu (site B intact), and B-Glu (site A intact) without added calcium or plus 5 mM CaCl2 or 5 mM EDTA. Note the clear effect of calcium removal on the A-Glu mutant, indicating a role for the calcium ion(s) in site B.
Structure of OmpF–LPS Complexes in Solution.
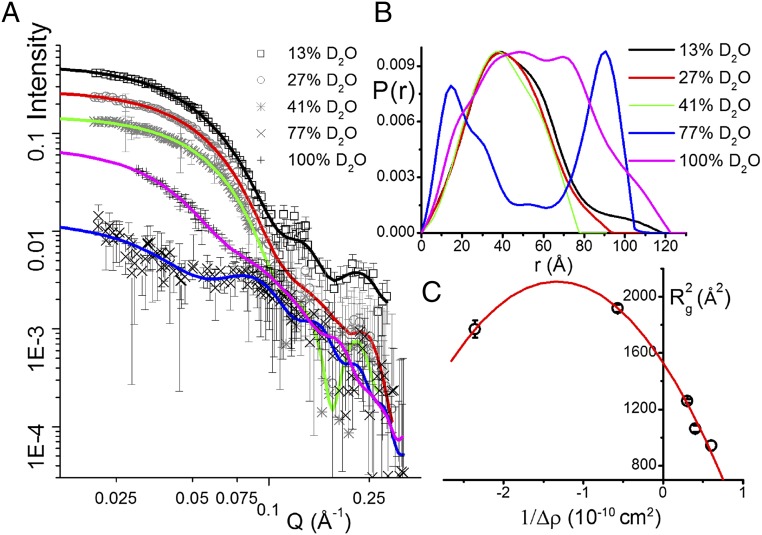
We performed dynamic light scattering (DLS) measurements on OmpF–LPS complexes in 1% SDS. This revealed a narrow distribution of radii slightly larger than OmpF alone (SI Appendix, Fig. S1) and is consistent with a population of isolated trimers. This pattern only changed to large aggregates when 10 or more molecules of LPS were added per trimer in calcium-containing solutions. Small-angle X-ray scattering (SAXS) was then used to more accurately measure the overall dimensions of OmpF–LPS–dodecylmaltoside micelles. Recently, it was shown that estimates of membrane protein structure can be derived from SAXS even when there is extensive scattering from detergent micelles (30). This enabled us to calculate a maximum size (Dmax) of about 130 Å for the OmpF–LPS complex (SI Appendix, Fig. S2). We then used neutron scattering, which can resolve different components of biological complexes (31). Neutron scattering is especially sensitive to the different neutron scattering length densities (nSLDs; analogous to a neutron refractive index) of hydrogen and its isotope deuterium. Thus, we prepared deuterated OmpF (d-OmpF) with an nSLD equal to that of 81% (vol/vol) D2O (the “match point”) (SI Appendix, Fig. S3A). We then formed complexes of d-OmpF and normal hydrogenous Ra-LPS (h-Ra-LPS), which has a match point of 27% (vol/vol) D2O (32). We then collected small-angle neutron scattering (SANS) data from the d-OmpF–h-Ra-LPS complexes in a range of D2O concentrations to highlight the separate contributions of protein and LPS to the complex (Fig. 4A). The measured the nSLD of the complexes has a match point of 70% (vol/vol) D2O, which most closely matches that expected for a complex with two LPSs per trimer (SI Appendix, Fig. S3 A and B). The scattering curves were used to calculate pairwise distance distributions [P(r) plots] (Fig. 4B). Small-angle scattering data collected from OmpF–LPS complexes in 13%, 27% (where LPS is completely invisible to neutrons), and 41% D2O were dominated by d-OmpF scattering and resembled previous d-OmpF data (32). By measuring SANS in 77% D2O, close to the contrast match point for d-OmpF, we obtained data dominated by LPS scattering. However, because each trimer may only contain six Ra-LPS molecules, which have a total mass of 27 kDa compared with the 120-kDa OmpF trimer, the scattering under these conditions is much weaker than when the protein dominates the signal (Fig. 4A and Table 1). Nevertheless, in this case, the LPS scattering profile was completely different and resulted in a P(r) plot with two peaks. This profile can be explained by scattering centers (LPS molecules) on the outside of the complex, separated by the diameter of the trimer, as seen previously with the complex of colicin N with OmpF (32). The first peak around 20 Å corresponds to distances within each group of bound LPS molecules at sites A and B. The second peak, centered at 90 Å, corresponds to the set of LPS–LPS distances across the trimer. We further analyzed the distribution of LPS using the Stuhrmann plot (33), the square of the radius of gyration () versus the inverse of the contrast between the scattering object and the solvent at different D2O concentrations (1/Δρ) (33) (Fig. 4C). Using the program MULCh, we fitted the data to the equation
where Rm is the Rg of an equivalent complex with a homogeneous scattering density, α relates to the difference in distribution of the scattering densities, and β relates to the difference in center of mass of the scattering densities (33). The result was the line in Fig. 4C with the following parameter values: = 1,534 ± 37 Å2, α = −863 ± 83, and β = 324 ± 51. The negative α-value indicates that the lower nSLD component (LPS) has a larger radius of gyration and is thus at the periphery of the complex. Finally, we computed theoretical scattering curves for a number of OmpF–LPS complexes using the program CRYSON (34) and compared them with our experimental data (SI Appendix, Fig. S3C). The data fitted OmpF models with Ra-LPS occupying either two (using the positions suggested by the OmpE36–LPS structure) or three (with an additional LPS next to site B) sites per trimer with similar statistics. The fits were improved when the sugar head groups were modeled as bent away from the trimer (SI Appendix, Fig. S3C).
Fig. 4.
SANS data indicate that LPS binds at the periphery of OmpF in SDS solution. (A) Log/log plot of scattering data for deuterated (d-)OmpF in complex with hydrogenous Ra-LPS after size-exclusion chromatography. Q (momentum transfer) is a product of the scattering angle and neutron wavelength (6 Å) (Methods). Fitted lines were generated by the program BayesApp to calculate the P(r) vs. distance (r) plots (see Table 1 for fitting parameters and SI Appendix, Fig. S8 for an enlargement of the panel). P(r) is the real-space pair-distance function, which describes the distribution of pairs of scattering centers within the complex (Methods). The error bars in A represent the range of intensity values about the plotted data point and are the result of data reduction and averaging procedures within the program GRASP. (B) P(r) plot calculated from data in A using the same color scheme shows that at 13%, 27%, and 41% D2O, where LPS scattering is minimal, the plots resemble free OmpF (32). At 77% D2O, when the d-OmpF scattering is minimal, the plot describes small groups of scattering centers separated by about 90 Å. This corresponds to groups of LPS arranged at fixed sites around the trimer, as in the case of OmpF–colicin complexes (32). (C) The line of best fit to the Stuhrmann plot, the square of the radius of gyration () versus the inverse of the contrast (1/Δρ) (Methods), has a negative value of α (Results), a result most easily appreciated from the apex of the parabola being at negative values of 1/Δρ. This indicates that the low-nSLD LPS has a larger Rg than the high-nSLD OmpF and is likely to be situated at the periphery of the complex.
Table 1.
