Abstract
The first ever genome-wide association study (GWAS) of ulcerative colitis in genetically distinct north Indian population identified two novel genes namely CFB and SLC44A4. Considering their biological relevance, we investigated allelic/genetic heterogeneity in these genes among ulcerative colitis cohorts of north Indian, Japanese and Dutch origin using high-density ImmunoChip case–control genotype data. Comparative linkage disequilibrium profiling and test of association were performed. Of the 28 CFB SNPs, similar strength of association was observed for rs4151657 (novel ulcerative colitis GWAS SNP) in north Indians (P=1.73 × 10−10) and Japanese (P=2.02 × 10−12) but not in the Dutch. Further, a three-marker haplotype was shared between north Indians and Japanese (P<10−8), but a different five-marker haplotype was associated (P=2.07 × 10−6) in the Dutch. Of the 22 SLC44A4 SNPs, rs2736428 (novel ulcerative colitis GWAS SNP) was found significantly associated in north Indians (P=4.94 × 10−10) and Japanese (P=3.37 × 10−9), but not among the Dutch. These results suggest (i) apparent allelic heterogeneity in CFB and genetic heterogeneity in SLC44A4 across different ethnic groups; (ii) shared ulcerative colitis genetic etiological factors among Asians; and finally (iii) re-exploration of GWAS findings together with high-density genotyping/sequencing and trans-ethnic fine mapping approaches may help identify shared and population-specific risk variants and enable to explain missing disease heritability.
Introduction
Ulcerative colitis (UC), a subtype of inflammatory bowel disorder (IBD), is a complex autoimmune disorder of severe medical consequences. Multiple genetic along with environmental and immunological factors and their interactions contribute to susceptibility to the disease.1 This condition is emerging as an important health problem in India with an incidence rate of 6.02/105 persons/year and a crude prevalence rate of 44.3/105 individuals, which is comparable to the west, where incidence is 3–15/105/year and prevalence is 50–80/105. But these statistics are much higher than other Asian countries like Japan and Korea, with incidence rates of 1.95/105/year and 1.23/105/year, respectively, and prevalence rates of 5.5–18.12/105 and 7.57/105, respectively.2
Over the preceding years, several potential UC associated loci were identified, initially via genome-wide linkage scans and thereafter by genome-wide association studies (GWASs) and their meta-analysis revealing new insights into UC pathogenesis.3, 4, 5, 6, 7, 8, 9 However, most of these studies were primarily carried out in European populations. Recently, International Inflammatory Bowel Disease Genetics Consortium (IIBDGC) conducted a trans-ancestry study using new genotype array, called Immunochip. The chip was designed to densely genotype overlapping risk loci among common immune-mediated diseases. This study substantially increased the number of known genetic risk loci for IBD to 200.10 The non-European UC GWAS performed to date also identified novel susceptibility loci11 and revealed shared UC risk loci between European and non-European cohorts.12 These UC-specific studies also confirmed the long-established association between UC and the classical human leukocyte antigen (HLA) locus, which contains genes encoding antigen-presenting proteins, and plays a crucial role in the regulation of the adaptive immune system.
Our first ever GWAS on UC from the genetically distinct north Indian (NI) population identified seven novel susceptibility genes namely CFB, SLC44A4, 3.8-1/HCG26, MSH5, NOTCH4, HSPA1L and BAT2 from the extended HLA region and were shown to be HLA independent based on conditional regression analysis.13 Of these seven novel genes, the two top significant hits, namely CFB (rs4151657; P=5.10 × 10−14) and SLC44A4 (rs2736428; P=4.86 × 10−11), were selected for further analysis.
Complement activation can occur via three pathways: classical, alternative or the lectin pathway. CFB (Complement factor B; 6141 bp) encodes a secreted protein that is involved in the alternative pathway of complement activation and is expressed mainly by liver and mononuclear phagocytes.14, 15 The complement system has an important role to play in the body and is involved in lysis of pathogens, opsonization, inflammation and immune clearance,16 thus warranting perfect regulation. Improper regulation of the complement system has been implicated in a number of autoimmune and inflammatory disorders.17 Variations within CFB have been previously associated with age-related macular degeneration18 and atypical hemolytic uremic syndrome,19 suggesting its potential role in inflammatory disorders. A recent study20 showed overexpression of CFB mRNA in inflamed versus normal colonic mucosa of IBD patients, suggesting its role in IBD pathogenesis by inappropriate activation of the complement system, contributing to chronic inflammation, one of the hallmarks of UC. This confirms the role of CFB in UC etiology and further supports our novel GWAS findings. Based on this knowledge, complete exon resequencing of CFB in 50 NI UC cases to identify novel UC associated variant(s) revealed five reported SNPs, one non-synonymous in exon 1 (rs4151667 T>A), two adjacent non-synonymous in exon 2 (rs12614 C>T, rs641153 G>A) and two synonymous (rs1048709 G>A in exon 3 and rs4151669 G>A in exon 4), all of which were in the same haplotype block (D′=1) with the GWAS index SNP rs4151657 within intron 10. Of these, rs12614 was predicted to be the most damaging on the basis of in silico analysis and was taken forward for functional analysis. The % alternate pathway activity assessed in the 52 UC case sera samples with 29 wild-type homozygous (CC) and 23 heterozygous and homozygous variant (CT+TT) genotypes of rs12614 revealed significantly (P=0.01) lower activity in the latter group.13 These findings correlate to lower hemolytic activity of variant CFB which is consistent with the autoimmune nature of the disease, resultant lower efficiency of clearance of pathogens and thus increased susceptibility to infections and consequently disease development.
Next, an extensive investigation of structural and regulatory variants within SLC44A4 (solute carrier family 44, member 4; 15855 bp) was undertaken,21 which revealed possible functional relevance of this gene in UC biology. The protein encoded by this gene, also named TPPT (thiamine pyrophosphate transporter), is a transmembrane thiamine pyrophosphate transporter expressed mainly in the colon. It has been suggested that TPPT plays an important role in the uptake of thiamine pyrophosphate generated in the colon by gut microbiota, thus contributing to thiamine nutrition, especially of the colonocytes.22 It has been observed that chronic fatigue in IBD is a consequence of mild thiamine deficiency.23
Given the biological relevance of CFB and SLC44A4 in UC pathogenesis as exemplified by our work, the present study evaluated allelic heterogeneity in these two genes across three genetically divergent populations namely NI, Japanese and Dutch to (a) corroborate our GWAS findings and (b) identify population-specific signals by utilizing high-density ImmunoChip genotype data generated as a part of the IIBDGC project.
Subjects and methods
ImmunoChip genotype data and quality control
Genotype data for a total of 28 SNPs within CFB (~6 kb) and 22 SNPs within SLC44A4 (~16 kb) were retrieved from the total genotype data generated on an Illumina Infinium ImmunoChip platform, a custom-made chip with 196 524 markers used in a recently completed trans-ethnic ImmunoChip study.10 Sample quality control (QC) for the Indian and Japanese study samples was done using PLINK v1.07 (http://pngu.mgh.harvard.edu/purcell/plink/).24 Samples with ambiguous sex, missing genotype rate ≥0.02 and outlying heterozygosity rate (threshold=mean±4 SD) were removed. Sample QC for Dutch study samples are detailed elsewhere.10
Study participants
Indian UC patients and controls were self-reported north Indians, recruited from Dayanand Medical College and Hospital, Ludhiana, Punjab state. These were a subset of the larger cohort previously used for the GWAS as detailed elsewhere.13 Similarly, Japanese UC patients were recruited from the Kyushu University with 25 affiliated hospitals. Controls were collected from the Midosuji and other related Rotary Clubs and the BioBank Japan project. All these samples were used in previous studies.11, 25 Dutch UC patients were recruited from the outpatient IBD clinic at the Department of Gastroenterology and Hepatology, University of Groningen and University Medical Center Groningen, the Netherlands. Control DNA samples were derived from healthy blood donors. All these samples were used in previous studies.9 All the three sample sets have been included in the recent ImmunoChip analysis.10 Briefly, UC subjects were diagnosed according to standard clinical diagnostic criteria. The controls were age, sex and ethnicity matched healthy unrelated blood donors with no history of chronic inflammatory autoimmune or infectious diseases. Informed consent was obtained from each participant, and approval for the study was obtained from the ethical committees of respective institutions.