Scattering parameters for the SANS data calculated using BayesApp
| D2O, % | I0 × 103 | Rg, Å | Dmax, Å |
| Deuterated samples (d-OmpF–h-LPS) | |||
| 13 | 501 ± 0.866 | 35.48 ± 0.11 | 116.2 ± 1.71 |
| 27 | 276 ± 0.434 | 32.6 ± 0.08 | 91.99 ± 2.05 |
| 41 | 153 ± 0.234 | 30.68 ± 0.07 | 76.05 ± 1.68 |
| 77 | 12.7 ± 0.162 | 42.06 ± 0.46 | 101.01 ± 1.6 |
| 100 | 73.3 ± 0.94 | 43.78 ± 0.33 | 119.56 ± 1.65 |
| Hydrogenated samples (h-OmpF–h-LPS) | |||
| 100 | 376 ± 1.12 | 36.84 ± 0.24 | 121.7 ± 7.99 |
Structure Determination of the OmpE36–LPS Complex.
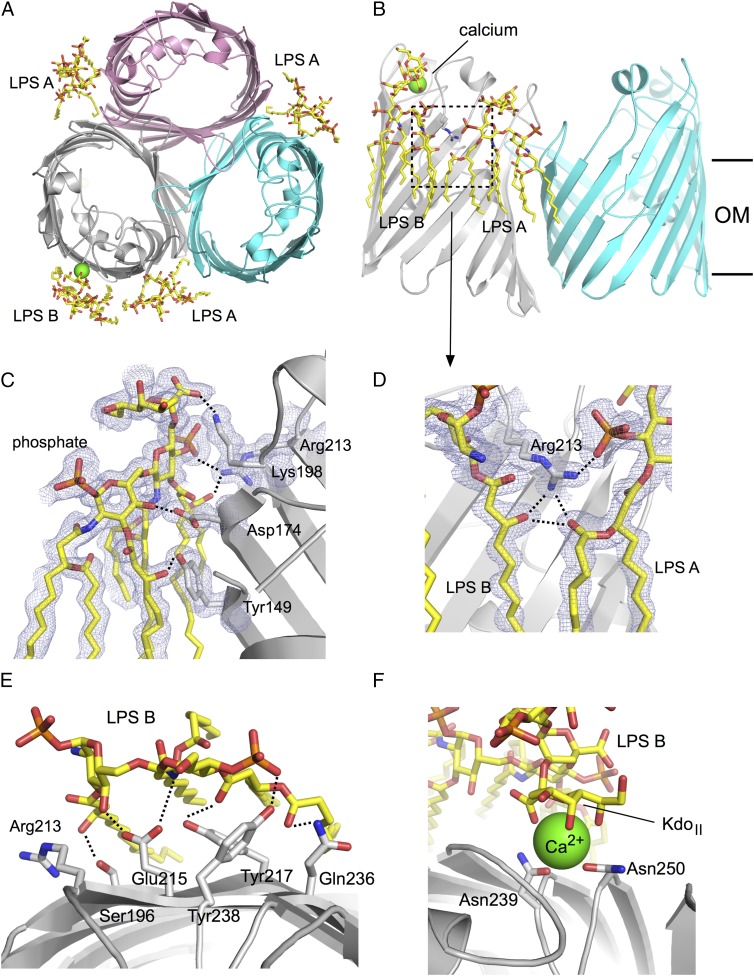
To provide support for the biochemical data, we overexpressed and purified several enterobacterial porins from the OM of E. coli with the goal of obtaining a structure with bound LPS. We initially focused our efforts on E. coli OmpF, but did not succeed in obtaining well-diffracting crystals. We then extended our focus to the OmpF and OmpC orthologs from other, closely related enteric bacteria, including those from Klebsiella pneumoniae and Enterobacter cloacae. The OmpC ortholog from E. cloacae (OmpE36) gave well-diffracting crystals. The structure was solved by molecular replacement with E. coli OmpC as a search model [Protein Data Bank (PDB) ID code 2J1N; 66% sequence identity] using data to 1.45-Å resolution. There are two porin trimers in the asymmetric unit, arranged as a double layer of 2D crystals (SI Appendix, Fig. S4). Inspection of the initial difference maps showed clear density for large molecules bound to the outside surface of the trimer, which could be unambiguously assigned to LPS (Fig. 5 and SI Appendix, Fig. S4).
Fig. 5.
X-ray crystal structure of the E. cloacae OmpE36–LPS complex. (A) Cartoon view of the OmpE36 trimer from the top, with the bound LPS molecules indicated by stick models. The bound calcium ion is shown as a green sphere. (B) Side view showing LPS A and LPS B binding. The approximate boundaries of the hydrophobic core of the OM are indicated by horizontal lines. (C) Close-up of LPS B, with polar interactions between the LPS and OmpE36 indicated by dashed lines. (D) Close-up of the boxed region in B, showing the interactions of the central Arg213 with both LPS A and LPS B. For clarity, some LPS B acyl chains have been removed. (E) View from the extracellular side highlighting interactions between LPS B and OmpE36. (F) Close-up of the calcium ion bridging LPS B and OmpE36.
A total of four LPS molecules are bound to each OmpE36 trimer (Fig. 5A). One LPS molecule (LPS A) is bound in each groove between two monomers, corresponding to site A (Fig. 5A). In addition, one OmpE36 monomer per trimer contains an additional LPS molecule at a position corresponding to site B (LPS B; Fig. 5A and SI Appendix, Fig. S4). This particular LPS molecule is sandwiched by a symmetry-related protein molecule (SI Appendix, Fig. S4), providing a possible explanation for why there is only one OmpE36 monomer with two molecules of bound LPS. In addition to LPS A and B, there is one other LPS molecule within the asymmetric unit. This molecule (denoted LPS C) is relatively poorly ordered and also bridges to a crystallographic symmetry mate (SI Appendix, Fig. S4). We have previously observed the ability of LPS to promote 2D crystallization of E. coli OmpF trimers, and the bridging positions of the site B (and possibly site C) LPS molecule help explain this behavior (35).
For the LPS A and B molecules, complete density is visible for the lipid A moiety as well as for both Kdo (3-deoxy-d-manno-oct-2-ulosonic acid) residues, that is, for Re-LPS. Beyond the KdoI moiety, LPS B has clear density for GMH (l-glycero-d-manno-heptopyranose) and part of the Rc moieties (Fig. 1A and SI Appendix, Fig. S5), but beyond GMH the density is of insufficient quality to allow the building of a reliable model. For GMH itself, density is visible only for the manno-pyranose ring. For LPS A, no model can be built beyond the Kdo residues. The likely reason for the relatively poor density is that beyond the Kdo moieties, the LPS sugars bend away from the protein and hence do not form many interactions. This is supported by the improvement of fits of SANS data to model complexes when the Ra-LPS head groups are bent away from the protein in the detergent micelle (SI Appendix, Fig. S3C). Interestingly, the density in the lipid A region reveals that both LPS A and B are heptaacylated (SI Appendix, Fig. S5), which is in contrast to the hexaacylated LPS that is thought to dominate in E. coli and which is also observed in the FhuA structures.
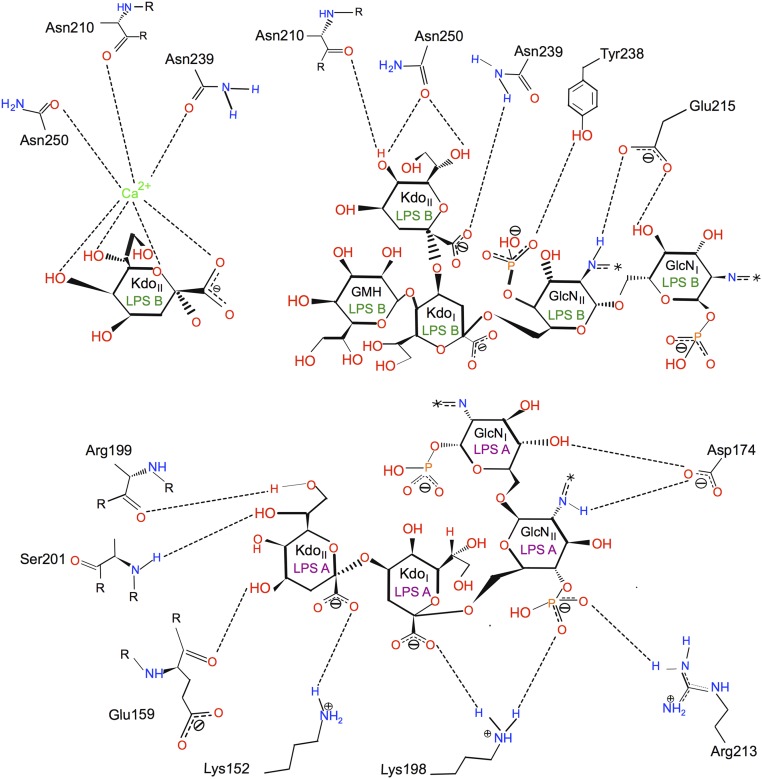
In addition to numerous van der Waals interactions between the lipid A acyl chains and the hydrophobic exterior of the barrel, LPS A and B make a number of polar interactions with residues of OmpE36, including both salt bridges and hydrogen bonds. For LPS A, salt bridges are present between Lys198 and the GlcNII-4P of lipid A and the carboxylate of KdoI, as well as between Arg213 (OmpF K210) and GlcNII-4P (Fig. 6). The configuration of donors and acceptors suggests that these salt bridges are dynamic and not all present all of the time. This may explain why, despite few clearly resolved salt bridges, the removal of lysine and arginine residues still reduces LPS binding to OmpF. A final, presumably stable, salt bridge is present between Lys152 and the carboxylate of KdoII. Hydrogen bonds are present between the Asp174 side chain and GlcNI and GlcNII and between the peptide backbone of several residues (Glu159, Arg199, and Ser201) and KdoII (Fig. 6). For the LPS B molecule, density consistent with a metal molecule is present between the LPS and OmpE36. Analysis with the CheckMyMetal server (36) assigns this metal with high confidence as a calcium ion. Because no metals were added during purification and crystallization, the calcium must have been copurified from the E. coli OM. The ligands of the calcium are the side chains of Asn239 and Asn250 as well as the backbone carbonyl of Asn210. In addition, four functional groups on KdoII provide ligands to the calcium (Figs. 5F and 6). The calcium ion most likely stabilizes the LPS–porin interaction substantially. In addition to the calcium-mediated interactions, there are also hydrogen bonds between the calcium-liganding residues and KdoII, between Tyr238 and GlcNII-4P, and last between Glu215 and GlcNI and GlcNII (Fig. 6). Thus, the three Asn residues have dual roles in that they hydrogen-bond to the KdoII directly but also coordinate the calcium ion that interacts with KdoII. Our work therefore reveals the structural basis of the previously observed calcium-dependent LPS–porin binding.
Fig. 6.
Two-dimensional map showing interactions between OmpE36 residues and LPS A and B. The figure was generated using PoseView (66). GlcNI and GlcNII are the reducing and nonreducing 2-amino-2-deoxy-α-d-glucopyranose residues of lipid A (glucosamine), respectively. Acyl chains of lipid A are indicated by asterisks.
Discussion
The high-resolution X-ray crystal structure of the porin OmpE36 from E. cloacae defines two binding sites for LPS molecules per porin monomer on the exterior surface of the barrel. The crystal structure confirms that the areas of density previously observed by electron microscopy correspond to LPS (8, 18). Likewise, the neutron contrast matching enabled us to observe solely the LPS scattering in the OmpF–LPS complexes, and the resulting data fit to a peripheral distribution of groups of LPS molecules around the trimer. The fitted distances between the scattering centers fit well to measured distances in the crystal structure between LPS A–B (∼20 Å) and LPS A–A (∼90 Å). The structure also agrees with our biochemical and biophysical data for E. coli OmpF, which is not surprising given the high sequence similarity between both porins (SI Appendix, Figs. S6 and S7). Taking the sequence and structural similarity argument further, we predict that LPS binding sites are conserved in gram-negative bacterial porins. Why then has no porin structure with bound LPS been reported previously despite a large body of structural work? One possible reason is that, based on SDS/PAGE and previous FFE analysis (8), preparations of porins are heterogeneous with respect to their bound LPS. This is even the case for LPS added in vitro (Fig. 1E), and will likely be exacerbated with protein isolated from the OM. Moreover, the extended ladders observed on SDS/PAGE, when using samples equilibrated with large excesses of LPS, suggest that there may be more than two sites per monomer. Another reason likely lies in crystal-packing constraints. The LPS–OmpE36 structure reveals that whereas LPS A is present in all three grooves between porin monomers, LPS B is only observed at one of three possible sites. This LPS molecule is sandwiched by a monomer from a crystallographic symmetry-related neighbor, likely stabilizing it within the crystal. As can be appreciated from SI Appendix, Fig. S4, the crystal packing is not compatible with LPS being present at the other two B site positions. Thus, the exceptionally large size of the LPS head group creates tight constraints on the possible packing within the crystal, which together with preparation inhomogeneity provides an explanation for why porin–LPS complexes have proved difficult to crystallize previously. Indeed, crystallization of several other porins from Enterobacter and Klebsiella spp. purified by us following the same procedure has not yet yielded structures with bound LPS.
Comparison of LPS Binding in FhuA and OmpE36/OmpF.