Statistical analyses
Firstly, LD was estimated in each of the three populations using Haploview 4.2 (http://www.broadinstitute.org/haploview/haploview).26 We next performed single SNP and haplotypic association analyses using PLINK v1.07 (http://pngu.mgh.harvard.edu/purcell/ plink/).24 Sliding window haplotypes were generated using UNPHASED 3.1.5.27 P-values for individual marker and sliding window haplotypes were represented graphically using Graphical Assessment of Sliding P-values (GrASP v0.82 beta) (http://research.nhgri.nih.gov/ GrASP/)28 to present and assess P-values from multiple tests.
In silico analysis of SNPs
SIFT (http://sift.jcvi.org/);29 PolyPhen2 (http://genetics.bwh.harvard.edu/pph2/);30 PolyMiRTS (http://compbio.uthsc.edu/miRSNP/)31, 32 and RegulomeDB (http://regulomedb.org/)33 were used for in silico characterization of SNPs analyzed in this study.
The association data for NI, Japanese and Dutch populations have been submitted to GWAS central database (Submission ID: HGVST 1840) available at the URL http://www.gwascentral.org/study/HGVST1840.
Results
CFB
ImmunoChip genotype data for 28 CFB SNPs (Table 1) obtained for NI (897 cases and 896 controls), Japanese (719 cases and 3263 controls) and Dutch (1729 cases and 1350 controls) UC case–control cohorts were tested for allelic and haplotypic association separately and population-wise results are presented below.
Table 1. Association status of CFB SNPs with UC in north Indian, Japanese and Dutch populations.
| North Indian | Japanese | Dutch | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MAF | P-value | OR (95% CI) | MAF | P-value | OR (95% CI) | MAF | P-value | OR (95% CI) | ||||||
| SNP | Positiona(Build hg19/GRCh38.p2) | Regionb | Cases | Controls | Cases | Controls | Cases | Controls | ||||||
| rs9332730 C>G | Chr6:g.31944232 | 5′ near gene | Failed HWE | Not called | Failed HWE | |||||||||
| rs4151648 G>A | Chr6:g.31945298 | 5′ near gene | Monomorphic | Monomorphic | Monomorphic | |||||||||
| rs4151667 T>A Leu9His | Chr6:g.31946247 | Exon 1 | 0.05 | 0.06 | 0.71 | 0.95 (0.71–1.26) | 0.02 | 0.03 | 0.02 | 0.61 (0.39–0.94) | 0.05 | 0.05 | 0.44 | 1.1 (0.87–1.39) |
| rs12614 G>A Arg32Trp | Chr6:g.31946402 | Exon 2 | 0.17 | 0.23 | 6.57 × 10−5 | 0.72 (0.61–0.84) | 0.05 | 0.07 | 0.01 | 0.73 (0.57–0.93) | 0.09 | 0.1 | 0.06 | 0.85 (0.72–1) |
| rs13194698 G>A | Chr6:g.31946896 | Intron 2 | 0.02 | 0.05 | 4.66 × 10−6 | 0.42 (0.29–0.62) | 0.002 | 0.007 | 0.03 | 0.3 (0.09–0.97) | 0.01 | 0.01 | 0.64 | 0.89 (0.56–1.42) |
| rs11754061 T>G Ser118Ala | Chr6:g.31947060 | Exon 3 | Monomorphic | Monomorphic | Not in 1000 genome | |||||||||
| rs4151650 C>T Tyr135Tyr | Chr6:g.31947113 | Exon 3 | Monomorphic | Monomorphic | MAF<0.001 | |||||||||
| rs1048709 G>A Arg150Arg | Chr6:g.31947158 | Exon 3 | 0.09 | 0.12 | 0.006 | 0.74 (0.6–0.92) | 0.22 | 0.27 | 0.0004 | 0.78 (0.69–0.9) | 0.19 | 0.21 | 0.06 | 0.89 (0.78–1) |
| rs4151669 G>A Pro168Pro | Chr6:g.31947367 | Exon 4 | 0.05 | 0.05 | 0.88 | 0.98 (0.73–1.31] | 0.01 | 0.02 | 0.23 | 0.73 (0.44–1.22) | 0.05 | 0.05 | 0.48 | 1.09 (0.86–1.38) |
| rs4151670 G>A Tyr224Tyr | Chr6:g.31947755 | Exon 5 | 0.02 | 0.04 | 0.005 | 0.56 (0.37–0.85) | 0.002 | 0.006 | 0.05 | 0.33 (0.1–1.07] | 0.02 | 0.02 | 0.99 | 1 (0.68–1.47] |
| rs4151651 G>A Gly252Ser | Chr6:g.31947837 | Exon 5 | 0.006 | 0.005 | 0.49 | 1.38 (0.55–3.43) | Monomorphic | 0.04 | 0.03 | 0.13 | 1.24 (0.94–1.63) | |||
| rs4151652 G>A | Chr6:g.31948169 | Intron 6 | Monomorphic | Monomorphic | 0.002 | 0.001 | 0.38 | 1.82 (0.47–7.06) | ||||||
| rs17201431 A>G | Chr6:g.31948236 | Intron 6 | 0.01 | 0.006 | 0.01 | 2.52 (1.21–5.26] | 0.006 | 0.003 | 0.11 | 1.85 (0.85–4.03) | 0.03 | 0.02 | 0.61 | 1.09 (0.79–1.51) |
| rs512559 A>G | Chr6:g.31948285 | Intron 6 | Monomorphic | Monomorphic | 0.03 | 0.03 | 0.31 | 1.16 (0.87–1.55) | ||||||
| rs537160 G>A | Chr6:g.31948623 | Intron 7 | 0.11 | 0.13 | 0.14 | 0.86 (0.7–1.05) | 0.26 | 0.3 | 0.001 | 0.81 (0.71–0.92) | 0.33 | 0.38 | 4.29 × 10−5 | 0.8 (0.72–0.89) |
| rs1803306 G>T Met365Ile | Chr6:g.31948888 | Exon 8 | Monomorphic | Monomorphic | Not in 1000 genome | |||||||||
| rs4151654 A>G | Chr6:g.31949139 | Intron 8 | Monomorphic | Monomorphic | MAF<0.001 | |||||||||
| rs541862 A>G | Chr6:g.31949174 | Intron 8 | 0.13 | 0.14 | 0.3 | 0.91 (0.75–1.09) | 0.08 | 0.08 | 0.52 | 0.93 (0.76–1.15) | 0.09 | 0.08 | 0.19 | 1.13 (0.94–1.36) |
| rs2072634 G>A Val455Val | Chr6:g.31949514 | Exon 10 | 0.11 | 0.11 | 0.76 | 1.03 (0.84–1.27) | 0.08 | 0.08 | 0.91 | 0.99 (0.8–1.22) | 0.03 | 0.03 | 0.71 | 1.06 (0.78–1.44) |
| rs4151656 A>C | Chr6:g.31949564 | Intron 10 | Monomorphic | Not called | Monomorphic | |||||||||
| rs4151657 A>G (NI GWAS INDEX SNP) | Chr6:g.31949763 | Intron 10 | 0.41 | 0.31 | 1.73 × 10−10 | 1.56 (1.36–1.79) | 0.52 | 0.42 | 2.02 × 10−12 | 1.5 (1.34–1.69) | 0.36 | 0.32 | 0.002 | 1.19 (1.07–1.32) |
| rs4151659 A>G Lys565Glu | Chr6:g.31950687 | Exon 13 | Monomorphic | Monomorphic | 0.008 | 0.008 | 0.78 | 0.92 (0.52–1.63) | ||||||
| rs1270942 T>C | Chr6:g.31951083 | Intron 14 | Monomorphic | Monomorphic | Not called | |||||||||
| rs4151660 T>G Asp651Glu | Chr6:g.31951241 | Exon 15 | Monomorphic | Monomorphic | MAF0.001 | |||||||||
| rs4151661 C>A | Chr6:g.31951537 | Intron 16 | Monomorphic | Monomorphic | Not in 1000 genome | |||||||||
| rs4151662 C>G | Chr6:g.31951542 | Intron 16 | Monomorphic | Monomorphic | Monomorphic | |||||||||
| rs2072633 G>A | Chr6:g.31951801 | Intron 17 | 0.25 | 0.28 | 0.06 | 0.86 (0.74–1) | 0.4 | 0.47 | 1.3 × 10−6 | 0.75 (0.67–0.84) | 0.43 | 0.47 | 0.003 | 0.86 (0.78–0.95) |
| rs4151672 G>A | Chr6:g.31952053 | 3' UTR | 0.25 | 0.28 | 0.82 | 0.97 (0.72–1.29) | 0.02 | 0.03 | 0.02 | 0.6 (0.39–0.93) | 0.05 | 0.05 | 0.56 | 1.07 (0.85–1.36) |
Example of description of variants – rs9332730: human genome reference sequence hg19 chr6:g.31944232 C>G.