The determination of the OmpE36–LPS structure allows a comparison with FhuA–LPS, for which 11 coordinate files are present in the PDB that all show one LPS molecule bound to the same site. For FhuA, basic residues dominate the polar interactions with the bound LPS. No fewer than six different lysine and arginine residues interact with the LPS inner core via salt bridges and hydrogen bonds, whereas only a few noncharged residues of FhuA interact with the LPS molecule. The dominance of basic residues in LPS binding formed the basis of the proposed four-residue LPS-binding motif (20) that allowed prediction of LPS binding sites on E. coli OmpF (18). Despite the successful prediction, however, there are substantial differences in LPS binding between the two proteins. First, the dominance of basic residues in LPS binding is much less pronounced in OmpE36 compared with FhuA. Three basic residues interact with LPS A in OmpE36 (Fig. 6), which in E. coli OmpF may be reduced to two (SI Appendix, Figs. S6 and S7). By contrast, only one basic residue (Arg213) interacts with LPS B in OmpE36 (Fig. 5). In OmpF, Lys210 is at the same position as Arg213 but, because the lysine cannot interact with both LPS A and B at the same time, it is not clear how much Lys210 contributes to the binding of each of the LPS molecules in OmpF. Molecular dynamics simulations could likely illuminate this issue. Other than the possible involvement of Lys210, only one other basic residue likely interacts with LPS B in OmpF. This is Arg235, which occupies the same position as Tyr238 in OmpE36 and is likely to interact with GlcNII-4P (Fig. 6 and SI Appendix, Fig. S7). The second difference in the mode of LPS binding between the two structures is the calcium-mediated interaction in OmpE36–LPS not present in the FhuA–LPS structure. The original FhuA–LPS complex structure [PDB ID code 2FCP (19)] contained a putative nickel metal ion bound to one of the phosphates of LPS that did not contribute to the binding to FhuA. However, other FhuA–LPS structures do not show any metal-mediated interactions between the protein and LPS, and we can therefore conclude that divalent metals do not play a role in the FhuA–LPS interaction. The final major difference in LPS binding between the two OMPs lies in the role of the KdoII moieties. In FhuA–LPS, KdoII points away from the protein and contributes just one interaction to the binding to FhuA (a salt bridge between Arg384 and the KdoII carboxylate). In sharp contrast, in both LPS A and B, the KdoII forms the interaction hub with OmpE36 (Fig. 6). This is especially true for LPS B, for which KdoII mediates a number of metal-mediated and direct interactions with the porin (Fig. 6). It is therefore clear that OMPs can interact with LPS in different ways, and an expanded database incorporating the LPS–OmpE36 data should enable a better prediction of LPS binding sites on OMPs.
Role of Metal in LPS Binding.
The site B LPS reveals the molecular basis of the calcium-dependent porin–LPS interaction (Fig. 1C) (8). The extended ladders of OmpF–LPS observed on SDS/PAGE when using samples equilibrated with large excesses of LPS occur only in the presence of calcium, indicating (i) that there may be additional B-type sites for OmpF–LPS interactions and (ii) that calcium mediates LPS–LPS interactions leading to an additional row of associated lipids surrounding the complex. The reduction of the size of the LPS–OmpF ladder by EDTA treatment is consistent with only the A sites being occupied by LPS in the absence of calcium, as suggested by the metal-independent binding of LPS A from the OmpE36 structure. Magnesium has also been shown to stabilize LPS–LPS interactions and thus divalent cations are critical for both LPS–LPS (28) and LPS–porin interactions, explaining why EDTA can destabilize the OM sufficiently for small proteins such as lysozyme to penetrate into the periplasm. The importance of divalent metal ions for stabilization of the LPS monolayer is a logical consequence of the presence of many negatively charged phosphate groups (28), which without neutralization would be repulsive and render the OM highly unstable.
The Essential Role for LPS in OM Structure and Function.
The intricate interactions between OmpF and LPS revealed here explain how these very different molecules combine to maintain OM integrity. The porin–LPS interface is likely to be rigid and thus regions of high porin density, observed by electron microscopy (37, 38) and atomic force microscopy (AFM) (39), are likely to be robust nonfluid structures, with the LPS having very limited diffusion. In an AFM study, Schabert and Engel (40) showed that the addition of loosely bound LPS changed 2D crystals from rectangular to hexagonal, whereas EM (24) and AFM (39) studies of bacterial outer membranes revealed quasicrystalline arrangements of porins and LPS in vivo. Furthermore, OmpF stabilized by the polymeric detergent amphipol can be rapidly induced to form dense 2D arrays by the addition of excess LPS (35). These data, combined with the specific interactions revealed here, explain how porins and LPS form tight assemblies on the surfaces of gram-negative bacteria that preserve the necessary stability and impermeability of the OM. Light microscopy on live cells using fluorescently labeled outer-membrane proteins has provided the best insights into the arrangement of stable assemblies within the outer membrane. In one report it was shown that whereas trimeric porins form relatively immobile regions, monomeric proteins such as BtuB show the faster diffusion expected of individual proteins (41). More recently, it was shown that assemblies of trimeric OMPs containing trapped BAM complexes are created near the cell midline and stay intact over several cell-division cycles to finally end up at the cell poles (42). The trimeric porins are likely to form stable ordered protein–LPS complexes at the heart of these long-lived assemblies.
Although a barrier for large and relatively hydrophobic antibiotics such as macrolides, the OM is also a target for antimicrobial molecules such as polymyxin and colicins. Polymyxins are strongly cationic lipopeptides that are likely able to replace the divalent cations bound to LPS, thus gaining access to the core of the membrane. The large antibacterial protein colicin N was recently shown to use the inner core region of LPS as its specific OM receptor, and thus must be able to access the regions of the LPS molecule close to the hydrophobic core (43). In fact, colicin N is the only molecule known to displace tightly bound LPS from the outside of the OmpF trimer (18).
Data gathered over many years have indicated an important role for LPS in the in vivo maturation of porins (7, 23, 25, 44–46). Here we provide support for these observations by showing that the successful in vivo OM biogenesis of OmpF requires the presence of an intact LPS B site at the periphery of the trimer. By contrast, the successful production of functional LPS-free OmpF by folding into detergent micelles suggested that LPS was not an essential component for OmpF in vitro (17, 47). OMP biogenesis is mediated by the BAM complex, which accepts unfolded proteins from periplasmic chaperones (48) and mediates the formation of β-barrels in the OM via a mechanism that is not yet well-understood (49–51). How LPS binding at site B affects OmpF biogenesis is therefore not clear, but the fact that this site is not required for in vitro folding suggests that the specific LPS B–OmpF interaction stabilizes an intermediate during the BAM-mediated biogenesis of porin trimers in the OM. Future work will be required to establish the extent to which LPS specifically interacts with other OMPs and whether such interactions, analogous to OmpF, directly affect OMP biogenesis in vivo. Such data will enable us to better understand the stability and dynamics of the OM, in particular its role as a barrier to antibiotics.
Methods
OmpF Production.
OmpF mutants were expressed from the plasmid pMS119 encoding the desired mutant ompF gene and purified from the OM of E. coli BZB1107 (ompF::Tn5) as described previously (32). Transformed cells were grown at 37 °C in LB (Luria–Bertani) medium supplemented with 100 µg/mL ampicillin, 30 µg/mL kanamycin, and 0.05% (vol/vol) glucose to inhibit the expression of another OMP, LamB. PhoE porin expression is suppressed by the phosphate present in the medium. The parental strain is naturally devoid of OmpC. When the OD600 of the cell culture reached 0.6, isopropyl β-d-1-thiogalactopyranoside (final concentration of 1 mM) was added to induce protein expression and cells were grown for a further 3 h. Cells were harvested by centrifugation at 8,000 × g for 10 min at 4 °C.
Production of Deuterated OmpF.