Exons are numbered according to NG_008191.1.
NI UC cohort
CFB coverage on ImmunoChip, QC and LD profile
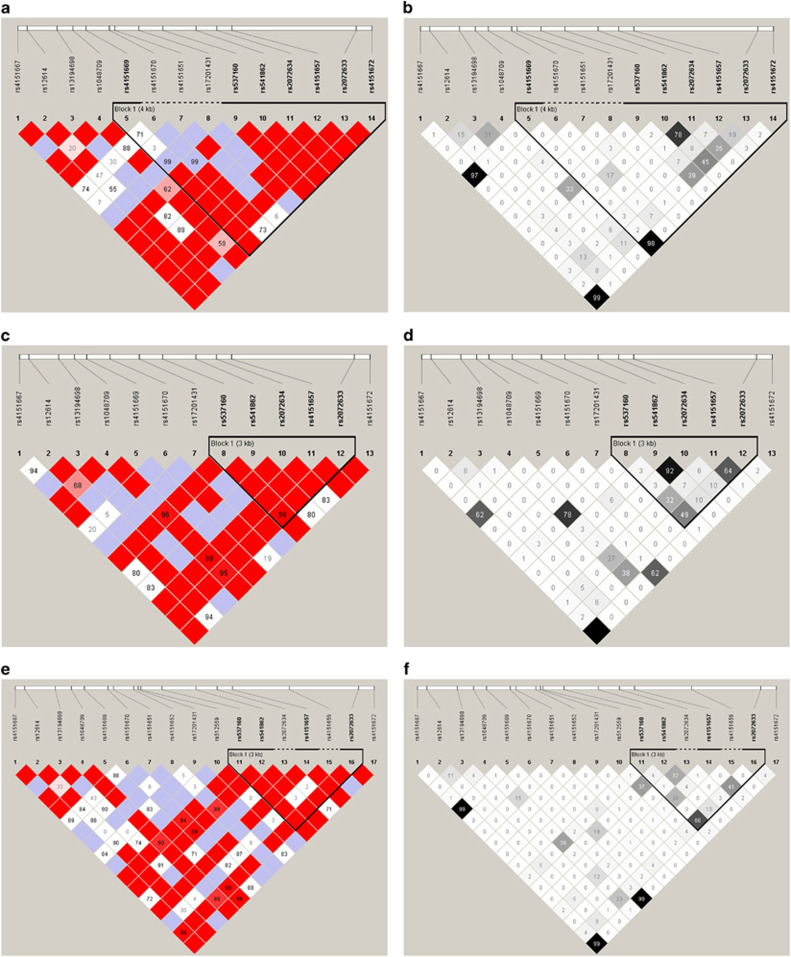
Of the 28 SNPs, 13 were monomorphic and one deviated from Hardy–Weinberg Equilibrium (HWE) (P=2.08 × 10−7). Of the 14 remaining SNPs, each with HWE P>10−3, MAF >0.001 and ≥99.7% genotyping efficiency (Table 1) three exonic SNPs namely rs4151667, rs4151669 and rs4151672 were in LD (r2>0.9) with each other and an intronic SNP rs541862 was in LD (r2=0.78) with rs2072634, an exonic SNP (Figure 1). These 14 SNPs were taken forward for analysis.
Figure 1.
LD plots of CFB SNPs analyzed in NI, Japanese and Dutch populations. LD plot of CFB in NI displaying (a) D′ values and (b) r2 values, LD plot of CFB in Japanese displaying (c) D′ values and (d) r2 values, LD plot of CFB in Dutch displaying (e) D′ values and (f) r2 values.
Allelic association
Of the 14 SNPs, the Indian GWAS index SNP rs4151657 (intron 10) was the most significant (unadjusted P=1.73 × 10−10), and five others namely rs12614 (exon 2), rs13194698 (intron 2), rs1048709 (exon 3), rs4151670 (exon 5) and rs17201431 (intron 6) were nominally associated at P≤0.05 (Table 1).
Haplotypic association
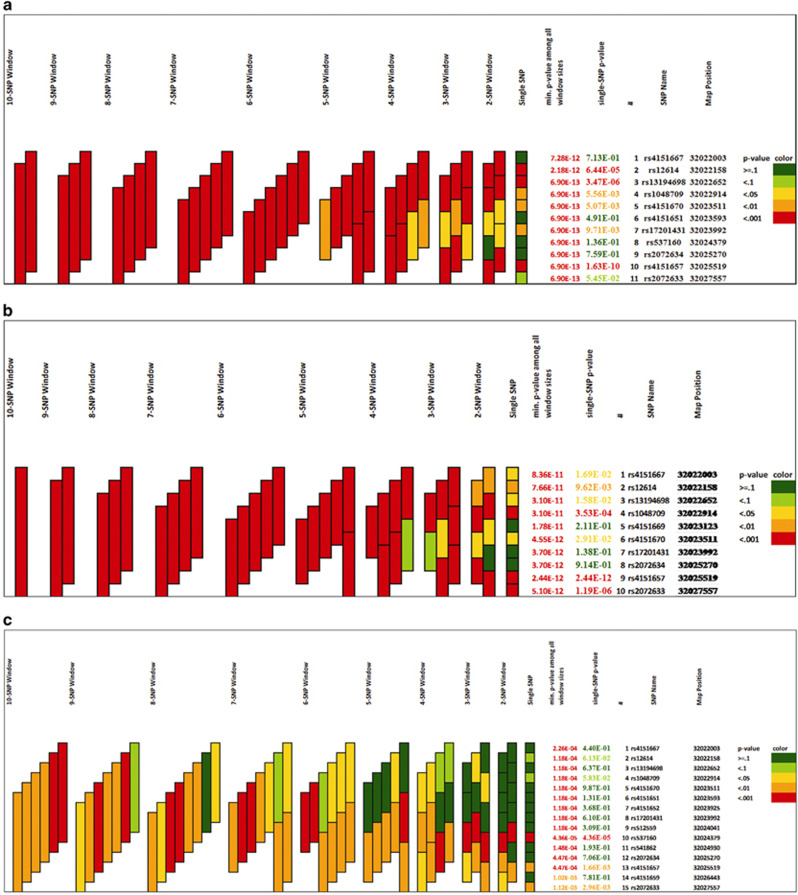
Of the three exonic SNPs in LD namely rs4151667, rs4151669 and rs4151672, only rs4151667 was used as proxy as it was non-synonymous and damaging on in silico predictions and of rs541862 and rs2072634 in LD, rs2072634 which was exonic was retained. Using these 11 markers and 1–11 marker sliding window haplotypes constructed on PLINK, 66 sliding windows and a total of 280 haplotypes with minimum frequency ≥0.01 were generated (Supplementary Table S1). The threshold P-value of <1.8 × 10−4 was set after Bonferroni correction was applied. A number of haplotypes were found significantly associated. A four marker haplotype (rs17201431–rs537160–rs2072634–rs4151657) was the smallest haplotype (A–G–G–G), encompassing the GWAS index SNP rs4151657 that was most significantly associated (P=4.4 × 10−11). Of the 11 marker haplotypes that were generated, one predisposing haplotype (T–G–G–G–G–G–A–G–G–G–G) with frequency 0.42 in cases and 0.31 in controls (377 cases and 278 controls), containing the predisposing alleles of GWAS index SNP rs4151657 and all SNPs except rs17201431 showing allelic association was found to be significantly associated (P=2.7 × 10−11). Global P-values of 1–11 marker sliding window haplotypes generated using UNPHASED 3.1.5 and graphed using GrASP v0.82 beta, keeping a minimum haplotype frequency threshold of 0.001 are presented in Supplementary Table S2 and Figure 2. Of the 11 marker haplotypes generated, the same haplotype as shown above (T–G–G–G–G–G–A–G–G–G–G) was found significantly associated (P=2.4 × 10−10). rs12614, rs13194698 and rs4151657 seem to be the main contributors within CFB, as can be seen from Figure 2.
Figure 2.
CFB haplotype associations illustrated using graphical assessment of sliding P-values (GrASP v0.82 beta) in (a) NI, (b) Japanese and (c) Dutch populations.
Japanese UC cohort
CFB coverage on immunochip, QC and LD profile
Of the 28 SNPs in CFB on ImmunoChip, 2 were not called and 13 were monomorphic/uninformative in the Japanese UC cohort. Of the 13 remaining SNPs, each with MAF >0.001, HWE P>10−3 and ≥99.7% genotyping efficiency (Table 1), the two exonic SNPs rs4151667 and rs4151672 were in LD (r2=1) with each other, an exonic SNP rs1048709 was in LD (r2=0.78) with an intronic SNP rs537160, and an intronic SNP rs541862 was in LD (r2>0.9) with an exonic SNP rs2072634, which is slightly different from the NI pattern (Figure 1). These 13 SNPs were taken forward for analysis.
Allelic association
NI UC GWAS index SNP rs4151657 came up significantly associated (P=2.02 × 10−12) along with eight other SNPs namely rs4151667 (exon 1), rs12614 (exon 2), rs13194698 (intron 2), rs1048709 (exon 3), rs4151670 (exon 5), rs537160 (intron 7), rs2072633 (intron 17) and rs4151672 (3’UTR) showing nominal association (P≤0.05) (Table 1).
Haplotypic association
Of the two exonic SNPs namely rs4151667 and rs4151672 in LD, rs4151667 was retained as it is non-synonymous and damaging on in silico predictions; of rs1048709 and rs537160 in LD, rs1048709 was retained as it is exonic and damaging on in silico predictions; and of rs541862 and rs2072634 in LD, rs2072634 was retained as it is exonic. 1–10 marker sliding window haplotypes constructed on PLINK generated 55 sliding windows and a total of 187 haplotypes with minimum frequency ≥0.01 (Supplementary Table S3). The threshold P-value of <2.7 × 10−4 was set after Bonferroni correction was applied. A number of haplotypes were found significantly associated. A three-marker haplotype (rs17201431–rs2072634–rs4151657) was the smallest haplotype (A–G–G), encompassing the GWAS index SNP rs4151657 that showed most significant association (P=9.6 × 10−13). Of the 10 marker haplotypes that were generated, the same predisposing haplotype (T–C–C–G–G–C–T–C–C–G) that was found associated in NI was found associated in Japanese population (P=2.6 × 10−11) as well, with frequency 0.53 in cases and 0.43 in controls (384 cases and 1407 controls). 1–10 marker sliding window haplotypes generated using UNPHASED 3.1.5 and GrASP v0.82 beta, keeping a minimum haplotype frequency threshold of 0.001, revealed the same pattern of association (Supplementary Table S2 and Figure 2). From Figure 2, it is apparent that the three SNPs namely rs1048709, rs4151657 and rs2072633 are the main drivers for association of this region to UC.
Dutch UC cohort
CFB coverage on ImmunoChip, QC and LD profile
Of the 28 CFB SNPs on ImmunoChip three were not in 1000 Genome, one failed HWE, one failed heterogeneity, and three were monomorphic and three had MAF<0.001 in Dutch UC cohort. Of the remaining 17 SNPs which were analyzed further, each with MAF >0.001, HWE P>10−3 and ≥99.8% genotyping efficiency (Table 1), three exonic SNPs namely rs4151667, rs4151669 and rs4151672 were in LD (r2=0.99) with each other (Figure 1).
Allelic association
India GWAS index SNP rs4151657 showed only a nominal association (P=0.002), along with two other intronic SNPs, namely rs537160 (P=4.29 × 10−5) and rs2072633 (P=0.003, Table 1).
Haplotypic association
Of the three exonic SNPs namely rs4151667, rs4151669 and rs4151672 which were in LD, only rs4151667 was retained as it was non-synonymous and damaging on in silico predictions. 1–15 marker sliding window haplotypes constructed on PLINK generated 120 sliding windows and a total of 695 haplotypes with minimum frequency ≥0.01 (Supplementary Table S4). Some haplotypic combinations withstood the Bonferroni corrected P-value threshold of <7.2 × 10−5. A five-marker haplotype (rs4151651–rs4151652–rs17201431–rs512559–rs537160) was the smallest haplotype (G–G–A–A–A) showing most significant association (P=2.07 × 10−6). It was a protective haplotype with a frequency of 0.3 in cases and 0.35 in controls. 1–15 marker sliding window haplotypes generated using UNPHASED 3.1.5 and GrASP v0.82 beta, keeping a minimum haplotype frequency threshold of 0.001 revealed similar pattern of association (Supplementary Table S2 and Figure 2). As can be seen from Figure 2, rs537160 seems to be the only contributor within this region to UC.
In silico analysis of CFB SNPs
SIFT and POLYPHEN2 prediction of the four missense SNPs namely rs4151667 (exon 1), rs12614 (exon 2), rs4151651 (exon 5) and rs4151659 (exon 13) showed the first two to be damaging (Supplementary Table S5). The 3′ UTR SNP rs4151672 was checked on PolymiRTS Database 3.0, and the reference allele C was found to disrupt two conserved miRNA sites and variant allele T was found to create a new miRNA site, and thus possibly functional. Checking all SNPs on RegulomeDB, most SNPs were predicted to be near DNA features or regulatory elements like transcription factor-binding sites and also affect protein binding. Of note, three SNPs namely rs1048709 (exon 3), rs17201431 (intron 6) and rs2072633 (intron 17), which showed allelic association in either of the three populations (Table 1), were predicted to have cis-eQTL effects on a number of HLA genes (Supplementary Table S6), which are in the vicinity of CFB, which may suggest the role of CFB via HLA genes.
SLC44A4
ImmunoChip genotype data for 22 SLC44A4 SNPs (Table 2) obtained for NI (897 cases and 896 controls), Japanese (724 cases and 3271 controls) and Dutch (1729 cases and 1350 controls) UC case–control cohorts were tested for allelic and haplotypic association separately and population-wise results are presented below.