The deuterated OmpF was produced from E. coli BE3000 (15, 32). Cells were first adapted onto an H2O-based, solid minimal medium plate followed by growth on an 85% D2O minimal medium plate (52). Once colonies grew on the plate (which normally took a few days, due to the slower growth of the cells), selected larger colonies were grown in 50 mL 85% D2O minimal liquid medium. Once growth was established overnight, these cells were inoculated at a 1:20 ratio into 2 × 50 mL fresh 85% D2O minimal liquid medium. This step was repeated three times to increase the initial growth rate. Cells were harvested by centrifugation at 8,000 × g at room temperature and resuspended in 10 mL fresh 85% D2O minimal liquid medium. Ten milliliters of cell culture was inoculated into 1.5 L minimal medium in a 2-L Minifors fermenter (INFORS) at 37 °C using glucose as a carbon source. During cell growth, a pH of 7.3 was maintained by adding 10% (wt/vol) NaOH. The measurement of OD600 was performed to monitor cell growth. When the OD600 reached at least 10.0, the cells were harvested by centrifugation at 8,000 × g for 10 min at 4 °C.
Purification of OmpF.
Hydrogenous and deuterated forms of OmpF were purified using the same procedures. The cell pellet was resuspended in 20 mM sodium phosphate (pH 6.8) and supplemented with DNase (10 µg/mL) and RNase (10 µg/mL). The cells were lysed by sonication on ice for 15 min. To remove unbroken cells and cell debris, the suspension was centrifuged at 3,000 × g for 15 min. After removing the pellet containing unbroken cells and cell debris, the supernatant was centrifuged at 40,000 rpm for 1 h in a 45 Ti rotor on a Beckman L7-80 ultracentrifuge to isolate the membrane pellet. The membrane pellet obtained was resuspended in wash buffer [20 mM Tris, 2% (wt/vol) SDS, pH 7.4] and then homogenized at 55 °C for 1 h. The solution was centrifuged at 40,000 rpm for 1 h in a 45 Ti rotor on a Beckman L7-80 ultracentrifuge and the supernatant was discarded. This wash step was repeated twice. The extraction of OmpF from the remaining membrane pellet was carried out by incubating and homogenizing the pellet in wash buffer with additional 500 mM NaCl. After incubation for 1 h at room temperature, the extract was centrifuged at 40,000 rpm for 1 h in a 45 Ti rotor on a Beckman L7-80 ultracentrifuge. The extraction was repeated once to increase the yield of OmpF. The supernatant containing SDS-solubilized OmpF was dialyzed against 5 mM NaHCO3, 0.1% (wt/vol) SDS overnight at 37 °C. Precipitation of OmpF (to concentrate or change detergent) was achieved by mixing cold ethanol, prechilled at −80 °C, with OmpF samples to give a final ethanol concentration of 90% (vol/vol). This solution was then incubated at −20 °C overnight. To isolate the precipitated OmpF pellet, the solution was centrifuged at 17,000 × g for 1 h. The OmpF pellet was dried under a stream of air. OmpF was recovered by resuspension in 20 mM Tris⋅HCl (pH 7.5), 300 mM NaCl, 2% (vol/vol) octyl-polyoxyethylene (POE).
Preparation of in Vitro Folded OmpF.
OmpF inclusion bodies were expressed and folded as described previously (17) with slight modification. The purified OmpF from inclusion bodies was folded by a 20-fold dilution in 50 mM Tris⋅HCl (pH 8.0), 1 mM DTT, 0.1 mM EDTA containing a mixture of 1% (wt/vol) DDG (n-dodecyl-β-d-glucopyranoside) and 0.4% (wt/vol) DDM (n-dodecyl-β-d-maltoside). After a stationary incubation at 37 °C for at least 3 d, the sample was precipitated by mixing 1:9 with cold ethanol (see above) and then the folded trimeric OmpF was recovered by resuspending in 20 mM Tris⋅HCl (pH 7.4), 0.5% (vol/vol) octyl-POE. To completely exchange the folding detergents to another detergent, buffer exchange was achieved by anion-exchange chromatography using a 1-mL HiTrap Q Sepharose column equilibrated with 20 mM Tris⋅HCl (pH 7.4), 0.5% (vol/vol) octyl-POE. The refolded OmpF was eluted using a linear salt gradient.
Preparation of LPS.
Samples of LPS, Smooth (E. coli 055:B5), Ra (E. coli EH-100), Rc (E. coli J5), Rd (E. coli F-583; Rd2), and Re (Salmonella minnesota strain Re595) were from Sigma-Aldrich. Each was dissolved in 20 mM Tris⋅HCl (pH 7.4) to give a final concentration of 2 mg/mL. The LPS solution was sonicated in a water bath for 20 min and temperature-cycled six times between 4 and 70 °C. The resulting solution was kept at 4 °C overnight before use.
Dynamic Light Scattering.
DLS measurements were performed with Zetasizer Nano (Malvern Instruments). Fifty microliters of protein at 0.1–0.5 mg/mL in a 45-µL quartz glass cuvette (Hellma; 105.251-QS) was measured at 25 °C in triplicate. Size measurements and data analysis were performed by Zetasizer software.
X-Ray Crystal Structure Determination of E. cloacae OmpE36.
Genomic DNA of E. cloacae was obtained from Basilea Pharmaceutica. The ompE36 gene including the signal sequence was amplified using PCR, digested with NcoI and XbaI, and ligated with the arabinose-inducible vector pBAD24 containing an ampicillin resistance gene. The ligated product was electroporated into DH5α-competent cells and plated on LB-ampicillin plates (100 μg/mL) for overnight incubation at 37 °C. Screening for positive clones was done using colony PCR, and clones were confirmed by DNA sequencing (Eurofins MWG). Porin-deficient E. coli omp8-competent cells (ΔompA ΔompC ΔompF ΔlamB) (53) were transformed with one of the positive clones, and protein was expressed using 0.1% arabinose for induction (37 °C, 3 h). The cells were harvested by centrifugation [1,914 × g for 30 min (Avanti J-26 XP Centrifuge, Beckman Coulter Inc.)] and lysed with a cell disrupter (0.75 kW; Constant Systems; one pass at 23 kpsi). Total membranes were collected by ultracentrifugation using a 45 Ti rotor (Beckman Coulter Inc.; 45 min; 42,000 rpm). The resulting membrane pellet was extracted twice with 0.5% N-lauroylsarcosine (sarkosyl) detergent (in 20 mM Hepes, pH 7.5) followed by ultracentrifugation using a 45 Ti rotor (Beckman; 30 min, 42,000 rpm) to solubilize and remove inner-membrane proteins, followed by an overnight extraction at 4 °C in 1% lauryldimethylamine-oxide (LDAO) in 10 mM Hepes, 50 mM NaCl (pH 7.5) to solubilize the OMPs. LDAO extraction was followed by ultracentrifugation using a 50.2 Ti rotor (Beckman; 30 min, 50,000 rpm) and the supernatant was subjected to Resource Q anion-exchange chromatography in 0.2% LDAO at pH 7.5 (10 mM Hepes, 50 mM NaCl). After elution, the protein was further purified by gel-filtration chromatography in 10 mM Hepes, 100 mM LiCl, 0.4% C8E4 (pH 7.5). The purified protein was concentrated to 24 mg/mL. Crystal trays were set up using 10 mg/mL and 15 mg/mL concentrations and the crystal hits obtained in each case were optimized to obtain good-quality crystals. The optimized crystal condition for OmpE36 was 0.4 M ammonium sulfate, 0.1 M Mes, 10% (wt/vol) PEG 3350 (pH 6.5). Crystals were harvested, cryoprotected with glycerol (∼20%), and flash-frozen in liquid nitrogen. X-ray diffraction data were collected at the Diamond Light Source and processed using XDS (54). The crystal structure was solved using data to 1.45-Å resolution by molecular replacement using MolRep (55) with E. coli OmpC (PDB ID code 2J1N) as the search model. The refinement was done in Refmac5 (55), and Coot (56) was used for model (re)building. The data collection and refinement statistics are summarized in SI Appendix, Table S1.