Table 2. Association status of SLC44A4 SNPs with UC in north Indian, Japanese and Dutch populations.
| North Indian | Japanese | Dutch | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MAF | P-value | OR (95% CI) | MAF | P-value | OR (95% CI) | MAF | P-value | OR (95% CI) | ||||||
| SNP | Positiona (Build hg19/GRCh38.p2) | Regionb | Cases | Controls | Cases | Controls | Cases | Controls | ||||||
| rs9461727 C>A | Chr6:g.31863944 | Intron 20 | 0.01 | 0.02 | 0.13 | 0.66 (0.38–1.14) | 0.003 | 0.005 | 0.22 | 0.53 (0.19–1.5) | 0.002 | 0.002 | 0.94 | 1.04 (0.36–3) |
| rs6915800 G>A Arg493Cys | Chr6:g.31865883 | Exon 14 | Monomorphic | 0 | 0.002 | 0.08 | 0 | Monomorphic | ||||||
| rs4947332 G>A | Chr6:g.31866420 | Intron 13 | 0.02 | 0.05 | 2.31 × 10−5 | 0.46 (0.32–0.66) | 0.01 | 0.02 | 0.03 | 0.55 (0.31–0.95) | 0.03 | 0.03 | 0.45 | 0.89 (0.67–1.2) |
| rs693906 G>C | Chr6:g.31867387 | Intron 13 | 0.08 | 0.1 | 0.22 | 0.87 (0.69–1.09) | 0.06 | 0.07 | 0.13 | 0.83 (0.65–1.06) | 0.14 | 0.16 | 0.006 | 0.82 (0.71–0.94) |
| rs11965547 G>A | Chr6:g.31868374 | Intron 13 | 0.13 | 0.13 | 0.45 | 0.93 (0.76–1.13) | 0.22 | 0.26 | 0.005 | 0.82 (0.72–0.94) | 0.11 | 0.12 | 0.22 | 0.91 (0.77–1.06) |
| rs521977 C>A | Chr6:g.31869050 | Intron 13 | 0.1 | 0.11 | 0.35 | 0.91 (0.73–1.12) | 0.07 | 0.09 | 0.03 | 0.79 (0.64–0.96) | 0.28 | 0.32 | 0.0002 | 0.81 (0.73–0.91) |
| rs660594 A>G | Chr6:g.31869473 | Intron 12 | 0.31 | 0.35 | 0.04 | 0.86 (0.75–0.99) | NOT CALLED | 0.44 | 0.49 | 0.0002 | 0.83 (0.75–0.91) | |||
| rs577272 A>G | Chr6:g.31870186 | Intron 11 | 0.33 | 0.38 | 0.007 | 0.83 (0.72–0.95) | 0.31 | 0.36 | 0.0001 | 0.78 (0.69–0.89) | 0.44 | 0.49 | 0.0004 | 0.83 (0.75–0.92) |
| rs644827 G>A Val326Met | Chr6:g.31870664 | Exon 11 | 0.33 | 0.38 | 0.008 | 0.83 (0.72–0.95f | 0.31 | 0.36 | 0.0001 | 0.78 (0.69–0.89) | 0.44 | 0.49 | 0.0004 | 0.83 (0.75–0.92) |
| rs644774 A>G | Chr6:g.31870713 | Intron 10 | 0.33 | 0.38 | 0.008 | 0.83 (0.72–0.95) | 0.31 | 0.36 | 0.0001 | 0.79 (0.7–0.89) | 0.44 | 0.49 | 0.0004 | 0.83 (0.75–0.92) |
| rs494620 G>A Tyr271Tyr | Chr6:g.31870936 | Exon 10 | 0.48 | 0.41 | 1.25 × 10−5 | 1.34 (1.18–1.53) | 0.36 | 0.43 | 2.68 × 10−6 | 0.75 (0.67–0.85) | 0.41 | 0.37 | 0.002 | 1.18 (1.06–1.31) |
| rs2242665 A>G Ile187Val | Chr6:g.31871532 | Exon 8 | 0.33 | 0.38 | 0.008 | 0.83 (0.72–0.95) | 0.31 | 0.36 | 0.0001 | 0.79 (0.69–0.89) | 0.44 | 0.49 | 0.0004 | 0.83 (0.75–0.92) |
| rs2242664 A>G Gly179Gly | Chr6:g.31871554 | Exon 8 | 0.33 | 0.38 | 0.008 | 0.83 (0.72–0.95) | 0.31 | 0.36 | 0.0001 | 0.79 (0.69–0.89) | 0.44 | 0.49 | 0.0004 | 0.83 (0.75–0.92) |
| rs3132442 A>G | Chr6:g.31871717 | Intron 7 | 0.34 | 0.38 | 0.007 | 0.83 (0.72–0.95) | 0.31 | 0.36 | 0.0001 | 0.79 (0.69–0.89) | 0.46 | 0.51 | 0.0002 | 0.83 (0.75–0.91) |
| rs3130481 C>G | Chr6:g.31871979 | Intron 7 | 0.34 | 0.38 | 0.007 | 0.83 (0.72–0.95) | 0.31 | 0.36 | 0.0001 | 0.78 (0.69–0.89) | 0.46 | 0.51 | 0.0002 | 0.83 (0.75–0.91) |
| rs3130482 A>C | Chr6:g.31872005 | Intron 7 | 0.34 | 0.38 | 0.007 | 0.83 (0.72–0.95) | 0.31 | 0.36 | 0.0001 | 0.78 (0.69–0.89) | 0.46 | 0.51 | 0.0002 | 0.83 (0.75–0.91) |
| rs614549 A>G | Chr6:g.31872848 | Intron 7 | 0.44 | 0.37 | 1.32 × 10−5 | 1.35 (1.18–1.54) | 0.42 | 0.49 | 1.4 × 10−7 | 0.73 (0.65–0.82) | 0.36 | 0.32 | 0.002 | 1.18 (1.06–1.31) |
| rs12661281 A>T Asp123Val | Chr6:g.31874821 | Exon 6 | 0.12 | 0.08 | 0.0002 | 1.52 (1.22–1.9) | 0.1 | 0.13 | 7.8 × 10−5 | 0.68 (0.57–0.83) | 0.16 | 0.14 | 0.03 | 1.18 (1.02–1.36) |
| rs2736428 G>A (NI GWAS INDEX SNP) | Chr6:g.31876147 | Intron 2 | 0.39 | 0.29 | 4.94 × 10−10 | 1.56 (1.35–1.79) | 0.42 | 0.51 | 3.37 × 10−9 | 0.71 (0.63–0.79) | 0.33 | 0.29 | 0.002 | 1.19 (1.07–1.33) |
| rs6907185 A>G | Chr6:g.31876907 | Intron 2 | Failed HWE | 0.002 | 0.006 | 0.05 | 0.33 (0.1–1.07) | 0.07 | 0.07 | 0.36 | 1.1 (0.9–1.34) | |||
| rs9267658 G>A | Chr6:g.31878208 | Intron 1 | 0.04 | 0.04 | 0.67 | 0.93 (0.67–1.29) | MAF<0.001 | 0.13 | 0.16 | 0.01 | 0.83 (0.72–0.96) | |||
| rs9267659 G>A | Chr6:g.31878457 | Intron 1 | 0.11 | 0.12 | 0.48 | 0.93 (0.76–1.14) | 0.07 | 0.09 | 0.03 | 0.79 (0.64–0.98] | 0.21 | 0.22 | 0.36 | 0.94 (0.83–1.07] |
Example of description of variants – rs9461727: human genome reference sequence hg19 chr6:g.31863944 C>A.
Exons are numbered according to NG_023058.1.
NI UC cohort
SLC44A4 coverage on ImmunoChip, QC and LD profile
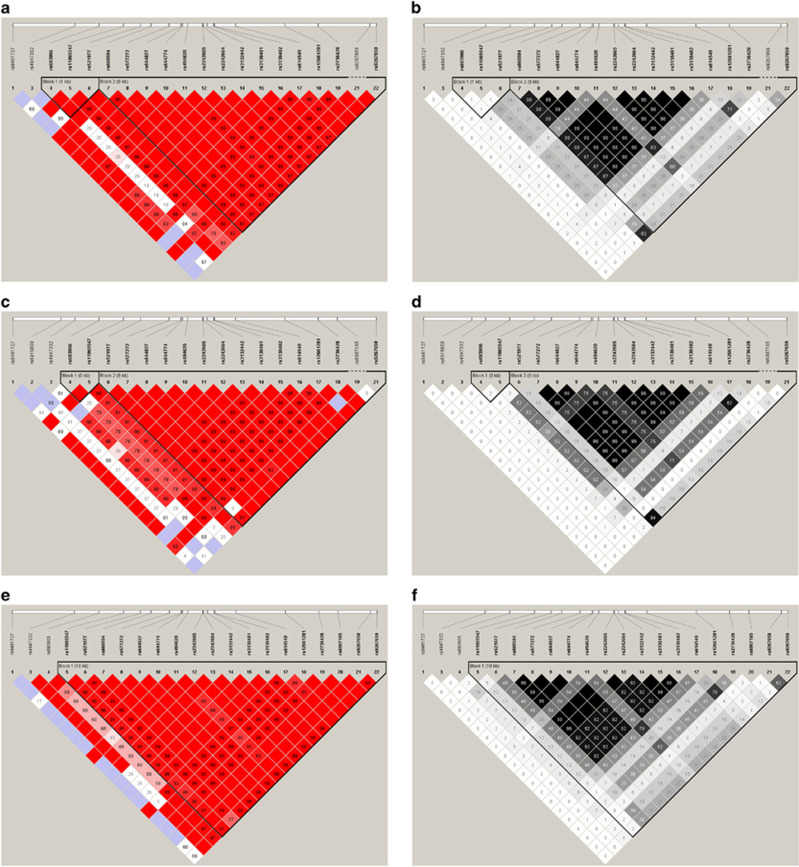
Of the 22 SNPs, one was monomorphic and one deviated from Hardy–Weinberg equilibrium (HWE) (P=7 × 10−4). The 20 remaining SNPs, each with HWE P>10−3, MAF>0.001 and ≥99.9% genotyping efficiency (Table 2), were taken forward for analysis. Nine SNPs namely rs660594, rs577272, rs644827, rs644774, rs2242665, rs2242664, rs3132442, rs3130481 and rs3130482 were in LD (r2≥0.88) with each other; rs494620 was in LD (r2=0.83) with rs614549 and rs521977 and rs9267659 were also in LD (r2=0.82) with each other (Figure 3).21
Figure 3.
LD plots of SLC44A4 SNPs analyzed in NI,21 Japanese and Dutch populations. LD plot of SLC44A4 in NI displaying (a) D′ values and (b) r2 values, LD plot of SLC44A4 in Japanese displaying (c) D ′ values and (d) r2 values, LD plot of SLC44A4 in Dutch displaying (e) D′ values and (f) r2 values.
Allelic association
The NI GWAS index SNP rs2736428 (intron 2) was the most significantly associated (P=4.94 × 10−10), while 13 others were nominally associated at P≤0.05, namely rs4947332 (intron 13), rs660594 (intron 12), rs577272 (intron 11), rs644827 (exon 11), rs644774 (intron 10), rs494620 (exon 10), rs12661281 (exon 6), rs2242665 and rs2242664 (exon 8), rs3132442, rs3130481, rs3130482 and rs614549 (intron 7) (Table 2).
Haplotypic association
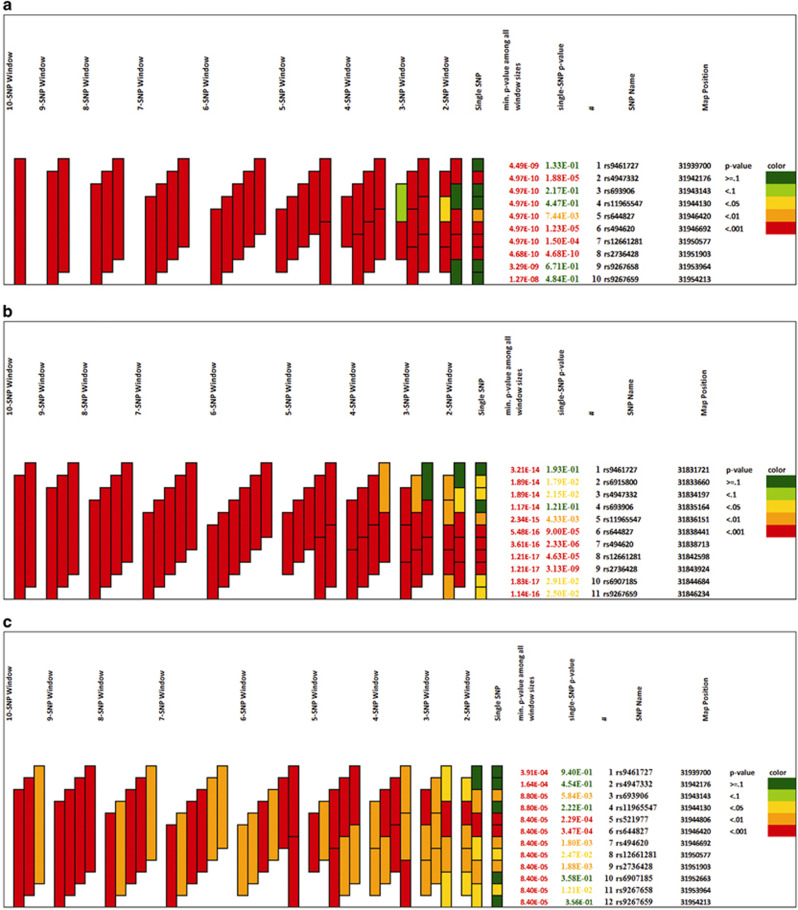
Of the nine SNPs in LD as mentioned above, rs644827 was selected as proxy as it was an exonic missense variant and seemed more damaging than others on in silico predictions. Of rs494620 and rs614549 in LD, rs494620 was retained as it was exonic; and of the two intronic SNPs rs521977 and rs9267659 in LD, rs9267659 was retained as it seemed more likely to have regulatory effects as predicted on RegulomeDB. 1–10 marker sliding window haplotypes constructed on PLINK generated 55 sliding windows and a total of 308 haplotypes with minimum frequency ≥0.01 (Supplementary Table S7). The threshold P-value of <1.6 × 10−4 was set after Bonferroni correction was applied. A number of haplotypes were found significantly associated. A six marker haplotype (rs9461727–rs4947332–rs693906–rs11965547–rs644827–rs494620) was the smallest haplotype (C–G–G–G–G–A) showing most significant association (P=5.97 × 10−11) with a frequency of 0.42 in cases and 0.32 in controls. 1–10 marker sliding window haplotypes generated using UNPHASED 3.1.5 and GrASP v0.82 beta, keeping a minimum haplotype frequency threshold of 0.001 revealed rs4947332, rs494620, rs12661281 and rs2736428 to be the main drivers for association (Supplementary Table S8 and Figure 4).
Figure 4.
SLC44A4 haplotype associations illustrated using graphical assessment of sliding P-values (GrASP v0.82 beta) in (a) NI, (b) Japanese and (c) Dutch population.
Japanese UC cohort
SLC44A4 coverage on immunochip, QC and LD profile
Of the 22 SNPs, one was not called and one had MAF<0.001. The 20 remaining SNPs, each with HWE P>10−3, MAF >0.001 and 100% genotyping efficiency (Table 2) were taken forward for analysis. Eight SNPs namely rs577272, rs644827, rs644774, rs2242665, rs2242664, rs3132442, rs3130481 and rs3130482 were in LD (r2≥0.99) with each other; NI GWAS index SNP rs2736428 was in LD (r2=0.92) with rs614549 and rs521977 and rs9267659 were also in LD (r2=0.94) with each other (Figure 3).
Allelic association
The NI GWAS index SNP rs2736428 (intron 2) was the most significantly associated (P=3.37 × 10−9), while 16 others showed nominal (P≤0.05) to moderate association (P≤10−5) (Table 2).
Haplotypic association
Of the eight SNPs in LD as mentioned above, rs644827 was selected as proxy as it was an exonic missense variant and seemed more damaging than others on in silico predictions. Of rs2736428 and rs614549 in LD, rs2736428 was retained as it showed more significant association; of the two intronic SNPs rs521977 and rs9267659 in LD, rs9267659 was retained as it seemed more likely to have regulatory effects as predicted on RegulomeDB. 1–11 marker sliding window haplotypes constructed on PLINK generated 66 sliding windows and a total of 316 haplotypes with minimum frequency ≥0.01 (Supplementary Table S9). The threshold P-value of <1.6 × 10−4 was set after Bonferroni correction was applied. A number of haplotypes were found significantly associated. A five-marker haplotype (rs11965547–rs644827–rs494620–rs12661281–rs2736428) was the smallest haplotype (G–G–A–A–A) showing most significant association (P=9.91 × 10−20) with a frequency of 0.47 in cases and 0.35 in controls. 1–11 marker sliding window haplotypes generated using UNPHASED 3.1.5 and GrASP v0.82 beta, keeping a minimum haplotype frequency threshold of 0.001 revealed rs644827, rs494620, rs12661281 and rs2736428 to be the main drivers for association (Supplementary Table S8 and Figure 4).