Small-Angle Neutron Scattering Sample Preparation and Data Collection.
LPS was removed from in vivo folded d-OmpF using size-exclusion chromatography (SEC) on a Superose 12 column equilibrated with 20 mM Tris (pH 7.5), 300 mM NaCl, 10 mM EDTA, 0.5% octyl-POE. The purified d-OmpF was then used to form complexes by incubation with Ra-LPS at a 1:5 d-OmpF:LPS molar ratio at 37 °C overnight. To remove unbound LPS, the samples were again passed through the same Superose 12 column using the same buffer without EDTA. After SEC, the proteins in the fractions were precipitated by prechilled ethanol at a 1:9 ratio (90% ethanol in total). The protein pellet was resuspended with 50 mM potassium phosphate (pH 7.5), 150 mM NaCl containing 0.5% hydrogenous/deuterated (h/d) SDS in 13%, 27%, 41%, 77%, and 100% D2O and dialyzed against the same buffer; 50 mM sodium phosphate buffer (pH 7.4), 300 mM NaCl was made at two different H2O:D2O ratios [13% (vol/vol) and 100% (vol/vol) D2O]. Hydrogenous SDS (h-SDS) was added to 13% D2O buffer (which has the same scattering length density as h-SDS and is thus the lowest SLD we can use) to give a final concentration of 0.5% (wt/vol), whereas 5% (wt/vol) h-SDS and 95% (wt/vol) deuterated SDS were added to 100% D2O buffer to give a final concentration of 0.5% (wt/vol) h/d-SDS with the same SLD as 100% D2O. These two buffers were used in SANS and as stock solutions to prepare 27%, 41%, and 77% D2O buffer in which the SLD of SDS was also matched to the solvent.
SANS experiments were conducted on the D22 beamline at the Institut Laue-Langevin (ILL) at 20 °C using 1-mm quartz rectangular cuvettes. The data were collected using the following instrument configuration: Sample detector distances were 11.2, 5.6, and 1.5 m and the collimation lengths were 11.2, 5.5, and 2.8 m, respectively. This configuration covers the momentum transfer (Q = 4πsinθ/λ, where 2θ is the scattering angle and λ is the neutron wavelength, in this case 6 Å) range between 0.01 and 0.35 Å. The data reduction was carried out using standard protocols implemented in GRASansP software (https://www.ill.eu/instruments-support/instruments-groups/groups/lss/grasp/home/). Neutron scattering contrasts for d/h OmpF–LPS complexes were calculated using program SASSIE (https://sassie-web.chem.utk.edu/sassie2/) (57). Experimental scattering parameters of d/h OmpF–LPS complexes were determined using the Bayesian approach for indirect Fourier transformation of scattering curves obtained in reciprocal space (www.bayesapp.org/) (58, 59). The match points of both protein and LPS were determined as √I0 vs. the D2O fraction in solution (I0 normalized to the sample concentration). A Stuhrmann plot of the square of the radius of gyration () versus the inverse of the contrast between the scattering object and the solvent at different D2O concentrations (Δρ) (33) was obtained using the routine implemented in the program MULCh smb-research.smb.usyd.edu.au/NCVWeb/ (60).
Small-Angle X-Ray Scattering.
SAXS data for dodecylmaltoside-solubilized OmpF–LPS complexes were collected at 20 °C on beamline BM29 (61) at the European Synchrotron Radiation Facility (ESRF). Scattering curves were recorded at a wavelength of 1.008 Å at a sample–detector distance of 2.85 m covering the momentum transfer range 0.05 < Q < 0.45 Å−1. The sample was gel-filtered before SAXS, and the final concentration of samples was between 0.4 and 1 mg/mL. Initial data processing and averaging were carried out according to ref. 62. The scattering curves were converted to the real-space pair-distribution function P(r) using a Bayesian approach (www.bayesapp.org/) (58, 59). The detergent shell of solubilized in vitro folded OmpF and OmpF–LPS complexes was modeled using the program Memprot (30).
Supplementary Material
Acknowledgments
We are grateful to Drs. Anne Martel (ILL) and Christoph Mueller-Dieckmann (ESRF) for their help with small-angle scattering data collection and treatment. We also thank Andrew Dale for his help with analyzing mutants, and Dr. Andrew Whitten for advice on SANS analysis. We thank the Institut Laue-Langevin for beam time on D22 (Proposal 8-03-777/795) and the ESRF for the grant of beam time allocation MX1463 (beamline BM29). This work was supported by a Royal Thai Government Scholarship (to W.A.) and the Wellcome Trust (Grant 093581). M.P. is employed as a PhD student in the EU Marie Curie network (ITN) “Translocation,” project number 607694. The research of B.v.d.B. leading to these results has received support from the Innovative Medicines Initiative’s Joint Undertaking under Grant Agreement 115525, resources that are composed of financial contributions from the European Union’s Seventh Framework Programme (FP7/2007–2013) and European Federation of Pharmaceutical Industries and Associations companies in kind contribution.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The crystallography, atomic coordinates, and structure factors reported in this paper have been deposited in the Protein Data Bank in Europe (PDBe ID code 5FVN).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1602382113/-/DCSupplemental.