Dutch UC cohort
SLC44A4 coverage on ImmunoChip, QC and LD profile
Of the 22 SNPs, only one was monomorphic. The 21 remaining SNPs, each with HWE P>10−3, MAF >0.001 and ≥99.8% genotyping efficiency (Table 2), were taken forward for analysis. Nine SNPs namely rs660594, rs577272, rs644827, rs644774, rs2242665, rs2242664, rs3132442, rs3130481 and rs3130482 were in LD (r2≥0.9) with each other; NI GWAS index SNP rs2736428 and rs494620 were in LD (r2=0.78) with rs614549 (Figure 3).
Allelic association
Unlike in the other two populations detailed above, the NI GWAS index SNP rs2736428 (intron 2) showed only nominal association (P≤0.05) along with 15 other SNPs (Table 2).
Haplotypic association
rs644827 which is non-synonymous was selected as proxy out of the nine SNPs in LD and out of rs494620 and rs614549 in LD, rs494620 which is exonic was retained. 1–12 marker sliding window haplotypes constructed on PLINK generated 78 sliding windows and a total of 491 haplotypes with minimum frequency ≥0.01 (Supplementary Table S10). Only three haplotypes, namely rs11965547–rs521977 (G–C), rs4947332–rs693906–rs11965547–rs521977 (G–G–G–C) and rs9461727–rs4947332–rs693906–rs11965547–rs521977 (C–G–G–G–C) (P=~10−5), all three common haplotypes with a frequency ~0.6 in cases and ~0.5 in controls crossed the Bonferroni corrected P-value threshold (P≤10−4). 1–12 marker sliding window haplotypes were also generated using UNPHASED 3.1.5 and GrASP v0.82 beta (Supplementary Table S8 and Figure 4).
In silico analysis of SLC44A4 SNPs
The three missense SNPs namely rs12661281, rs2242665 and rs644827 were predicted to be benign on both SIFT and POLYPHEN2.21 RegulomeDB predicted most of the SNPs to be within transcription factor-binding motifs and affect protein binding. Some SNPs were also found to have cis-eQTL effects on a number of HLA genes (Supplementary Table S11).21
Discussion
CFB, a component of the alternate pathway of complement system, emerged as a novel susceptibility gene in the first ever GWAS on UC among NI.13 There is evidence for circulating immune complexes and enhanced production of components of the complement system in IBD in Caucasian populations, suggesting increased complement activation in such patients.34, 35, 36, 37 SLC44A4, a thiamine pyrophosphate transporter, was another of our NI UC GWAS top hits.13 However, neither CFB nor SLC44A4 have been identified in any of the larger Caucasian GWAS,3, 4, 5, 6, 7 their meta-analysis8, 9 and more recently in ImmunoChip analysis,10 or in the non-European UC cohorts studied to date.11, 12 Needless to say, such a striking difference across ethnic groups may be due to inherent statistical limitations of GWAS, which mainly relies on single SNP analysis, incomplete coverage of functional common or rare variants, poor representation of appropriate proxies on commercial genotyping arrays due to population-specific LD patterns, among others factors like allelic/genetic heterogeneity, varying environmental components like gut microbiome influenced by geographical location, lifestyle factors such as diet, smoking, etc. leaving much of the disease heritability unexplained. Keeping in view the biological significance of CFB and SLC44A4, we attempted to identify allelic heterogeneity in these two genes by comparing three populations namely NI, Japanese and Dutch of different ethnic origin.
Of the 28 CFB SNPs present on the ImmunoChip, 14, 13 and 17 were retained after stringent QC in NI, Japanese and Dutch, respectively, while approximately 40% of CFB SNPs present on the ImmunoChip were monomorphic/uninformative (Table 1) in all the three study populations reiterating the need to have population-specific commercial arrays, which will undoubtedly contribute to the black box of missing disease heritability and partially explain non-replication of European findings in other ethnically distinct populations. The reported NI UC GWAS index SNP rs4151657 within CFB consistently showed strong allelic association in the Japanese as well (P=2.02 × 10−8), but nominal in the Dutch (P=0.002, Table 1). It would be interesting to mention that a long-range haplotype in the MHC region (25–35 Mb), including CFB reflected strong association in the Japanese with UC, which they considered as one susceptibility locus.25 On the other hand, rs537160 was suggestively significant (P=4.29 × 10−5) in the Dutch cohort, which is indicative of allelic heterogeneity at CFB. Of the remaining nominally associated SNPs (P≤0.05) in any of the three populations, (a) none were common between NI and Dutch; (b) three exonic and one intronic SNPs were shared between NI and Japanese; and (c) two intronic SNPs were shared between Japanese and Dutch cohorts (Table 1). These promising findings suggest that trans-ethnic fine-mapping efforts using high-density genotyping/sequencing will undoubtedly restore the momentum of causal variant identification in complex disease research and may identify population-specific determinants. Genuine contribution of these alleles to UC may derive further support from the observed absence of LD between these markers in these two populations (Figure 1). Such allelic heterogeneity across distinct ethnic populations is not unexpected and, for example, has already been demonstrated for NOD2 in our previous study on UC patients from north India.38
Considering the associated SNP may not be the only or predominant determinant of the respective gene function and other SNPs in the gene, singly or in haplotypic combinations may contribute to the phenotype, we next estimated haplotypic diversity across the three populations. It may be mentioned that previous association studies have demonstrated high-risk haplotypes for various complex disorders, for example, a rare haplotype within CFH was found associated with age-related macular degeneration39 and haplotypes within STAT4 were found associated with systemic lupus erythematosus.40 Our haplotypic association results further reaffirm these findings. A minimal three-marker haplotype within CFB namely rs17201431–rs2072634–rs4151657 was shared across NI and Japanese (P<10−8), but a different five-marker haplotype namely rs4151651–rs4151652–rs17201431–rs512559–rs537160 was significantly associated (P=2.07 × 10−6) in the Dutch population after Bonferroni corrections. However, in NI and Japanese populations, the association seems to be driven mainly by rs4151657, the India GWAS index SNP and in the Dutch population, it is rs537160 (Figure 2), also identified in allelic association (Table 1). It is also noteworthy that the haplotypes associated in NI (0.41 in cases and 0.31 in controls) or Japanese (0.52 in cases and 0.42 in controls) and Dutch (0.3 in cases and 0.35 in controls) cohorts are rather common. As for the likely role of these two driver SNPs namely rs4151657 and rs537160, they may be involved in regulation of gene expression through transcription factor binding, as predicted by various in silico tools (Supplementary Table S6).
The NI UC GWAS index SNP rs2736428 within SLC44A4 was found significantly associated in Japanese (P=3.37 × 10−9) but only nominally associated (P=0.002) in the Dutch cohorts. Other than this, 11 out of the 22 SNPs within SLC44A4 showed nominal association in all three ethnic groups (Table 2), most of which were predicted to have regulatory effects (Supplementary Table S11). Allelic as well as haplotype associations revealed similar patterns across Indians and Japanese, but a different pattern was observed in the Dutch (Supplementary Table S8 and Figure 4), suggesting genetic heterogeneity across the two populations.
Taken together, our findings unequivocally demonstrate evidence of allelic heterogeneity in CFB and genetic heterogeneity in SLC44A4, biologically relevant genes for UC and utility of trans-ethnic studies. These observations reiterate the contemporary need for fine mapping of known loci and trans-ethnic comparisons for identification of common and unique risk variants. This in turn would have implications for predictive medicine and for further understanding of disease biology.
Acknowledgments
This work was supported by the Centre of Excellence in Genome Sciences and Predictive Medicine, Department of Biotechnology, Government of India (BT/01/COE/07/UDSC/2008) to BKT, AS and VM; VIDI grant (016.136.308) from the Netherlands Organization for Scientific Research (NWO) and the Broad Medical Research Program of The Broad Foundation (IBD-0318) to RKW; and the BioBank Japan Project from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) to MK. We acknowledge the International Inflammatory Bowel Disease Genetics Consortium (IIBDGC) for the Immunochip genotyping data. We thank Ms Anjali Dhyani for preparation of DNA samples and maintenance of the resource in the lab. Junior and senior research fellowship to AG from Council of Scientific and Industrial Research, New Delhi is gratefully acknowledged. We acknowledge infrastructure support provided to the Department of Genetics, UDSC, by the University Grants Commission, New Delhi under the Special Assistance Programme and Department of Science and Technology, New Delhi under FIST and DU-DST PURSE programmes.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
The authors declare no conflict of interest.