References
- 1.Saliba AE, Vonkova I, Gavin AC. The systematic analysis of protein-lipid interactions comes of age. Nat Rev Mol Cell Biol. 2015;16(12):753–761. doi: 10.1038/nrm4080. [DOI] [PubMed] [Google Scholar]
- 2.Nikaido H, Vaara M. Molecular basis of bacterial outer membrane permeability. Microbiol Rev. 1985;49(1):1–32. doi: 10.1128/mr.49.1.1-32.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pagès JM, James CE, Winterhalter M. The porin and the permeating antibiotic: A selective diffusion barrier in gram-negative bacteria. Nat Rev Microbiol. 2008;6(12):893–903. doi: 10.1038/nrmicro1994. [DOI] [PubMed] [Google Scholar]
- 4.Rosenbusch JP. Characterization of the major envelope protein from Escherichia coli. Regular arrangement on the peptidoglycan and unusual dodecyl sulfate binding. J Biol Chem. 1974;249(24):8019–8029. [PubMed] [Google Scholar]
- 5.Schindler H, Rosenbusch JP. Matrix protein from Escherichia coli outer membranes forms voltage-controlled channels in lipid bilayers. Proc Natl Acad Sci USA. 1978;75(8):3751–3755. doi: 10.1073/pnas.75.8.3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schindler H, Rosenbusch JP. Matrix protein in planar membranes: Clusters of channels in a native environment and their functional reassembly. Proc Natl Acad Sci USA. 1981;78(4):2302–2306. doi: 10.1073/pnas.78.4.2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buehler LK, Kusumoto S, Zhang H, Rosenbusch JP. Plasticity of Escherichia coli porin channels. Dependence of their conductance on strain and lipid environment. J Biol Chem. 1991;266(36):24446–24450. [PubMed] [Google Scholar]
- 8.Holzenburg A, et al. Rapid isolation of OmpF porin-LPS complexes suitable for structure-function studies. Biochemistry. 1989;28(10):4187–4193. doi: 10.1021/bi00436a010. [DOI] [PubMed] [Google Scholar]
- 9.Hoenger A, Gross H, Aebi U, Engel A. Localization of the lipopolysaccharides in metal-shadowed reconstituted lipid-porin membranes. J Struct Biol. 1990;103(2):185–195. [Google Scholar]
- 10.Jap BK, Downing KH, Walian PJ. Structure of PhoE porin in projection at 3.5 Å resolution. J Struct Biol. 1990;103(1):57–63. doi: 10.1016/1047-8477(90)90086-r. [DOI] [PubMed] [Google Scholar]
- 11.Vakorina TI, et al. Interaction of porin from Yersinia pseudotuberculosis with different structural forms of endogenous lipopolysaccharide. Biochemistry (Mosc) 2003;68(9):976–983. doi: 10.1023/a:1026056411472. [DOI] [PubMed] [Google Scholar]
- 12.Weiss MS, Wacker T, Weckesser J, Welte W, Schulz GE. The three-dimensional structure of porin from Rhodobacter capsulatus at 3 Å resolution. FEBS Lett. 1990;267(2):268–272. doi: 10.1016/0014-5793(90)80942-c. [DOI] [PubMed] [Google Scholar]
- 13.Cowan SW, et al. Crystal structures explain functional properties of two E. coli porins. Nature. 1992;358(6389):727–733. doi: 10.1038/358727a0. [DOI] [PubMed] [Google Scholar]
- 14.Fairman JW, Noinaj N, Buchanan SK. The structural biology of β-barrel membrane proteins: A summary of recent reports. Curr Opin Struct Biol. 2011;21(4):523–531. doi: 10.1016/j.sbi.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garavito RM, Rosenbusch JP. Isolation and crystallization of bacterial porin. Methods Enzymol. 1986;125:309–328. doi: 10.1016/s0076-6879(86)25027-2. [DOI] [PubMed] [Google Scholar]
- 16.Parr TR, Jr, Poole K, Crockford GWK, Hancock REW. Lipopolysaccharide-free Escherichia coli OmpF and Pseudomonas aeruginosa protein P porins are functionally active in lipid bilayer membranes. J Bacteriol. 1986;165(2):523–526. doi: 10.1128/jb.165.2.523-526.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Visudtiphole V, Thomas MB, Chalton DA, Lakey JH. Refolding of Escherichia coli outer membrane protein F in detergent creates LPS-free trimers and asymmetric dimers. Biochem J. 2005;392(Pt 2):375–381. doi: 10.1042/BJ20051257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baboolal TG, et al. Colicin N binds to the periphery of its receptor and translocator, outer membrane protein F. Structure. 2008;16(3):371–379. doi: 10.1016/j.str.2007.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferguson AD, Hofmann E, Coulton JW, Diederichs K, Welte W. Siderophore-mediated iron transport: Crystal structure of FhuA with bound lipopolysaccharide. Science. 1998;282(5397):2215–2220. doi: 10.1126/science.282.5397.2215. [DOI] [PubMed] [Google Scholar]
- 20.Ferguson AD, et al. A conserved structural motif for lipopolysaccharide recognition by procaryotic and eucaryotic proteins. Structure. 2000;8(6):585–592. doi: 10.1016/s0969-2126(00)00143-x. [DOI] [PubMed] [Google Scholar]
- 21.Eren E, Murphy M, Goguen J, van den Berg B. An active site water network in the plasminogen activator Pla from Yersinia pestis. Structure. 2010;18(7):809–818. doi: 10.1016/j.str.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 22.Eren E, van den Berg B. Structural basis for activation of an integral membrane protease by lipopolysaccharide. J Biol Chem. 2012;287(28):23971–23976. doi: 10.1074/jbc.M112.376418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bolla JM, Lazdunski C, Pagès JM. The assembly of the major outer membrane protein OmpF of Escherichia coli depends on lipid synthesis. EMBO J. 1988;7(11):3595–3599. doi: 10.1002/j.1460-2075.1988.tb03237.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rachel R, Engel AM, Huber R, Stetter KO, Baumeister W. A porin-type protein is the main constituent of the cell envelope of the ancestral eubacterium Thermotoga maritima. FEBS Lett. 1990;262(1):64–68. [Google Scholar]
- 25.Laird MW, Kloser AW, Misra R. Assembly of LamB and OmpF in deep rough lipopolysaccharide mutants of Escherichia coli K-12. J Bacteriol. 1994;176(8):2259–2264. doi: 10.1128/jb.176.8.2259-2264.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Cock H, Tommassen J. Lipopolysaccharides and divalent cations are involved in the formation of an assembly-competent intermediate of outer-membrane protein PhoE of E.coli. EMBO J. 1996;15(20):5567–5573. [PMC free article] [PubMed] [Google Scholar]
- 27.Noinaj N, Rollauer SE, Buchanan SK. The β-barrel membrane protein insertase machinery from gram-negative bacteria. Curr Opin Struct Biol. 2015;31:35–42. doi: 10.1016/j.sbi.2015.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clifton LA, et al. An accurate in vitro model of the E. coli envelope. Angew Chem Int Ed Engl. 2015;54(41):11952–11955. doi: 10.1002/anie.201504287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Diedrich DL, Stein MA, Schnaitman CA. Associations of Escherichia coli K-12 OmpF trimers with rough and smooth lipopolysaccharides. J Bacteriol. 1990;172(9):5307–5311. doi: 10.1128/jb.172.9.5307-5311.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pérez J, Koutsioubas A. Memprot: A program to model the detergent corona around a membrane protein based on SEC-SAXS data. Acta Crystallogr D Biol Crystallogr. 2015;71(Pt 1):86–93. doi: 10.1107/S1399004714016678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Le Brun AP, Clifton LA, Holt SA, Holden PJ, Lakey JH. Deuterium labeling strategies for creating contrast in structure-function studies of model bacterial outer membranes using neutron reflectometry. Methods Enzymol. 2016;566:231–252. doi: 10.1016/bs.mie.2015.05.020. [DOI] [PubMed] [Google Scholar]