Supplementary Material
References
- Galvez J: Role of Th17 cells in the pathogenesis of human IBD. ISRN Inflamm 2014; 2014: 928461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sood A, Midha V, Sood N, Bhatia AS, Avasthi G: Incidence and prevalence of ulcerative colitis in Punjab, North India. Gut 2003; 52: 1587–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke A, Balschun T, Karlsen TH et al: Sequence variants in IL10, ARPC2 and multiple other loci contribute to ulcerative colitis susceptibility. Nat Genet 2008; 40: 1319–1323. [DOI] [PubMed] [Google Scholar]
- Silverberg MS, Cho JH, Rioux JD et al: Ulcerative colitis-risk loci on chromosomes 1p36 and 12q15 found by genome-wide association study. Nat Genet 2009; 41: 216–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Lee JC, Lees CW et al: Genome-wide association study of ulcerative colitis identifies three new susceptibility loci, including the HNF4A region. Nat Genet 2009; 41: 1330–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGovern DP, Gardet A, Torkvist L et al: Genome-wide association identifies multiple ulcerative colitis susceptibility loci. Nat Genet 2010; 42: 332–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke A, Balschun T, Sina C et al: Genome-wide association study for ulcerative colitis identifies risk loci at 7q22 and 22q13 (IL17REL). Nat Genet 2010; 42: 292–294. [DOI] [PubMed] [Google Scholar]
- Anderson CA, Boucher G, Lees CW et al: Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet 2011; 43: 246–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jostins L, Ripke S, Weersma RK et al: Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 2012; 491: 119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JZ, Van Sommeren S, Huang H et al: Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet 2015; 47: 979–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano K, Matsushita T, Umeno J et al: A genome-wide association study identifies three new susceptibility loci for ulcerative colitis in the Japanese population. Nat Genet 2009; 41: 1325–1329. [DOI] [PubMed] [Google Scholar]
- Yang SK, Hong M, Zhao W et al: Genome-wide association study of ulcerative colitis in Koreans suggests extensive overlapping of genetic susceptibility with Caucasians. Inflamm Bowel Dis 2013; 19: 954–966. [DOI] [PubMed] [Google Scholar]
- Juyal G, Negi S, Sood A et al: Genome-wide association scan in north Indians reveals three novel HLA-independent risk loci for ulcerative colitis. Gut 2014; 64: 571–579. [DOI] [PubMed] [Google Scholar]
- Garnier G, Ault B, Kramer M, Colten HR: Cis and trans elements differ among mouse strains with high and low extrahepatic complement factor B gene expression. J Exp Med 1992; 175: 471–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu LC, Morley BJ, Campbell RD: Cell-specific expression of the human complement protein factor B gene: evidence for the role of two distinct 5'-flanking elements. Cell 1987; 48: 331–342. [DOI] [PubMed] [Google Scholar]
- Kindt TJ, Goldsby RA, Osborne BA, Kuby J (eds): Immunology, 6th edn. New York, USA: W. H. Freeman and Company, 1992. [Google Scholar]
- Carroll MV, Sim RB: Complement in health and disease. Adv Drug Deliv Rev 2011; 63: 965–975. [DOI] [PubMed] [Google Scholar]
- Gold B, Merriam JE, Zernant J et al: Variation in factor B (BF) and complement component 2 (C2) genes is associated with age-related macular degeneration. Nat Genet 2006; 38: 458–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goicoechea de Jorge E, Harris CL, Esparza-Gordillo J et al: Gain-of-function mutations in complement factor B are associated with atypical hemolytic uremic syndrome. Proc Natl Acad Sci USA 2007; 104: 240–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostvik AE, Granlund A, Gustafsson BI et al: Mucosal toll-like receptor 3-dependent synthesis of complement factor B and systemic complement activation in inflammatory bowel disease. Inflamm Bowel Dis 2014; 20: 995–1003. [DOI] [PubMed] [Google Scholar]
- Gupta A, Thelma BK: Identification of critical variants within SLC44A4, an ulcerative colitis susceptibility gene identified in a GWAS in north Indians. Genes Immun 2016; 17: 105–109. [DOI] [PubMed] [Google Scholar]
- Nabokina SM, Inoue K, Subramanian VS, Valle JE, Yuasa H, Said HM: Molecular identification and functional characterization of the human colonic thiamine pyrophosphate transporter. J Biol Chem 2014; 289: 4405–4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantini A, Pala MI: Thiamine and fatigue in inflammatory bowel diseases: an open-label pilot study. J Altern Complement Med 2013; 19: 704–708. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K et al: PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y, Yamazaki K, Umeno J et al: HLA-Cw*1202-B*5201-DRB1*1502 haplotype increases risk for ulcerative colitis but reduces risk for Crohn's disease. Gastroenterology 2011; 141: 864–871. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ: Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005; 21: 263–265. [DOI] [PubMed] [Google Scholar]
- Dudbridge F: Likelihood-based association analysis for nuclear families and unrelated subjects with missing genotype data. Hum Hered 2008; 66: 87–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathias RA, Gao P, Goldstein JL et al: A graphical assessment of p-values from sliding window haplotype tests of association to identify asthma susceptibility loci on chromosome 11q. BMC Genet 2006; 7: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC: Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 2009; 4: 1073–1081. [DOI] [PubMed] [Google Scholar]
- Adzhubei IA, Schmidt S, Peshkin L et al: A method and server for predicting damaging missense mutations. Nat Methods 2010; 7: 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya A, Ziebarth JD, Cui Y: PolymiRTS Database 3.0: linking polymorphisms in microRNAs and their target sites with human diseases and biological pathways. Nucleic Acids Res 2014; 42: D86–D91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebarth JD, Bhattacharya A, Chen A, Cui Y: PolymiRTS Database 2.0: linking polymorphisms in microRNA target sites with human diseases and complex traits. Nucleic Acids Res 2012; 40: D216–D221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle AP, Hong EL, Hariharan M et al: Annotation of functional variation in personal genomes using RegulomeDB. Genome Res 2012; 22: 1790–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahrenstedt O, Knutson L, Nilsson B, Nilsson-Ekdahl K, Odlind B, Hallgren R: Enhanced local production of complement components in the small intestines of patients with Crohn's disease. N Engl J Med 1990; 322: 1345–1349. [DOI] [PubMed] [Google Scholar]
- Potter BJ, Brown DJ, Watson A, Jewell DP: Complement inhibitors and immunoconglutinins in ulcerative colitis and Crohn's disease. Gut 1980; 21: 1030–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson HJ, Potter BJ, Jewell DP: Humoral immune system in inflammatory bowel disease: I. Complement levels. Gut 1977; 18: 749–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen H, Petersen PH, Svehag SE: Circulating immune complexes in ulcerative colitis—II. Correlation with serum protein concentrations and complement conversion products. Clin Exp Immunol 1978; 31: 81–91. [PMC free article] [PubMed] [Google Scholar]
- Juyal G, Amre D, Midha V, Sood A, Seidman E, Thelma BK: Evidence of allelic heterogeneity for associations between the NOD2/CARD15 gene and ulcerative colitis among North Indians. Aliment Pharmacol Ther 2007; 26: 1325–1332. [DOI] [PubMed] [Google Scholar]
- Raychaudhuri S, Iartchouk O, Chin K et al: A rare penetrant mutation in CFH confers high risk of age-related macular degeneration. Nat Genet 2011; 43: 1232–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namjou B, Sestak AL, Armstrong DL et al: High-density genotyping of STAT4 reveals multiple haplotypic associations with systemic lupus erythematosus in different racial groups. Arthritis Rheum 2009; 60: 1085–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.