- 32.Clifton LA, et al. Low resolution structure and dynamics of a colicin-receptor complex determined by neutron scattering. J Biol Chem. 2012;287(1):337–346. doi: 10.1074/jbc.M111.302901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stuhrmann HB, Fuess H. Neutron small-angle scattering study of hen egg-white lysozyme. Acta Crystallogr A. 1976;32:67–74. [Google Scholar]
- 34.Svergun DI, et al. Protein hydration in solution: Experimental observation by X-ray and neutron scattering. Proc Natl Acad Sci USA. 1998;95(5):2267–2272. doi: 10.1073/pnas.95.5.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arunmanee W, Harris JR, Lakey JH. Outer membrane protein F stabilised with minimal amphipol forms linear arrays and LPS-dependent 2D crystals. J Membr Biol. 2014;247(9–10):949–956. doi: 10.1007/s00232-014-9640-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng H, et al. Validation of metal-binding sites in macromolecular structures with the CheckMyMetal web server. Nat Protoc. 2014;9(1):156–170. doi: 10.1038/nprot.2013.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoenger A, Pagès JM, Fourel D, Engel A. The orientation of porin OmpF in the outer membrane of Escherichia coli. J Mol Biol. 1993;233(3):400–413. doi: 10.1006/jmbi.1993.1520. [DOI] [PubMed] [Google Scholar]
- 38.Hoenger A, Ghosh R, Schoenenberger CA, Aebi U, Engel A. Direct in situ structural analysis of recombinant outer membrane porins expressed in an OmpA-deficient mutant Escherichia coli strain. J Struct Biol. 1993;111(3):212–221. doi: 10.1006/jsbi.1993.1051. [DOI] [PubMed] [Google Scholar]
- 39.Jarosławski S, Duquesne K, Sturgis JN, Scheuring S. High-resolution architecture of the outer membrane of the gram-negative bacteria Roseobacter denitrificans. Mol Microbiol. 2009;74(5):1211–1222. doi: 10.1111/j.1365-2958.2009.06926.x. [DOI] [PubMed] [Google Scholar]
- 40.Schabert FA, Engel A. Reproducible acquisition of Escherichia coli porin surface topographs by atomic force microscopy. Biophys J. 1994;67(6):2394–2403. doi: 10.1016/S0006-3495(94)80726-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spector J, et al. Mobility of BtuB and OmpF in the Escherichia coli outer membrane: Implications for dynamic formation of a translocon complex. Biophys J. 2010;99(12):3880–3886. doi: 10.1016/j.bpj.2010.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rassam P, et al. Supramolecular assemblies underpin turnover of outer membrane proteins in bacteria. Nature. 2015;523(7560):333–336. doi: 10.1038/nature14461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnson CL, et al. The antibacterial toxin colicin N binds to the inner core of lipopolysaccharide and close to its translocator protein. Mol Microbiol. 2014;92(3):440–452. doi: 10.1111/mmi.12568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kloser A, Laird M, Deng M, Misra R. Modulations in lipid A and phospholipid biosynthesis pathways influence outer membrane protein assembly in Escherichia coli K-12. Mol Microbiol. 1998;27(5):1003–1008. doi: 10.1046/j.1365-2958.1998.00746.x. [DOI] [PubMed] [Google Scholar]
- 45.Koplow J, Goldfine H. Alterations in the outer membrane of the cell envelope of heptose-deficient mutants of Escherichia coli. J Bacteriol. 1974;117(2):527–543. doi: 10.1128/jb.117.2.527-543.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Patel GJ, Kleinschmidt JH. The lipid bilayer-inserted membrane protein BamA of Escherichia coli facilitates insertion and folding of outer membrane protein A from its complex with Skp. Biochemistry. 2013;52(23):3974–3986. doi: 10.1021/bi400103t. [DOI] [PubMed] [Google Scholar]
- 47.Watanabe Y, Sano Y, Takagi T. Refolding of Escherichia coli outer membrane protein porin. Prog Biophys Mol Biol. 1996;65(Suppl 1):54. [Google Scholar]
- 48.Bulieris PV, Behrens S, Holst O, Kleinschmidt JH. Folding and insertion of the outer membrane protein OmpA is assisted by the chaperone Skp and by lipopolysaccharide. J Biol Chem. 2003;278(11):9092–9099. doi: 10.1074/jbc.M211177200. [DOI] [PubMed] [Google Scholar]
- 49.Han L, et al. Structure of the BAM complex and its implications for biogenesis of outer-membrane proteins. Nat Struct Mol Biol. 2016;23(3):192–196. doi: 10.1038/nsmb.3181. [DOI] [PubMed] [Google Scholar]
- 50.Gu Y, et al. Structural basis of outer membrane protein insertion by the BAM complex. Nature. 2016;531(7592):64–69. doi: 10.1038/nature17199. [DOI] [PubMed] [Google Scholar]
- 51.Bakelar J, Buchanan SK, Noinaj N. The structure of the β-barrel assembly machinery complex. Science. 2016;351(6269):180–186. doi: 10.1126/science.aad3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Artero JB, Härtlein M, McSweeney S, Timmins P. A comparison of refined X-ray structures of hydrogenated and perdeuterated rat gammaE-crystallin in H2O and D2O. Acta Crystallogr D Biol Crystallogr. 2005;61(Pt 11):1541–1549. doi: 10.1107/S0907444905028532. [DOI] [PubMed] [Google Scholar]
- 53.Prilipov A, Phale PS, Van Gelder P, Rosenbusch JP, Koebnik R. Coupling site-directed mutagenesis with high-level expression: Large scale production of mutant porins from E. coli. FEMS Microbiol Lett. 1998;163(1):65–72. doi: 10.1111/j.1574-6968.1998.tb13027.x. [DOI] [PubMed] [Google Scholar]
- 54.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53(Pt 3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 56.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 57.Curtis JE, Raghunandan S, Nanda H, Krueger S. SASSIE: A program to study intrinsically disordered biological molecules and macromolecular ensembles using experimental scattering restraints. Comput Phys Commun. 2012;183(2):382–389. [Google Scholar]
- 58.Hansen S. Simultaneous estimation of the form factor and structure factor for globular particles in small-angle scattering. J Appl Crystallogr. 2008;41(2):436–445. [Google Scholar]
- 59.Hansen S. BayesApp: A web site for indirect transformation of small-angle scattering data. J Appl Crystallogr. 2012;45(3):566–567. [Google Scholar]
- 60.Whitten AE, Cai S, Trewhella J. MULCh: Modules for the analysis of small-angle neutron contrast variation data from biomolecular assemblies. J Appl Crystallogr. 2008;41(1):222–226. [Google Scholar]
- 61.Pernot P, et al. New beamline dedicated to solution scattering from biological macromolecules at the ESRF. J Phys Conf Ser. 2010;247(1):012009. [Google Scholar]
- 62.Franke D, Kikhney A, Svergun D. Automated acquisition and analysis of small angle X-ray scattering data. Nucl Instrum Methods Phys Res A. 2012;689:52–59. [Google Scholar]
- 63.Heinrichs DE, Yethon JA, Whitfield C. Molecular basis for structural diversity in the core regions of the lipopolysaccharides of Escherichia coli and Salmonella enterica. Mol Microbiol. 1998;30(2):221–232. doi: 10.1046/j.1365-2958.1998.01063.x. [DOI] [PubMed] [Google Scholar]
- 64.Seydel U, Koch MHJ, Brandenburg K. Structural polymorphisms of rough mutant lipopolysaccharides Rd to Ra from Salmonella minnesota. J Struct Biol. 1993;110(3):232–243. doi: 10.1006/jsbi.1993.1026. [DOI] [PubMed] [Google Scholar]
- 65.Schrödinger 2010 The PyMOL Molecular Graphics System, Version 1.3r1. www.pymol.org.
- 66.Stierand K, Rarey M. Drawing the PDB: Protein-ligand complexes in two dimensions. ACS Med Chem Lett. 2010;1(9):540–545. doi: 10.1021/ml100164p. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.